Abstract
The worldwide epidemic of obesity has increased the urgency of developing a deeper understanding of physiological systems related to energy balance and energy storage, including the mechanisms controlling the development of fat cells (adipocytes). The differentiation of committed preadipocytes to adipocytes is controlled by PPARγ and several other transcription factors 1, but the molecular basis for preadipocyte determination is not understood. Using a novel method for the quantitative analysis of transcriptional components, we identified the zinc-finger protein Zfp423 as a factor enriched in preadipose versus non-preadipose fibroblasts. Ectopic expression of Zfp423 in non-adipogenic NIH 3T3 fibroblasts robustly activates expression of PPARγ in undifferentiated cells and permits cells to undergo adipocyte differentiation under permissive conditions. ShRNA-mediated reduction of Zfp423 expression in 3T3-L1 cells blunts preadipocyte PPARγ expression and diminishes the ability of these cells to differentiate. Furthermore, both brown and white adipocyte differentiation is strikingly impaired in Zfp423-deficient mouse embryos. Zfp423 regulates PPARγ expression, in part, through amplification of the BMP signaling pathway, an effect dependent on the SMAD binding capacity of Zfp423. This study identifies Zfp423 as a transcriptional regulator of preadipocyte determination.
Keywords: adipogenesis, PPARγ, preadipocytes, brown adipose tissue, white adipose tissue, cell fate determination, BMP signaling, SMAD signaling, Zfp423
There has been great progress in unraveling the transcriptional pathways controlling adipocyte differentiation, the process in which committed preadipose fibroblasts undergo a morphological and biochemical transition into mature adipocytes in response to appropriate cues 1. However, little is known about how the determined preadipose state is controlled, particularly the transcriptional mechanisms that distinguish preadipocytes from non-adipogenic fibroblasts. There have been significant barriers to the discovery of genes that control preadipocyte determination. First, preadipose commitment appears to be a quantitative trait, thus genes controlling this cellular state may be controlled quantitatively rather than qualitatively 2. Second, while molecular markers to allow for the enrichment of a preadipocyte population from adipose tissue now exist 3, 4, markers specific for the purification of preadipose fibroblasts and non-adipogenic fibroblasts from adipose tissue are lacking. Additionally, immortalized preadipose cell lines have disparate histories and are notoriously aneuploid, potentially confounding the appropriate designation of cells as non-preadipose or preadipose 5, 6.
In order to identify the molecular mechanisms controlling preadipocyte commitment, we employed the strategy summarized in Supplementary Figure 1. First, we derived 42 new clonal sublines of Swiss 3T3 fibroblasts, and evaluated their adipogenic potential. Oil-Red O staining of accumulated lipids following the induction of differentiation revealed six highly adipogenic preadipose cell lines, as well as six cell lines with little adipogenic potential (<10% of cells) (Fig. 1a,b). Next, we devised a novel real-time PCR platform to quantitatively determine the expression of all or nearly all transcription regulators (~1800) encoded by the mouse genome. This new transcription-factor primer array, which we refer to as Quanttrx, is based on the work by Gray and colleagues, which previously described the derivation of in situ hybridization probes targeting the mRNAs encoding murine transcriptional components 7. We improved this methodology by building a high-throughput platform for quantitative real-time PCR, utilizing newly designed real-time PCR primers that amplify a more comprehensive set of all known or predicted murine transcriptional components (See Methods and Supplementary Figure 1).
Figure 1. The C2H2 zinc finger protein Zfp423 is enriched in preadipocytes.

Oil-red-O (ORO) staining of preadipocyte cell lines (a) and non-adipogenic cells lines (b) at six days following the induction of adipogenesis with DMI. (c) Zfp423 expression in sub-confluent Swiss 3T3 subclones and existing preadipocyte and fibroblast cell lines. (n=3 replicates per cell line). In this, and other figures, bars represent mean ± standard deviation from the mean. (d) Western Blot of endogenous Zfp423 protein levels in fibroblast cell lines grown under non-differentiating conditions.
