Abstract
Breast carcinoma invasion is associated with prominent alterations in stromal fibroblasts. Carcinoma-associated fibroblasts (CAF) support and promote tumorigenesis, whereas normal mammary fibroblasts (NF) are thought to suppress tumor progression. Little is known about the difference in gene expression between CAF and NF or the patient-to-patient variability in gene expression. Paired CAF and NF were isolated from six primary human breast carcinoma specimens. RNA was extracted from low-passage cultures of CAF and NF and analyzed with Affymetrix Human Genome U133 Plus 2.0 arrays. The array data were examined with an empirical Bayes model and filtered according to the posterior probability of equivalent expression and fold difference in expression. Twenty-one genes (27 probe sets) were up-regulated in CAF, as compared to NF. Known functions of these genes relate to paracrine or intracellular signaling, transcriptional regulation, extracellular matrix and cell adhesion/migration. Ten genes (14 probe sets) were down-regulated in CAF, including the pluripotency transcription factor KLF4. Quantitative RT-PCR analysis of 10 genes validated the array results. Immunohistochemical staining for three gene products confirmed stromal expression in terms of location and relative quantity. Surprisingly, the variability of gene expression was slightly higher in NF than in CAF, suggesting inter-individual heterogeneity of normal stroma.
Keywords: breast neoplasms, tumor stroma, fibroblasts, invasion
Introduction
One of the hallmarks of cancer is the invasion of tumor cells into adjacent normal tissue (Hanahan and Weinberg, 2000). Invasive tumor cells interact with their microenvironment and remodel it into a neighborhood supportive of tumor growth and progression. The altered microenvironment is recognizable at the light microscopic level as desmoplasia and used by pathologists to diagnose invasion. The tumor bed, or stroma, contains a number of different cellular elements, which include inflammatory cells, endothelial cells and fibroblasts. Cancer associated fibroblasts (CAF) constitute a significant component of the tumor stroma. Since, in contrast to resting fibroblasts, many of these cells express muscle-specific actin, CAF are commonly referred to as myofibroblasts. CAF are not merely innocent bystanders but actively participate in reciprocal communication with the tumor cells, leading to accelerated tumor growth and progression (Erickson and Barcellos-Hoff, 2003; Orimo et al., 2005).
CAF are likely derived from resident fibroblasts and marrow-derived mesenchymal precursor cells, while their generation via epithelial mesenchymal transition of tumor cells is more controversial (Kalluri and Zeisberg, 2006; Karnoub et al., 2007; Mishra et al., 2008; Mueller and Fusenig, 2004), TGFβ and PDGF are thought to be the principal paracrine factors responsible for the induction (and recruitment) of CAF (Mueller and Fusenig, 2004). Compared to normal fibroblasts, CAF are phenotypically and functionally distinct. Some of these differences are reversible, while others persist when the fibroblasts are removed from the vicinity of carcinoma cells. Stable gene expression changes in CAF vs. normal fibroblasts (NF) may be due to epigenetic (Hu et al., 2005) and possibly genetic alterations (Kurose et al., 2002; Patocs et al., 2007).
Global gene expression profiling of breast carcinomas has revealed a considerable degree of heterogeneity between individual tumors (Perou et al., 2000). These studies have led to new classifications and risk stratification of breast carcinomas based on their gene expression signatures (Massague, 2007; van 't Veer et al., 2002). The samples used for those analyses were whole tissues, including tumor cells and stroma, but the transcriptional pattern is generally attributed primarily to the carcinoma cells. Most recently, global gene expression in the stromal compartment (fibroblasts, inflammatory cells, endothelium) has been shown to powerfully predict clinical outcome (Finak et al., 2008) and therapy response (Farmer et al., 2009). A variety of differences have been identified between breast carcinoma stroma and normal mammary stroma, primarily resulting in increased expression of cytokines, extracellular matrix molecules and proteases (Casey et al., 2008; Iacobuzio-Donahue et al., 2002; Singer et al., 2008; Singer et al., 2002; Tang et al., 2004). Our understanding of differential gene expression specifically in stromal fibroblasts is incomplete and information on potential inter-individual heterogeneity of gene expression in CAF and NF is currently lacking.
The purpose of the present study was to address this knowledge gap by identifying genes differentially expressed in CAF vs. matched NF and by analyzing the heterogeneity of gene expression profiles in the two cell types. Using an empirical Bayes model and defined inclusion criteria, the transcription of 21 genes was found to be up-regulated in CAF, whereas the expression of 10 genes was down-regulated. Surprisingly, the variability of gene expression in NF exceeded that of CAF. This finding suggests an unexpected inter-individual heterogeneity of gene expression in normal mammary fibroblasts and a relative synchronization of gene expression in CAF.
Results
Characteristics of Fibroblasts from Human Tissue Samples
Fibroblasts were isolated from six breast carcinomas and from adjacent normal breast tissue. The histology of all tissues was validated by frozen section analysis of a tissue piece taken from the samples prior to tissue mincing (Fig. 1A, B). Five infiltrating ductal carcinomas and one mucinous carcinoma representing a spectrum of different grades (two grade 3, three grade 2 and one grade 1) were obtained. The fibroblasts were grown in selective culture medium until examination by phase contrast microscopy indicated that all epithelial elements had disappeared. Labeling with anti-vimentin and anti-pancytokeratin antibodies confirmed the purity of the fibroblast cultures (Fig. 1C, D). Several cultures were also labeled with an antibody to CD31, which failed to reveal any endothelial cell contamination (not shown). For further characterization of fibroblast cultures, immunoflourescence labeling for the fibroblast marker FSP1 and the myofibroblast marker α-smooth muscle actin (αSMA) was performed. CAF and NF were universally positive for FSP1. The frequency of αSMA expression was higher in CAF than in NF in four of the six cases (Fig. 1E, F).
