Abstract
Extensive changes in plant transcriptome and metabolome have been observed by numerous research groups after transferring plants from optimal conditions to sulfur (S) deficiency. Despite intensive studies and recent important achievements, like identification of SLIM1/EIL3 as a major transcriptional regulator of the response to S-deficiency, many questions concerning other elements of the regulatory network remain unanswered. Investigations of genes with expression regulated by S-deficiency stress encoding proteins of unknown function might help to clarify these problems. This study is focused on the UP9C gene and the UP9-like family in tobacco. Homologs of these genes exist in other plant species, including a family of four genes of unknown function in Arabidopsis thaliana (LSU1-4), of which two were reported as strongly induced by S-deficit and to a lesser extent by salt stress and nitrate limitation. Conservation of the predicted structural features, such as coiled coil region or nuclear localization signal, suggests that these proteins might have important functions possibly mediated by interactions with other proteins. Analysis of transgenic tobacco plants with silenced expression of UP9-like genes strongly argues for their significant role in regulation of plant response to S-deficit. Although our study shows that the UP9-like proteins are important components of such response and they might be also required during other stresses, their molecular functions remain a mystery.
Keywords: Sulfate assimilation, sulfur deficit, tobacco, Arabidopsis, transcription regulators, gene expression
INTRODUCTION
Several factors, including decreased atmospheric pollution with industrial gases, reduction of sulfur (S) content in fuel, and use of fertilizers rich in nitrogen (N) and phosphorus (P) but free from S, resulted in S becoming in many geographical regions the major limiting factor in plant production (Schnug et al., 2004). The importance of a sufficient supply of S for appropriate development and performance of various crops (Haneklaus et al., 2005; Scherer, 2001) and for food quality (Muttucumaru et al., 2006) has gradually become evident.
Regulation of gene expression in response to S source depletion is an important aspect of S metabolism in plants. For several plant species, including maize (Petrucco et al., 1996), spinach (Prosser et al., 2001), tobacco (Migge et al., 2000), and duckweed (Kopriva et al., 2002), research in this field concentrated on a limited number of genes with known metabolic or enzymatic functions. Such an approach was helpful in demonstrating the linkage between the S, P, and N nutrition, but was not sufficient to identify the regulatory circuits involved in plant response to S nutrition status. Most of the high-throughput transcriptome analyses were performed in Arabidopsis thaliana using micro- and macroarrays. The authors noticed and discussed a cross-talk between various pathways induced by S-deficiency, including auxin and jasmonate biosynthesis pathways (Hirai et al., 2003; Nikiforova et al., 2003), activation of defense response through oxidative stress (Maruyama-Nakashita et al., 2003), and involvement of O-acetyl-L-serine (OAS) as a general regulator for global gene expression under sulfur-nutrition stress (Hirai et al., 2003). A transcriptional regulator of plant sulfur response and metabolism, SLIM1/EIL3 that belongs to the EIL family, has been identified in A. thaliana (Maruyama-Nakashita et al., 2006). Mutants with impaired function of SLIM1 not only have pleiotropic effects on transcription of many genes regulated by S-deficit, but they also have changed level of relevant primary and secondary metabolites, including cysteine, glutathione, OAS, methionine, and glucosinolates.
It is generally accepted that adjusting plant metabolism to the conditions of S-deficit is a complex process involving a network of factors, such as transcriptional and posttranscriptional regulators, miRNA, and signaling molecules (for a recent review, see Lewandowska and Sirko (2008)). The list of known factors influencing response to S-deficit is still too incomplete to explain all aspects of plant response to it. There must be several other (not yet identified) transcriptional or posttranscriptional regulators involved in this process. We could expect to find such regulators among proteins with known function but previously not associated with response to S-deficit, as well as among proteins with unknown functions. For example, Kasajima and co-workers (2007) reported that the expression of some S-deficiency-responsive genes, such as sulfate transporter (SULTR2;2) and adenosine-5′-phosphosulfate reductase 1 (APR1) was up-regulated in the normal conditions in the A. thaliana mutants with impaired function of BIG gene. The detailed role of BIG is unknown; however, the protein was previously recognized as being involved in such processes as light-signaling pathway, auxin transport, cytokinin, and ethylene sensitivity, giberellin signaling (Kanyuka et al., 2003; Lopez-Bucio et al., 2005; Sponsel et al., 1997) and no reports about its role in S-deficiency response had been published before. The second example might be a cyclophilin CYP20-3 (also known as ‘ROC4’) and its recently proposed role in regulatory mechanism linking cysteine biosynthesis to light, redox, and stress. The interaction of CYP20-3 with the chloroplast izoform of serine acetyltransferase (SAT) appeared to be required for a fully functional enzyme and for normal accumulation of thiol compounds in response to stress (Dominguez-Solis et al., 2008).
On the other hand, genes encoding proteins of unknown function, although reported in various studies as strongly regulated during S-deficit at the transcription level in Arabidopsis and other plant species, have not been further investigated. Several such genes were found among Nicotiana tabacum genes previously identified in our laboratory as regulated during short-term S-deficit (Wawrzynska et al., 2005). The UP9A and UP9C genes, which belong to this group, are up-regulated in all plant organs in response to S-deficit (Lewandowska et al., 2005). The major aims of this study were (1) preliminary characterization of UP9C protein, its sub-cellular localization in plants, and identification of its protein partners, and (2) examination of the effects of UP9C silencing in transgenic tobacco plants grown in optimal and S-deficient conditions.
RESULTS
A UP9-Like Family Exists in Tobacco and in Other Plant Species
The UP9 gene has been identified in our laboratory as strongly up-regulated at the level of transcription during early stages of S-starvation (Wawrzynska et al., 2005). Further studies revealed that, in fact, two types of sequences, UP9A and UP9C, corresponded to a gene previously denoted by us as UP9 (Lewandowska et al., 2005). After database searches using a coding region of UP9C, it was possible to identify multiple tobacco UP9-like ESTs that could be grouped into six clusters according to the sequence of their coding regions. Organization of these genes within the tobacco genome and their expression is obscure, in contrast to A. thaliana, where it is clear that four UP9-like genes are located in pairs of the direct neighbors on two chromosomes (III and V). The A. thaliana genes were recently named LSU1–LSU4 (Low Sulfur Upregulated) and, according to the GENEVESTIGATOR database (Zimmermann et al., 2004), at least two of them, LSU1 (At3g49580) and LSU2 (At5g24660), are strongly expressed under S-deprivation (4.3- and 13.6-fold up-regulation, respectively). Families of UP9-like genes exist also in other plant species, such as rice, poplar, tomato, and potato.
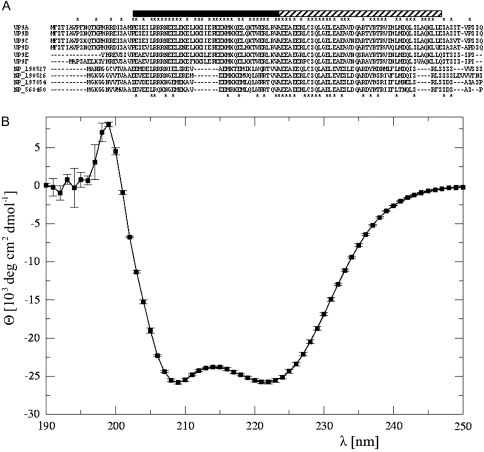
Multiple sequence alignment of the UP9-like proteins from A. thaliana (four members) and tobacco (six members) are shown in Figure 1A. Computer analyses of the secondary structure of UP9-like proteins using GOR4 program predicted the extensive regions being in a form of α-helix. The predicted structure was experimentally verified by us by monitoring of the circular dichroism (CD) spectra of the recombinant UP9C produced in Escherichia coli cells (Figure 1B). The observed curve was characterized by two well resolved minima at 209 and 220 nm with molar ellipticity of –25 900 and –25 800 (deg cm2 dmol−1), respectively. Such form of CD spectra indicated that the UP9C protein is almost fully α-helical (Yang et al., 1986). The de-convolution procedure, performed by the K2d program (Andrade et al., 1993), confirmed that the dominant contributions to the CD spectrum came from α-helical (∼0.82) and random coil (∼0.17) conformations. The contributions from extended β secondary structures were extremely small.
Figure 1.
Amino Acid Sequence Alignment of UP9-Like Proteins from Tobacco and Arabidopsis (A) and Circular Dichroism Spectrum of Recombinant UP9C (B).
(A) All tobacco UP9-like ESTs identified after a database similarity search with a coding region of UP9C were grouped into six clusters (arbitrary denoted UP9A-F) according to the sequence of their coding regions: UP9A (GB# DQ444223, DV999485, FG623584, BP535499), UP9B (GB# BP533592, FG638291, FG637038), UP9C (GB# AY547446, FG641107, EB445189), UP9D (GB# FG636997), UP9E (GB# BP531586, EB430507), UP9F (GB# BY31337). The alignment of six open reading frames deduced from each of the tobacco EST group and of four homologs from A. thaliana (NP_190527, NP_190526, NP_197854, and NP_56450) was performed using the MAFFT. The asterisks above the sequences indicate residues identical in six tobacco proteins, while the asterisks below the sequences indicate the residues identical in all aligned proteins. The stripped bar above alignment shows the alpha-helical region predicted by GOR4 and its filled part indicates a two-stranded coiled coil detected by MULTICOIL.
