Abstract
The ability to adjust growth and development to the availability of mineral nutrients in the soil is an essential life skill of plants but the underlying signaling pathways are poorly understood. In Arabidopsis thaliana, shortage of potassium (K) induces a number of genes related to the phytohormone jasmonic acid (JA). Using comparative microarray analysis of wild-type and coi1-16 mutant plants, we classified transcriptional responses to K with respect to their dependence on COI1, a central component of oxylipin signaling. Expression profiles obtained in a short-term experiment clearly distinguished between COI1-dependent and COI1-independent K-responsive genes, and identified both known and novel targets of JA-COI1-signaling. During long-term K-deficiency, coi-16 mutants displayed de novo responses covering similar functions as COI1-targets except for defense. A putative role of JA for enhancing the defense potential of K-deficient plants was further supported by the observation that plants grown on low K were less damaged by thrips than plants grown with sufficient K.
Keywords: Hormonal regulation, nutrition, secondary metabolism/natural products, transcriptome analysis, plant–insect interactions, Arabidopsis
INTRODUCTION
Potassium (K) is an essential macronutrient for all living organisms due to vital functions in enzyme activation, protein synthesis, solute transport and osmoregulation (Marschner, 1995; Oria-Hernandez et al., 2005). In the field, the demand for K of young, fast-growing crops is high and therefore K supply can become yield-limiting even when fertilizers are applied (Syers, 1998; Dobermann et al., 1999). K-deficiency impacts not only on crop yield, but also on crop quality, taste, mechanical properties and stress resistance (Marschner, 1995; Laegreid et al., 1999; Amtmann, 2009).
High and low-affinity K transport proteins facilitate uptake of K from the soil and its reallocation between different tissues (Epstein et al., 1963; Maathuis and Sanders, 1996; for review, see Amtmann et al., 2004; Ashley et al., 2006). Regulation of these transporters has been an area of intensive research (reviewed in Véry and Sentenac, 2003; Amtmann and Blatt, 2009), and the recent discovery of the Ca/CBL/CIPK pathway (Li et al., 2006; Xu et al., 2006) has further enhanced our understanding of how plants adjust K uptake to the external K supply. Such control mechanisms endow plants with the capacity for efficient cellular and whole-plant K homeostasis (Walker et al., 1996), thereby allowing them to cope with transient fluctuations of external K.
If K-deficiency persists, plants have to initiate a much wider adaptive response, which involves re-prioritization of growth, development and metabolism to ensure maximal seed production under nutrient-limited conditions. Field and glasshouse trials have produced a vast amount of data on the physiological consequences of K-deficiency (reviewed in Kafkafi et al., 2001), but it is not known which of these reflect unavoidable damage and which have adaptive function. Even less is known about the underlying regulatory mechanisms. A role of the ethylene pathway in K-perception and signaling in the roots is now emerging (Shin and Schachtman, 2004; Amtmann et al., 2005; Shin et al., 2005; Schachtman and Shin, 2006; Amtmann and Armengaud, 2007; Pandey et al., 2007; Jung et al., 2009) but we still lack essential information about the signaling pathways that integrate adaptive responses to K-deficiency in the leaves.
Previous microarray analysis of Arabidopsis thaliana plants subjected to changes in external K supply indicated that the phytohormone jasmonic acid (JA) could also be involved in plant responses to K-deficiency (Armengaud et al., 2004). K-responsiveness at the transcript level was found for enzymes involved in JA biosynthesis (e.g. lipoxygenase, LOX2) and known targets of JA-signaling (e.g. vegetative storage protein, VSP2; see Figure 3 in Armengaud et al., 2004). A search with Genevestigator (Zimmermann et al., 2004) shows that most of these genes respond also to treatments with methyl-jasmonate (MeJA) or the JA precursor OPDA (see Supplemental Figure A in SI4). The transcriptional profile suggested a reversible increase in JA levels during K-deficiency (Amtmann et al., 2004), and this was subsequently confirmed by others (Cao et al., 2006). JA is known to integrate plant responses to developmental and environmental cues, such as senescence, wounding, and defense (Creelman and Mullet, 1997), but had not previously been linked to nutrient stress. From the transcript profiles and the existing knowledge of JA-dependent processes, we developed a model in which JA links a K-deficiency signal to a number of physiological responses (see Figure 5 in Armengaud et al., 2004), including growth inhibition (Staswick et al., 1992), nutrient recovery from senescent tissues (He et al., 2002), production of organic cations (Perez-Amador et al., 2002), as well as control of ion transport and stomatal closure (Evans, 2003; Munemasa et al., 2007). While this scheme provided a useful working model to test possible roles of JA in plant adaptation, it lacked direct evidence for JA-dependence of the underlying transcript responses to K.
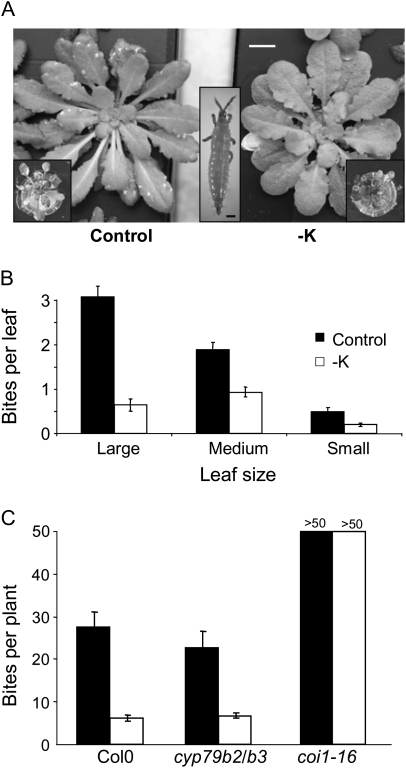
Figure 3.
K-Deficiency Reduced Thrips Damage on A. thaliana.
(A) Typical appearance of control (left) or K-starved (right) Arabidopsis thaliana Col0 wild-type plants exposed to thrips (Frankliniella sp., middle inset, bar is 100 μm). Insect bites are visible as white spots on the leaves of control plants. Bar = 1 cm.
(B) Numbers of insect bites on leaves of control (black bars) or K-starved (white bars) wild-type plants. Forty plants were analyzed for each condition. Average numbers of bites for different leaf sizes (large: >25 mm2, medium: 10–25 mm2, small: <10 mm2 leaf surface) are shown. All differences are significant at p < 0.001 (t-test).
(C) Numbers of insect bites on leaves of control (black bars) or K-starved (white bars) wild-type and mutant plants. Mutants were defective for JA-dependent indolic gucosinolate production (cyp79b2/b3) or JA-signaling through COI1 (coi1-16).
A color version of this figure is available from the Supplemental Material (SI6).
The JA-signaling pathway involved in responses to MeJA treatment, wounding and biotic stress has been studied extensively (Balbi and Devoto, 2008). A central role was assigned to COI1, a F-box protein that determines the substrate specificity of the E3 ubiquitin ligase SCFC°I1 complex (Xie et al., 1998; Devoto et al., 2005; Devoto and Turner, 2005; Yan et al., 2009). This complex targets proteins that act as repressors of JA-induced transcriptional responses for degradation, and thus constitutes a central component of JA-signaling (Staswick, 2008). Recently, it was shown that COI1 itself is a receptor for Ile-JA (Yan et al., 2009).
In this study, we have used microarrays to evaluate which transcriptional responses of A. thaliana plants to varying external K supply require the presence of a functional COI1 gene by comparing transcriptional responses in wild-type with those in coi1-16 mutants. Clearly, microarray analysis can only be a first step towards unraveling the role of COI1 in the complex regulatory network underlying plant responses to K. Nevertheless, the analysis clearly showed that the number of genes responding to K-treatment was reduced in coi1-16 mutants. Based on a quantitative comparison of transcript changes between wild-type and coi1-16 plants K-responsive genes were assigned into four classes of transcript profiles with respect to external K supply and COI1-dependence. While many genes responded to K in a COI1-dependent manner, the function of COI1 in plant adaptation to K-stress seems to be redundant because coi1-16 mutants are not affected in their growth under long-term K-starvation. However, experiments with herbivorous insects indicate that a necessary function of COI1 in low K is apparent when K-deficiency is accompanied by biotic stress.
RESULTS
Physiological and Developmental Phenotype of coi1-16 Plants on Low K
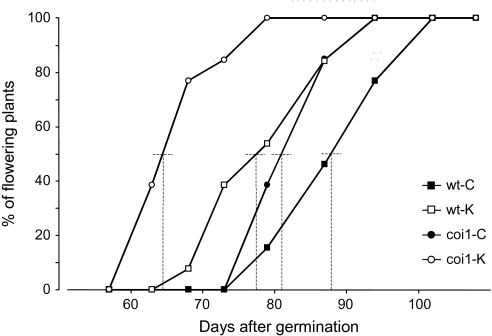
To investigate the role of COI1 in plant responses to K-deficiency, we analyzed the phenotype of coronatine-insensitive (coi) 1–16 mutants (Ellis and Turner, 2002). There was no indication that coi1-16 plants were more severely affected by K-starvation than wild-type plants. The relative reduction in fresh weight caused by low K was even slightly less in coi1-16 mutants than in wild-type (Table 1). No significant differences between K-deficient wild-type and coi1-16 mutant plants were detected concerning water or K content (Table 1). However, coi1-16 mutants flowered earlier than the wild-type in control medium, and this phenotype was enhanced on low K (Figure 1). While wild-type plants flowered approximately 10 d earlier in low K than in control medium, coi1-16 plants flowered more than 20 d earlier in low K than in control medium.
Table 1.
Fresh Weight, Water Content, and Leaf K-Concentration in Wild-Type and coi1-16 Mutants.
| Fresh weight (% control) | wt | coi1-16 |
| K-deficiency3 | 45.3 ± 5.2 | 55.1 ± 0.8 [0.1] |
| K re-supply4 | 104.4 ± 5.3 | 106.2 ± 4.7 [0.4] |
| Water content (%) | wt | coi1-16 |
| Control plants | 92.7 ± 0.4 | 92.4 ± 0.2 [0.4] |
| K-deficient | 90.8 ± 0.6 | 90.5 ± 0.3 [0.4] |
| K re-supplied | 91.1 ± 0.2 | 91.4 ± 0.2 [0.1] |
| K-concentration (% DW) | wt | coi1-16 |
| Control plants | 5.9 ± 0.5 | 5.3 ± 0.1 [0.2] |
| K-deficient | 1.1 ± 0.1 | 1.0 ± 0.1 [0.4] |
| K re-supplied | 2.2 ± 0.3 | 2.3 ± 0.1 [0.4] |
Figure 1.
Onset of Flowering in a Population of Wild-Type and coi1-16 A. thaliana Plants.
Plants were grown hydroponically in control or low-K medium. At each time point, the number of plants that flowered was counted. For each genotype and condition, the time point at which 50% of the plants flowered is indicated with a dashed line.