We performed Quanttrx assays on proliferating adipogenic and non-adipogenic fibroblasts in culture, prior to the growth arrest period that precedes the induction of adipose differentiation (Supplementary Fig. 1). This was to ensure that these data would not include genes whose expression was primarily linked to the differentiation process per se. We identified five genes whose expression was enriched in non-adipogenic fibroblasts, and four genes whose expression was enriched in preadipose fibroblasts (Supplementary Fig. 2, Data Set 1). One of the genes enriched in preadipocytes was PPARγ, a dominant and essential regulator of both brown and white adipocyte differentiation 8-10 (Supplementary Fig. 2b). Analysis of PPARγ expression levels in these cell lines with isoform-specific primers indicated that PPARγ2 mRNA levels, but not PPARγ1 levels, were more abundant in preadipose fibroblasts than in non-adipogenic fibroblasts (Supplementary Fig. 3). However, the expression of well-characterized differentiation-dependent regulators of PPARγ expression did not correlate with the adipogenic potential of these cell lines (Supplementary Fig. 4). As anticipated and discussed above, the correlation between these genes and the adipogenic potential of the fibroblasts was not perfect. For most of these genes the adipogenic clone Swiss 27 resembled that of a non-adipogenic fibroblast, while the gene expression pattern of the non-adipogenic clone Swiss 30 more closely resembled the pattern found in preadipose cells (Supplementary Fig. 2a,b).
We focused on components enriched in a majority of preadipocytes, which might serve as initiators of preadipocyte commitment. Ectopic expression of PLZF or Foxa2 in non-adipogenic fibroblasts did not render cells competent to undergo adipogenesis (data not shown). On the contrary, gain of function experiments described below led us to study extensively the large C2H2 zinc-finger protein, Zfp423, which was initially isolated from a yeast two-hybrid screen for interacting partners of the EBF family of transcription factors in the olfactory epithelium 11. With the exception of the two consistently outlying clones (Swiss 27 and Swiss 30), levels of Zfp423 mRNA were higher in all preadipose cell lines when compared to the non-adipogenic fibroblasts (Fig. 1c). 3T3-L1 preadipocytes exhibit the greatest adipogenic potential in the response to the standard hormonal cocktail and have by far the highest mRNA and protein levels of Zfp423 (Fig. 1c,d). Conversely, we did not detect either Zfp423 mRNA or protein in NIH 3T3 fibroblasts, cells with the least potential for adipogenesis under our experimental conditions (Fig. 1c,d). In most preadipocyte cell lines examined, the expression of Zfp423 is not significantly regulated during their differentiation into fat cells; therefore, the expression of Zfp423 is not necessarily linked to the differentiation step of adipogenesis (Supplementary Fig. 5). In mice, Zfp423 mRNA levels were enriched in white and brown adipose tissue, as well as in the brain where Zfp423 exhibits numerous functions (Supplementary Fig. 6). Analysis of fractionated adipose tissue indicates that Zfp423 mRNA is abundantly expressed in both the stromal-vascular fraction (SVF) of the tissues that contain committed preadipocytes, and in the mature adipocytes (Supplementary Fig.7). On the contrary, we did not detect appreciable Zfp423 mRNA expression in primary myoblasts, or C2C12 myoblasts (data not shown), suggesting that Zfp423 was not a general regulator of precursor specification in mesenchymal lineages.
To evaluate the role of Zfp423 in preadipocyte commitment, we first ectopically expressed Zfp423 in non-adipogenic NIH 3T3 fibroblasts; these cells do not express detectable Zfp423 and have very little potential for adipogenesis. Retroviral expression of Zfp423 resulted in a 12-fold elevation of PPARγ2 mRNA levels in the undifferentiated state (Fig. 2a), while the expression of PPARγ1 remained undetectable (Supplementary Fig. 3). The 12-fold elevation of PPARγ2 expression brought the total mRNA levels of PPARγ2 to the levels observed in most preadipocyte cell lines (Supplementary Fig 3). On the other hand, the expression of most known upstream regulators of PPARγ expression was not significantly altered (Fig. 2a). In response to pro-differentiative cell culture conditions, Zfp423-expressing cells underwent robust adipocyte differentiation, giving rise to lipid accumulating cells that express molecular markers of differentiated adipocytes (Fig. 2b,c). These data indicate that Zfp423 can regulate PPARγ gene expression in undifferentiated fibroblasts and can render these cells competent to undergo adipocyte differentiation.
Figure 2. Zfp423 regulates preadipocyte PPARγ gene expression and adipocyte differentiation in vitro.