(B) The mean values of three independent measurements with SD-indicated molar ellipticity (deg cm2 dmol−1) of 64 μM UP9C protein in a light spectrum between 190 and 250 nm are shown.
The MULTICOIL program assigned the coiled coil region between residues 24 and 63 of UP9C as two-stranded, which suggested that this region might be involved in dimerization of the protein. In fact, the UP9C–UP9C homodimers formation was observed in the yeast two-hybrid experiment (Lewandowska et al., 2005). Moreover, formation of mixed heterodimers consisting of UP9C from N. tabacum and the product of LSU1 (At3g49580) gene from A. thaliana was also observed in the yeast two-hybrid system (not shown).
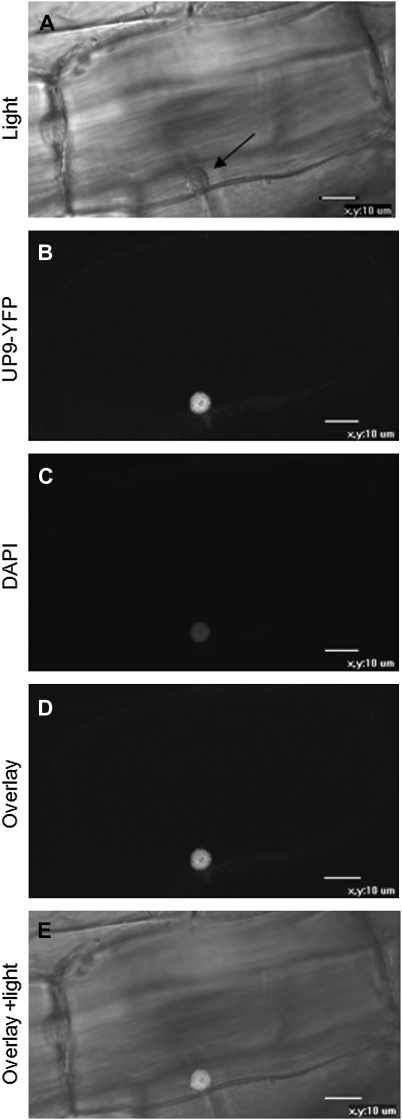
The potential bipartite nuclear localization signal was found using the MOTIFSCAN program in the region of residues 16–33 of UP9C. Accordingly, the PSORT program predicted nuclear or cytosol-nuclear localization of this protein. The predicted sub-cellular localization of UP9C protein was experimentally verified by us in transgenic tobacco seedlings, where the nuclear location of the UP9C–EYFP fusion protein was observed (Figure 2). It is necessary to mention that the UP9C–EYFP protein was found in the nucleus independently of the S-nutrition status of the plant material (not shown).
Figure 2.
Sub-Cellular Localization of the UP9C–EYFP Fusion Protein in the Root of 3-Week-Old Transgenic Tobacco.
(A) Light microscopy image showing the cell nucleus marked by an arrow.
(B) Fluorescence microscopy image showing the nuclear localization of the UP9C–EYFP fusion protein.
(C) Fluorescence microscopy image showing the nucleus stained with DAPI.
(D) Overlay of (B) and (C).
(E) Overlay of (A), (B), and (C). Scale bar = 10 μm.
Identification of Tobacco Proteins Interacting with UP9C
No clue about the potential role of UP9-like proteins could be found in the literature. Therefore, in search of a function of the UP9-like proteins, we decided to identify the protein partners of UP9C using the yeast two-hybrid system and the respective cDNA library prepared from N. tabacum grown for 2 d in S-deficient conditions. The screen resulted in identification of 17 independent clones submitted (GB#: GU066878–GU066894). The relative large number of clones and a huge variety of identified proteins were not surprising because of the helical structure of UP9C; however, it is necessary to verify whether all detected interactions take place in plants. Most of the identified clones encode proteins of unknown function but some are recognized as coding for described proteins, such as transcription factors AP2-like (GB# GU066884), jasmonate ZIM-domain protein 1 (JAZ1) involved in biosynthesis of jasmonic acid (GB# GU066894), and ACC oxidase involved in ethylene biosynthesis (GB# GU066878 and GU066879). A proved nuclear location of UP9C is in an agreement with identification of numerous clones encoding proteins with a possible localization in the nucleus, such as GB# GU066880, GU066884, GU066888, and GU066894. On the other hand, ability of UP9C to interact with proteins of a possible non-nuclear location might suggest a probable function of UP9C also in the other cellular compartments. Detection of the UP9C–EYFP fusion inside the nucleus does not exclude its presence in the other compartments, particularly in the cytosol, because the UP9C localization might depend on multiple factors, such as posttranslational modifications and interaction with other proteins.
Effects of Constitutive Ectopic Overexpression of the UP9C Gene in Antisense Orientation
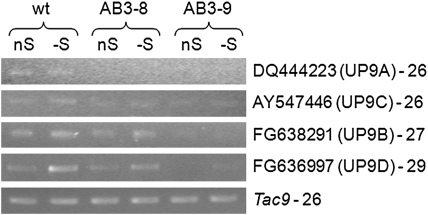
The binary plasmid pAB3 enabling overexpression of UP9C in antisense orientation was used for generation of the transgenic tobacco lines with down-regulated expression of UP9C. The T1 progeny of two independently obtained ‘antisense’ AB3 lines (AB3-8 and AB3-9) were used in most experiments, although some results were additionally confirmed in other antisense lines, such as AB3-1, AB3-3, AB3-6, and AB3-12. To verify the effects of antisense expression of UP9C, the level of mRNA corresponding to the four closest homologs (UP9A–UP9D) was monitored in mature leaves of the parental line and the transgenic lines AB3-8 and AB3-9 grown in S-sufficient and S-deficient conditions (Figure 3). The expression of both UP9C and UP9A was apparently silenced in both transgenic lines, while the silencing of two other UP9 family members encoding UP9B and UP9D was much more effective in AB3-9 than in AB3-8.
Figure 3.
Evaluation of Expression Levels of UP9-Like Transcripts in Mature Leaves of the Analyzed Plant Lines.
The primers used in semiquantitative RT–PCR (sqRT–PCR) were designated to the non-coding sequences of the indicated EST clones and they are listed in Table 2. The GB#, the name of the protein cluster, and the number of cycles are marked next to the corresponding expression profile; wt, the parental line (LA Burley 21).
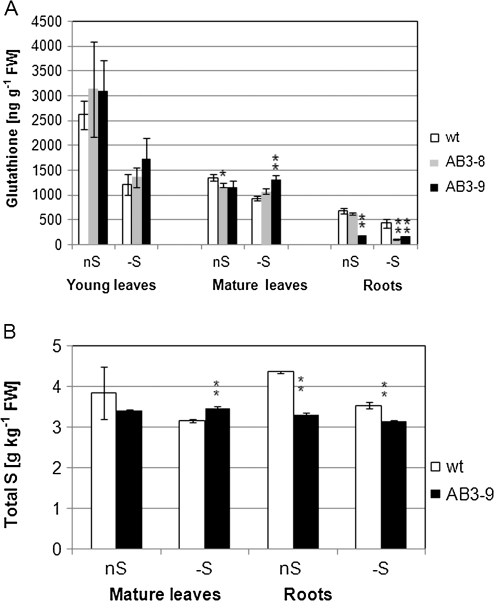
It has been shown previously that growth in S-deficient medium results in reduction of non-protein thiols in most plant organs (Nikiforova et al., 2003; Wawrzynska et al., 2005). Interestingly, silencing of the UP9-like genes affects thiols level in mature leaves and roots of AB3 lines but rather not in young leaves (Figure 4A). The level of total glutathione in mature leaves of AB3 lines grown in S-sufficient conditions was slightly lower than that of the parental line (88 and 85% for AB3-8 and AB3-9, respectively) but, more spectacularly, in AB3 lines grown in S-deficient conditions, thiols remained on the same level (or were even slightly increased) as compared to S-sufficient conditions. This apparent lack of response to S limitation in the mature leaves of AB3 lines (confirmed in several other independent AB3 lines; results not shown) was in a contrast to the reduction of thiols in the mature leaves of the parental line grown in S-deficient conditions. Glutathione measurements in the roots revealed further significant differences between AB3 lines and the parental line. In the roots of AB3-8 grown in S-sufficient conditions, glutathione level was as in the parental line, while in S-deficient conditions, it was very low and similar to that in AB3-9 (27% of that in the parental line; Figure 4A).
Figure 4.
Contents of Glutathione (A) and Total S (B) in the Analyzed Tobacco Lines.
Plants were grown in the optimal AB medium (nS) or for 2 d in S-deficient conditions in the AB-S medium (–S). The statistically significant differences between the transformants and the parental line (wt) grown in the same conditions are marked with either a single (for p < 0.05) or double (for p < 0.01) asterisks.
(A) The material was combined from five plants for each analyzed group; the mean data from five independent extractions with standard deviations (SD) are shown.