Reduced Number of K-Responsive Transcripts in coi1-16 Plants
To investigate the requirement of a functional JA-COI1-signaling pathway for transcriptional responses to K, we repeated microarray experiments previously carried out with wild-type plants (Armengaud et al., 2004) with coi1-16 mutant plants. As before, two treatments were applied (Armengaud et al., 2004). In a long-term-starvation experiment, plants were grown from germination for 2 weeks on a medium that was not supplied with K (‘K-free’ medium; see Methods). In short-term re-supply experiments, K-starved plants were supplied with K (or K-free medium as control) for 6 h. Shoot material was harvested from three separately grown plant batches. RNA was isolated, labeled, and hybridized to A. thaliana microarrays (University of Arizona) as before.
For each treatment, genes were ranked in two lists (one for up and one for down-regulation) according to the strength and significance of their response across the replicate experiments using Rank Products (Breitling et al., 2004). Detailed information on all genes that showed K-dependent transcript changes in coi1-16 is provided in the Supplementary Information SI1. As expected, if JA plays an important role in K-signaling, the total number of K-responsive genes was smaller in coi1-16 than in wild-type plants (Table 2). The effect was particularly evident for genes up-regulated during K-starvation and down-regulated upon K re-supply, which was indeed the transcriptional profile most clearly related to JA in wild-type plants (Armengaud et al., 2004).
Table 2.
Numbers of K-Responsive Genes in Wild-Type (wt) and coi1-16 Mutant Plants
| K-deficiency (14 d) |
K re-supply (6 h) |
|||||||
| Down |
Up |
Down |
Up |
|||||
| FDR1(%) | wt | coi1-16 | wt | coi1-16 | wt | coi1-16 | wt | coi1-16 |
| <0.01 | 1 | 0 | 19 | 5 | 3 | 1 | 4 | 3 |
| <0.1 | 8 | 2 | 45 | 19 | 12 | 2 | 13 | 17 |
| <1 | 17 | 21 | 99 | 51 | 56 | 10 | 50 | 86 |
False discovery rate.
‘Loss’ and ‘Gain’ of Transcriptional Regulation by K in coi1-16 Mutants
For comparison of transcript changes in response to the K treatments between wild-type and coi1-16 genotypes, we employed an algorithm that was previously developed by our group to provide a means for quantitative and statistically testable two-factor comparison, in this case nutrient supply (factor 1) and genotype (factor 2). Vector Analysis (Breitling et al., 2005) has several advantages over other methods for comparative microarray data analysis. In particular, it does not require pre-assignment of transcripts into K-‘regulated’ and ‘non-regulated’ genes, thereby avoiding wrong interpretation of transcript changes lying closely at either side of a cut-off value (e.g. 1.9-fold and 2.1-fold) as being ‘different’, or of those lying above the cut-off but displaying large differences (e.g. two-fold and 20-fold) as being ‘the same’.
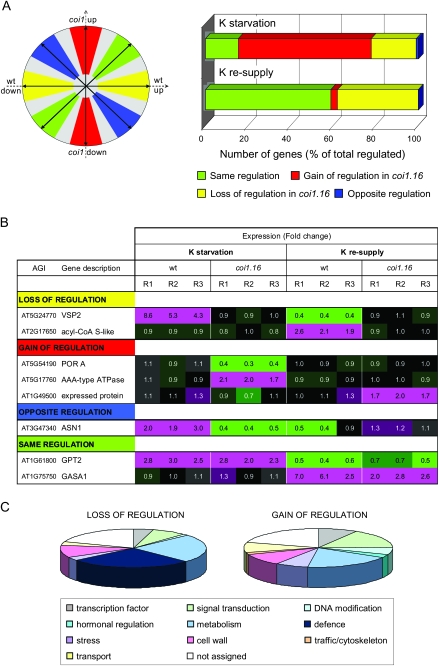
For each gene, fold-changes of transcript levels measured in coi1-16 were plotted against wild-type fold-changes (nine pair-wise comparisons from three replicates). Based on the direction of the resulting sum vector, the obtained transcriptional responses were assigned into four main classes (Figure 2A), namely ‘same regulation’ (up or down-regulated in both genetic backgrounds), ‘loss of regulation’ (up or down-regulated in wild-type but unchanged in coi1-16), ‘gain of regulation’ (unchanged in wild-type but up or down-regulated in coi1-16), and ‘opposite regulation’ (up-regulated in wild-type and down-regulated in coi1-16 or vice versa). Statistical evaluation of the transcript behavior was based on three parameters: the average length of the replicate vectors (l) as a measure of overall strength of the response, the length of the sum vector (p) as a measure of the consistency of the response across replicates, and the angle between the sum-vector and the closest prototypic vector (a) as a measure of the clarity of the response. Figure 2A shows the relative number of genes identified for each class after applying a common set of statistical constraints (see Methods). The analysis was carried out separately for long-term K-starvation and short-term K-re-supply.
Figure 2.
Effect of COI1 on K-Dependent Gene Expression.
(A) Comparison of transcriptional profiles between wild-type and coi1-16 mutant plants. The scheme on the left shows the assignment of sum vectors obtained by Vector Analysis into regulatory classes. For each gene, fold-changes of transcript levels measured in coi1-16 in response to K-starvation or K re-supply are plotted against the respective wild-type fold-changes resulting in nine vectors (from three replicate experiments). The direction of the sum vector allows assignment of the obtained transcriptional responses into four regulatory groups, namely ‘same regulation’ (green), ‘loss of regulation’ (yellow), ‘gain of regulation’ (red), and ‘opposite regulation’ (blue). Prototypic sum vectors ideally representing each regulatory class are drawn. Genes with sum vectors in gray areas were excluded from further analysis based on inconsistency of the response or ambiguity of the classification. The chart on the right shows the relative number of genes identified for each class (same color-coding as on the left) after applying a common set of statistical constraints (see Methods). Upper bar: K-starvation (104 transcripts in total). Lower bar: K re-supply (129 transcripts in total). For individual genes, see Tables 3 and 4, and SI2.
(B) Examples of expression profiles within the four regulatory classes. Transcripts were identified as belonging to a particular regulatory class (same color coding as in (A)) for K-starvation, re-supply, or both treatments. R1–R3 denote replicate experiments of the same treatment, fold-changes are given in boxes, up-regulation is shown in pink, down-regulation in green.
(C) Functional categories of genes displaying loss or gain of regulation by K in coi1-16 mutants. For individual genes, see SI2. Note that defense-related genes were not represented among genes that gained responsiveness to K in coi1-16.
Interestingly, the effect of COI1 disruption on K-dependent transcriptional changes differed considerably between the two K treatments. In response to K re-supply, most of the transcripts showed either the same regulation in the two genotypes (56%) or a loss of K-dependence in coi1-16 mutants (36%). By contrast, after long-term starvation, only 15% of all K-regulated genes showed the same response in coi1-16 and wild-type, while the majority of transcripts (65%) displayed changes in coi1-16 but not in wild-type plants (‘gain of regulation’). Figure 2B shows examples of transcriptional profiles represented in the different regulatory classes. All genes assigned to a particular regulatory class are listed in Tables 3 (K-starvation) and 4 (K re-supply), and their transcriptional profiles are presented in the Supplementary File SI2.
Table 3.
Transcriptional Response in coi1-16 Mutants Compared to wt1: K-Starvation
| AGI | Name | Description | Statistics2 |
Average fold change3 |
|||
| a | l | p | wt | coi | |||
| LOSS OF REGULATION | |||||||
| At1g26420 | BBE | FAD-linked oxidoreductase family | 2.1 | 0.6 | 0.0 | 1.8 | 1.0 |
| At4g15100 | SCPL30 | Serine carboxypeptidase S10 family | 5.0 | 0.5 | 0.1 | 1.9 | 1.1 |
| At5g24420 | 6PGL | 6-phosphogluconolactonase-like | 8.1 | 1.7 | 0.0 | 5.2 | 1.3 |
| At1g76790 | O-methyltransferase, family 2 | 9.2 | 0.5 | 0.3 | 1.7 | 0.9 | |
| At4g08870 | ARGAH2 | Putative arginase | 9.2 | 1.2 | 0.0 | 3.6 | 1.2 |
| At1g04710 | Putative acetyl-CoA acyltransferase | 9.4 | 0.7 | 0.4 | 2.0 | 1.1 | |
| At5g05210 | Putative protein | 12.3 | 0.8 | 0.2 | 2.3 | 1.2 | |