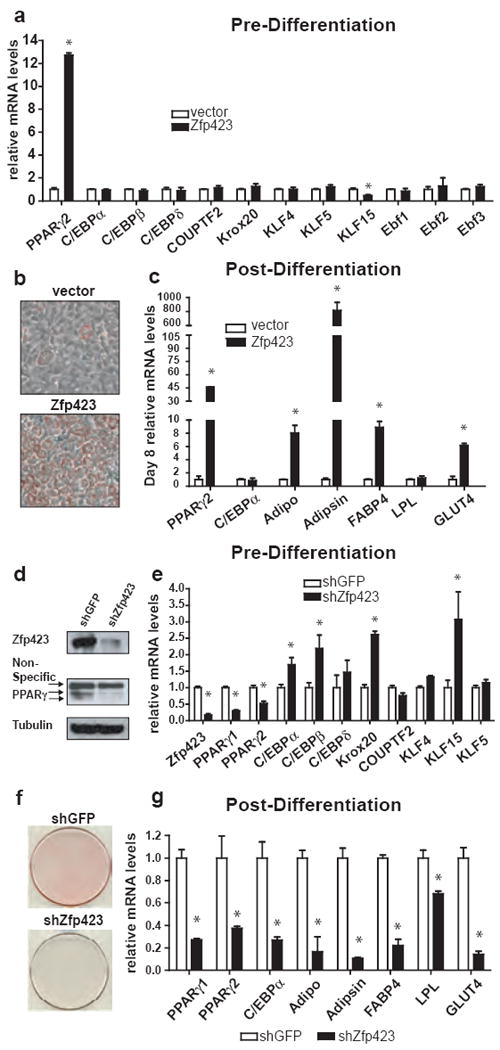
(a) Transcription factor gene expression in undifferentiated NIH 3T3 fibroblasts ectopically expressing Zfp423. (b) ORO staining of control and Zfp423-expressing cells 8 days following the induction of adipocyte differentiation. (c) Expression of adipocyte selective genes in the differentiated cultures shown in (b). (d) Western blot of Zfp423 and PPARγ protein levels in 3T3-L1 preadipocytes expressing shGFP (control) or shRNA against Zfp423 (shZfp423). Tubulin protein levels serve as a loading control. (e) Transcription factor gene expression in undifferentiated preadipocytes expressing shGFP or shZfp423. (f) ORO staining six days after the induction of adipocyte differentiation in 3T3-L1 cells expressing shGFP or shZfp423. (g) Expression of adipocyte selective mRNAs in cultures shown in (f). * denotes p<0.05 in Student’s T-test. n=3 replicates
We investigated the requirement of Zfp423 in preadipocytes by utilizing specific short-hairpin RNA (shRNA) sequences targeting Zfp423. Retroviral delivery of Zfp423 shRNA into 3T3-L1 preadipocytes greatly reduced the expression of Zfp423 in undifferentiated cells (Fig. 2d,e). Likewise, both PPARγ1 and PPARγ2 expression were significantly lower in Zfp423 knock-down preadipocytes, when compared to control cells (Fig 2d,e). In contrast, known upstream activators of PPARγ transcription were either unaltered or even upregulated in Zfp423 deficient preadipocytes (Fig. 2e). In response to pro-differentiative cell culture conditions, Zfp423-deficient 3T3-L1 cells failed to undergo significant adipocyte differentiation, as indicated by the presence of fewer lipid accumulating cells (Fig. 2f) and lower expression of adipocyte-selective genes (Fig. 2g). As Zfp423 is also abundantly expressed in the SVF of brown adipose tissue (BAT) (Supplementary Fig. 7), we knocked down Zfp423 expression in immortalized BAT precursor cells and found that PPARγ expression in these cells was also Zfp423 dependent (Supplementary Fig. 8). In addition, PRDM16, a BAT-selective regulator of PPARγ expression and activity 12, 13, was modestly but significantly decreased in Zfp423- deficient brown preadipocytes. (Supplementary Fig. 8).