(B) The material was combined from five plants for each analyzed group; the mean of three assays with SD values are shown.
The level of total S monitored in the parental line and the more efficiently ‘silenced’ line, AB3-9, revealed more statistically significant differences (Figure 4B). The level of total S in AB3-9 grown in S-sufficiency was lower than in the parental line and it was not reduced (in the leaves) or reduced to a low extent (in the roots) by S limitation. In the leaves of AB3-9, about 88 and 110% of S present in the leaves of the parental line in S-sufficient and S-deficient conditions, respectively, was found. The reduction of S content in the roots of AB3-9 in comparison to the parental line was more spectacular than in the leaves (76 and 89% in S-sufficient and S-deficient conditions, respectively).
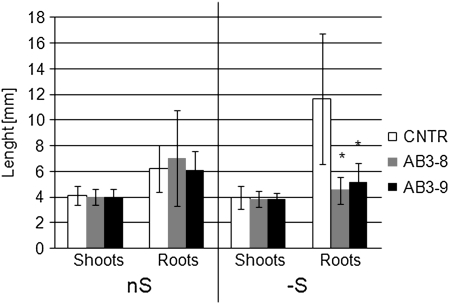
The observed biochemical discrepancy had rather a small influence on the performance of AB3 plants in non-stressing conditions, since, during vegetative growth in optimal, S-sufficient conditions, the transgenic plants looked rather similar to the parental line. Further experiments revealed that the roots of the seedlings of AB3 lines grown in S-deficient conditions were significantly shorter than those of the control line (Figure 5).
Figure 5.
Comparison of Root and Shoot Lengths in Seedlings Grown for 3 Weeks on AB Plates Maintained in a Vertical Position in Either the Optimal (nS) or S-Deficient (–S) AB-S Medium.
The average data (with SD indicated) were calculated from 30–50 measured seedlings, depending on the line. The statistically significant (p < 0.05) differences between the transformants and the control (CNTR) are marked with asterisks. Similar results were obtained for two control lines, parental line (not shown), and a tobacco line transformed with GFP expression cassette (CNTR).
Misregulation of UP9-Like Genes Affects Expression of Other Genes
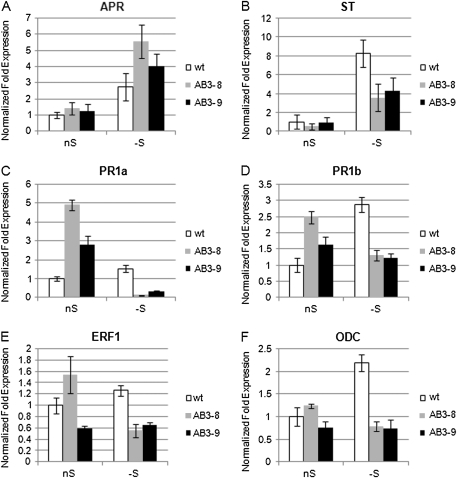
Expression of the two genes known to respond to S-deficit and encoding crucial enzymes of sulfate assimilation, ST encoding group 1 sulfate transporter (GB# AJ745720) and APR (GB# AY648056) encoding APS reductase, was tested in the parental and AB3 plants grown in S-sufficient and S-deficient medium (Figure 6A and 6B). The analyses were limited to mature leaves of the tested plants, because the most spectacular changes in non-protein thiol levels had been observed in these parts of the plants (see Figure 4A). Both tested genes responded to S-deficit in all lines. However, in both AB3 lines, the level of ST transcript was about two-fold lower as compared to the parental line in S-deficient condition. In contrast, the level of APR transcript was about 1.5–2-fold more abundant in AB3 lines than in the parental line in S-deficient conditions.
Figure 6.
The Expression Levels of the Selected Genes in Mature Leaves of the Analyzed Tobacco Lines.
In all real time RT–PCR (qRT–PCR) reactions, both Tac9 and UBQ2 served as references, except for the qRT–PCR shown in (A), in which only Tac9 was used as a reference gene. nS, optimal conditions; –S, 2 d of S-deficit. Each experiment was performed at least two times in triplicate using two independently isolated RNA templates, with similar results. The presented data are from one representative experiment performed in triplicate with SD indicated.
Some of the UP9C interacting partners identified during yeast two-hybrid search were related to the action and synthesis of plant hormones. Besides, plant hormones might be involved in mediating the root growth phenotype of AB3 seedlings in S-deficient medium (see Figure 5). To further investigate the problem of influence of misregulation of the UP9-like genes on the level of plant hormones, the expression of genes known to respond to ethylene (ET) and jasmonic acid (JA) was monitored. The following genes were selected: PR1a (GB# D90196) and PR1b (GB# X66942) coding for pathogenesis-related acidic and basic proteins, respectively, and ERF1 (GB# D38123) encoding the ethylene-responsive transcription factor 1. The levels of expression of these genes in optimal conditions and after treatment for 2 d with S-deficiency are shown in Figure 6C–6E. All three genes were induced in the parental line during S-starvation, while in the antisense lines, they were usually down-regulated in S-deficient conditions. On the other hand, in the S-sufficient conditions, significantly raised expression of PR1a and PR1b in both AB3 lines and ERF1 in AB3-8 line in comparison to the parental line was observed.
Since ethylene production is connected with the synthesis of polyamines, which are involved in response to environmental stresses, the expression of genes encoding proteins related with polyamine synthesis, such as S-adenosylmethionine synthetase (SAMS, GB# AY445582), S-adenosylmethionine decarboxylase (SAMDC, GB# U91924), ornithine decarboxylase (ODC, GB# AF127242), and arginine decarboxylase (ADC, GB# AF321137), was also monitored. The level of expression of SAMS, SAMDC, and ADC in both AB3 lines remained similar to that of the parental line (not shown), while the expression of ODC in S-deficient conditions was strongly altered (reduced) in both AB3 lines (Figure 6F). The observed pattern of ODC expression might suggest that in the parental line, the level of polyamines would raise in the S-deficient conditions, while in the AB3 lines, such an increase would not take place.
Metabolite Profiles in Antisense Tobacco Transformants
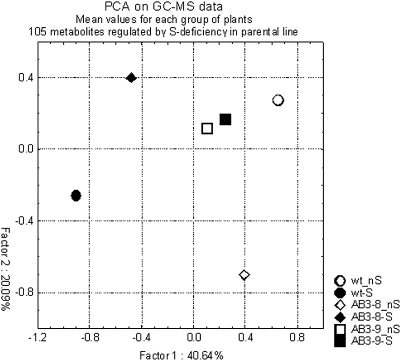
The profile of polar metabolites was examined in the mature leaves of AB3-8, AB3-9, and parental line grown in S-deficient and optimal conditions. Initial data used for analysis included 559 peaks of metabolites detected in the measured samples. Among these peaks, only about 10% were identified as annotated compounds. Comparison of the data for the parental line grown in the optimal conditions and the parental line grown in S-deficiency allowed identification of metabolites that had significantly different levels in these two conditions of growth. Depending on the selected borderline significance (p-value), the number of peaks regulated by S-deficiency in the parental line was different and reached 27 for p < 0.05, 47 for p < 0.1, and 105 for p < 0.2. The largest set of regulated metabolites (66 up- and 39 down-regulated in S-deficient medium) was chosen for the subsequent factorial analysis, which revealed that both AB3 lines (particularly AB3-9) independently from growth conditions have a tendency to cluster rather with the wild-type plants grown in S-sufficient conditions than with the wild-type plants grown in S-deficient conditions (Figure 7). This observation might suggest that AB3 lines have problems with adjusting their metabolism to the conditions of S-deficiency. Comparison of the regulation of the selected set of the metabolites in the parental line and the AB3 lines is shown in Table 1. It is shown as an X-fold ratio of the metabolite level in S-deficiency versus its level in optimal conditions of the same line. A list of peaks representing metabolites apparently up-regulated during S-deficit in a ‘UP9-dependent way’ includes glutamine, lysine, threonine, asparagine, spermidine, serine, citric, aspartic, and malic acids. Unfortunately, huge differences between replicas hindered more sophisticated analysis of the metabolite data. This variation might result from technical problems with preparation of the plant tissues for the assay.
Figure 7.
Principal Component Analysis of Metabolite Profiling Data for Parental Line (wt) and the Transgenic Lines Grown Either in the Optimal Medium (nS) or in the S-Deficient Medium (–S).
Table 1.