| At1g26400 | FAD-linked oxidoreductase family | 12.5 | 1.1 | 0.5 | 2.7 | 1.2 | |
| At2g39030 | Expressed protein | 12.7 | 0.9 | 0.4 | 2.7 | 1.2 | |
| At4g24350 | Putative protein | 12.9 | 0.6 | 0.6 | 1.6 | 0.9 | |
| At2g39330 | Putative myrosinase-binding protein | 1.2 | 1.2 | 0.0 | 3.1 | 1.0 | |
| At5g24770 | VSP2 | Vegetative storage protein 2 | 1.6 | 1.7 | 0.0 | 6.1 | 0.9 |
| At5g44420 | PDF1.2 | Plant defensin protein | 2.7 | 1.1 | 0.4 | 5.9 | 1.1 |
| At1g79720 | Putative aspartyl protease | 4.1 | 0.8 | 0.2 | 2.1 | 1.1 | |
| At2g26010 | PDF1.3 | Plant defensin protein, putative | 4.8 | 1.5 | 0.0 | 5.6 | 1.2 |
| At5g24780 | VSP1 | Vegetative storage protein 1 | 6.8 | 2.3 | 0.0 | 10.2 | 1.3 |
| At2g43530 | Putative trypsin inhibitor | 8.7 | 0.6 | 0.0 | 1.8 | 1.1 | |
| At5g28510 | Glycosyl hydrolase family 1 | 4.5 | 2.1 | 0.0 | 7.6 | 1.2* | |
| At1g52400 | BG1 | Glycosyl hydrolase family 1 | 10.3 | 2.1 | 0.3 | 8.7 | 1.5 |
| At4g30620 | Putative protein | 4.5 | 0.7 | 0.4 | 2.3 | 1.0 | |
| At2g23440 | Unknown protein | 5.2 | 1.1 | 0.2 | 3.2 | 1.1 | |
| At5g52110 | Putative protein | 10.8 | 0.7 | 0.6 | 2.0 | 1.2 | |
| GAIN OF REGULATION | |||||||
| At5g61590 | ERF/AP2/B3 | Ethylene responsive element binding | 1.6 | 0.5 | 0.2 | 1.0 | 0.6 |
| At3g52270 | Transcription initiation factor IIF | 3.9 | 0.7 | 0.1 | 1.0 | 0.6 | |
| At5g26930 | GAtA zinc finger protein | 10.9 | 0.6 | 0.2 | 0.9 | 0.6 | |
| At3g61830 | ARF18 | Auxin response factor-like protein | 14.3 | 0.7 | 0.4 | 0.8 | 0.4 |
| At3g45390 | LRK1 | Receptor-like protein kinase | 0.6 | 0.5 | 0.5 | 1.0 | 0.6 |
| At1g10660 | Unknown protein | 0.6 | 0.5 | 0.2 | 1.0 | 0.6 | |
| At1g25390 | Wall-associated kinase, putative | 2.2 | 0.8 | 0.8 | 1.0 | 0.5 | |
| At3g08510 | PCL2 | Phospholipase C | 2.3 | 0.5 | 0.1 | 1.0 | 0.7 |
| At4g39890 | RABH1C | GTP-binding protein, putative | 2.5 | 0.8 | 0.2 | 1.0 | 0.5 |
| At1g11350 | Serine/threonine kinase, putative | 2.5 | 0.5 | 0.4 | 1.0 | 0.7 | |
| At4g19110 | Kinase-like protein | 3.6 | 0.6 | 0.5 | 1.0 | 0.5 | |
| At2g29800 | Hypothetical protein | 5.5 | 0.6 | 0.0 | 1.1 | 0.6 | |
| At4g23190 | CRK11 | Serine/threonine kinase-like protein | 6.8 | 0.7 | 0.2 | 1.1 | 0.6 |
| At4g13490 | Putative protein | 13.4 | 0.6 | 1.0 | 0.9 | 0.6 | |
| At3g61260 | Putative DNA-binding protein | 2.6 | 0.6 | 0.6 | 1.0 | 0.6 | |
| At4g00850 | GIF3 | A. thaliana cDNA T45454 | 2.7 | 0.6 | 0.1 | 1.0 | 0.6 |
| At5g24490 | 30S | Putative protein | 5.3 | 1 | 0.4 | 1.1 | 0.5 |
| At1g13270 | MAP1C | Methionine aminopeptidase I | 1.2 | 0.6 | 0.9 | 1.0 | 0.6 |
| At5g54190 | PORA | Protochlorophyllide oxidoreductase | 2.8 | 1 | 0.3 | 1.1 | 0.4 |
| At3g29320 | Glucan phosphorylase, putative | 3.7 | 0.5 | 0.2 | 1.0 | 0.6 | |
| At5g28840 | GME | Epimerase/dehydratase-like protein | 3.8 | 0.6 | 0.6 | 1.0 | 0.6 |
| At1g21100 | O-methyltransferase 1, putative | 4.9 | 1 | 0.6 | 0.9 | 0.4 | |
| At4g27440 | PORB | Protochlorophyllide reductase | 5.4 | 0.9 | 0.1 | 1.1 | 0.4 |
| At1g61520 | LHCA3 | Light-harvesting chlorophyll a/b | 6.2 | 0.5 | 0.3 | 0.9 | 0.6 |
| At1g09350 | Putative galactinol synthase | 6.6 | 0.7 | 0.2 | 1.0 | 0.6 | |
| At3g19820 | DWF1 | Cell elongation protein, Dwarf1 | 9.8 | 0.7 | 0.3 | 0.9 | 0.6 |
| At5g65780 | Branched-chain aa aminotransferase | 12.2 | 0.5 | 0.5 | 1.1 | 0.7 | |
| At4g08670 | Putative lipid transfer protein | 14.9 | 0.5 | 0.9 | 0.9 | 0.6 | |
| At3g23175 | Expressed protein | 0.1 | 0.6 | 0.7 | 1.0 | 0.6 | |
| At1g06460 | ADC31.2 | Heat shock protein, putative | 9.2 | 0.7 | 0.7 | 1.1 | 0.5 |
| At4g21580 | NADPH quinone oxidoreductase | 11.8 | 0.5 | 0.6 | 1.1 | 0.7 | |
| At4g35770 | Senescence-associated protein 1 | 13.2 | 0.7 | 0.7 | 1.2 | 0.5 | |
| At3g23920 | BMY7 | Glycosyl hydrolase family 14 | 0.3 | 0.5 | 0.2 | 1.0 | 0.6 |
| At1g47705 | F16N3.33, putative peroxidase | 5.2 | 0.6 | 0.3 | 1.0 | 0.6 | |
| At2g16890 | Putative glucosyltransferase | 9 | 0.8 | 0.3 | 1.1 | 0.5 | |
| At3g08670 | Hypothetical protein | 12.3 | 0.7 | 0.1 | 1.2 | 0.5 | |
| At5g55230 | MAP65-1 | Putative protein | 9.4 | 0.6 | 0.4 | 0.9 | 0.6 |
| At2g24710 | GLR2.3 | Putative ligand-gated ion channel | 4.7 | 0.6 | 0.8 | 1.0 | 0.6 |
| At3g46560 | TIM9 | Small zinc finger-like protein TIM9 | 6.4 | 0.6 | 0.4 | 1.1 | 0.6 |
| At2g28180 | AtCHX8 | Hypothetical protein | 8.8 | 0.6 | 0.9 | 0.9 | 0.6 |
| At4g18200 | Putative protein | 10.7 | 0.9 | 0.4 | 0.9 | 0.5 | |
| At2g41560 | Potential Ca2+-ATPase, isoform 4 | 13.1 | 0.5 | 0.8 | 0.9 | 0.6 | |
| At2g29680 | CDC6 | Putative CDC6 protein | 2.9 | 0.8 | 0.0 | 1.0 | 0.5 |
| At3g57785 | Expressed protein | 0.4 | 0.8 | 0.2 | 1.0 | 0.5 | |
| At1g17690 | Expressed protein | 1 | 1 | 0.2 | 1.0 | 0.4 | |
| At1g24060 | Hypothetical protein | 2.2 | 0.5 | 0.8 | 1.0 | 0.6 | |
| At3g42190 | Putative protein | 2.9 | 0.6 | 0.0 | 1.0 | 0.6 | |
| At1g53870 | Expressed protein | 4 | 0.9 | 0.0 | 1.1 | 0.4 | |
| At3g24250 | Hypothetical protein | 5.1 | 0.7 | 0.1 | 1.1 | 0.5 | |
| At1g73140 | Hypothetical protein | 6 | 0.5 | 0.6 | 1.1 | 0.6 | |
| At2g15890 | Expressed protein | 6.5 | 1.3 | 0.6 | 1.1 | 0.4 | |
| At1g78460 | Hypothetical protein | 9.6 | 0.5 | 0.9 | 1.1 | 0.6 | |
| At1g80720 | Expressed protein | 10.4 | 0.7 | 0.3 | 1.1 | 0.5 | |
| At3g19460 | Expressed protein | 11.2 | 0.8 | 0.2 | 0.8 | 0.5 | |
| At2g31110 | Hypothetical protein | 14.8 | 0.6 | 0.2 | 1.2 | 0.6 | |
| At4g00150 | SCL6 | Scarecrow-like 6 (SCL6) | 10.7 | 0.5 | 1.0 | 0.9 | 1.7 |
| At5g10380 | Putative protein | 10.8 | 1 | 0.6 | 1.3 | 2.9 | |
| At3g42270 | Putative protein | 14 | 0.5 | 0.2 | 1.1 | 1.6 | |
| At5g44390 | BBE | FAD-linked oxidoreductase family | 11 | 0.6 | 0.0 | 1.1 | 1.7 |
| At2g17480 | AtMLO8 | Similar to Mlo proteins H. vulgare | 10.8 | 0.5 | 0.6 | 1.1 | 1.7 |
| At2g43570 | Glycosyl hydrolase family 19 | 5.6 | 0.6 | 0.8 | 1.1 | 1.7 | |
| At4g08410 | Extensin-like protein | 14 | 0.6 | 0.1 | 1.1 | 1.6 | |
| At1g76930 | AtEXT4 | Expressed protein | 14.7 | 0.7 | 0.5 | 1.2 | 1.8 |
| At5g17760 | BCS1-like protein | 0.9 | 0.7 | 0.8 | 1.0 | 2.0 | |
| At3g26470 | Expressed protein | 2.1 | 0.6 | 0.9 | 1.0 | 1.8 | |
| OPPOSITE REGULATION | |||||||
| At3g47340 | ASN1 | Asparagine synthetase | 1 | 1.2 | 0.6 | 2.3 | 0.4 |
| SAME REGULATION | |||||||
| At3g60390 | HAT3-TF | Homeobox-leucine zipper protein | 7.5 | 0.6 | 0.5 | 0.7 | 0.7 |
| At1g26680 | B3-TF | Hypothetical protein | 7.8 | 1.7 | 0.3 | 0.4 | 0.3 |
| At2g42600 | PPC2 | Phosphoenolpyruvate carboxylase | 0.6 | 0.6 | 0.0 | 0.7 | 0.7 |
| At4g04955 | ALN | Expressed protein | 9.7 | 0.6 | 0.2 | 0.6 | 0.7 |
| At5g40780 | LHT1 | Amino acid permease | 12.4 | 0.8 | 0.3 | 0.7 | 0.5 |
| At3g04070 | NAM-TF | NAM-like protein | 6.7 | 0.5 | 0.8 | 1.5 | 1.4 |
| At3g15950 | TSA1-Like | Unknown protein | 13.4 | 0.6 | 0.3 | 1.8 | 1.4 |
| At3g55190 | Lipase-like protein | 12.1 | 0.5 | 0.5 | 1.3 | 1.6 | |
| At4g37990 | ELI3-2 | Cinnamyl-alcohol dehydrogenase | 0.7 | 0.6 | 0.5 | 1.5 | 1.5 |
| At2g37770 | Aldo/keto reductase family | 1.5 | 0.7 | 0.8 | 1.8 | 1.6 | |
| At1g61800 | GPT2 | Gluc-6-P/P-translocator precursor | 4.3 | 1.3 | 0.1 | 2.8 | 2.4 |
| At5g06320 | NHL3 | Harpin-induced protein-like | 1.3 | 0.9 | 0.7 | 1.8 | 1.9 |
| At2g43510 | TI1 | Putative trypsin inhibitor | 5.6 | 1.1 | 0.3 | 2.2 | 1.9 |
| At3g51860 | CAX3 | Ca2+/H+-exchanging protein-like | 6.6 | 1.3 | 1.0 | 2.5 | 2.8 |
| At1g54570 | Expressed protein | 8.1 | 0.8 | 0.9 | 1.6 | 1.9 | |
| At1g19180 | JAZ1 | Expressed protein | 14.1 | 1.0 | 0.7 | 2.3 | 1.7 |
Regulatory classes determined with vector analysis (Breitling et al., 2005).
Statistical parameters: angle (a), average vector length (l), consistency p-value in % (p).
From three replicate experiments.
Note that this gene is down-regulated by low K in pen2 (see SI5).