Figure 1.
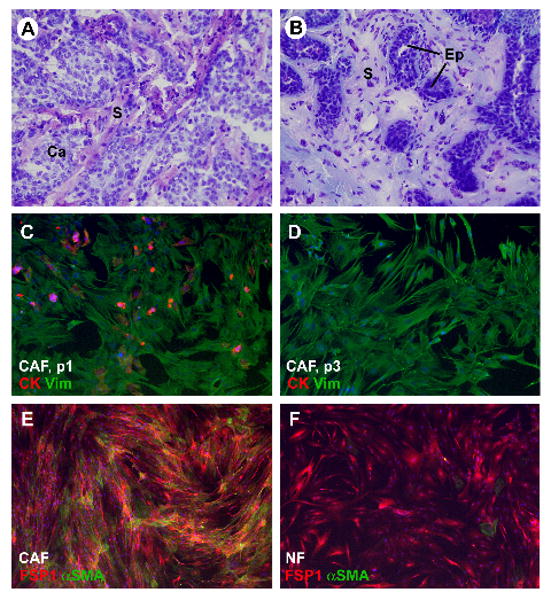
Characterization of fibroblasts isolated from human breast tissue samples: A) Hematoxylin-eosin-stained frozen section of tissue piece taken from infiltrating ductal carcinoma sample prior to tissue mincing. B) Hematoxylin-eosin-stained frozen section of tissue piece taken from adjacent normal mammary tissue sample prior to mincing. C) Carcinoma-associated fibroblasts (CAF) from human breast carcinoma sample at passage 1 (p1). The cells were immunofluorescence-labeled for epithelial marker cytokeratin (red signal) and the mesenchymal marker vimentin (green signal). Note a few epithelial cells and epithelial cell fragments among the fibroblasts. D) Carcinoma-associated fibroblasts (CAF) from the same human breast carcinoma sample as shown in panel “C” at passage 3 (p3). The cells were immunofluorescently labeled for epithelial marker cytokeratin (red signal) and mesenchymal marker vimentin (green signal). At this stage, no epithelial cells are left. E) CAF labeled for FSP1 (red signal) and αSMA (green signal). F) Normal fibroblasts (NF) labeled for FSP1 (red signal) and αSMA (green signal). Abbreviations: Ca = carcinoma; S = stroma). Original magnification: 200× for all images. Blue signal in panels C – F: nuclear label DAPI
Differential Gene Expression in CAF vs. NF
The gene expression profiles generated with Affymetrix arrays from the six CAF samples and their matched NF controls were examined by unsupervised cluster analysis. In some patients, CAF and matched NF clustered closely together (e.g. cases 8 and 2) whereas in others (e.g. cases 6 and 7), CAF and NF showed distinctly different expression profiles (Fig. 2). This result indicates that NF and CAF are not easily distinguishable by unsupervised protocols.
Figure 2.
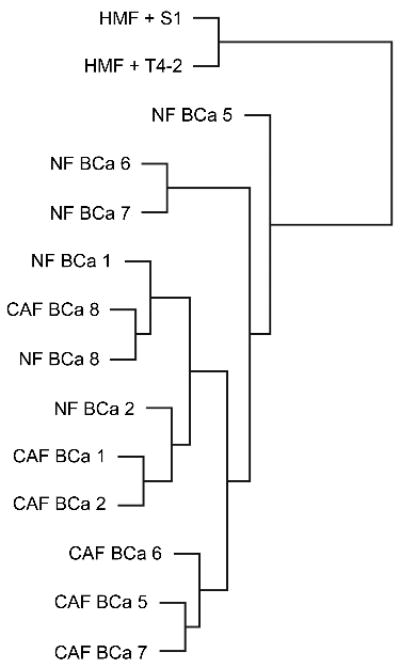
Cluster analysis of gene expression: Gene expression data obtained with Affymetrix arrays from CAF and NF isolated from all six cases (BCa 1, 2, 5, 6, 7, 8) were subjected to unsupervised cluster analysis. Also included were results from a human mammary fibroblast cell line (HMF) grown in 3D co-culture with S1 normal mammary epithelial cells or with malignant T4-2 cells.
Also included in this analysis were immortalized human mammary fibroblasts (HMF) in 3D co-culture with T4-2 carcinoma cells or S1 mammary epithelial cells. T4-2 and S1 cells stem from the same patient and are commonly used as a model system to compare transformed and normal mammary epithelial cells. HMF gene expression segregated into a discrete cluster – whether grown in co-culture with T4-2 carcinoma cells or with S1 cells (Fig. 2). This suggests that immortal HMF are distinct from primary NF and do not become “CAF-like” when exposed to carcinoma cell paracrine factors. The difference in gene expression between HMF co-cultured with T4-2 tumor cells vs. HMF cells co-cultured with S1 epithelial cells likely reflects short-term induction by carcinoma cell-derived secreted factors. Gene expression in HMF was not further considered in this study.
Using empirical Bayes modeling, we ranked genes which were most consistently differentially expressed in CAF vs. NF. With a cut-off value of 0.005 for the posterior probability of equivalent expression and after eliminating genes with less than two-fold overexpression, we found that 21 genes (27 probe sets) were overexpressed in CAF compared to NF (Table 1). The known functions of these genes fall into the categories of paracrine signaling, intracellular signaling, transcription regulation, extracellular matrix production and cell adhesion/migration. Using the same posterior probability cut-off, we identified ten genes (14 probe sets) which were expressed at least two-fold higher in NF than in CAF (Table 2). Interestingly, two of these genes (AKR1C1 and AKR1C2) are thought to be involved in steroid hormone metabolism and polycyclic aromatic hydrocarbon detoxification. Other genes in this group have purported roles in transcription, migration and cell signaling.