Metabolites Identified as Regulated by S-Deficiency in the Parental Line (wt) and the Ratio (X-Fold) of their Mean Values as Indicated.
| Peak no. | Annotation | X-fold | X-fold | X-fold | |||
| wt –S/nS | t-test | AB3-8 –S/nS | t-test | AB3-9 –S/nS | t-test | ||
| 4250 | n/a | 21.40 | 0.06 | 0.08 | 0.25 | 1.54 | 0.51 |
| 414 | n/a | 20.33 | 0.06 | 0.07 | 0.25 | 1.78 | 0.40 |
| 1797 | n/a | 19.59 | 0.07 | 0.07 | 0.25 | 1.91 | 0.33 |
| 168 | n/a | 14.42 | 0.06 | 0.08 | 0.25 | 1.49 | 0.44 |
| 27 | Glutamine | 4.88 | 0.07 | 0.37 | 0.49 | 1.04 | 0.85 |
| 316 | n/a | 4.84 | 0.09 | 0.18 | 0.29 | 1.10 | 0.83 |
| 648 | n/a | 3.56 | 0.05 | 0.33 | 0.24 | 1.22 | 0.72 |
| 1406 | n/a | 3.09 | 0.04 | 0.31 | 0.16 | 0.91 | 0.72 |
| 588 | Lysine | 3.09 | 0.03 | 0.35 | 0.14 | 0.85 | 0.51 |
| 4074 | n/a | 2.99 | 0.10 | 1.95 | 0.35 | 0.60 | 0.52 |
| 2018 | n/a | 2.97 | 0.13 | 1.13 | 0.82 | 0.70 | 0.64 |
| 3147 | n/a | 2.93 | 0.11 | 1.63 | 0.49 | 0.65 | 0.54 |
| 1559 | Ornithine | 2.74 | 0.04 | 4.48 | 0.00 | 2.16 | 0.12 |
| 2940 | n/a | 2.70 | 0.11 | 1.54 | 0.53 | 0.55 | 0.50 |
| 3463 | n/a | 2.69 | 0.12 | 0.55 | 0.55 | 0.43 | 0.38 |
| 3373 | n/a | 2.48 | 0.04 | 1.24 | 0.80 | 0.10 | 0.02 |
| 4794 | n/a | 2.32 | 0.19 | 1.10 | 0.93 | 0.93 | 0.91 |
| 584 | n/a | 2.29 | 0.04 | 0.63 | 0.40 | 0.85 | 0.79 |
| 121 | n/a | 2.22 | 0.04 | 0.82 | 0.50 | 1.39 | 0.21 |
| 4603 | n/a | 2.09 | 0.03 | 1.53 | 0.28 | 1.26 | 0.61 |
| 95 | Threonine | 2.08 | 0.03 | 0.54 | 0.44 | 1.36 | 0.20 |
| 574 | n/a | 2.06 | 0.05 | 0.65 | 0.23 | 1.52 | 0.48 |
| 2167 | n/a | 2.04 | 0.06 | 1.89 | 0.09 | 2.23 | 0.17 |
| 1020 | n/a | 1.99 | 0.01 | 1.54 | ND | 0.23 | ND |
| 793 | n/a | 1.92 | 0.12 | 0.76 | 0.37 | 1.33 | 0.13 |
| 759 | n/a | 1.87 | 0.04 | 1.38 | 0.30 | 1.73 | 0.08 |
| 3378 | n/a | 1.85 | 0.16 | 0.46 | 0.43 | 0.44 | 0.33 |
| 1435 | n/a | 1.82 | 0.13 | 1.00 | 1.00 | 1.00 | 0.97 |
| 1060 | n/a | 1.80 | 0.01 | 1.29 | 0.46 | 1.13 | 0.71 |
| 4924 | n/a | 1.79 | 0.13 | 0.42 | 0.20 | 0.82 | 0.72 |
| 224 | Alanine | 1.77 | 0.18 | 1.27 | 0.63 | 1.81 | 0.15 |
| 941 | n/a | 1.75 | 0.19 | 0.71 | 0.60 | 0.14 | 0.01 |
| 326 | n/a | 1.72 | 0.04 | 0.71 | 0.52 | 0.99 | 0.97 |
| 229 | Spermidine | 1.69 | 0.04 | 0.61 | 0.37 | 0.96 | 0.89 |
| 2608 | n/a | 1.69 | 0.08 | 1.24 | 0.39 | 1.32 | 0.06 |
| 2574 | n/a | 1.69 | 0.15 | 1.28 | 0.57 | 1.39 | 0.49 |
| 2421 | n/a | 1.66 | 0.14 | 1.99 | 0.19 | 0.76 | 0.50 |
| 112 | Citric acid | 1.63 | 0.00 | 0.87 | 0.61 | 1.03 | 0.86 |
| 738 | Aspartic acid | 1.60 | 0.08 | 0.65 | 0.38 | 1.26 | 0.33 |
| 151 | n/a | 1.60 | 0.01 | 1.07 | 0.80 | 1.12 | 0.58 |
| 2967 | n/a | 1.59 | 0.13 | 2.39 | 0.18 | 1.48 | 0.32 |
| 413 | n/a | 1.58 | 0.06 | 0.83 | 0.48 | 1.19 | 0.63 |
| 2354 | n/a | 1.57 | 0.16 | 1.01 | 0.97 | 0.77 | 0.55 |
| 3670 | n/a | 1.57 | 0.16 | 0.40 | 0.04 | 2.07 | 0.06 |
| 1529 | n/a | 1.55 | 0.14 | 1.90 | 0.02 | 2.16 | 0.00 |
| 497 | n/a | 1.52 | 0.04 | 1.70 | 0.14 | 1.26 | 0.39 |
| 115 | Serine | 1.52 | 0.12 | 0.38 | 0.34 | 1.07 | 0.78 |
| 806 | n/a | 1.51 | 0.16 | 0.75 | 0.57 | 0.55 | 0.39 |
| 1244 | n/a | 1.51 | 0.11 | 0.65 | 0.25 | 1.22 | 0.37 |
| 147 | n/a | 1.50 | 0.13 | 0.70 | 0.29 | 1.02 | 0.95 |
| 1303 | n/a | 1.48 | 0.13 | 1.75 | 0.32 | 1.68 | 0.22 |
| 535 | Threonine | 1.47 | 0.08 | 1.07 | 0.60 | 1.23 | 0.05 |
| 825 | n/a | 1.43 | 0.19 | 0.62 | 0.14 | 1.41 | 0.05 |
| 161 | Asparagine | 1.42 | 0.13 | 0.60 | 0.37 | 1.10 | 0.53 |
| 1729 | n/a | 1.41 | 0.19 | 1.49 | 0.21 | 1.27 | 0.05 |
| 53 | n/a | 1.38 | 0.01 | 1.42 | 0.22 | 1.05 | 0.81 |
| 445 | n/a | 1.36 | 0.02 | 1.03 | 0.86 | 0.95 | 0.66 |
| 288 | n/a | 1.31 | 0.13 | 1.39 | 0.20 | 1.10 | 0.33 |
| 40 | n/a | 1.28 | 0.17 | 0.77 | 0.54 | 1.39 | 0.22 |
| 798 | n/a | 1.26 | 0.02 | 0.92 | 0.49 | 1.02 | 0.81 |
| 730 | n/a | 1.26 | 0.10 | 1.15 | 0.62 | 1.00 | 0.97 |
| 21 | n/a | 1.23 | 0.06 | 1.03 | 0.85 | 0.96 | 0.73 |
| 26 | Malic acid | 1.20 | 0.06 | 0.92 | 0.56 | 1.01 | 0.94 |
| 81 | Putrescine | 1.19 | 0.13 | 0.93 | 0.52 | 1.29 | 0.01 |
| 160 | n/a | 1.19 | 0.06 | 0.67 | 0.00 | 0.87 | 0.05 |
| 20 | Threonic acid-1,4-lactone | 1.17 | 0.12 | 0.71 | 0.01 | 0.90 | 0.09 |
| 131 | n/a | 0.89 | 0.10 | 0.91 | 0.11 | 1.17 | 0.01 |
| 9 | n/a | 0.87 | 0.08 | 0.90 | 0.09 | 1.19 | 0.01 |
| 1555 | n/a | 0.87 | 0.10 | 0.97 | 0.87 | 0.96 | 0.85 |
| 178 | n/a | 0.86 | 0.04 | 0.70 | 0.00 | 1.55 | 0.00 |
| 1295 | Fumaric acid | 0.80 | 0.05 | 0.86 | 0.11 | 0.81 | 0.02 |
| 470 | Galactonic acid | 0.80 | 0.18 | 0.78 | 0.18 | 1.12 | 0.67 |
| 428 | n/a | 0.79 | 0.20 | 0.52 | 0.03 | 1.66 | 0.01 |
| 2088 | n/a | 0.76 | 0.20 | 0.65 | 0.26 | 1.19 | 0.54 |
| 1413 | n/a | 0.76 | 0.14 | 0.69 | 0.31 | 0.76 | 0.38 |
| 278 | Trehalose | 0.75 | 0.16 | 0.77 | 0.31 | 0.78 | 0.31 |
| 754 | n/a | 0.74 | 0.15 | 0.21 | 0.00 | 1.55 | 0.07 |
| 4215 | n/a | 0.72 | 0.10 | 1.31 | 0.19 | 1.04 | 0.92 |
| 360 | n/a | 0.71 | 0.15 | 0.49 | 0.21 | 0.57 | 0.21 |
| 248 | n/a | 0.71 | 0.16 | 0.43 | 0.01 | 1.03 | 0.93 |
| 2726 | n/a | 0.70 | 0.15 | 0.54 | 0.06 | 1.75 | 0.05 |
| 258 | n/a | 0.69 | 0.13 | 0.25 | 0.07 | 2.35 | 0.02 |
| 1106 | n/a | 0.69 | 0.19 | 0.32 | 0.07 | 1.40 | 0.44 |
| 911 | n/a | 0.66 | 0.14 | 0.78 | 0.53 | 0.91 | 0.62 |
| 848 | n/a | 0.66 | 0.17 | 0.89 | 0.68 | 0.64 | 0.26 |
| 172 | n/a | 0.65 | 0.03 | 1.02 | 0.88 | 1.40 | 0.11 |
| 842 | n/a | 0.64 | 0.14 | 1.14 | 0.75 | 0.62 | 0.29 |
| 1297 | Ethanolamine | 0.64 | 0.13 | 1.35 | 0.11 | 0.93 | 0.63 |
| 2212 | n/a | 0.64 | 0.16 | 0.47 | 0.09 | 5.00 | 0.00 |
| 76 | n/a | 0.63 | 0.16 | 0.28 | 0.09 | 0.99 | 0.98 |
| 45 | n/a | 0.63 | 0.10 | 0.42 | 0.01 | 1.65 | 0.05 |
| 2028 | n/a | 0.61 | 0.17 | 0.60 | 0.24 | 1.58 | 0.09 |
| 3912 | n/a | 0.61 | 0.10 | 0.29 | 0.06 | 1.53 | 0.28 |
| 930 | n/a | 0.59 | 0.11 | 0.35 | 0.00 | 1.06 | 0.57 |
| 2087 | n/a | 0.57 | 0.11 | 0.33 | 0.01 | 1.13 | 0.74 |
| 801 | n/a | 0.54 | 0.16 | 0.46 | 0.13 | 0.43 | 0.12 |
| 418 | n/a | 0.52 | 0.17 | 0.88 | 0.85 | 2.14 | 0.29 |
| 649 | n/a | 0.52 | 0.01 | 1.35 | 0.21 | 0.91 | 0.66 |
| 4292 | n/a | 0.50 | 0.08 | 0.30 | 0.04 | 0.99 | 0.98 |
| 509 | n/a | 0.48 | 0.20 | 0.63 | 0.58 | 1.22 | 0.82 |
| 2977 | n/a | 0.46 | 0.14 | 0.29 | 0.14 | 0.51 | 0.29 |
| 134 | n/a | 0.38 | 0.02 | 0.75 | 0.47 | 0.80 | 0.59 |
| 1985 | n/a | 0.34 | 0.03 | 1.03 | 0.94 | 0.79 | 0.58 |
| 1815 | n/a | 0.24 | 0.01 | 1.63 | 0.41 | 1.52 | 0.40 |
| 975 | n/a | 0.21 | 0.11 | 3.73 | ND | 1.13 | 0.90 |
The statistical significances of differences between the two compared groups are shown as the p-values of the t-test; n/a, not annotated, ND, impossible to determine because of low number of measurements in one or both groups.