As a complementary approach, we evaluated the ability of primary embryonic fibroblasts (MEFs) from Zfp423-deficient mice to undergo adipogenesis 14. Cultures of wild-type primary MEFs contained undetectable basal levels of PPARγ mRNA (data not shown); therefore, the effect of Zfp423 deficiency on basal PPARγ gene expression could not be explored in this model. However, differentiated cultures of primary MEFs isolated from embryonic day (E) 13.5 Zfp423 knockout embryos had significantly fewer lipid-laden adipocytes when compared to differentiated cultures from wild type controls (Supplementary Fig. 9a). Expression of adipocyte genes in these cultures was also reduced (Supplementary Fig. 9b). In addition, those adipocytes that did arise in the cultures of Zfp423 knockout MEFs appeared smaller than those found in wild type cultures, with less overall lipid accumulation (Supplementary Fig. 9a). The defect in adipogenesis likely lies upstream or at the level of PPARγ activation since ectopic expression of PPARγ in Zfp423 knockout MEFs stimulates adipogenesis (data not shown). Taken together, these results clearly indicates that Zfp423 functions as a critical regulator of PPARγ expression in the preadipocyte state, and is required for normal morphological and molecular differentiation of adipocytes.
Next, we evaluated the requirement of Zfp423 in the regulation of the broader preadipocyte gene expression program, particularly genes related to the committed state. To determine an expression signature that defines a committed 3T3 preadipocyte, we performed global expression profiling of several adipogenic and non-adipogenic cell lines using gene arrays. Unsupervised sample clustering indicated that all four adipogenic clones examined exhibited a closer gene expression pattern to one another than to non-adipogenic cells (Supplementary Fig. 10). From the expression data we derived a gene expression signature consisting of 48 genes that characterizes the determined preadipose state (See Methods, Supplementary Table 1).
To determine if Zfp423 regulates the molecular identity of committed preadipocytes, we analyzed the expression of all 48 genes plus the additional transcriptional components identified by the Quanttrx assays in 3T3-L1 preadipocytes expressing control or the Zfp423 shRNA. Of the 8 transcriptional components assayed in Zfp423 deficient preadipocytes, 3 genes (PPARγ2, Pou3f3, and Hmx1) were significantly altered in a direction consistent with a non-adipogenic fibroblast phenotype (Supplementary Fig. 11a). In addition, while their observed differences were not statistically significant, Jazf1 and Satb1 also appeared to be upreguated in shZfp423-expressing preadipocytes (Supplementary Fig. 11a). This suggests the presence of a Zfp423-dependent transcriptional hierarchy in preadipose fibroblasts. Strikingly, of the 32 genes whose expression is normally enriched in non-adipogenic fibroblasts, 19 were Zfp423 dependent (Supplementary Fig. 11b). Of the 13 genes enriched in preadipocytes, expression of 10 of these genes were Zfp423-dependent, with 7 exhibiting lower preadipocyte gene expression in the absence of Zfp423 (Supplementary Fig 11c). Taken together, these data demonstrate that a major portion of the preadipose gene expression program is dependent on Zfp423.
Numerous studies have implicated the BMP/SMAD signaling cascade in the early events of adipogenesis, particularly in the induction of PPARγ 15-19. Interestingly, Zfp423 was previously identified as a BMP-dependent transcriptional coactivator of SMAD proteins 20. Thus, we first asked whether the SMAD interaction domain of Zfp423 is required for its pro-adipogenic actions under the standard adipocyte differentiation conditions. In addition, we asked whether Zfp423, via the SMAD interaction domain, mediates the pro-adipogenic response to BMPs. We compared cells expressing full length Zfp423 to cells expressing a mutant of Zfp423 that lacks the well-characterized SMAD binding domain (Zfp423 ΔSBD) 20 (Fig. 3a). When stable NIH 3T3 cell lines expressing either full-length Zfp423 or Zfp423 ΔSBD were studied (Fig. 3b), we observed that the basal expression of PPARγ2 in undifferentiated fibroblasts was similarly induced by both proteins (Fig. 3c). Furthermore, both the full-length protein and Zfp423 mutant were equally capable of inducing adipogenesis, as shown by the comparable expression of adipocyte-selective mRNAs in differentiated cultures and similar morphological appearance of lipid-accumulating fat cells (Fig. 3d). Thus, the SMAD binding domain of Zfp423 is not required for the adipogenic activity of Zfp423 under these typical cell culture conditions.
Figure 3. Zfp423 amplifies the pro-adipogenic actions of BMP proteins through its SMAD protein interaction domain.
