Abstract
Previous studies of CD8+ T cell immunodominance after primary virus infection of F1 mice compared with their inbred parents have generally concluded that no dramatic changes occur. Here we re-visit this issue using vaccinia virus (VACV), which has a large genome, a recently defined immunodominance hierarchy in mice and is a candidate vector for vaccines. We found that immunogenicity of VACV peptides defined using inbred mice was highly variable in F1 progeny: some peptides were equally immunogenic in F1 and inbred, while others elicited responses that were reduced by more than 90% in F1 mice. Further, the dominance of a peptide in the relevant inbred parent did not predict whether or not it would be poorly immunogenic in F1 mice. This result held using F1 hybrids of MHC-congenic mice, suggesting that MHC differences alone were responsible. It was also extended to foreign epitopes expressed by a recombinant VACV vaccine. F1 mice were less able to mount responses to the poorly immunogenic peptides when used as a sole immunogen, ruling out immunodomination. In addition, conserved TCR Vβ usage between inbred and F1 mice did not always correlate with strong responses in F1 mice. However direct estimation of naïve precursor numbers showed that these were reduced in F1 compared with inbred mice for specificities that were poorly immunogenic in the hybrids. These data have implications for our understanding of the extent to which MHC diversity alters the range of epitopes that are immunogenic in outbred populations.
Introduction
Immunodominance, or the unequal recognition of different epitopes from the same antigen, is a basic characteristic of CD8+ T cell responses and as such has relevance for our understanding of adaptive immunity to infection and for vaccine design. The mechanisms underlying immunodominance are complex but fall into two main categories, those related to antigen processing and presentation and those related to the responding CD8+ T cell population (1, 2). Multiple factors that influence the amount of peptide-MHC available to prime CD8+ T cells can limit the immunogenicity of one epitope relative to others. These include (but are not limited to) protein abundance and time of expression (3–7), ability of the antigenic peptide to be generated from a protein (8–12) and the binding affinity (both the on and off rates) of the peptide for its presenting MHC (12–16). On the responding T cell side, abundance and possibly clonal diversity of precursors in the naïve repertoire are likely to be most important (3, 12, 13, 16–22), but other factors such as avidity (23, 24), IFN-γ expression, including kinetics of expression (25, 26) and regulatory environment (27) may also play roles. Finally it has been reported that presence or absence of dominant epitopes or prior priming of responses to individual epitopes affects the entire hierarchy through the phenomenon of immunodomination (28–35). Of all these various factors, two recent papers suggest that MHC binding and frequency of precursors in the naïve TCR repertoire are likely to be the best predictors of immunodominance (13, 18). Much of this knowledge has been gained using model virus infections in inbred mice but there is evidence that immunodominance is also a characteristic of human CD8+ T cell responses (36–40).
The relationship between antigen diversity and immunodominance has not been very well characterized. This is important for any attempt to relate studies done using inbred mice, especially C57Bl/6 that have only two available restriction elements, to outbred populations such as humans with up to six restriction elements and therefore three times greater epitope diversity. The possibility that adding new restriction elements, as would be the case in the F1 progeny of two inbred strains, might compromise CD8+ T cell responses restricted by another element was first described in the late 1970s (41, 42). Further investigation suggested that these data were consistent with suppression of clones that cross-reacted with self in F1 mice resulting in large gaps in the T cell repertoire that could be seen at the level of a whole virus (43). However, this early work was not able to investigate responses at the individual epitope level, nor were strictly quantitative assays available to measure T cell responses. More recently, comparisons of the dominance hierarchies established using inbred parents with those in F1 progeny have been published for influenza A, lymphocytic choriomeningitis (LCMV), respiratory syncitial and murine cytomegaloviruses (MCMV) (13, 25, 44–47).