Table 1.
Genes up-regulated in CAF compared to NF
| Affy Probeset | Gene Symbol | Fold Difference (CAF/NF) | Gene Name |
|---|---|---|---|
| 220979_s_at | ST6GALNAC5 | 12.2 | ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| 219090_at | SLC24A3 | 6.3 | solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 |
| 223484_at | C15orf48 | 5.7 | chromosome 15 open reading frame 48 |
| 209909_s_at | TGFB2 | 5.6 | transforming growth factor, beta 2 |
| 202718_at | IGFBP2 | 5.4 | insulin-like growth factor binding protein 2 |
| 207426_s_at | TNFSF4 | 5.2 | tumor necrosis factor (ligand) superfamily, member 4 |
| 228109_at | RASGRF2 | 4.5 | Ras protein-specific guanine nucleotide-releasing factor 2 |
| 217428_s_at | COL10A1 | 4.4 | collagen, type X, alpha 1(Schmid metaphyseal chondrodysplasia) |
| 203397_s_at | GALNT3 | 4.3 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) |
| 209604_s_at | GATA3 | 4.2 | GATA binding protein 3 |
| 226701_at | GJA5 | 4.1 | gap junction protein, alpha 5, 40kDa |
| 202796_at | SYNPO | 3.7 | synaptopodin |
| 204653_at | TFAP2A | 3.6 | transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| 206796_at | WISP1 | 3.1 | WNT1 inducible signaling pathway protein 1 |
| 205286_at | TFAP2C | 2.9 | transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| 47550_at | LZTS1 | 2.7 | leucine zipper, putative tumor suppressor 1 |
| 213541_s_at | ERG | 2.6 | v-ets erythroblastosis virus E26 oncogene homolog (avian) |
| 219655_at | C7orf10 | 2.6 | chromosome 7 open reading frame 10 |
| 236738_at | LOC401097 | 2.5 | similar to LOC166075 |
| 203821_at | HBEGF | 2.3 | heparin-binding EGF-like growth factor |
| 202625_at | LYN | 2.1 | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
Table 2.
Genes down-regulated in CAF compared to NF
| Affy Probeset | Gene Symbol | Fold Difference (CAF/NF) | Gene Name |
|---|---|---|---|
| 238178_at | NA | 0.261 | NA |
| 204151_x_at | AKR1C1 | 0.375 | aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase) |
| 209699_x_at | AKR1C2 | 0.379 | aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; 3-alpha hydroxysteroid dehydrogenase, type III) |
| 232668_at | C8orf72 | 0.386 | chromosome 8 open reading frame 72 |
| 209355_s_at | PPAP2B | 0.388 | phosphatidic acid phosphatase type 2B |
| 229377_at | GRTP1 | 0.412 | growth hormone regulated TBC protein 1 |
| 220266_s_at | KLF4 | 0.437 | Kruppel-like factor 4 (gut) |
| 202948_at | IL1R1 | 0.445 | interleukin 1 receptor, type I |
| 1561574_at | SLIT3 | 0.475 | slit homolog 3 (Drosophila) |
| 213895_at | EMP1 | 0.477 | epithelial membrane protein 1 |
NA, not annotated
Validation of Gene Expression Data
The expression levels of ten genes (7 with increased expression in CAF; 3 with decreased expression in CAF) were validated by qRT-PCR. Sufficient amounts of RNA were available from five of the six patients. In all but one of the tissue pairs, the expression arrays and qRT-PCR yielded concordant results (Fig. 3). A review of the outlier case did not reveal any unusual histologic features in either normal or tumor tissue. To determine whether the differences between CAF and NF extend to the protein level, we examined the expression of three of these genes (WISP1, TGFB2 and KLF4) by immunohistochemistry in an independent set of 26 breast carcinomas and adjacent normal tissue. Due to some tissue section loss during epitope retrieval, 24 sample pairs remained for the WISP1 and TGFB2 analysis and 25 for the KLF4 analysis. WISP1 labeling was found in CAF in the majority of tumors but only in one sample of NF (Fig. 4). TGFB2 labeling was elevated in CAF compared to NF. Conversely, and consistent with the RNA data, KLF4 was more abundant in NF of normal terminal duct lobular units than in CAF. Some of the breast carcinoma sections included an area with a wound healing response due to a prior biopsy procedure. Interestingly, TGFB2 and WISP1 expression in wound site myofibroblasts was elevated, similar to CAF, but KLF4 expression was also high, thus resembling NF (not shown).
Figure 3.
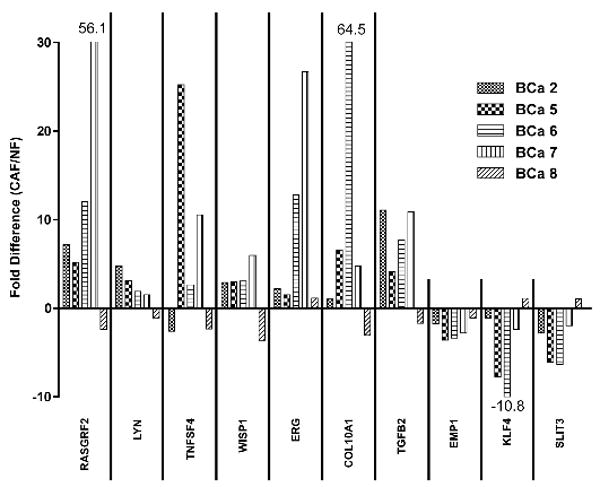
Validation of mRNA levels: The mRNA levels of ten genes (7 overexpressed in CAF, 3 overexpressed in NF) were analyzed by qRT-PCR in CAF and NF from five breast cancer patients. The results are expressed as fold difference between CAF and NF. In all but one case (BCa 8), the results were congruent with the cumulative Affymetrix data.