(a) Schematic illustrating the protein domains and zinc-finger (ZF) motifs of full length Zfp423 and Zfp423 lacking the SMAD binding domain (Zfp423 ΔSBD). ZF 14-20 serves as the well-characterized SMAD interaction domain. (b) Western blot of ectopic full length Zfp423 or Zfp423 ΔSBD protein expression in NIH 3T3 cells. Tubulin protein levels serve as a loading control. (c) PPARγ2 mRNA levels in undifferentiated cells expressing Zfp423 or Zfp423 ΔSBD. (d) Expression of adipocyte selective mRNAs in cultures following differentiation with DMI and Rosiglitazone and ORO staining of differentiated cultures. (e) PPARγ2 mRNA levels in undifferentiated NIH 3T3 cells expressing a control vector or full length Zfp423 following 48 hours of incubation with increasing doses of BMP4. (f) PPARγ2 gene expression in undifferentiated, BMP4 treated cells expressing control, full length Zfp423, or Zfp423 ΔSBD. (g) Expression of adipocyte selective mRNAs in cultures following BMP4-induced differentiation, and oil-red O staining of lipid accumulation in differentiated cultures. * denotes p<0.05 in Student’s T-test. n=3 replicates.
We also treated proliferating control or Zfp423-expressing cells with BMP4 and measured PPARγ2 mRNA levels prior to the induction of differentiation (Fig. 3e). Relatively high concentrations of BMP4 (>25 ng/mL) were required to induce PPARγ2 expression in control fibroblasts (Fig. 3e). In sharp contrast, NIH 3T3 cells ectopically expressing wild-type Zfp423 showed a significant leftward shift of the BMP dose-response curve, with Zfp423-expressing cells much more sensitive to low concentrations of BMP4 (>100-fold induction at 12 ng/mL) (Fig. 3e). To determine if the SMAD interaction domain of Zfp423 is required for this effect of the BMP proteins, we compared PPARγ mRNA levels in BMP-treated control, Zfp423-expressing, and Zfp423 ΔSBD expressing cell lines. Indeed, Zfp423 mutant-expressing cells failed to respond to the BMP signal to further enhance PPARγ gene expression (Fig. 3f). In addition, while cells expressing full-length Zfp423 were capable of undergoing BMP-driven adipogenesis, cells expressing Zfp423 ΔSBD or the empty viral vector did not undergo significant adipocyte differentiation when stimulated with low doses of BMP4 (Fig. 3g). Similar results were observed when we treated these same cell lines with BMP2 (Supplementary Figure 12). These results indicate that the SMAD-binding domain of Zfp423 is not necessary for its activity in preadipocyte commitment under basal culture conditions, but is absolutely required for Zfp423 modulation of adipogenic activity induced by BMPs.
To determine if Zfp423 regulates adipose cell development in vivo, we examined the formation of adipocytes in Zfp423-deficient mice 14. Zfp423 knock-out mice exhibit large defects in the development of the midline structures of the brain 14, 21 and most of these Zfp423-/- mice died within 24 hours of birth 14, 21, 22. While the few recovered postnatal Zfp423-/- mice exhibited noticeable defects in adipose mass (data not shown), we could not exclude the possibility that alterations in energy balance, secondary to CNS abnormalities, leads to reduced energy storage in adipose tissues. Thus, we focused our analysis here on E18.5 of mouse development, where Zfp423-/- embryos are found in normal mendelian ratio and are grossly indistinguishable from littermate controls.
Transverse sections of the thoracic region of E18.5 mice revealed abundant interscapular BAT in all Zfp423+/+ mice examined (Fig. 4a). In contrast, the interscapular BAT in all Zfp423-/- embryos examined was much smaller and appeared grossly abnormal when compared to the BAT found in wild-type mice (Fig. 4b,c). Furthermore, remnant BAT from mutant mice contained large amounts of interspersed connective tissue when compared to controls (Fig. 4d,e). FABP4 antibody staining indicated that the while the remaining BAT in Zfp423 knockout mice expressed FABP4 (Supplementary Fig. 13a-c), UCP1 expression was nearly absent in most of the remnant BAT of knockout embryos, suggesting poor differentiation of the remaining adipose cells (Supplementary Fig. 13d-h). Within the mesodermal lineages, these histological abnormalities appeared specific to the adipose tissue, as muscle and connective tissues surrounding the BAT in the Zfp423-/- embryos appeared grossly normal (data not shown).
Figure 4. Impaired brown and white adipocyte differentiation in Zfp423 deficient embryos.