Many genes were quickly down-regulated upon re-supply of K in the wild-type (Armengaud et al., 2004). This response was significantly attenuated in coi1-16 mutants in the case of at least 45 genes (Table 4 and Supplementary File SI2), including vegetative storage protein VSP2 (At5g24770), the cytochrome P450 CYP79B3 (At2g22330, involved in glucosinolate production (Hull et al., 2000), and a number of myrosinase-associated and myrosinase-binding proteins (e.g. MBP1, At1g52040), as well as several stress and defense-related transcripts such as the dehydration-responsive NAC transcription factor RD26 (At4g27410), an aspartyl protease (At1g79720), and a disease resistance protein (At3g25020, Yamaguchi-Shinozaki et al., 1992). COI1-dependence was also found for down-regulation of genes encoding the cation co-transporter CAX7 (At5g17860), several metabolic enzymes (ARGAH2, At4g08870; MGD3, At2g11810; PEPCK, At4g37870; GSTU4, At2g29460), as well as regulatory proteins (putative caltracin At2g46600, serine threonine protein kinase At5g15080, protein phosphatase 2C At5g59220, growth regulating factor AtGRF2, At4g37740; Kim et al., 2003). Up-regulation of AGP17 (At2g23130) encoding an arabinogalactan protein (Sun et al., 2005) and an acyl-CoA synthetase-like protein (At2g17650) by K re-supply was also lost in the coi1-16 mutant. By contrast, the response to K of ADC2 (At4g34710, SPE2), encoding arginine decarboxylase required for polyamine biosynthesis, was unchanged in coi1-16, despite the fact that ADC2 expression has been reported to be JA-dependent (Perez-Amador et al., 2002). This confirms the notion that JA also employs COI1-independent-signaling pathways (Devoto et al., 2005).
Table 4.
Transcriptional Response in coi-16 Mutants Compared to wt1: K Re-Supply.
| AGI | Name | Description | Statistics2 |
Average fold change3 |
|||
| a | l | p | wt | coi | |||
| LOSS OF REGULATION | |||||||
| At4g27410 | RD26 | Putative protein | 1.8 | 0.6 | 0.0 | 0.6 | 1.0 |
| At3g28650 | CHP-rich zinc finger protein | 2.4 | 0.7 | 0.3 | 0.6 | 1.0 | |
| At4g37740 | GRF2 | Putative protein | 4.8 | 0.7 | 0.3 | 0.5 | 1.0 |
| At2g04840 | Hypothetical protein | 0.1 | 0.8 | 0.4 | 0.5 | 1.0 | |
| At4g22250 | Hypothetical protein | 8.8 | 0.5 | 0.7 | 0.6 | 1.1 | |
| At5g15080 | Serine/threonine protein kinase | 12.0 | 0.8 | 0.7 | 0.5 | 0.9 | |
| At5g59220 | PP2C | Protein phosphatase 2C | 13.1 | 0.7 | 0.0 | 0.5 | 0.9 |
| At2g46600 | Putative caltractin | 14.2 | 0.9 | 0.2 | 0.4 | 0.8 | |
| At4g03920 | Putative protein | 2.8 | 0.6 | 0.6 | 0.6 | 1.0 | |
| At3g44860 | Methyltransferase-related | 2.3 | 1.0 | 0.3 | 0.3 | 1.1 | |
| At4g08870 | ARGAH2 | Putative arginase | 7.2 | 0.9 | 0.2 | 0.4 | 0.9 |
| At4g37870 | PEPCK | Phosphoenolpyruvate carboxykinase | 8.0 | 0.7 | 0.9 | 0.5 | 0.9 |
| At1g26570 | UGD1 | UDP-glucose dehydrogenase | 8.7 | 0.7 | 0.4 | 0.5 | 0.9 |
| At2g11810 | MGD3 | Monogalactosyldiacylglycerol synth | 8.9 | 0.9 | 0.0 | 0.4 | 0.9 |
| At3g44870 | Methyltransferase-related | 13.5 | 0.8 | 0.1 | 0.4 | 1.2 | |
| At2g22330 | CYP79B3 | Putative cytochrome P450 | 14.2 | 0.6 | 0.0 | 0.5 | 0.9 |
| At1g54020 | Myrosinase-associated protein | 1.9 | 1.1 | 0.0 | 0.3 | 1.0 | |
| At1g52040 | MBP1 | Myrosinase-binding protein | 5.4 | 0.9 | 0.1 | 0.5 | 1.1 |
| At5g38540 | Myrosinase binding protein-like | 9.8 | 0.8 | 0.2 | 0.5 | 0.9 | |
| At3g21380 | Unknown protein | 11.9 | 0.5 | 0.3 | 0.6 | 0.9 | |
| At5g61820 | Putative protein | 12.5 | 0.5 | 0.6 | 0.6 | 0.9 | |
| At5g24770 | VSP2 | Vegetative storage protein 2 | 3.2 | 0.9 | 0.2 | 0.4 | 1.0 |
| At1g64160 | Disease resistance response protein | 5.8 | 0.6 | 0.3 | 0.6 | 1.0 | |
| At1g72260 | Thi2.1 | Thionin | 6.0 | 0.7 | 0.2 | 0.5 | 0.9 |
| At2g43530 | Putative trypsin inhibitor | 9.6 | 0.5 | 0.3 | 0.6 | 0.9 | |
| At1g20440 | Hypothetical protein | 14.0 | 0.9 | 0.6 | 0.4 | 0.8 | |
| At3g25020 | Disease resistance protein family | 14.5 | 0.7 | 0.2 | 0.5 | 0.9 | |
| At5g57625 | Putative pathogenesis-related protein | 1.2 | 0.6 | 0.7 | 0.6 | 1.0 | |
| At2g29460 | GSTU4 | Glutathione transferase, putative | 2.9 | 0.6 | 0.0 | 0.5 | 1.0 |
| At1g79720 | Putative aspartyl protease | 6.8 | 0.6 | 0.3 | 0.6 | 1.1 | |
| At2g33380 | RD20 | RD20 protein | 8.7 | 0.6 | 0.0 | 0.5 | 0.9 |
| At1g11580 | PME | Pectin methylesterase, putative | 1.2 | 0.8 | 0.0 | 0.5 | 1.0 |
| At1g24070 | CSLA10 | Glucosyltransferase, putative | 11.5 | 0.8 | 0.0 | 0.5 | 0.9 |
| At2g43570 | Glycosyl hydrolase family 19 | 12.1 | 0.5 | 0.9 | 0.6 | 0.9 | |
| At2g17500 | Expressed protein | 3.1 | 0.6 | 0.0 | 0.6 | 1.0 | |
| At5g17860 | CAX7 | Putative sodium–calcium exchanger | 14.3 | 0.5 | 0.4 | 0.6 | 0.9 |
| At1g70350 | Hypothetical protein | 1.1 | 0.5 | 0.7 | 0.6 | 1.0 | |
| At1g17620 | Expressed protein | 1.4 | 0.5 | 0.4 | 0.6 | 1.0 | |
| At5g66650 | Putative protein | 1.7 | 0.5 | 0.9 | 0.6 | 1.0 | |
| At1g74800 | Hypothetical protein | 2.6 | 0.7 | 0.2 | 0.5 | 1.0 | |
| At3g53630 | Putative protein | 2.8 | 0.5 | 0.3 | 0.6 | 1.0 | |
| At4g18610 | Putative protein | 8.4 | 1.0 | 0.3 | 0.3 | 0.9 | |
| At1g17380 | Expressed protein | 8.7 | 0.9 | 0.4 | 0.4 | 0.9 | |
| At1g70700 | Hypothetical protein | 12.7 | 0.7 | 0.3 | 0.6 | 1.2 | |
| At1g06140 | Hypothetical protein | 14.4 | 0.5 | 0.6 | 0.6 | 1.1 | |
| At2g17650 | Acyl-CoA synthetase like protein | 3.1 | 0.8 | 0.2 | 2.2 | 1.0 | |
| At2g23130 | AGP17 | Arabinogalactan-protein (AgP17) | 11.0 | 0.6 | 0.1 | 1.8 | 1.1 |
| At4g23820 | Polygalacturonase, putative | 11.3 | 0.5 | 0.7 | 1.6 | 1.1 | |
| At2g26520 | Expressed protein | 14.7 | 1.0 | 0.1 | 2.7 | 1.3 | |
| GAIN OF REGULATION | |||||||
| At1g26680 | B3-TF | Hypothetical protein | 2.4 | 1.1 | 0.3 | 1.1 | 0.3 |
| At4g09760 | Choline kinase gmCK2p-like protein | 6.9 | 0.5 | 0.3 | 0.9 | 0.6 | |
| At3g61830 | ARF18 | Auxin response factor-like protein | 12.1 | 0.6 | 0.1 | 0.9 | 0.5 |
| At1g49500 | Expressed protein | 9.5 | 0.6 | 0.8 | 1.1 | 1.8 | |
| SAME REGULATION | |||||||
| At1g63840 | Putative RINg zinc finger protein | 1.2 | 0.5 | 0.2 | 0.7 | 0.7 | |
| At1g67970 | HSFA8 | Heat shock transcription factor | 6.1 | 0.5 | 0.2 | 0.7 | 0.7 |
| At2g17040 | NAM-TF | NAM (no apical meristem)-like | 8.6 | 0.5 | 0.6 | 0.6 | 0.7 |
| At5g58350 | WNK4 | MAP kinase | 0.2 | 0.5 | 0.9 | 0.7 | 0.7 |
| At2g41100 | TCH3 | Calmodulin-like protein | 1.0 | 0.8 | 0.9 | 0.6 | 0.6 |
| At1g08450 | CRT3 | Calreticulin, putative | 4.6 | 0.6 | 0.8 | 0.7 | 0.7 |
| At1g65800 | ARK2 | Receptor kinase, putative | 7.2 | 0.6 | 0.3 | 0.7 | 0.7 |
| At2g31880 | Leucine-rich repeat protein kinase | 12.4 | 0.7 | 0.4 | 0.6 | 0.7 | |
| At1g09070 | SRC2 | Expressed protein | 14.7 | 1.0 | 0.8 | 0.4 | 0.6 |
| At5g48660 | Putative protein | 9.6 | 0.5 | 0.6 | 0.7 | 0.8 | |
| At5g37600 | GSR1 | Glutamine synthetase | 0.5 | 0.6 | 0.7 | 0.7 | 0.7 |
| At4g34230 | CAD5 | Cinnamyl alcohol dehydrogenase | 0.9 | 0.5 | 0.9 | 0.7 | 0.7 |
| At3g14990 | 4-methyl-5(β-hydroxyethyl)-thiazole | 3.4 | 0.6 | 0.1 | 0.7 | 0.7 | |
| At3g22890 | APS1 | ATP sulfurylase, putative | 3.7 | 0.6 | 1.0 | 0.7 | 0.7 |
| At5g64000 | SAL2 | 3(2),5-bisphosphate nucleotidase | 4.7 | 0.7 | 0.7 | 0.6 | 0.6 |