Figure 4.
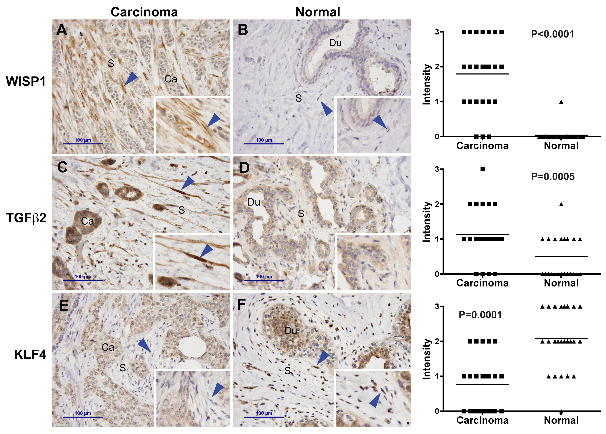
Validation of protein levels by immunohistochemistry: Paraffin sections from breast carcinoma and adjacent normal mammary gland tissue were immuno-labeled for the protein products of genes that had been found to be overexpressed in CAF (WISP1, TGFb2) or in NF (KLF4). Blue arrowheads: stromal fibroblasts. Scale bar indicates 100 μm. The labeling intensity in fibroblasts of tumor or adjacent normal stroma was scored manually on a scale from 0 (no detectable labeling) to 3 (strong labeling). The horizontal lines indicate the means.
Analysis of Variability of Gene Expression
Next, we compared the heterogeneity of gene expression in CAF vs. NF. Unexpectedly, the estimated probability of NF gene expression variance being higher than CAF gene expression variance was 0.547 with a 95% confidence interval of 0.543 to 0.551 (p<0.0001). This result indicates that gene expression is more variable in the NF samples than the CAF samples. To examine the possibility that the increased heterogeneity was due to a single sample, which might have skewed the distribution, we performed a leave-one-out procedure on all samples. The conclusion of increased variability in the NF samples was maintained in this analysis, indicating that this result was not caused by an outlier sample. To determine the amount of heterogeneity attributable to assay variability, we isolated RNA from two independent cultures of a single, new NF isolate and performed Affymetrix array analysis. The median ratio of the sample variance of gene expression in this set of duplicates to the sample variance in the original set of NF was 0.30, suggesting that the heterogeneity was not primarily caused by assay noise. Furthermore, it is unlikely that assay noise would be different in CAF vs. NF.
Next, we performed F-tests and ranked the q-values of individual genes to identify those that show a significant difference in variance between CAF and NF (q<0.05). The list of these genes (Table 3) included three with known functions: a metalloprotease thought to be involved in inflammation, a mucin and a transcription factor. These results should be interpreted with caution as the F-test relies heavily on an assumption of normal data (in this case, log-normal).
Table 3.
Genes with a significant difference in variance between NF and CAF
| Affy Probeset | Gene Symbol | Gene Name |
|---|---|---|
| 216910_at | XPNPEP2 | X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound |
| 217117_x_at | MUC3A | mucin 3A, cell surface associated |
| 217926_at | HSPC023 | HSPC023 pseudogene |
| 233739_at | NA | NA |
| 236471_at | NFE2L3 | nuclear factor (erythroid-derived 2)-like 3 |
NA, not annotated
Discussion
Using primary cultures of fibroblasts from human tissue samples, we identified sets of genes that are differentially expressed in fibroblasts from breast carcinomas compared to fibroblasts from adjacent normal breast tissue. Surprisingly, there is essentially no overlap with respect to CAF-specific genes described by different investigators (Allinen et al., 2004; Casey et al., 2008; Iacobuzio-Donahue et al., 2002; Mercier et al., 2008; Singer et al., 2008). COL10A1 was the only gene identified both by us and Casey and co-workers in a very recent study (Casey et al., 2008). The apparent discrepancies may be caused by differences in the cell isolation techniques, the analysis platforms and statistical methods. For example, Allinen and coworkers used affinity purification of dispersed cells, followed by serial analysis of gene expression and pooled the results for myofibroblasts and myoepithelial cells (Allinen et al., 2004). Since myoepithelial cells are not a stromal component, this approach would be expected to yield a list of genes distinct from those expressed by CAF alone. Casey's group and Finak isolated RNA from microdissected stroma and analyzed gene expression with the Affymetrix and Agilent platforms, respectively (Casey et al., 2008; Finak et al., 2008). Unavoidably, microdissection captures endothelial cells and hematopoietic cells in addition to fibroblasts. Despite these technical differences, all studies identify genes involved in similar functions, namely extracellular matrix constituents and paracrine and intracellular signaling molecules. In our study, the fibroblasts were isolated to apparent purity by mechanical means and short-term culture under conditions favoring fibroblast growth. It is likely that this passage of the cells ex vivo masked some of the originally existing differences in gene expression. Notably, a very different set of genes was differentially expressed in mammary fibroblasts after 5 days of 3D co-culture with S1 mammary epithelial cells or T4-2 carcinoma cells. In this experimental setting, all of the differences are expected to be due to short-term, paracrine induction.