DISCUSSION
In our earlier studies, two UP9-like genes (GB# AY547446 and DQ444223) were identified as up-regulated by S-deficit (Lewandowska et al., 2005; Wawrzynska et al., 2005). Due to the amphidiploid nature of N. tabacum, we suspected that they corresponded to genes located on the matching chromosomes of the genome S and T (Volkov et al., 1999). However, they can also be genes from different chromosomal loci on one genome. Subsequent analysis has shown that at least six distinct UP9-like proteins exist in N. tabacum (Figure 1) and that other UP9-like genes might be also up-regulated in S-deficient conditions (Figure 3). The differences in the sequences outside coding regions of the EST clones classified into one cluster indicate that either they represent separate genes (despite encoding the identical proteins) or that there exist differences between the cultivars in the non-coding sequences of individual UP9-like counterparts. In A. thaliana, there are four genes (LSU1–LSU4) encoding homologs of UP9. They are organized in two tandems located in two genome positions, one on chromosome III (3 kb apart) and the second on chromosome V (2.5 kb apart). Two of these genes (the upstream genes of each tandem) were reported as strongly up-regulated by S-deficit (Maruyama-Nakashita et al., 2005; Nikiforova et al., 2005) and the regulation of at least one of them (LSU1, At3g49580) is dependent on SLIM1 (Maruyama-Nakashita et al., 2006).
The predicted structure of UP9-like proteins is largely in the form of coiled coils. The proteins containing such motifs can form multiple interactions and fulfill a variety of functions due to their ability to undergo dynamic rearrangements (Parry et al., 2008). The UP9-like proteins can form dimers (Lewandowska et al., 2005). Most probably, the interaction is mediated through the coiled coil region; however, solution characterization and crystallographic studies might be necessary to establish the factual three-dimensional structure and verify the localization of the oligomerization domains. Moreover, in the yeast two-hybrid experiment performed in our laboratory, a range of plant proteins was identified as potential partners of UP9C. Their further analysis might help to decipher the role of UP9-like proteins.
Antisense mRNA has frequently been used in plants to lower gene expression through posttranscriptional gene silencing (Chintapakorn and Hamill, 2003; Halitschke and Baldwin, 2003; Siritunga and Sayre, 2003). Transgenic tobacco lines obtained in our study had apparently misregulated expression of UP9-like genes. It should be noted that the expression of the entire family was not completely abolished by the antisense construct and a substantial expression of some members, particularly in AB3-8, was present (Figure 3). Molecular and biochemical assays (Figures 4 and 6) as well as metabolite profiling (Figure 7 and Table 1) revealed that many changes in the level of metabolites that were characteristic for plants grown in S-deficient medium did not occur in the AB3 plants. It is necessary to mention that interpretation of metabolite data is somewhat problematical because of the incomplete peaks identification and a low statistical significance of the results (large differences between the replicas). Nevertheless, some results of metabolite profiling are in agreement with the gene expression data. For example, the level of spermidine, increased during S-deficit in the parental line but not in the AB3 lines (Table 1), correlates very well with the expression of the ODC gene (Figure 6F) encoding ornithine decarboxylase that is involved in the pathway of polyamines biosynthesis. However, additional experiments using the knockout plants and multiple knockouts are needed to properly assess the function of UP9-like proteins in tobacco and/or LSU1-like proteins in Arabidopsis.
The transcriptional regulator from A. thaliana, SLIM1/EIL3, was shown to affect expression of many genes regulated by S-deficit, including LSU1 (At3g49580), one of the UP9 homologs (Maruyama-Nakashita et al., 2006). Recent experiments performed in our laboratory revealed that a tobacco protein NtEIL2 that belongs to the same family as SLIM1 activates the promoter of UP9C gene in S-deficient conditions (Wawrzynska et al., 2009). Therefore, some effects observed in slim1 mutants could be due to the lack of appropriate level of UP9-like regulator. UP9-like proteins seem to be a part of a regulatory network that is in charge of an adequate whole-plant response to S-deficit.
One of the striking effects of UP9 family silencing in AB3 lines is their failure to diminish the level of S-containing metabolites in mature leaves (source tissue) in response to S-deprivation (Figure 4). A possible explanation for this phenomenon might be either a defect in a long-distance roots-to-shoots signaling of S status or a defect in adjusting the metabolism in the shoots to the conditions of S-deficit encountered by the roots. It is still uncertain how S-deficiency stress is sensed and transmitted on a long distance within plants. Usually, the ability to detect and adapt to environmental changes relies on complex molecular machinery that integrates the action of hormonal and non-hormonal signals (Chico et al., 2008; Lin et al., 2009). Some effects observed in the ‘antisense’ lines might be explained if in these lines occurred problems with controlling either the level or the function of plant hormones. The expression of three arbitrary selected genes (PR1a, PR1b, and ERF1) induced by the stress hormones was reduced in AB3 lines grown in S-deficient conditions (Figure 6). This result indicates that at least one of these hormones might be limiting for gene expression in AB3 lines in S-deficient conditions. On the other hand, the elevated expression of these genes in AB3 in S-sufficient conditions is difficult to explain. Perhaps, somehow, the silencing of UP9-like genes causes a stress situation both in optimal and in S-deficient conditions of growth. It is worth emphasizing that in the S-sufficient conditions, the level of thiols in the mature leaves was significantly lower in AB3 than in the parental line (Figure 4). It means that the appropriate level (or balance) of UP9-like proteins might be needed also in the optimal (S-sufficient) conditions. This phenomenon could be further clarified only after establishing the function of individual members of the UP9-like family.
The function of UP9C and its complexes with other proteins is still elusive. We propose that UP9C, through its multiple protein–protein interactions, might be involved in precise and rapid adjustment of plant metabolism to S-deficit on multiple levels, presumably through influencing the level and/or action of plant hormones. The UP9C involvement could be either indirect, through interactions with transcription factors (JAZ1, AP2-like), or direct, through interactions with enzymes, such as ACC oxidase. Binding of UP9C might affect the stability of its partners and/or their ability to interact with other proteins. On the other hand, sub-cellular localization of UP9C and its ability to interact with other proteins can be regulated by additional factors, such as by redox potential and posttranslational modifications (phosphorylation, ubiquitination). Such modifications could also affect possible dimerization or heterodimerization with other members of the UP9-like family, whose function can be partially overlapping but also potentially dependent on a composition of a dimer. Despite additional studies being needed to assess the precise biological role of UP9-like proteins, this study shows that at least part of the specificity of response to the S-deficiency stress is provided by one or more members of the UP9-like family of proteins.