For influenza A virus, Belz etal (44) showed that the Db-restricted NP366 and PA224 are co-dominant in H-2b- inbred mice, but the latter loses this status in H-2b×d F1 mice, inducing around ten-fold fewer CD8+ T cells in F1 compared to inbred mice. Data published by Chen etal (2) also show reduced responses to PA224 in H-2b×d F1 mice, albeit to a less obvious degree and the authors concluded that in the case of influenza virus infection, responses to all epitopes were reduced fairly equally in F1 mice. Two studies using LCMV have likewise concluded that immunodominance hierarchies established in inbred strains are preserved relatively well in their F1 progeny (13, 25). Although the most recent work suggested that responses to some of the less dominant epitopes tended to be compromised in F1 mice (13). If these data are viewed differently and a direct comparison of the size of CD8+ T cell responses to each epitope in inbred and F1 mice is made, it can be seen that the extra restriction elements in the F1 mice have variable effects on different epitopes: Responses to the Ld-restricted NP118 and NP314 are almost as high in H-2b×d F1 as in H-2d inbred mice, whereas responses to the Db-restricted GP33 and GP276 in the same F1 mice are approximately a half and a quarter, respectively, of the magnitude seen in inbred H-2b mice. In a third model using a virus with a small genome, a similar situation was found for two epitopes of respiratory syncitial virus, namely Kd-restricted M282 and Db-restricted M187. Whereas M282 elicited similar, very large numbers of CD8+ T cells in the lungs of infected H-2d and H-2b×d F1 mice, M187 elicited very high responses in H-2b, but around 75% fewer in H-2b×d F1 mice (47). MCMV is the only virus with a larger genome for which studies of this type have been published. Here, marked differences were seen in F1 versus inbred mice, but these were linked to genes outside the MHC. Indeed, genes other than MHC dictate immunodominance hierarchies and total CD8+ T cell responses to MCMV in inbred as well as F1 mice. This, in addition to the persistent nature of MCMV makes further investigation of the phenomenon difficult in this model. Finally, none of these studies have found holes in the repertoire that are large enough to explain the original observations made in the 1970s (41, 44).
Of the more recent papers, only the work with influenza virus made an attempt to define a mechanism for the reduced responses for an epitope in F1 mice. It was concluded that the reduction of responses to the PA224 epitope of influenza A virus in H-2b×d F1 compared with H-2b parent was due to the loss in the F1 mice of PA224-specific CD8+ T cell clones sharing a Vβ7 chain in their TCR that dominate responses to this epitope in H-2b mice. Presumably these clones are deleted in the thymus during negative selection in H-2b×d F1 mice and, because they comprise a large fraction of the PA224-specific cells, their loss leads to a substantial reduction in the total number of precursors able to recognize this epitope. This type of mechanism has been confirmed for the loss of a dominant public TCR in human responses to the Epstein-Barr Virus, HLA B8-restricted FLRGRAYGL peptide, where individuals also express HLA-B*4402 (48). Given these precedents, one might predict that epitopes to which responses are characterized by limited clonal diversity are more likely to be prone to greatly reduced immunogenicity in the context of some non-presenting MHC molecules.
Recently we and others have mapped a large number CD8+ T cell epitopes for vaccinia virus (VACV)4 (49–52) allowing immunodominance to be studied in this system where the virus is around ten times larger than influenza virus and LCMV. This model is not complicated by the persistence of virus and we have already found some surprising differences in the drivers of immunodominace between VACV and influenza/LCMV (53). Immunodominance in VACV infections is also of particular interest because this virus is being used as a vector for recombinant vaccines. CD8+ T cell responses to VACV-based vaccines are dominated by those targeting the vector and it is reasonable to assume that finding ways to focus this response away from the vector and towards the recombinant antigen would improve this class of vaccines (54, 55). Here we compare CD8+ T cell responses elicited by several epitopes from VACV and a VACV-based multi-epitope vaccine in H-2b, H-2d and F1 H-2b×d mice and find that responses to a surprising number of epitopes are compromised in F1 mice. This observation is then explored further and the implications discussed.