Our data show that considerable differences in gene expression are maintained ex vivo. This finding is consistent with work by other investigators who observed differential gene expression in mammary fibroblasts after brief in vitro passage (Singer et al., 2007). Persistent changes in fibroblast gene expression could be explained by genetic or epigenetic alterations. The concept of genetic alterations in breast carcinoma stroma is the subject of considerable controversy (Campbell et al., 2008; Patocs et al., 2007; Qiu et al., 2008). Epigenetic changes, on the other hand, have been convincingly documented in stromal fibroblasts by methylation-specific digital karyotyping (Fiegl et al., 2006; Hu et al., 2005).
The list of genes overexpressed in CAF compared to NF affords some interesting insights into the dynamics of the carcinoma microenvironment. For example, the transcription of Wnt-1 inducible signaling pathway protein 1 (WISP1) is elevated in CAF and WISP1 protein is detected in breast cancer stroma by IHC. The mammary gland-targeted expression of Wnt-1 in mice causes tumors and interestingly, WISP1, is aberrantly expressed in the mammary tumor stroma of these animals, consistent with the paracrine induction by epithelial cell-derived Wnt-1 (Kim et al., 2008; Pennica et al., 1998). WISP1 expression in CAF suggests active paracrine Wnt-1 signaling in human breast cancer. Apart from serving as a marker for Wnt-1 signaling, WISP1 may also have direct roles in tumorigenesis. WISP1 expression has been associated with aggressive features in breast carcinomas (Xie et al., 2001) and the protein inhibits apoptotic pathways (You et al., 2002).
Abnormal ECM production is one of the defining characteristics of tumor stroma (Hanahan and Weinberg, 2000). The transcripts of ECM constituents have been consistently identified in CAF global gene expression studies (Allinen et al., 2004; Casey et al., 2008; Finak et al., 2008; Singer et al., 2002). Thus, we were not surprised to discover a collagen among the genes up-regulated in CAF. Collagen type-X (COL10A), however, is an unusual collagen to be found in the mammary gland, since it is primarily produced by chondrocytes in the epiphyseal growth plate (Sutmuller et al., 1997). The role of collagen type-X in mammary gland stroma is unknown.
TGF-beta isoforms are well-characterized as promoters of ECM production in wound healing and in tumors (Chang et al., 2007). Nakagawa and co-workers identified increased levels of TGFβ2 in CAF of colon carcinoma metastases to the liver (Nakagawa et al., 2004). TGFβ2 is secreted by breast carcinoma cells (de Jong et al., 1998) and fibroblasts (Fig. 4) and likely contributes to aberrant ECM synthesis in breast tumors. The finding that heparan sulfate binding epidermal growth factor-like factor (HB-EGF) expression is elevated in CAF of breast carcinomas is interesting, since this molecule has the ability to stimulate mitogenesis of both fibroblasts and carcinoma cells. Thus, HB-EGF may have dual functions as an autocrine and a paracrine factor in breast cancer (Duque et al., 2001; Narita et al., 2007).
The transcription of several genes is consistently lower in CAF than in NF. Notably, AKR1C1 and AKR1C2, two closely related genes involved in progesterone metabolism, are expressed at lower levels in CAF. The loss of these enzymes in breast carcinomas has previously been reported, but expression in normal tissues had been attributed to the epithelial compartment (Ji et al., 2004; Lewis et al., 2004). Expression silencing of these two enzymes in mammary epithelial cells resulted in elevated progesterone concentrations in vitro (Ji et al., 2004). Loss of expression of AKR1C1 and AKR1C2 in stromal fibroblasts may alter the hormonal milieu in breast carcinomas. KLF4 was also expressed at high levels in normal mammary gland stroma and lost in CAF. KLF4 is a bifunctional transcription factor that can either activate or repress gene transcription depending on the target gene. Recent work demonstrating that the forced expression of KLF4 together with three other transcription factors reverses the differentiation of mature skin fibroblasts and reverts them into pluripotent stem (iPS) cells has garnered much attention (Takahashi et al., 2007). It is tempting to speculate that the down-regulation of KLF4 participates in the differentiation of resting fibroblasts to myofibroblasts. The inverse relationship between KLF4 and WISP1 is intriguing in view of the finding by Zhang and co-workers that KLF4 can inhibit Wnt signaling (Zhang et al., 2006).
One of the unexpected findings of our study is the observation that gene expression profiles of normal mammary fibroblasts are more heterogeneous than those of CAF. This may indicate that the mammary stroma indeed varies between individuals, lending support to the hypothesis that the ability of the stroma to act as a barrier to cancer development or to promote tumorigenesis may also be variable (Mueller and Fusenig, 2004). Conversely, the heterogeneous gene expression of NF relative to CAF may be a reflection of the uniformity of gene expression in CAF. This latter interpretation is consistent with the conclusion by Allinen et al. (Allinen et al., 2004) that myofibroblasts isolated from different infiltrating ductal carcinomas are highly similar.
In summary, we identified distinct differences in gene expression between CAF and NF, which are maintained in short-term cell culture. Altered gene expression in fibroblasts likely contributes to carcinoma growth and progression by enhancing ECM production, promoting stromal-epithelial paracrine signaling and altering steroid hormone metabolism.