In addition, it is possible to speculate that an elevated glutathione pool in mature leaves of AB3 lines grown in S-deficient conditions (accompanied by increased expression of APR) might result from the occurrence of an intracellular oxidative stress. A strong induction of APR (and SAT2.1 encoding chloroplast izoform of SAT) transcript and increased glutathione pool have been observed in the cat2 (catalase-deficient) mutant of Arabidopsis in the conditions of growth that were generating production of hydrogen peroxide (Queval et al., 2009).
METHODS
DNA Manipulations and Plasmid Construction
Conventional techniques were used for DNA manipulation and transformation (Sambrook et al., 1989). All primers are listed in Table 2. To obtain the plasmid-containing UP9C in the reverse orientation, a PCR fragment of about 350 bp was cloned into vector pRTL2 (Carrington et al., 1991). In the resulting plasmid, the UP9C gene is under the control of the CaMV 35S promoter and terminator, in the reverse orientation, thus enabling expression of an antisense transcript of UP9C. The entire cassette (1.5 kb) was then inserted into the HindIII site of plant vector pGreenII0029 (Hellens et al., 2000). The resulting plasmid, pAB3, also has the nptII gene present within the T-DNA (kanamycin selection marker). To obtain the N-terminal fusion of UP9C to the fluorescent tag EYFP (UP9C–EYFP), a fragment of about 350 bp containing the coding region of UP9C lacking the stop codon amplified by PCR using attB-tailed gene-specific primers was cloned into pDONR221 (Gateway® Technology, Invitrogen). In the next step, the fragment was re-cloned into the plant binary vector pH7YWG2 (Karimi et al., 2005). The resulting plasmid, pUP9C–EYFP, contains the UP9C–EYFP sequence under the control of the CaMV 35S promoter and terminator, and the hygromycin selection marker within the T-DNA. The pJKU1 plasmid, used for the yeast two-hybrid experiment, was obtained after cloning of the appropriate PCR product (0.35 kb) into the PstI/EcoRI sites of the pGBT9 vector (Clontech).
Table 2.
Oligonucleotides Used in the Study.
| Purpose | GB# of the target | Sequence |
| orf for ‘antisense’ constructs | AY547446 (UP9C) | 5′-CGGGATCCATGTTTTCGACAATTGCT-3’ |
| 5′-GCAAGCTTGGTACCTCATTGGGAACTGGGAAC-3’ | ||
| orf for UP9C–EYFP fusion | AY547446 (UP9C) | 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCAATGTTTTCGACAATTGCTGT-3’ |
| 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTTGGGAACTGGGAACGGTAA-3’ | ||
| orf for Y2H | AY547446 (UP9C) | 5′-CGGAATTCTTTTCGACAATTGCTGTGC-3’ |
| 5′-GCCTGCAGAGCGTCAAACTCCATTACCC-3’ | ||
| sqRT–PCR | DQ444223 (UP9A) | 5′-ACCACAAATCAATCTCCTACCT-3’ |
| 5′-ACGTGCTCTACCTAGTTTGAA-3’ | ||
| AY547446 (UP9C) | 5′-ACCACAAATCAGTTTCCTGCATT-3’ | |
| 5′-ACGTGCTCTACCTAGTTTGAA-3’ | ||
| FG638291 (UP9B) | 5′-TCAACTACTACTATTTCCTTCAACATG-3’ | |
| 5′-ACCTCCATTAATCCGAGTGCCTAA-3’ | ||
| FG636997 (UP9D) | 5′-TGCCTCAGTTTTAATAGTAGCAATTTT-3’ | |
| 5′-AATACAAACAGGCCGCCACCAA-3’ | ||
| X69885 (Tac9) | 5′-CCTCCCACATGCTATTCTCC-3’ | |
| 5′-AGAGCCTCCAATCCAGACAC-3’ | ||
| qRT-PCR | X69885 (Tac9) | 5′-GTGTCTGGATTGGAGGCTCT-3’ |
| 5′-ATGAAGCAAACCTGCTGGAA-3’ | ||
| DQ138111 (UBQ2) | 5′-AGGTTGATGATTCCGGTAAGG-3’ | |
| 5′-CACACTCAGCATTGGGACAC-3’ | ||
| AY648056 (APR) | 5′-TTCCCTGACGCTGTTGAAG-3’ | |
| 5′-CCACCACCATCTTCCCTCT-3’ | ||
| AJ745720 (ST) | 5′-CCATTATCCTCTCGACGTTA-3’ | |
| 5′-TCAAACCAGAAATTGCACCA-3’ | ||
| AF321137 (ADC) | 5′-TGGAGGTGGGCTTGGAATT-3’ | |
| 5′-TGAACAACTGCGGAGGCAT-3’ | ||
| AF127242 (ODC) | 5′-TGATTGGCTGGTTTTTCCTAA-3’ | |
| 5′-AGGTGGTTCATCAGCTTGGA-3’ | ||
| U91924 (SAMD) | 5′-TGGCAAGGGTGGTTCCAT-3’ | |
| 5′-TCTTTCCAGCAGCCCTTCA-3’ | ||
| AY445582 (SAMS) | 5′-ACCGGATGGCAAGACACAAG-3’ | |
| 5′-TCCTGGGCGATCTGCTCA-3’ | ||
| D90196 (PR1a) | 5′-AGTCATGGGATTTGTTCTCTT-3’ | |
| 5′-ACGGCAAGAGTGGGATATTA-3’ | ||
| X66942 (PR1b) | 5′-TTGGTGGTATTATGGAGGTGTG-3’ | |
| 5′-ACTGCCGTTGACTCATCACCT-3’ | ||
| D38123 (ERF1) | 5′-TAGGGGAAGGCATTACAGAG-3’ | |
| 5′-TCGTATGTTCCAAGCCAAA-3’ |
RT–PCR Analysis
Total plant RNA was used as a template for synthesis of the first-strand cDNA. The 20-μl RT reaction contained 5 μg RNA, 2 pmoles of specific reverse primer or oligo(dT) primers, 1 mM dNTP Mix, 20 units RiboLock™ Ribonuclease Inhibitor, and 1 μl of the RevertAid™ H Minus M-MuLV Reverse Transcriptase (MBI Fermentas) in the buffer supplied by the manufacturer. The RNA and primers were preheated to 70°C for 5 min and chilled on ice before adding the 5× reaction buffer for Reverse Transcriptase, dNTP Mix, Ribonuclease inhibitor, and RNase-free water to 19 μl. Then, the mixture was incubated for 5 min at 37°C. Next, the reverse transcriptase was added and the RT reaction was incubated for 1 h at 42°C. The reverse transcription was stopped by heating for 10 min at 70°C. The reaction mixture was diluted (usually 20 or 100 times) and 1 μl of the diluted reaction mixture was then used for the PCR with the specific primers designed for selected cDNAs (Table 2). A variable number (25–35) of cycles (30 s denaturation at 94°C, 30 s annealing at 50°C, and 1 min extension at 72°C) were then performed for semiquantitative RT–PCR (sqRT–PCR). Actin (Tac9) cDNA amplification using the same temperature profile for 25–30 cycles (dependent on the dilution of the first-strand cDNA) served as the internal control. DNA samples were separated by electrophoresis on 1% agarose gels, and the images of ethidium-bromide-stained bands were obtained with a BioRad Imaging System.
For real-time PCR (qRT–PCR), the first-strand cDNA synthesis was the same as for sqRT–PCR, with one exception—oligo(dT) primers (Invitrogen) were used instead of a specific primer. The reactions were performed in an iQ5 thermocycler (BioRad). The pairs of primers used in qRT–PCR reactions are listed in Table 2. Each experiment was performed at least twice in triplicate using two independently isolated RNA templates. The primers were designed to have similar annealing temperatures of about 50–53°C; the efficiency of the primers was analyzed before main experiments in a temperature gradient range from 49 to 55°C (a typical cycle: 15 s denaturation at 94°C, 15 s annealing at 52°C, and 15 s extension at 72°C). Tac9 and UBQ2 were used as internal controls.
Plant Culture Media
The MS medium used for plant cultivation in sterile (in vitro) conditions was prepared according to the Murashige and Skoog protocol (Murashige and Skoog, 1962) using concentrated components from Sigma macro- and micro-elements (10× conc.), MS Vitamins (1000× conc.), 3% sucrose; for solid media, 0.8 g of agar (Difco Oxoid) per 100 ml was added and the pH was adjusted to 5.8 with 1 M KOH. Media were autoclaved for 15 min at 121°C. For tobacco hydroponic culture, the AB medium devised in our laboratory was used (Wawrzynska et al., 2005).