| At4g05020 | NDB2 | A. thaliana cDNA W43435 | 6.4 | 0.8 | 0.6 | 0.6 | 0.6 |
| At2g29370 | Putative tropinone reductase | 11.8 | 0.5 | 0.1 | 0.7 | 0.6 | |
| At5g19550 | ASP2 | Aspartate aminotransferase 2 | 12.6 | 0.5 | 0.8 | 0.8 | 0.7 |
| At4g34710 | ADC2 | Arginine decarboxylase SPE2 | 9.9 | 1.7 | 0.2 | 0.3 | 0.4 |
| At1g69930 | GSTU11 | Glutathione transferase, putative | 4.3 | 0.5 | 0.9 | 0.7 | 0.7 |
| At5g06320 | NHL3 | Harpin-induced protein-like | 5.0 | 0.9 | 0.9 | 0.5 | 0.6 |
| At1g11910 | Putative aspartic proteinase | 9.0 | 0.7 | 0.2 | 0.6 | 0.7 | |
| At1g01470 | LEA14 | Hypothetical protein | 9.2 | 1.1 | 0.4 | 0.4 | 0.5 |
| At3g25010 | Disease resistance protein family | 12.2 | 0.8 | 0.1 | 0.5 | 0.6 | |
| At3g10980 | SAG20 | Unknown protein | 14.0 | 0.6 | 0.7 | 0.6 | 0.7 |
| At3g52400 | SYP122 | Syntaxin SYP122 | 5.3 | 0.8 | 0.3 | 0.6 | 0.6 |
| At1g19370 | Expressed protein | 7.1 | 0.6 | 0.4 | 0.6 | 0.7 | |
| At2g22500 | Mitochondrial carrier protein family | 8.5 | 0.6 | 0.4 | 0.6 | 0.7 | |
| At5g52760 | Expressed protein; protein | 9.9 | 0.5 | 0.6 | 0.7 | 0.6 | |
| At1g61800 | GPT2 | Gluc-6-P/P-translocator precursor | 10.8 | 0.9 | 0.8 | 0.5 | 0.6 |
| At5g39520 | Expressed protein | 10.9 | 0.6 | 0.6 | 0.7 | 0.7 | |
| At1g48610 | Regulatory protein HAL3B | 2.9 | 0.7 | 0.9 | 1.5 | 1.6 | |
| At3g60320 | bZIP protein | 11.1 | 0.6 | 0.5 | 1.4 | 1.7 | |
| At4g17980 | NAM-TF | NAM (no apical meristem)-like | 11.5 | 0.5 | 0.7 | 1.3 | 1.6 |
| At5g14260 | Putative protein | 2.1 | 0.5 | 0.8 | 1.4 | 1.5 | |
| At2g31680 | RABA5D | GTP-binding protein, putative | 2.2 | 0.8 | 0.6 | 1.8 | 1.8 |
| At4g03110 | RNA-binding CELF protein, putative | 5.3 | 0.6 | 0.1 | 1.4 | 1.6 | |
| At2g31010 | Putative protein kinase | 5.5 | 0.7 | 0.4 | 1.5 | 1.6 | |
| At5g24240 | Ubiquitin | 8.3 | 1.1 | 0.3 | 2.4 | 1.9 | |
| At4g00060 | Hypothetical protein | 10.9 | 0.6 | 0.8 | 1.4 | 1.6 | |
| At1g64940 | CYP89A6 | Cytochrome p450, putative | 11.6 | 0.7 | 0.6 | 1.9 | 1.5 |
| At5g23970 | Acyltransferase family | 12.8 | 0.7 | 0.8 | 1.8 | 1.5 | |
| At1g63290 | D-ribulose-5-phosphate-3-epimerase | 0.3 | 0.8 | 0.3 | 1.7 | 1.7 | |
| At4g25050 | ACP4 | Acyl carrier-like protein | 6.2 | 0.8 | 0.9 | 1.6 | 1.8 |
| At1g72610 | GLP1 | Germin-like protein | 8.5 | 0.8 | 0.5 | 1.6 | 1.9 |
| At5g65730 | Xyloglucan endo-transglycosylase | 13.3 | 0.6 | 0.8 | 1.6 | 1.4 | |
| At4g33970 | Extensin-like protein; protein | 13.9 | 0.7 | 0.4 | 1.7 | 1.4 | |
| At1g73840 | Hydroxyproline-rich glycoprot | 14.8 | 0.6 | 0.8 | 1.3 | 1.6 | |
| At3g26520 | TIP1.2 | Gamma tonoplast intrinsic prot | 0.6 | 0.6 | 0.4 | 1.6 | 1.6 |
| At3g16240 | TIP2.1 | Delta tonoplast integral protein | 3.3 | 0.9 | 0.6 | 1.9 | 1.8 |
| At1g49510 | Unknown protein | 0.0 | 0.5 | 0.1 | 1.4 | 1.4 | |
| At3g13720 | PRA1 | Expressed protein | 0.5 | 0.5 | 0.5 | 1.4 | 1.5 |
| At4g20260 | Endomembrane-associated protein | 1.1 | 0.6 | 0.4 | 1.6 | 1.6 | |
| At3g28460 | Unknown protein | 1.1 | 0.7 | 0.4 | 1.6 | 1.7 | |
| At3g07470 | Expressed protein | 1.3 | 0.7 | 0.1 | 1.7 | 1.7 | |
| At5g57320 | Villin | 4.0 | 0.7 | 0.4 | 1.7 | 1.5 | |
| At1g61740 | Unknown protein | 4.4 | 0.7 | 0.9 | 1.6 | 1.7 | |
| At5g18050 | Auxin-induced protein-like | 5.2 | 0.7 | 0.2 | 1.6 | 1.7 | |
| At3g52130 | 5B protein like protein | 5.6 | 0.6 | 0.7 | 1.6 | 1.5 | |
| At5g14920 | Putative protein | 5.8 | 0.5 | 0.4 | 1.4 | 1.5 | |
| At3g13500 | Hypothetical protein | 5.8 | 0.6 | 0.3 | 1.4 | 1.6 | |
| At1g69040 | ACR4 | ACT Domain Repeat Protein | 5.9 | 0.7 | 0.3 | 1.5 | 1.6 |
| At2g46740 | Hypothetical protein | 6.8 | 0.9 | 0.8 | 1.8 | 2.1 | |
| At3g25930 | Expressed protein | 7.2 | 0.6 | 0.4 | 1.4 | 1.6 | |
| At4g24170 | Putative protein | 8.4 | 0.6 | 0.2 | 1.6 | 1.4 | |
| At3g63390 | Putative protein | 9.0 | 0.7 | 0.8 | 1.9 | 1.5 | |
| At5g64160 | Expressed protein | 10.5 | 0.5 | 0.3 | 1.4 | 1.5 | |
| At4g16830 | Nuclear antigen homolog | 11.0 | 0.7 | 0.4 | 1.5 | 1.8 | |
| At1g35617 | Hypothetical protein | 11.3 | 0.7 | 0.3 | 1.5 | 1.8 | |
| At1g68960 | Hypothetical protein | 11.7 | 0.6 | 0.3 | 1.4 | 1.8 | |
| At1g62510 | Similar to 14Kd proline-rich | 11.9 | 0.5 | 0.4 | 1.6 | 1.4 | |
| At1g75750 | GASA1 | Expressed protein | 12.6 | 2.1 | 0.7 | 5.2 | 2.5 |
| At5g52570 | Putative beta-carotene hydroxylase | 13.0 | 0.5 | 0.6 | 1.3 | 1.5 | |
| At5g13470 | Putative protein | 13.2 | 0.6 | 0.5 | 1.8 | 1.4 | |
| At2g45180 | Expressed protein | 13.6 | 1.1 | 0.8 | 2.7 | 1.8 | |
| At3g49900 | Putative protein | 14.9 | 0.6 | 0.4 | 1.4 | 1.6 | |
Regulatory classes determined with vector analysis (Breitling et al., 2005).
Statistical parameters: angle (a), average vector length (l), consistency p-value in % (p).
From three replicate experiments.
During long-term starvation (Table 3 and Supplementary File SI2), ‘loss of regulation’ in coi1-16 plants was again apparent for well known downstream targets of JA that were up-regulated in wild-type plants, namely VSP1 (At5g24780), PDF1.2a (At5g44420), and PDF1.3 (At2g26010 (Staswick, 1984; Penninckx et al., 1998). However, in this experiment, the majority of genes ‘gained’ K responsiveness in coi1-16 (unchanged in wt but up or down-regulated in coi1-16; Figure 2A). ‘Gain of regulation’ was displayed by genes encoding transcription factors (e.g. ethylene-responsive ERF, At5g61590; Nakano et al., 2006), and auxin-responsive ARF18, At3g61830 (Okushima et al., 2005), and regulatory proteins (e.g. RABH1C, At4g39890, and PLC2, At3g08510; Otterhag et al., 2001; Vernoud et al., 2003), as well as metabolic enzymes (e.g. BMY7, At3g23920, and MAP1C, At1g13270; Ross et al., 2005; Sparla et al., 2006) and transporters (e.g. GLR2.3, At2g24710, and ACA4, At2g41560; Geisler et al., 2000).
‘Gain of Regulation’ in coi1-16 Mutants under Long-Term K-Stress Excludes Defense-Related Genes
The large number of ‘gain-of-regulation’ transcripts in long-term-starved coi1-16 plants suggests that if given enough time, the mutants initiate new responses to K-stress. Since some of these might compensate for the lack of JA-related responses, their functional spectrum could provide a clue to the physiological role of JA in plant adaptation to low K. We therefore compared predicted functions of genes ‘gaining’ responsiveness during long-term starvation with those of genes displaying ‘loss of regulation’ upon short-term K re-supply (likely to be direct targets of JA-signaling). Although there was no evidence for gene-by-gene replacement (e.g. replacement of one family member by another), similar functional categories were represented by ‘loss’ and ‘gain-of-regulation’ genes, suggesting that the latter may serve a compensatory role in coi1-16 mutants (Figure 2C and Supplementary File SI2). The only functional category represented among ‘loss-of-regulation’ but not among ‘gain-of-regulation’ genes was ‘defense’. The fact that COI1-dependent induction of defense-related genes could be lost without causing a physiological phenotype indicates that it is not required for plant adaptation to K-deficiency under sterile laboratory conditions.