Materials and Methods
Tissue Samples
Tissue was obtained with approval from the Health Sciences Institutional Review Board of the University of Wisconsin – Madison. Fresh surgical specimens (mastectomies and excisional biopsies) were available from six patients with invasive breast carcinomas. Approximately 500 mm3 each were taken from grossly recognizable tumor and adjacent normal breast tissue. In compliance with the IRB protocol, no further patient information was obtained. H&E stained frozen sections were prepared from each tissue sample to confirm benignity or malignancy and to obtain information about histological subtype and histopathological grade.
Fibroblast Isolation
To isolate fibroblasts from tumor and adjacent normal tissue, we followed a modification of a protocol developed by Allinen and coworkers (Allinen et al., 2004). The tissue was minced and digested for 2 hours at 37°C in DMEM (Mediatech) with 10% fetal bovine serum (Gemini Bio-Products) supplemented with penicillin-streptomycin (Mediatech), 2 mg/ml collagenase I (Sigma), and 2 mg/ml hyaluronidase (Sigma). After centrifugation and washing with Hank's balanced salt solution (HBSS; HyClone), cells were trypsinized, resuspended in complete media and plated in plastic dishes.
Culture and Characterization of Primary Fibroblasts
CAF and NF were routinely maintained in DMEM and 10% fetal bovine serum at 37°C in a humidified atmosphere containing 5% CO2. RNA was extracted from confluent cultures as soon as epithelial cells had disappeared. This criterion was met typically after one to three passages. The purity of the fibroblasts was assessed in parallel cultures by immunolabeling for the epithelial cell marker pan-cytokeratin (mouse monoclonal antibody, dilution 1:100; Lab Vision) and the endothelial cell marker von Willebrand factor (rabbit polyclonal antibody, dilution 1:1000; Dako) and the mesenchymal marker vimentin (mouse monoclonal antibody, dilution 1:100; Lab Vision). For further characterization, the fibroblast cultures were labeled with antibodies to α-smooth muscle actin (mouse monoclonal antibody αSMA, dilution 1:100, Sigma) and fibroblast specific protein FSP1/S100A4 (rabbit polyclonal, dilution 1:50, Abcam).
Microarray Analysis
Total RNA from CAF and NF was isolated using the RNeasy Micro kit according to the manufacturer's instructions (Qiagen). Total RNA was eluted in a final volume of 20 μl (H2O) and stored at -80°C until further processing. The Ambion MessageAmp II-Biotin Enhanced kit was used to synthesize double-stranded cDNA and produce biotin-labeled cRNA from 500 ng of total RNA. After fragmentation, 10 μg of cRNA were hybridized at 45°C for 16 h to Affymetrix HG_U133 Plus 2.0 oligonucleotide arrays containing probes to more than 47,000 transcripts.
Statistical Analysis
The microarray data were normalized using robust multi-array average (RMA) (Irizarry et al., 2003). Using the resulting normalized intensities, an empirical Bayes method was applied (Kendziorski et al., 2003). The lognormal-normal model was found to be reasonable and provided an estimate for each gene of the posterior probability of equivalent expression (PP-EE). Genes were ranked according to this value and the results were further filtered according to the magnitude of change in expression: only genes that were at least 2-fold up or down regulated were considered. To evaluate differences in variability of expression, an F-test for equal variances of CAF and NF log-intensities was performed for each gene. To account for multiple testing issues, q-values were calculated (Storey and Tibshirani, 2003) to provide a local expected false discovery rate for a list of genes with the most significant difference in variance. All computations were performed using R language and environment (R-Development-Core-Team, 2008). Immunohistochemistry scores were considered as continuous variables and the scores from tumor and normal tissue stroma were compared by Wilcoxon matched-pairs signed-ranks test using InStat software (Graphpad Software, www.graphpad.com).
Semiquantitative Real-Time PCR (qRT-PCR)
qRT-PCR was performed to validate the differential gene expression pattern revealed by microarray analysis. Seven genes which were up-regulated (RASGRF2, LYN, TNFSF4, WISP1, ERG, COL10A1, TGFB2) and three that were down-regulated (EMP1, KLF4, and SLIT3) in CAF were examined. Aliquots from the same total RNA samples used for the microarray analysis were reverse-transcribed into cDNA using the RT2 First Strand Kit (SuperArray). qRT-PCR reactions were run on a LightCycler instrument (Roche) with gene-specific primer sets (SuperArray) and RT2 SYBR Green PCR Master Mix (SuperArray) following the manufacturer's instructions. Peptidylprolyl isomerase A (PPIA), TATA box binding protein (TBP), transferrin receptor (TFRC), and ubiquitin C (UBC) mRNA levels were used as internal controls for normalizing different samples.
Immunohistochemistry
Immunolabeling was performed on a Lab Vision Autostainer 360. Five μm thick paraffin sections were subjected to heat-induced epitope retrieval with EDTA, pH 8.0 (WISP1, TGFB2) or citrate, pH 6.0 (KLF4). After inhibition of endogenous peroxidase and blocking, the slides were incubated with primary antibody for 60 minutes. The following primary antibodies were used: Goat anti-WISP1/CCN4 (1:50; R&D Systems), mouse anti-TGFβ2 (1:500; Abcam) and goat anti-KLF4 (1:100; R&D Systems). This was followed by incubation with an appropriate horseradish peroxidase-based detection reagent (Biocare) and DAB+ chromogenic substrate (DAKO). The slides were counterstained with hematoxylin and coverslipped. The labeling intensity of fibroblasts within carcinoma stroma and stroma of adjacent normal terminal duct lobular units (TDLU) was scored visually on a scale ranging from 0 to 3 (0 = negative; 1 = weakly positive; 2 = moderately positive; 3 = strongly positive).