Plant Material, Growth Conditions, and Genetic Transformation
Transgenic UP9C–EYFP (overexpressing UP9C–EGFP fusion) and AB3 lines (overexpressing UP9C in antisense orientation) used in this study were derivative of Nicotiana tabacum cv. LA Burley 21 (Legg et al., 1970). Agrobacterium-mediated genetic transformation of 3-week-old tobacco seedlings was performed as previously described (Wawrzynski et al., 2006). Seeds of the self-pollinated primary tobacco transformants (T0) with confirmed expression of the transgenes were collected and used for the subsequent experiments. Surface-sterilized seeds of AB3 lines from T1 generation were germinated in vitro at 23°C with a photoperiod of 18 h light/6 h dark on solid MS plates (with addition of 100–200 μg ml−1 kanamycin for the transgenic lines). Three-week-old plantlets were transferred to the AB liquid medium (Wawrzynska et al., 2005) and cultivated hydroponically in a growth chamber (16 h light, 22°C/8 h darkness, 18°C), with weekly changes of the medium. Two-month-old plants were divided into two groups. The control group was transferred to the optimal medium (AB) while the other was transferred to the sulfur-free medium (AB-S) containing 1 mM MgCl2 instead of 1 mM MgSO4. Unless specified otherwise, after 2 d, five plants from each group were harvested and fractionated into four parts, namely young developing leaves, mature leaves, stems and roots, which were then pooled. Tobacco UP9C–EGFP seedlings of T1 were grown in vitro for 3–4 weeks in liquid half-strength MS medium with 50 μg ml−1 hygromycin with a constant rotation (240 rpm) at 22°C with a photoperiod of 18 h light/6 h dark.
Assay of Non-Protein Thiols and Chemical Elements
The total amount of both reduced and oxidized forms of glutathione was determined by the high-performance liquid chromatography (HPLC) method after subsequent extraction, reduction, and derivatization as described by Williams et al. (2002). The amount of reduced thiol groups present in the low-molecular weight compounds was measured as previously described (Wawrzynski et al., 2006) according to the modified Ellman's test (Ellman, 1959).
For quantification of chemical elements, dried plant material was homogenized and, for each sample, 35–70 mg were processed and elemental contents analyzed by inductively coupled plasma optical emission spectroscopy using an IRIS Advantage Duo ER/S (Thermo Jarrell Ash) as described previously (Becher et al., 2004).
Analysis of Circular Dichroism Spectrum
The recombinant UP9C protein with the His-tag at the N-terminus was overproduced in Escherichia coli DH5α. The His-tag was removed by digestion with thrombin (Amersham Biosciences, Vienna, Austria) and the recombinant UP9C protein was refolded in non-denaturing conditions. All measurements were carried out in a 1-mm path-length cell at 298 K in 10 mM TRIS buffer with 5% glycerol and 100 mM NaCl, pH 7.0. Circular dichroism spectra were recorded using an Aviv-202 CD spectrometer equipped with a HP 89100A temperature control unit. The concentration of protein was determined with a Biospectrometer (Eppendorf) using a 6970 molar extinction coefficient calculated by the PROTPARAM program (www.expasy.ch/tools/protparam.html). The spectrum was de-convoluted to the contribution from different secondary structure forms by the K2d program (Andrade et al., 1993).
Yeast Two-Hybrid Experiment
The experiment with the cDNA library from N. tabacum plants grown for 2 d in S-deficient medium was prepared using the BD Machmaker Library Construction Kit (BD-Biosciences-Clontech, Mountain View, CA, USA) as recommended by the manufacturer. The pJKU1 plasmid was used as a bait. The number of analyzed transformants was approximately 2 × 105. The positive prey clones, after retransformation, were confirmed for their ability to activate the three reporter genes, HIS3, ADE2, and lacZ, when co-transforming yeasts with a plasmid containing the coding region of UP9C fused to the DNA fragment encoding the binding domain of GAL4 as bait.
Confocal Microscopy Analysis of Protein Localization
Three to four-week-old seedlings were used in experiment without fixing, stained with DAPI (1 μg ml−1, infiltrated with syringe, after 7 s washed three times, 5 min each with 10 ml dH2O) and visualized using an Eclipse TE2000-E (Nikon) confocal microscope. For the negative control, the seedlings of the wild-type plants were used. The seedlings containing an unfused EGFP construct were analyzed as positive controls. Data for negative and positive control are not shown.
Metabolite Analysis by GC–MS
Material extraction, derivatization, and metabolite analysis were performed as described earlier (Nikiforova et al., 2005). For each group, five samples were independently prepared from the mature leaves combined from five plants. Groups of parental plants were grown in duplicate. Thus, data for the AB3 lines are from five measurements of the independently extracted samples, while data for the control line are from 10 measurements of the independently extracted samples (five for each of the two combined sets of the material from five plants).
Statistical Analysis of Metabolite Data
Data analysis was performed using Statistica 6.0 (StatSoft Polska Sp. z o.o., Poland). When the ratio of the means of two groups of plants was compared, the t-test was used to evaluate the significance of the results. The principal component analysis (PCA) was implemented by using the covariance matrix of the normalized data.
Other In Silico Analysis and Accession Numbers
Similarity searches were performed with BLASTP at NCBI (www.ncbi.nlm.nih.gov/BLAST/) or with BLASTN using the TGI Database of plant EST at DFCI (http://compbio.dfci.harvard.edu/tgi/plant.html). Translation of nucleotide sequences was generated at EMBL–EBI (www.ebi.ac.uk/services/index.html). Multiple sequence alignment was generated by MAFFT using the E-INS-i strategy (http://align.bmr.kyushu-u.ac.jp/mafft/online/server/) (Katoh et al., 2005). The following programs were accessed through the ExPASy Proteomic Server (www.expasy.ch/): GOR4 (Combet et al., 2000) for secondary structure prediction, MULTICOIL (Wolf et al., 1997) for predicting the coiled coil regions, PSORT (Horton and Nakai, 1997) for predicting the localization, SMART (Letunic et al., 2004) and MOTIFSCAN (Falquet et al., 2002) for identification of the protein domains and patterns. The expression of A. thaliana genes was checked using the GENEVESTIGATOR program (Zimmermann et al., 2004) at the Swiss Federal Institute of Technology, Zurich (www.genevestigator.ethz.ch/).
FUNDING
This work was supported by the Polish Ministry of Science and Higher Education (grant No. N N302 119435).
Acknowledgments
The authors acknowledge Joachim Kopka and Alexander Erban (MPI-MP, Golm-Potsdam) for assistance with getting and analyzing GC–MS data, Ute Krämer (MPI-MP, Golm-Potsdam) for mineral elements analysis, and Anna Anielska-Mazur (IBB PAS, Warsaw) for help with microscope analysis. No conflict of interest declared.
References
- Andrade MA, Chacon P, Merelo JJ, Moran F. Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Protein Eng. 1993;6:383–390. doi: 10.1093/protein/6.4.383. [DOI] [PubMed] [Google Scholar]
- Becher M, Talke IN, Krall L, Kramer U. Cross-species microarray transcript profiling reveals high constitutive expression of metal homeostasis genes in shoots of the zinc hyperaccumulator Arabidopsis halleri. Plant J. 2004;37:251–268. doi: 10.1046/j.1365-313x.2003.01959.x. [DOI] [PubMed] [Google Scholar]
- Carrington JC, Freed DD, Leinicke AJ. Bipartite signal sequence mediates nuclear translocation of the plant potyviral NIa protein. Plant Cell. 1991;3:953–962. doi: 10.1105/tpc.3.9.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chico JM, Chini A, Fonseca S, Solano R. JAZ repressors set the rhythm in jasmonate signaling. Curr. Opin. Plant Biol. 2008;11:486–494. doi: 10.1016/j.pbi.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Chintapakorn Y, Hamill JD. Antisense-mediated down-regulation of putrescine N-methyltransferase activity in transgenic Nicotiana tabacum L. can lead to elevated levels of anatabine at the expense of nicotine. Plant Mol. Biol. 2003;53:87–105. doi: 10.1023/B:PLAN.0000009268.45851.95. [DOI] [PubMed] [Google Scholar]
- Combet C, Blanchet C, Geourjon C, Deleage G. NPS@: network protein sequence analysis. Trends Biochem. Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- Dominguez-Solis JR, He Z, Lima A, Ting J, Buchanan BB, Luan S. A cyclophilin links redox and light signals to cysteine biosynthesis and stress responses in chloroplasts. Proc. Natl Acad. Sci. U S A. 2008;105:16386–16391. doi: 10.1073/pnas.0808204105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellman GL. Tissue sulfhydryl groups. Arch Biochem Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- Falquet L, Pagni M, Bucher P, Hulo N, Sigrist CJ, Hofmann K, Bairoch A. The PROSITE database, its status in 2002. Nucleic Acids Res. 2002;30:235–238. doi: 10.1093/nar/30.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halitschke R, Baldwin IT. Antisense LOX expression increases herbivore performance by decreasing defense responses and inhibiting growth-related transcriptional reorganization in Nicotiana attenuata. Plant J. 2003;36:794–807. doi: 10.1046/j.1365-313x.2003.01921.x. [DOI] [PubMed] [Google Scholar]