Plants in Low-K Conditions Are Less Prone to Thrips Attack
The large number of defense-related genes induced by K-starved A. thaliana plants raises the question whether increased JA-production protects K-deficient plants against additional biotic stress. Thrips (Frankliniella sp.) are among the most vicious insect pests affecting cultivated plants. In a recent study, it was shown that numbers and feeding of Frankliniella occidentalis on A. thaliana and Brassica rapa were reduced by pre-treating plants with JA, whereas coi1-1 plants were hypersensitive to thrips (Abe et al., 2009). We therefore investigated whether K nutrition had an effect on thrips attack of hydroponically grown A. thaliana plants and whether this effect was dependent on COI1. Plants were initially grown in K-sufficient control medium to eliminate any effects related to plant size and leaf surface, subsequently exposed to a medium lacking K to induce a JA response, and finally transferred to a growth chamber infested with thrips (Frankliniella sp.; Figure 3A). To quantify thrips attack, we counted insect bites, visible as chlorotic spots on the leaves (Figure 3A). Numbers of bites were assessed separately for leaves within three size classes. Wild-type and coi1-16 mutant plants in low-K and control medium were grown in parallel to ensure identical thrips exposure and accompanying conditions. Over an observation time of 2 weeks, K-starved wild-type plants contracted significantly lower numbers of bites than K-sufficient plants, independent of leaf size (Figure 3B). For coi1-16 plants, thrips-inflicted damage was lethal within a few days (insets in Figure 3A) in both control and low-K plants, and individual bites could no longer be distinguished (Figure 3C). Mutants deleted in CYP79B2 and B3, two genes required for the (JA-dependent) production of indolic glucosiolates, showed similar levels of thrips attack and a similar difference between K-sufficient and K-starved plants as wild-type plants (Figure 3C), suggesting that neither the high susceptibility of coi1-16 plants nor the low susceptibility of K-starved wild-type plants were due to changes of indolic glucosinolate levels.
DISCUSSION
Phenotype of coi1-16 Mutants on Low K
Despite the differences in gene expression between coi1-16 and wild-type plants on low K, there was no striking difference in growth, water or K content. In agreement with this finding, no genes encoding known K-transporters or regulators of these featured among COI1-dependent K-responsive genes. We conclude that JA/COI1-signaling does not play a major role in regulating uptake or root-shoot allocation of K. However, coi1-16 plants did flower earlier than wild-type plants, particularly on low K. In accordance with existing knowledge that (1) JA is an important signal for senescence (Reinbothe et al., 2009), (2) K-deficiency induces senescence and the expression of senescence-related genes (Armengaud et al., 2004; Cao et al., 2006), and (3) this induction is inhibited by salicylic acid (probably through antagonistic function with JA), early flowering in coi1-16 mutants could indicate defective nutrient recovery from senescent leaves, which is particularly important under long-term nutrient shortage. The fact that wild-type plants also flowered earlier in low-K conditions (albeit to a lesser extent than coi1-16) supports a linkage between early flowering and K-deficiency. This issue should be investigated in the future by fine-mapping K-concentration in different tissues and at different developmental stages in both genotypes.
Treatments and K Status of the Plants
As before, two treatments were applied to assess transcriptional responses of the plants to external K supply. In a long-term starvation experiment, plants were grown from germination for 2 weeks on a medium that was not supplied with K (but contained traces of K at the beginning of the experiment; see Methods). This experiment was designed to reveal long-term adaptive responses of the plants to K-deficiency but will also, to a minor extent, reflect secondary changes caused by deficiency symptoms. We have reported before that the K-starved plants had significantly lower root and shoot K-concentrations than the control plants but showed no visible symptoms until day 12, probably due to effective K-uptake and redistribution (Armengaud et al., 2004, 2009). Over the last 2 d before harvest, the plants developed K-deficiency symptoms in the form of leaf chlorosis, decrease in shoot growth rate, and arrest of lateral root growth, indicating that whole-plant K levels had reached a critical level. Indeed, cytoplasmic K in epidermal root cells had fallen to low mM concentrations (Armengaud et al., 2009). We also previously recorded changes in the metabolite spectrum (especially with respect to reducing sugars, organic acids, and glutamate/glutamine ratio) but not the total protein or chlorophyll content (Armengaud et al., 2009), suggesting that the plants had successfully adjusted their growth, photosynthetic rate, and metabolism to the limited nutrient supply (Amtmann and Armengaud, 2009; Tschoep et al., 2009). A short-term K re-supply experiment (6 h K re-supply to K-deficient plants) was designed to identify early signals and responses directly linked to the K-stimulus. As reported before, during this period of time, root K-concentrations increased but there was no change in growth, shoot K-concentration or visible symptoms (Armengaud et al., 2004). The fact that many transcripts, metabolites and enzymes displayed opposite changes in response to long-term starvation and short-term re-supply (Armengaud et al., 2004, 2009) indicated not only that these changes were indeed directly related to the K supply, but also that the K-starved plants were still capable of quickly reversing their responses to K-deficiency. The experimental design also ensured that measured transcript responses were not linked to changes in other ions in the medium, as these differed between the treatments (Armengaud et al., 2004).
Genotypic Differences
To assess the dependency of transcript changes on a functional JA-COI1-signaling pathway, we compared transcript profiles of coi1-16 mutants to those previously obtained for Col0 wild-type plants. It should be noted that coi1-16 mutants differ from Col0 wild-type in two other genes: GL1 (Ellis and Turner, 2002) and PEN2 (Westphal et al., 2008). It could therefore be possible that some of the identified differences between coi1-16 and wild-type are in fact caused by gl1 or pen2. A previous microarray study of A. thaliana plants subjected to MeJA, herbivorous insects, and wounding investigated the contribution of gl1 to the gl1coi1 transcriptome (Reymond et al., 2004), and found it to be generally small. Comparison of our data with their data shows a good overlap not only between transcript responses to K-treatments and herbivore attack, but also between COI1-dependence of these changes (Supplemental Table SI3). For all genes that had significantly lower response to both K and herbivores in the coi1 genotype than in the wild-type (marked with ** in SI3), the difference between gl1 mutant and wild-type (if any) was considerably smaller than between gl1/coi1 mutant and wild-type, and can therefore be assigned to COI1. The good overlap between our results and those from Reymond and colleagues (who used a different coi1 mutant from the one used here) is particularly reassuring in the light of the recent finding that the coi1-16 mutant contains a hitherto unidentified mutation in PEN2 (Westphal et al., 2008). PEN2 is a glycosyl hydrolase that converts 4-methoxyindol-3-ylmethylglucosinolate to downstream products with antifungal properties (Bednarek et al., 2009). We carried out a qPCR analysis for a number of K-responsive genes (selected for their COI1 (in-)dependence and possible relation to glucosinolate biosynthesis in general and PEN2 in particular). As shown in Supplemental Figure SI4, in all cases but one, the response to K was similar between pen2 mutants and wild-type and different between pen2 and coi1-16 mutants (Supplemental Figure SI5). The only gene for which a significant effect of the pen2 mutation on its response to low K was found was At5g28510, encoding a glucosyl hydrolase (marked with * in Table 3) closely related to PEN2. The response of another gene of the same family (BG1, AtAt1g52400) did not differ between pen2 and wild-type. We conclude that the effects of GL1 and PEN2 on K-responsiveness are minor compared to COI1. Nevertheless, future studies investigating specific genes listed in Tables 3 and 4 should confirm their COI1 dependence in other coi1 lines.
Transcript Profiles Identify Known and Novel Targets of JA-Signaling
A central role of JA-signaling through COI1 in plant responses to varying K supply was apparent in the fact that the total number of K-responsive transcripts was significantly smaller in coi1-16 than in wild-type plants. Based on a quantitative comparison of transcript changes between coi1-16 and wild-type plants, we assigned all transcripts into four main categories, and those abiding to strict statistical constraints are shown in Tables 3 and 4 (see Methods for statistical cut-offs). The observation that several known JA/COI1 targets had lost responsiveness to K-starvation (e.g. VSP1 and PDFs; Table 3) and/or K re-supply (e.g. VSP2 and Thi2.1; Table 4) in coi1-16 validated our experimental approach. We also explored how the transcripts listed in Tables 3 and 4 responded to other conditions using Genevestigator (Zimmermann et al., 2004). Most of the ‘loss-of-regulation’ transcripts but very few ‘same-regulation’ transcripts displayed a strong response to OPDA and MeJA treatments (Supplemental Figure SI4, B–E). This adds further support to the notion that the categories identified here separate well between genes that respond to K through the oxylipin-COI1-signaling pathway and those that do not.
Particular suitability of the short-term K re-supply experiment to identify novel K-dependent COI1 targets was evident in the fact that the transcriptional profiles of coi1-16 plants showed a clear separation between JA/COI1-dependent and JA/COI1-independent K-regulated transcripts (Figure 2). More than half of the genes that were previously identified as K-regulated in the wild-type displayed the same response to K re-supply in coi1-16 plants (Figure 2A), which indicates that the loss of responsiveness in other genes was indeed due to a lack of COI1 function rather than a general problem with transcriptional regulation. Novel targets of JA/COI-signaling identified in the re-supply experiment (Table 4) include genes with function in transport, primary metabolism, and cell wall composition, which are likely to be related to previously reported reversible changes in primary metabolism (pyruvate kinase), membrane potential, and growth during plant adaptation to low K (Armengaud et al., 2009), as well as a number of regulatory proteins (Table 4). Identification of genes that were significantly independent of COI1 in their response to K (‘same regulation’) are also interesting, as they will include upstream components of the JA/COI1 pathway and/or components of parallel signaling pathways. For example, we identified COI1-independent up-regulation by low K of JAZ1 (Table 3), a repressor of JA-dependent transcription that is regulated by JA via protein degradation (Thines et al., 2007). Transcriptional induction of JAZ1 is likely to exert a negative feedback regulation during transient JA responses; however, fast induction of JAZ1 after wounding and herbivore attack is COI1-dependent (Chung et al., 2008).
‘Gain’ of Regulation in coi1-16 Mutants during Long-Term K-Deficiency
Long-term K-starvation produced very different transcript profiles. Here, the majority of genes ‘gained’ K-responsiveness in the coi1-16 background. In most cases, this involved a transcriptional down-regulation that did not occur in wild-type plants. A function of COI1 in repressing K-regulation of these genes is unlikely, as such function should be similarly apparent during short-term re-supply (as ‘gain of regulation’ with opposite changes in the same transcripts), which was not the case. It is more likely that the coi1-16 plants induce new responses to K-starvation because they experience a different physiological state under K-deficiency from wild-type plants. This raises the question of whether the K-regulated genes in coi1-16 mutants functionally compensate for the lack of JA-mediated responses, and thus account for the absence of a growth phenotype in low K. ‘Gain-of-regulation’ genes covered indeed similar functional categories as direct COI1-targets identified in the K re-supply experiment (e.g. metabolism, cell wall modification and transport; Figure 2C) and included a number of ethylene and auxin-responsive genes, indicating that coi1-1 mutants mobilize alternative pathways.