Acknowledgments
We thank Dr. Christina Kendziorski for help with the array analyses and Dr. Korise Rasmusson for her assistance with the manuscript preparation. Maret Bauer was supported by a scholarship from the Dr. Mildred Scheel Stiftung. This research was funded by National Institutes of Health grant RO1 CA107012 and a grant from the Wisconsin Partnership program.
Contributor Information
M Bauer, Department of Pathology and Laboratory Medicine, University of Wisconsin-Madison, Madison WI, 53792, USA.
G Su, Department of Pathology and Laboratory Medicine, University of Wisconsin-Madison, Madison WI, 53792, USA.
C Casper, Department of Biostatistics and Medical Informatics, University of Wisconsin-Madison, Madison WI, 53792, USA.
R He, Department of Pathology and Laboratory Medicine, University of Wisconsin-Madison, Madison WI, 53792, USA.
W Rehrauer, Department of Pathology and Laboratory Medicine, University of Wisconsin-Madison, Madison WI, 53792, USA.
A Friedl, Department of Pathology and Laboratory Medicine, University of Wisconsin-Madison, Madison WI, 53792, USA.
References
- Allinen M, Beroukhim R, Cai L, Brennan C, Lahti-Domenici J, Huang H, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Campbell IG, Qiu W, Polyak K, Haviv I. Breast-cancer stromal cells with TP53 mutations. N Engl J Med. 2008;358:1634–5. doi: 10.1056/NEJMc086024. author reply 1636. [DOI] [PubMed] [Google Scholar]
- Casey T, Bond J, Tighe S, Hunter T, Lintault L, Patel O, et al. Molecular signatures suggest a major role for stromal cells in development of invasive breast cancer. Breast Cancer Res Treat. 2008 doi: 10.1007/s10549-008-9982-8. [DOI] [PubMed] [Google Scholar]
- Chang CF, Westbrook R, Ma J, Cao D. Transforming growth factor-beta signaling in breast cancer. Front Biosci. 2007;12:4393–401. doi: 10.2741/2396. [DOI] [PubMed] [Google Scholar]
- de Jong JS, van Diest PJ, van der Valk P, Baak JP. Expression of growth factors, growth inhibiting factors, and their receptors in invasive breast cancer. I: An inventory in search of autocrine and paracrine loops. J Pathol. 1998;184:44–52. doi: 10.1002/(SICI)1096-9896(199801)184:1<44::AID-PATH984>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Duque JL, Adam RM, Mullen JS, Lin J, Richie JP, Freeman MR. Heparin-binding epidermal growth factor-like growth factor is an autocrine mediator of human prostate stromal cell growth in vitro. J Urol. 2001;165:284–8. doi: 10.1097/00005392-200101000-00080. [DOI] [PubMed] [Google Scholar]
- Erickson AC, Barcellos-Hoff MH. The not-so innocent bystander: the microenvironment as a therapeutic target in cancer. Expert Opin Ther Targets. 2003;7:71–88. doi: 10.1517/14728222.7.1.71. [DOI] [PubMed] [Google Scholar]
- Farmer P, Bonnefoi H, Anderle P, Cameron D, Wirapati P, Becette V, et al. A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat Med. 2009;15:68–74. doi: 10.1038/nm.1908. [DOI] [PubMed] [Google Scholar]
- Fiegl H, Millinger S, Goebel G, Muller-Holzner E, Marth C, Laird PW, et al. Breast cancer DNA methylation profiles in cancer cells and tumor stroma: association with HER-2/neu status in primary breast cancer. Cancer Res. 2006;66:29–33. doi: 10.1158/0008-5472.CAN-05-2508. [DOI] [PubMed] [Google Scholar]
- Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008 doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hu M, Yao J, Cai L, Bachman KE, van den Brule F, Velculescu V, et al. Distinct epigenetic changes in the stromal cells of breast cancers. Nat Genet. 2005;37:899–905. doi: 10.1038/ng1596. [DOI] [PubMed] [Google Scholar]
- Iacobuzio-Donahue CA, Argani P, Hempen PM, Jones J, Kern SE. The desmoplastic response to infiltrating breast carcinoma: gene expression at the site of primary invasion and implications for comparisons between tumor types. Cancer Res. 2002;62:5351–7. [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Ji Q, Aoyama C, Nien YD, Liu PI, Chen PK, Chang L, et al. Selective loss of AKR1C1 and AKR1C2 in breast cancer and their potential effect on progesterone signaling. Cancer Res. 2004;64:7610–7. doi: 10.1158/0008-5472.CAN-04-1608. [DOI] [PubMed] [Google Scholar]
- Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–63. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- Kendziorski CM, Newton MA, Lan H, Gould MN. On parametric empirical Bayes methods for comparing multiple groups using replicated gene expression profiles. Stat Med. 2003;22:3899–914. doi: 10.1002/sim.1548. [DOI] [PubMed] [Google Scholar]
- Kim YC, Clark RJ, Ranheim EA, Alexander CM. Wnt1 expression induces short-range and long-range cell recruitments that modify mammary tumor development and are not induced by a cell-autonomous beta-catenin effector. Cancer Res. 2008;68:10145–53. doi: 10.1158/0008-5472.CAN-08-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurose K, Gilley K, Matsumoto S, Watson PH, Zhou XP, Eng C. Frequent somatic mutations in PTEN and TP53 are mutually exclusive in the stroma of breast carcinomas. Nat Genet. 2002;32:355–7. doi: 10.1038/ng1013. [DOI] [PubMed] [Google Scholar]