- Haneklaus S, Kerr CW, Schnug E. A chronicle of sulfur research in agriculture. In: Saito K, et al., editors. Sulfur Transport and Assimilation in Plants in the Post Genomic Era. Leiden, The Netherlands: Backhuys Publishers; 2005. pp. 249–256. [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 2000;42:819–832. doi: 10.1023/a:1006496308160. [DOI] [PubMed] [Google Scholar]
- Hirai MY, Fujiwara T, Awazuhara M, Kimura T, Noji M, Saito K. Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-L-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J. 2003;33:651–663. doi: 10.1046/j.1365-313x.2003.01658.x. [DOI] [PubMed] [Google Scholar]
- Horton P, Nakai K. Better prediction of protein cellular localization sites with the k nearest neighbors classifier. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1997;5:147–152. [PubMed] [Google Scholar]
- Kanyuka K, Praekelt U, Franklin KA, Billingham OE, Hooley R, Whitelam GC, Halliday KJ. Mutations in the huge Arabidopsis gene BIG affect a range of hormone and light responses. Plant J. 2003;35:57–70. doi: 10.1046/j.1365-313x.2003.01779.x. [DOI] [PubMed] [Google Scholar]
- Karimi M, De Meyer B, Hilson P. Modular cloning in plant cells. Trends Plant Sci. 2005;10:103–105. doi: 10.1016/j.tplants.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Kasajima I, Ohkama-Ohtsu N, Ide Y, Hayashi H, Yoneyama T, Suzuki Y, Naito S, Fujiwara T. The BIG gene is involved in regulation of sulfur deficiency-responsive genes in Arabidopsis thaliana. Physiol. Plant. 2007;129:351–363. [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopriva S, Suter M, von Ballmoos P, Hesse H, Krähenbühl U, Rennenberg H, Brunold C. Interaction of sulfate assimilation with carbon and nitrogen metabolism in Lemna minor. Plant Physiol. 2002;130:1406–1413. doi: 10.1104/pp.007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legg PD, Collins GB, Litton CC. Registration of LA Burley 21 tobacco germplasm. Crop Sci. 1970;10:212. [Google Scholar]
- Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, Schultz J, Ponting CP, Bork P. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 2004;32:D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowska M, Sirko A. Recent advances in understanding plant response to sulfur-deficiency stress. Acta Biochim. Polon. 2008;55:457–471. [PubMed] [Google Scholar]
- Lewandowska M, Wawrzynska A, Kaminska J, Liszewska F, Sirko A. Identification of novel proteins of Nicotiana tabacum regulated by short term sulfur starvation. In: Saito K, et al., editors. Sulfur Transport and Assimilation in Plants in the Post Genomic Era. Leiden, The Netherlands: Backhuys Publishers; 2005. pp. 153–156. [Google Scholar]
- Lin Z, Zhong S, Grierson D. Recent advances in ethylene research. J. Exp. Bot. 2009;60:3311–3336. doi: 10.1093/jxb/erp204. [DOI] [PubMed] [Google Scholar]
- Lopez-Bucio J, Hernandez-Abreu E, Sanchez-Calderon L, Perez-Torres A, Rampey RA, Bartel B, Herrera-Estrella L. An auxin transport independent pathway is involved in phosphate stress-induced root architectural alterations in Arabidopsis: identification of BIG as a mediator of auxin in pericycle cell activation. Plant Physiol. 2005;137:681–691. doi: 10.1104/pp.104.049577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Inoue E, Watanabe-Takahashi A, Yamaya T, Takahashi H. Transcriptome profiling of sulfur-responsive genes in Arabidopsis reveals global effects of sulfur nutrition on multiple metabolic pathways. Plant Physiol. 2003;132:597–605. doi: 10.1104/pp.102.019802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Tohge T, Saito K, Takahashi H. Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell. 2006;18:3235–3251. doi: 10.1105/tpc.106.046458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Watanabe-Takahashi A, Inoue E, Yamaya T, Takahashi H. Identification of a novel cis-acting element conferring sulfur deficiency response in Arabidopsis roots. Plant J. 2005;42:305–314. doi: 10.1111/j.1365-313X.2005.02363.x. [DOI] [PubMed] [Google Scholar]
- Migge A, Bork C, Hell R, Becker TW. Negative regulation of nitrate reductase gene expression by glutamine or asparagine accumulating in leaves of sulfur-deprived tobacco. Planta. 2000;211:587–595. doi: 10.1007/s004250000322. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962;15:473–493. [Google Scholar]
- Muttucumaru N, Halford NG, Elmore JS, Dodson AT, Parry M, Shewry PR, Mottram DS. Formation of high levels of acrylamide during the processing of flour derived from sulfate-deprived wheat. J. Agric. Food Chem. 2006;54:8951–8955. doi: 10.1021/jf0623081. [DOI] [PubMed] [Google Scholar]
- Nikiforova V, Freitag J, Kempa S, Adamik M, Hesse H, Hoefgen R. Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: interlacing of biosynthetic pathways provides response specificity. Plant J. 2003;33:633–650. doi: 10.1046/j.1365-313x.2003.01657.x. [DOI] [PubMed] [Google Scholar]
- Nikiforova VJ, Daub CO, Hesse H, Willmitzer L, Hoefgen R. Integrative gene-metabolite network with implemented causality deciphers informational fluxes of sulphur stress response. J. Exp. Bot. 2005;56:1887–1896. doi: 10.1093/jxb/eri179. [DOI] [PubMed] [Google Scholar]
- Parry DA, Fraser RD, Squire JM. Fifty years of coiled-coils and alpha-helical bundles: a close relationship between sequence and structure. J. Struct. Biol. 2008;163:258–269. doi: 10.1016/j.jsb.2008.01.016. [DOI] [PubMed] [Google Scholar]
- Petrucco S, Bolchi A, Foroni C, Percudani R, Rossi GL, Ottonello S. A maize gene encoding an NADPH binding enzyme highly homologous to isoflavone reductases is activated in response to sulfur starvation. Plant Cell. 1996;8:69–80. doi: 10.1105/tpc.8.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser IM, Purves JV, Saker LR, Clarkson DT. Rapid disruption of nitrogen metabolism and nitrate transport in spinach plants deprived of sulphate. J. Exp. Bot. 2001;52:113–121. [PubMed] [Google Scholar]
- Queval G, Thominet D, Vanacker H, Miginiac-Maslow M, Gakiere B, Noctor G. H2O2-activated up-regulation of glutathione in Arabidopsis involves induction of genes encoding enzymes involved in cysteine synthesis in the chloroplast. Mol. Plant. 2009;2:344–356. doi: 10.1093/mp/ssp002. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Frisch EF, Maniattis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Scherer HW. Sulphur in crop production—invited paper. Eur. J. Agron. 2001;14:81–111. [Google Scholar]
- Schnug E, Ernst WHO, Kraztz S, Knolle F, Haneklaus S. Aspects of ecotoxicology of sulphur in the Harz region—a guided excursion. Landbauforschung Voelkenrode. 2004;54:129–143. [Google Scholar]
- Siritunga D, Sayre RT. Generation of cyanogen-free transgenic cassava. Planta. 2003;217:367–373. doi: 10.1007/s00425-003-1005-8. [DOI] [PubMed] [Google Scholar]
- Sponsel VM, Schmidt FW, Porter SG, Nakayama M, Kohlstruk S, Estelle M. Characterization of new gibberellin-responsive semidwarf mutants of Arabidopsis. Plant Physiol. 1997;115:1009–1020. doi: 10.1104/pp.115.3.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov RA, Borisjuk NV, Panchuk II, Schweizer D, Hemleben V. Elimination and rearrangement of parental rDNA in the allotetraploid Nicotiana tabacum. Mol. Biol. Evol. 1999;16:311–320. doi: 10.1093/oxfordjournals.molbev.a026112. [DOI] [PubMed] [Google Scholar]
- Wawrzynski A, Kopera E, Wawrzynska A, Kaminska J, Bal W, Sirko A. Effects of simultaneous expression of heterologous genes involved in phytochelatin biosynthesis on thiol content and cadmium accumulation in tobacco plants. J. Exp. Bot. 2006;57:2173–2182. doi: 10.1093/jxb/erj176. [DOI] [PubMed] [Google Scholar]
- Wawrzynska A, Lewandowska M, Sirko A. Nicotiana tabacum EIL2 directly regulates expression of at least one tobacco gene induced by sulphur starvation. J. Exp. Bot. 2009 doi: 10.1093/jxb/erp356. doi: 10.1093/jxb/erp356. [DOI] [PubMed] [Google Scholar]
- Wawrzynska A, Lewandowska M, Hawkesford MJ, Sirko A. Using a suppression subtractive library-based approach to identify tobacco genes regulated in response to short-term sulphur deficit. J. Exp. Bot. 2005;56:1575–1590. doi: 10.1093/jxb/eri152. [DOI] [PubMed] [Google Scholar]
- Williams JS, Hall SA, Hawkesford MJ, Beale MH, Cooper RM. Elemental sulfur and thiol accumulation in tomato and defense against a fungal vascular pathogen. Plant Physiol. 2002;128:150–159. [PMC free article] [PubMed] [Google Scholar]
- Wolf E, Kim PS, Berger B. MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci. 1997;6:1179–1189. doi: 10.1002/pro.5560060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JT, Wu CS, Martinez HM. Calculation of protein conformation from circular dichroism. Methods Enzymol. 1986;130:208–269. doi: 10.1016/0076-6879(86)30013-2. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]