Defense-related genes constituted the only functional category of K-responsive genes that lost responsiveness to K re-supply in coi1-16 but did not feature among ‘gain-of-regulation’ transcripts in K-starved coi1-16 plants. Considering that this class was the largest class of K-regulated genes in the wild-type, it is surprising that the observed changes should be unnecessary for plant adaptation to low K. One possible explanation is that an increase in JA-mediated defense during K-deficiency has evolved to counteract their increased susceptibility to biological enemies (discussed below). In this case, the real advantage of a JA/COI1 response to K-deficiency would remain unnoticed in sterile laboratory conditions. In this context, it is interesting that many of the transcripts identified by Reymond et al. (2004) as regulated by herbivorous insects (S. littoralis and P. rapa) also responded to at least one of the K-treatments (Supplemental Table SI3). Considerably less overlap was found between K and MeJA treatment or wounding (Supplemental Table SI3).
‘Prophylactic’ Defense against Insects in K-Deficient Plants?
Investigating the effect of the K-deficiency-induced rise in JA on plant resistance against pathogens and pest is not an easy undertaking, as K-deficiency causes several changes in the plants that ease the attack (e.g. weakened skeleton and cell wall), improve the feeding quality (e.g. higher content of low-molecular sugars and nitrogen compounds), and increase the relative damage (smaller leaf size) (Amtmann et al., 2008). Even if partially offset by increased defense, one would therefore still expect to see more damage in K-deficient plants. We tried to eliminate the above factors by growing the plants first with sufficient K for a period of time before removing K from the growth medium. This yielded plants that were comparable in size but nevertheless induced a JA signal (enhanced LOX2 expression). After transfer to a thrips-infested growth chamber, the low-K plants suffered considerably less damage from the herbivorous insect than the control plants (Figure 3B). The observations suggest that the main function of JA/COI1 in K-deficient plants is to enhance their defense potential against herbivorous insects and other biological enemies. Such ‘prophylaxis’ could be advantageous, especially in small plants, which cannot afford to loose a substantial proportion of their leaf surface to herbivory. Unfortunately, insect damage of the coi1-16-mutants was so rapid and devastating that it was impossible to measure quantitative differences between control and K-deficient mutant plants. The difference in insect attack between control and low-K wild-type plants is clearly not due to JA-induced production of indolic glucosinolates (see up-regulation of CYP79B in K-deficient plants, Table 3) because it is still apparent in cyp79b2/b3 mutants that are defective in this pathway (Mikkelsen et al., 2003). More detailed experiments are now required to consolidate our hypothesis that K-deficiency induces a ‘prophylactic’ defense response via JA and COI1.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana (Col0 wild-type or mutants coi1-16) plants were grown on sterile vertical agar plates or hydroponically as described previously (Maathuis et al., 2003; Armengaud et al., 2004). The composition of the nutrient media was (in mM) 1.25 KNO3, 0.5 Ca(NO3)2, 0.5 MgSO4, 0.625 KH2PO4, NaH2PO4, 2 NaCl in the control medium, and 1.0 Ca(NO3)2, 0.5 MgSO4, 0.625 NaH2PO4, 1.375 NaCl in the –K (‘K-free’) medium. Both media contained the following micronutrients (in μM): 42.5 FeNaEDTA, 0.16 CuSO4, 45 H3BO3, 0.015 (NH4)6Mo7O24, 0.01 CoCl2, 0.38 ZnSO4, 1.8 MnSO4 (both media). Final concentrations of the altered ion in the two media, control (‘K-free’), were 1.875 (0) mM K, 0.5 (1) mM Ca2+, 1.25 (1) mM NO3– and 2 (1.375) mM Cl–. Plates contained 70 ml medium, supplemented with 3% sucrose and 1% agar (Type A, Sigma, Poole, UK). This agar contains a small amount of K (approx 80 μM), which is rapidly depleted by the growing plants. Changes in root and shoot K contents over the course of the long-term-starvation experiment and after K re-supply have been documented before (Armengaud et al., 2004, 2009). K re-supply to plants growing on plates with K-free medium consisted in replacing the condensed solution at the bottom of the Petri dishes with 5 ml K-free medium supplemented with 10 mM KCl. As a control, K-free medium was given instead. Plates were positioned vertically under a light source (16 h/d at 100 μE) at a constant temperature of 22°C.
For thrips experiments, plants were grown hydroponically in short days (9 h light at 200 μE) on control medium for 7 d and subsequently exposed to K-free medium for 4 weeks (control plants had a continuous supply of K) before being transferred to a growth chamber infested with thrips (Frankliniella sp.). Insect bites on leaves were counted 2 weeks later.
Microarray Experiments and Data Analysis
RNA extraction, reverse transcription, and direct Cy3 and Cy5 labeling of cDNA were performed as previously described (Armengaud et al., 2004). Glass arrays spotted with the Arabidopsis Genome Oligo Set version 1.0. (Qiagen) were obtained from D. Galbraith (University of Arizona). Array preparation, hybridization, washing, scanning (ScanArray Express scanner and software suite, Perkin Elmer, Warrington, UK), and signal quantification (QuantArray, Perkin Elmer, Warrington, UK) were carried out as described previously (Armengaud et al., 2004). Hybridization signals for control and treatment were quantile-normalized (Bolstad et al., 2003). Genes were sorted by their normalized expression ratio for each replicate in ascending and descending order. Rank products (RPs) across replicates were calculated for each gene (Breitling et al., 2004). Comparison between transcript changes in wild-type and coi1-16 mutants was performed using vector analysis (Breitling et al., 2005). For each gene expression, changes in the two genetic backgrounds were represented by a vector in a Cartesian plane. The length of the sum vector resulting from nine pair-wise comparisons across three replicate experiments was calculated using a Perl script (Breitling et al., 2005) and compared to 100 random permutations of the original dataset, thus generating a consistency p-value. To eliminate inconsistent responses, only transcripts yielding p-values less than 0.01 were considered. The overall strength of the response, l, was given by the average length of the nine individual vectors and only transcripts with l greater than 0.5 were chosen for further analysis. The angle between the sum vector and a prototypic vector was used for assignment into regulatory classes (Figure 2A and Supplementary Information SI2). To avoid ambiguous assignment, only those transcripts producing sum vectors that deviated by less than 15 degrees from the closest prototype were considered.
Microarray Data
A search engine based on AGI codes for expression profiles in roots and shoots of wild-type plants grown under different K-conditions is provided at www.brc.dcs.gla.ac.uk/∼rb106x/Arabidopsis_results.htm. Expression profiles for coi1-16 shoots are available at www.brc.dcs.gla.ac.uk/∼rb106x/coi_results.htm.
Supplementary Information
SI1 contains lists of K-responsive genes in coi1-16 mutants as identified by Rank Products. SI2 contains lists of genes assigned to four regulatory classes by Vector Analysis. Gene annotations are linked to TAIR and TIGR websites. SI3 contains a table showing comparison between our dataset and that generated by Reymond et al. (2004). SI4 shows the response to OPDA and MeJA treatments of the genes listed in Tables 3 and 4 (from Genevestigator). SI5 shows qPCR results for selected genes in Col0 wild-type, coi1-16 and pen2 mutants. SI6 is a color version of Figure 3.
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
This work was funded by the Biotechnology and Biological Sciences Research Council (BBSRC grants P17237 and D006775).
Acknowledgments
We thank John Turner (University of East Anglia) for supplying coi1-16 seeds, Barbara Halkier (University of Copenhagen) for supplying cyp79b2/b3 seeds, and Paul Schulze-Lefert (MPI, Cologne) for supplying pen2 seeds. We are grateful to Pawel Herzyk (University of Glasgow) for providing microarray scanning facilities and to Philip White (SCRI), Joel Milner (UoG), and Ari Sadanandom (UoG) for fruitful discussions. No conflict of interest declared.