- Lewis MJ, Wiebe JP, Heathcote JG. Expression of progesterone metabolizing enzyme genes (AKR1C1, AKR1C2, AKR1C3, SRD5A1, SRD5A2) is altered in human breast carcinoma. BMC Cancer. 2004;4:27. doi: 10.1186/1471-2407-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J. Sorting out breast-cancer gene signatures. N Engl J Med. 2007;356:294–7. doi: 10.1056/NEJMe068292. [DOI] [PubMed] [Google Scholar]
- Mercier I, Casimiro MC, Wang C, Rosenberg AL, Quong J, Minkeu A, et al. Human breast cancer-associated fibroblasts (CAFs) show caveolin-1 downregulation and RB tumor suppressor functional inactivation: Implications for the response to hormonal therapy. Cancer Biol Ther. 2008;7:1212–25. doi: 10.4161/cbt.7.8.6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra PJ, Mishra PJ, Humeniuk R, Medina DJ, Alexe G, Mesirov JP, et al. Carcinoma-associated fibroblast-like differentiation of human mesenchymal stem cells. Cancer Res. 2008;68:4331–9. doi: 10.1158/0008-5472.CAN-08-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller MM, Fusenig NE. Friends or foes - bipolar effects of the tumour stroma in cancer. Nat Rev Cancer. 2004;4:839–49. doi: 10.1038/nrc1477. [DOI] [PubMed] [Google Scholar]
- Nakagawa H, Liyanarachchi S, Davuluri RV, Auer H, Martin EW, De La Chapelle A, et al. Role of cancer-associated stromal fibroblasts in metastatic colon cancer to the liver and their expression profiles. Oncogene. 2004 doi: 10.1038/sj.onc.1208013. [DOI] [PubMed] [Google Scholar]
- Narita K, Chien J, Mullany SA, Staub J, Qian X, Lingle WL, et al. Loss of HSulf-1 expression enhances autocrine signaling mediated by amphiregulin in breast cancer. J Biol Chem. 2007;282:14413–20. doi: 10.1074/jbc.M611395200. [DOI] [PubMed] [Google Scholar]
- Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–48. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- Patocs A, Zhang L, Xu Y, Weber F, Caldes T, Mutter GL, et al. Breast-cancer stromal cells with TP53 mutations and nodal metastases. N Engl J Med. 2007;357:2543–51. doi: 10.1056/NEJMoa071825. [DOI] [PubMed] [Google Scholar]
- Pennica D, Swanson TA, Welsh JW, Roy MA, Lawrence DA, Lee J, et al. WISP genes are members of the connective tissue growth factor family that are up-regulated in wnt-1-transformed cells and aberrantly expressed in human colon tumors. Proc Natl Acad Sci U S A. 1998;95:14717–22. doi: 10.1073/pnas.95.25.14717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Qiu W, Hu M, Sridhar A, Opeskin K, Fox S, Shipitsin M, et al. No evidence of clonal somatic genetic alterations in cancer-associated fibroblasts from human breast and ovarian carcinomas. Nat Genet. 2008;40:650–5. doi: 10.1038/ng.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R-Development-Core-Team. A language and environment for statistical computing. R Foundation for Statistical Computing 2008 [Google Scholar]
- Singer CF, Gschwantler-Kaulich D, Fink-Retter A, Haas C, Hudelist G, Czerwenka K, et al. Differential gene expression profile in breast cancer-derived stromal fibroblasts. Breast Cancer Res Treat. 2007 doi: 10.1007/s10549-007-9725-2. [DOI] [PubMed] [Google Scholar]
- Singer CF, Gschwantler-Kaulich D, Fink-Retter A, Haas C, Hudelist G, Czerwenka K, et al. Differential gene expression profile in breast cancer-derived stromal fibroblasts. Breast Cancer Res Treat. 2008;110:273–81. doi: 10.1007/s10549-007-9725-2. [DOI] [PubMed] [Google Scholar]
- Singer CF, Kronsteiner N, Marton E, Kubista M, Cullen KJ, Hirtenlehner K, et al. MMP-2 and MMP-9 expression in breast cancer-derived human fibroblasts is differentially regulated by stromal-epithelial interactions. Breast Cancer Research & Treatment. 2002;72:69–77. doi: 10.1023/a:1014918512569. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutmuller M, Bruijn JA, de Heer E. Collagen types VIII and X, two non-fibrillar, short-chain collagens. Structure homologies, functions and involvement in pathology. Histol Histopathol. 1997;12:557–66. [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tang Y, Kesavan P, Nakada MT, Yan L. Tumor-stroma interaction: positive feedback regulation of extracellular matrix metalloproteinase inducer (EMMPRIN) expression and matrix metalloproteinase-dependent generation of soluble EMMPRIN. Mol Cancer Res. 2004;2:73–80. [PubMed] [Google Scholar]
- van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Xie D, Nakachi K, Wang H, Elashoff R, Koeffler HP. Elevated levels of connective tissue growth factor, WISP-1, and CYR61 in primary breast cancers associated with more advanced features. Cancer Res. 2001;61:8917–23. [PubMed] [Google Scholar]
- You Z, Saims D, Chen S, Zhang Z, Guttridge DC, Guan KL, et al. Wnt signaling promotes oncogenic transformation by inhibiting c-Myc-induced apoptosis. J Cell Biol. 2002;157:429–40. doi: 10.1083/jcb.200201110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Chen X, Kato Y, Evans PM, Yuan S, Yang J, et al. Novel cross talk of Kruppel-like factor 4 and beta-catenin regulates normal intestinal homeostasis and tumor repression. Mol Cell Biol. 2006;26:2055–64. doi: 10.1128/MCB.26.6.2055-2064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]