(a-c) Hematoxylin and eosin staining of transverse sections of the interscapular region of E18.5 Zfp423+/+ (a) and Zfp423-/- (b,c) embryos. (d,e) Higher magnification images of the interscapular BAT shown in (a,b). FABP4 staining of primitive white adipocytes in the subcutaneous region of E18.5 Zfp423+/+ mice (f) and E18.5 Zfp423-/- (g) mice (40x magnification) (h) Quantitation of FABP4+; lipid+; UCP1- cells in the subcutaneous regions of wild-type and Zfp423 knockout mice.
White fat cells first appear in the latest stages of embryogenesis, with the full expansion of white adipose tissue occurring postnatally, when the need for energy storage increases. We localized developing white adipocytes by FABP4 antibody staining of the subcutaneous regions in E18.5 embryos. We easily identified numerous FABP4+, lipid-laden cells in Zfp423+/+ mice at this stage. These adipocytes were indeed white adipocytes, as UCP1+ cells were not observed anywhere in the embryonic subcutaneous regions examined (data not shown). However, in the subcutaneous layer of littermate Zfp423-/- mice there was a great reduction in lipid-containing FABP4+ cells (Fig. 4f,g). On average, we identified 121 + 6 lipid-containing FABP4+ cells per mm2 of subcutaneous tissue in E18.5 Zfp423+/+ mice (~5 cells per 40x field). Zfp423-/- mice showed only an average of 32 ± 7 of lipid containing FABP4+ cells per mm2 of equivalent sections (<1 cell per 40x field) (Fig. 4h). These data reveal a genetic requirement for Zfp423 in the initial formation of both brown and white adipocytes in vivo.
Taken together, these data identify Zfp423 as a regulator of preadipose cell determination and suggest a model in which the regulation of the preadipocyte levels of PPARγ by Zfp423 (and likely other, unknown regulators) represents a crucial determinant of preadipocyte commitment (Supplementary Fig. 14). The differentiation signals, such as the standard hormonal cocktail used in our in vitro differentiation assays, function in large part to further stimulate the expression and activity of PPARγ in preadipocytes until a critical threshold to initiate adipocyte differentiation is achieved. While sensitizing cells to the BMP signals represents one mechanism by which Zfp423 regulates PPARγ expression, future experiments must delineate the additional mechanisms by which Zfp423 controls PPARγ expression, as well as the regulatory pathways acting upstream of, and parallel to, Zfp423 in preadipocyte determination.
Methods Summary
For adipocyte differentiation assays, confluent cultures of 3T3-L1 and Swiss 3T3 subclones were exposed to induction medium containing dexamethasone (1 μM), insulin (5 μg/ml), and isobutylmethylxanthine (0.5 mM) (DMI) and 10% FBS. 48 hours after induction, cells were maintained in DMEM containing insulin (5 μg/ml) and 10% FBS until ready for harvest. For NIH 3T3 cells, differentiation medium contained DMI, 6% FBS, and 1 μM rosiglitazone. After induction, cells were maintained in medium containing 6% FBS, insulin, and 1 μM rosiglitazone until ready for harvest. For BMP-induced adipogenesis of NIH 3T3 cell lines, cells were grown to confluence, and maintained at post-confluence, in medium containing 6 ng/mL BMP4 or 25 ng/mL BMP2 along with insulin and rosiglitazone. The derivation and genotyping of Zfp423 knockout mice has been previously described 14, 23. All animal experiments were performed according to procedures approved by the Dana-Farber Cancer Institute’s and Beth Isreal Deconess Medical Center’s Institutional Animal Care and Use Committee.
Supplementary Material
Acknowledgments
The authors are grateful to Drs. Sandra Kleiner and Shingo Kajimura for critical reading of the manuscript and to all members of the Spiegelman laboratory for useful discussions. We thank Dr. Bridget Wagner for technical assistance in utilizing robotic liquid handlers, Dr. Brian Seed’s laboratory for help with high-through query of Primer bank, and Mr. John Brestelli for performing high-throughput queries of the Primer 3 program. We are also grateful to Dr. David Bernlohr for the FABP4 antiserum. R.K.G is supported by the Ruth Kirstein NRSA (F32 DK079507-01), Z.A. is supported by K08 HL79172-01 (NHLBI) and the Smith Family Foundation Grant, P.S. is supported by NIH DK081605, and the research described in this study was supported by NIH DK31405 to B.M.S. and NIDCD R01DC008295 to R.R.R.
Footnotes
Author Contributions R.K.G and B.M.S. conceived and designed the experiments. R.K.G., Z.A., P.S., R.J.M., L.Y, and H.M.C., performed experiments. All authors analyzed the data. Y.A.R., H.K., and R.R.R. provided reagents and samples, and R.K.G and B.M.S. wrote the manuscript.