References
- Abe H, Shimoda T, Ohnishi J, Kugimiya S, Narusaka M, Seo S, Narusaka Y, Tsuda S, Kobayashi M. Jasmonate-dependent plant defense restricts thrips performance and preference. BMC Plant Biol. 2009;9:97. doi: 10.1186/1471-2229-9-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amtmann A. Learning from evolution: Thellungiella generates new knowledge on essential and critical components of abiotic stress tolerance in plants. Mol. Plant. 2009;2:3–12. doi: 10.1093/mp/ssn094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amtmann A, Armengaud P. The role of calcium sensor-interacting protein kinases in plant adaptation to potassium-deficiency: new answers to old questions. Cell. Res. 2007;17:483–485. doi: 10.1038/cr.2007.49. [DOI] [PubMed] [Google Scholar]
- Amtmann A, Armengaud P. Effects of N, P, K and S on metabolism: new knowledge gained from multi-level analysis. Curr. Opin. Plant Biol. 2009;12:275–283. doi: 10.1016/j.pbi.2009.04.014. [DOI] [PubMed] [Google Scholar]
- Amtmann A, Blatt MR. Regulation of macronutrient transport. New Phytologist. 2009;181:35–52. doi: 10.1111/j.1469-8137.2008.02666.x. [DOI] [PubMed] [Google Scholar]
- Amtmann A, Armengaud P, Volkov V. In: Potassium nutrition and salt stress. In Membrane Transport in Plants. Blatt MR, editor. Oxford: Blackwell Publishing; 2004. pp. 293–339. [Google Scholar]
- Amtmann A, Hammond JP, Armengaud P, White PJ. Nutrient sensing and signalling in plants: potassium and phosphorus. In: Callow JA, editor. Advances in Botanical Research Incorporating Advances in Plant Pathology. Academic Press; 2005. pp. 209–257. [Google Scholar]
- Amtmann A, Troufflard S, Armengaud P. The effect of potassium nutrition on pest and disease resistance in plants. Physiol. Plant. 2008;133:682–691. doi: 10.1111/j.1399-3054.2008.01075.x. [DOI] [PubMed] [Google Scholar]
- Armengaud P, Breitling R, Amtmann A. The potassium-dependent transcriptome of Arabidopsis reveals a prominent role of jasmonic acid in nutrient signaling. Plant Physiol. 2004;136:2556–2576. doi: 10.1104/pp.104.046482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armengaud P, Sulpice R, Miller AJ, Stitt M, Amtmann A, Gibon Y. Multilevel analysis of primary metabolism provides new insights into the role of potassium nutrition for glycolysis and nitrogen assimilation in Arabidopsis roots. Plant Physiol. 2009;150:772–785. doi: 10.1104/pp.108.133629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley MK, Grant M, Grabov A. Plant responses to potassium deficiencies: a role for potassium transport proteins. J. Exp. Bot. 2006;57:425–436. doi: 10.1093/jxb/erj034. [DOI] [PubMed] [Google Scholar]
- Balbi V, Devoto A. Jasmonate signalling network in Arabidopsis thaliana: crucial regulatory nodes and new physiological scenarios. New Phytologist. 2008;177:301–318. doi: 10.1111/j.1469-8137.2007.02292.x. [DOI] [PubMed] [Google Scholar]
- Bednarek P, et al. A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science. 2009;323:101–106. doi: 10.1126/science.1163732. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A. Vector analysis as a fast and easy method to compare gene expression responses between different experimental backgrounds. BMC Bioinformatics. 2005;6:181. doi: 10.1186/1471-2105-6-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S, Su L, Fang Y. Evidence for involvement of jasmonic acid in the induction of leaf senescence by potassium deficiency in Arabidopsis. Can. J. Bot. 2006;84:328–333. [Google Scholar]
- Chung HS, Koo AJ, Gao X, Jayanty S, Thines B, Jones AD, Howe GA. Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 2008;146:952–964. doi: 10.1104/pp.107.115691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creelman RA, Mullet JE. Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997;48:355–381. doi: 10.1146/annurev.arplant.48.1.355. [DOI] [PubMed] [Google Scholar]
- Devoto A, Turner JG. Jasmonate-regulated Arabidopsis stress signalling network. Physiol. Plant. 2005;123:161–172. [Google Scholar]
- Devoto A, Ellis C, Magusin A, Chang HS, Chilcott C, Zhu T, Turner JG. Expression profiling reveals COI1 to be a key regulator of genes involved in wound- and methyl jasmonate-induced secondary metabolism, defence, and hormone interactions. Plant Mol. Biol. 2005;58:497–513. doi: 10.1007/s11103-005-7306-5. [DOI] [PubMed] [Google Scholar]
- Dobermann A, Cassman KG, Mamaril CP, Shesby JE. Management of phosphorus, potassium, and sulfur in intensive irrigated lowland rice. Field Crop. Res. 1999;56:113–118. [Google Scholar]
- Epstein I, Rains DW, Elzam OE. Resolution of dual mechanisms of potassium absorption by barley roots. Proc. Natl Acad. Sci. U S A. 1963;49:684–692. doi: 10.1073/pnas.49.5.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans NH. Modulation of guard cell plasma membrane potassium currents by methyl jasmonate. Plant Physiol. 2003;131:8–11. doi: 10.1104/pp.014266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M, Frangne N, Gomes E, Martinoia E, Palmgren MG. The ACA4 gene of Arabidopsis encodes a vacuolar membrane calcium pump that improves salt tolerance in yeast. Plant Physiol. 2000;124:1814–1827. doi: 10.1104/pp.124.4.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Fukushige H, Hildebrand DF, Gan S. Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 2002;128:876–884. doi: 10.1104/pp.010843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull AK, Vij R, Celenza JL. Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc. Natl Acad. Sci. U S A. 2000;97:2379–2384. doi: 10.1073/pnas.040569997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JY, Shin R, Schachtman DP. Ethylene mediates response and tolerance to potassium deprivation in Arabidopsis. Plant Cell. 2009;21:607–621. doi: 10.1105/tpc.108.063099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafkafi U, Xu G, Imas P, Magen H, Tarchitzki J. Potassium and Chloride in Crops and Soils: The Role of Potassium Chloride Fertilizer in Crop Nutrition. Bern, Switzerland: International Potash Institute; 2001. [Google Scholar]
- Kim JH, Choi D, Kende H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 2003;36:94–104. doi: 10.1046/j.1365-313x.2003.01862.x. [DOI] [PubMed] [Google Scholar]
- Laegreid M, Bockman OC, Kaarstad O. Agriculture, Fertilizers and the Environment. Oxon, UK: CABI; 1999. [Google Scholar]
- Li L, Kim BG, Cheong YH, Pandey GK, Luan S. A Ca(2)+ signaling pathway regulates a K(+) channel for low-K response in Arabidopsis. Proc. Natl Acad. Sci. U S A. 2006;103:12625–12630. doi: 10.1073/pnas.0605129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maathuis F, Sanders D. Mechanisms of potassium absorption by higher plant roots. Physiol. Plant. 1996;96:158–168. [Google Scholar]
- Maathuis FJ, et al. Transcriptome analysis of root transporters reveals participation of multiple gene families in the response to cation stress. Plant J. 2003;35:675–692. doi: 10.1046/j.1365-313x.2003.01839.x. [DOI] [PubMed] [Google Scholar]
- Marschner H. Mineral Nutrition of Higher Plants. London: Academic Press; 1995. [Google Scholar]
- Mikkelsen MD, Petersen BL, Glawischnig E, Jensen AB, Andreasson E, Halkier BA. Modulation of CYP79 genes and glucosinolate profiles in Arabidopsis by defense signaling pathways. Plant Physiol. 2003;131:298–308. doi: 10.1104/pp.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munemasa S, Oda K, Watanabe-Sugimoto M, Nakamura Y, Shimoishi Y, Murata Y. The coi1 mutation reveals the hormonal signaling interaction between ABA and MeJA in Arabidopsis guard cells : specific impairment of ion channel activation and second messenger production. Plant Physiol. 2007 doi: 10.1104/pp.106.091298. PMID: 17220365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006;140:411–432. doi: 10.1104/pp.105.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell. 2005;17:444–463. doi: 10.1105/tpc.104.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oria-Hernandez J, Cabrera N, Perez-Montfort R, Ramirez-Silva L. Pyruvate kinase revisited: the activating effect of K+ J. Biol. Chem. 2005;280:37924–37929. doi: 10.1074/jbc.M508490200. [DOI] [PubMed] [Google Scholar]
- Otterhag L, Sommarin M, Pical C. N-terminal EF-hand-like domain is required for phosphoinositide-specific phospholipase C activity in Arabidopsis thaliana. FEBS Lett. 2001;497:165–170. doi: 10.1016/s0014-5793(01)02453-x. [DOI] [PubMed] [Google Scholar]
- Pandey GK, Cheong YH, Kim BG, Grant JJ, Li L, Luan S. CIPK9: a calcium sensor-interacting protein kinase required for low-potassium tolerance in Arabidopsis. Cell Res. 2007;17:411–421. doi: 10.1038/cr.2007.39. [DOI] [PubMed] [Google Scholar]
- Penninckx IA, Thomma BP, Buchala A, Metraux JP, Broekaert WF. Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell. 1998;10:2103–2113. doi: 10.1105/tpc.10.12.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Amador MA, Leon J, Green PJ, Carbonell J. Induction of the arginine decarboxylase ADC2 gene provides evidence for the involvement of polyamines in the wound response in Arabidopsis. Plant Physiol. 2002;130:1454–1463. doi: 10.1104/pp.009951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe C, Springer A, Samol I, Reinbothe S. Plant oxylipins: role of jasmonic acid during programmed cell death, defence and leaf senescence. FEBS J. 2009;276:4666–4681. doi: 10.1111/j.1742-4658.2009.07193.x. [DOI] [PubMed] [Google Scholar]
- Reymond P, Bodenhausen N, Van Poecke RMP, Krishnamurthy V, Dicke M, Farmer EE. A conserved transcript pattern in response to a specialist and a generalist herbivore. Plant Cell. 2004;16:3132–3147. doi: 10.1105/tpc.104.026120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross S, Giglione C, Pierre M, Espagne C, Meinnel T. Functional and developmental impact of cytosolic protein N-terminal methionine excision in Arabidopsis. Plant Physiol. 2005;137:623–637. doi: 10.1104/pp.104.056861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman DP, Shin R. Nutrient sensing and signaling: NPKS. Annu. Rev. Plant Biol. 2006;58:47–69. doi: 10.1146/annurev.arplant.58.032806.103750. [DOI] [PubMed] [Google Scholar]
- Shin R, Schachtman DP. Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proc. Natl Acad. Sci. U S A. 2004;101:8827–8832. doi: 10.1073/pnas.0401707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R, Berg RH, Schachtman DP. Reactive oxygen species and root hairs in Arabidopsis root response to nitrogen, phosphorus and potassium deficiency. Plant Cell Physiol. 2005;46:1350–1357. doi: 10.1093/pcp/pci145. [DOI] [PubMed] [Google Scholar]
- Sparla F, Costa A, Lo Schiavo F, Pupillo P, Trost P. Redox regulation of a novel plastid-targeted beta-amylase of Arabidopsis. Plant Physiol. 2006;141:840–850. doi: 10.1104/pp.106.079186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE. Novel regulation of vegetative storage protein genes. Plant Cell. 1984;2:1–6. doi: 10.1105/tpc.2.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE. JAZing up jasmonate signaling. Trends Plant Sci. 2008;13:66–71. doi: 10.1016/j.tplants.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Staswick PE, Su W, Howell SH. Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl Acad. Sci. U S A. 1992;89:6837–6840. doi: 10.1073/pnas.89.15.6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Xu J, Yang J, Kieliszewski MJ, Showalter AM. The lysine-rich arabinogalactan-protein subfamily in Arabidopsis: gene expression, glycoprotein purification and biochemical characterization. Plant Cell Physiol. 2005;46:975–984. doi: 10.1093/pcp/pci106. [DOI] [PubMed] [Google Scholar]
- Syers JK. Soil and Plant Potassium in Agriculture. London: International Fertiliser Society; 1998. [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448:661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- Tschoep H, Gibon Y, Carillo P, Armengaud P, Szecowka M, Nunes-Nesi A, Fernie AR, Koehl K, Stitt M. Adjustment of Growth and Central Metabolism to a Mild but Sustained Nitrogen-Limitation in Arabidopsis. Plant Cell Environ. 2009;32:300–318. doi: 10.1111/j.1365-3040.2008.01921.x. [DOI] [PubMed] [Google Scholar]
- Vernoud V, Horton AC, Yang Z, Nielsen E. Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 2003;131:1191–1208. doi: 10.1104/pp.013052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Véry AA, Sentenac H. Molecular mechanisms and regulation of K+ transport in higher plants. Annu. Rev. Plant Biol. 2003;54:575–603. doi: 10.1146/annurev.arplant.54.031902.134831. [DOI] [PubMed] [Google Scholar]
- Walker DJ, Leigh RA, Miller AJ. Potassium homeostasis in vacuolate plant cells. Proc. Natl Acad. Sci. U S A. 1996;93:10510–10514. doi: 10.1073/pnas.93.19.10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal L, Scheel D, Rosahl S. The coi1- 16 mutant harbors a second site mutation rendering PEN2 nonfunctional. Plant Cell. 2008;20:824–826. doi: 10.1105/tpc.107.056895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 1998;280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- Xu J, Li HD, Chen LQ, Wang Y, Liu LL, He L, Wu WH. A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell. 2006;125:1347–1360. doi: 10.1016/j.cell.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Koizumi M, Urao S, Shinozaki K. Molecular cloning and characterization of 9 cDNAs for genes that are responsive to desiccation to Arabidopsis thaliana: sequence analysis of one cDNA clone that encodes a putative transmembrane channel protein. Plant Cell Physiol. 1992;33:217–224. [Google Scholar]
- Yan JB, et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell. 2009;21:2220–2236. doi: 10.1105/tpc.109.065730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]