Complete microarray data is available at Gene Expression Omnibus (Accession # GSE19732) http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE19732.
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions.
Conflicting interests statement. The authors declare that they have no competing financial interests.
References
- 1.Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–73. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green H, Kehinde O. An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell. 1975;5:19–27. doi: 10.1016/0092-8674(75)90087-2. [DOI] [PubMed] [Google Scholar]
- 3.Rodeheffer MS, Birsoy K, Friedman JM. Identification of white adipocyte progenitor cells in vivo. Cell. 2008;135:240–9. doi: 10.1016/j.cell.2008.09.036. [DOI] [PubMed] [Google Scholar]
- 4.Tang W, et al. White fat progenitor cells reside in the adipose vasculature. Science. 2008;322:583–6. doi: 10.1126/science.1156232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green H, Kehinde O. Spontaneous heritable changes leading to increased adipose conversion in 3T3 cells. Cell. 1976;7:105–13. doi: 10.1016/0092-8674(76)90260-9. [DOI] [PubMed] [Google Scholar]
- 6.Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gray PA, et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–7. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- 8.Barak Y, et al. PPAR gamma is required for placental, cardiac, and adipose tissue development. Mol Cell. 1999;4:585–95. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- 9.Kubota N, et al. PPAR gamma mediates high-fat diet-induced adipocyte hypertrophy and insulin resistance. Mol Cell. 1999;4:597–609. doi: 10.1016/s1097-2765(00)80210-5. [DOI] [PubMed] [Google Scholar]
- 10.Rosen ED, et al. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol Cell. 1999;4:611–7. doi: 10.1016/s1097-2765(00)80211-7. [DOI] [PubMed] [Google Scholar]
- 11.Tsai RY, Reed RR. Cloning and functional characterization of Roaz, a zinc finger protein that interacts with O/E-1 to regulate gene expression: implications for olfactory neuronal development. J Neurosci. 1997;17:4159–69. doi: 10.1523/JNEUROSCI.17-11-04159.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kajimura S, et al. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;460:1154–8. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seale P, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–7. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng LE, Zhang J, Reed RR. The transcription factor Zfp423/OAZ is required for cerebellar development and CNS midline patterning. Dev Biol. 2007;307:43–52. doi: 10.1016/j.ydbio.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bowers RR, Kim JW, Otto TC, Lane MD. Stable stem cell commitment to the adipocyte lineage by inhibition of DNA methylation: role of the BMP-4 gene. Proc Natl Acad Sci U S A. 2006;103:13022–7. doi: 10.1073/pnas.0605789103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hata K, et al. Differential roles of Smad1 and p38 kinase in regulation of peroxisome proliferator-activating receptor gamma during bone morphogenetic protein 2-induced adipogenesis. Mol Biol Cell. 2003;14:545–55. doi: 10.1091/mbc.E02-06-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin W, et al. Schnurri-2 controls BMP-dependent adipogenesis via interaction with Smad proteins. Dev Cell. 2006;10:461–71. doi: 10.1016/j.devcel.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 18.Tang QQ, Otto TC, Lane MD. Commitment of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. Proc Natl Acad Sci U S A. 2004;101:9607–11. doi: 10.1073/pnas.0403100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tseng YH, et al. New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature. 2008;454:1000–4. doi: 10.1038/nature07221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hata A, et al. OAZ uses distinct DNA- and protein-binding zinc fingers in separate BMP-Smad and Olf signaling pathways. Cell. 2000;100:229–40. doi: 10.1016/s0092-8674(00)81561-5. [DOI] [PubMed] [Google Scholar]
- 21.Warming S, Rachel RA, Jenkins NA, Copeland NG. Zfp423 is required for normal cerebellar development. Mol Cell Biol. 2006;26:6913–22. doi: 10.1128/MCB.02255-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alcaraz WA, et al. Zfp423 controls proliferation and differentiation of neural precursors in cerebellar vermis formation. Proc Natl Acad Sci U S A. 2006;103:19424–9. doi: 10.1073/pnas.0609184103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng LE, Reed RR. Zfp423/OAZ participates in a developmental switch during olfactory neurogenesis. Neuron. 2007;54:547–57. doi: 10.1016/j.neuron.2007.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