Materials and Methods
Viruses and cell lines
VACV strain WR (Western Reserve) was grown and titrated in BHK-21 and BSC-1 cells respectively using standard methods. A recombinant VACV expressing a multi-epitope construct, murine-pt-rVV (56), was the kind gift of Andreas Suhrbier, Queensland Institute of Medical Research. All immortalized cell lines were maintained in Dulbecco’s Modified Eagle medium (DMEM) with glutamine and 10% fetal bovine serum (FBS) (D10) (Invitrogen). For use as stimulators in CD8+ T cell assays, 1–5 × 106 DC2.4 or P815 cells were infected with VACV at 5–10 PFU/cell in <200 µl of PBS at 37°C for 30–60 min with occasional shaking. After this initial incubation, 9 ml of D10 was added, and the incubation continued until a total time after infection of at least 4 h. Infected cells were spun and washed, and the appropriate number was added to T cell assays.
Synthetic peptides
Peptides were purchased from Genscript Corp. (Piscataway, NJ) or Mimotopes (Clayton, Vic Australia). Master stocks of peptides were made at 10 mg/ml in 100% DMSO and aliquots were stored at −70°C. Before use, peptides were diluted to required concentrations in serum-free DMEM.
Mice and infections
Specific pathogen-free C57BL/6 (H-2b), C57BL/10 (H-2b), DBA/2 (H-2d), C57BL/10.D2 (H-2d), C57BL/6×DBA/2 (referred to as BDF1; H-2b×d) and C57BL/10×C57BL/10.D2 (referred to as B10×D2F1 ;H-2b×d) were obtained from Animal Resource Centre (Perth, Australia). We used DBA/2 rather than the more commonly used BALB/c mice as the standard H-2d inbred strain because F1 hybrids between these and C57Bl/6 were a standard item offered by our supplier (BDF1 in their catalogue) and experiments found similar responses to H-2d-restricted CD8+ T cell epitopes in DBA/2 and BALB/c mice (not shown). Mice were housed, and experiments were done according to the relevant ethical requirements and under an approval from the ANU animal ethics and experimentation committee. Mice were infected i.p. with 1 × 106 PFU of VACV in 200 µl PBS.
Stimulations and intracellular staining of IFN-γ (ICS)
Mice were euthanized 7 days (acute response) or 3 months (memory response) after infection and spleens taken for analysis of CD8+ T cell responses by ICS as described previously (51). Briefly, splenocytes were plated at 1 × 106 cells/well into round-bottom 96-well plates. Synthetic peptides (Table 1) were added to a final concentration of 10−7 M and plates were incubated at 37°C and 5% CO2. After 1 h, 5 µg/ml brefeldin A (Sigma) was added and plates were incubated for another 3 h. Plates were spun at 4°C, medium was removed, and cells were resuspended in 50 µl of 1:150 diluted anti-CD8-PE (clone 53-6.7; BD Biosciences). After 30 min incubation on ice, cells were washed, resuspended in 50 µl of 1% paraformaldehyde, and incubated at room temperature for 20 min before another two washes and staining with 50 µl of 1:200 diluted anti-IFN-γ-APC (clone XMG1.2; BD Biosciences) overnight in PBS with 0.5% saponin (Sigma) at 4°C. Cells were washed three times before acquisition using a FACS LSR II (BD Biosciences). Analysis was done using Flowjo software (Tree Star Inc.). Events were gated for live lymphocytes on FCS × SSC followed by CD8+ T cells × IFN-γ. Backgrounds as determined using irrelevant peptides were usually in the order of 0.1% and were subtracted from the values presented for test samples.
Table 1.
Peptides used in the study
| Name | Description | Sequence | MHC Restriction |
|---|---|---|---|
| B820–27 | VACV-B8 | TSYKFESV | H-2Kb |
| K36–15 | VACV-K3 | YSLPNAGDVI | H-2Db |
| A47138–146 | VACV-A47 | AAFEFINSL | H-2Kb |
| A4288–96 | VACV-A42 | YAPVSPIVI | H-2Db |
| F226–34 | VACV-F2 | SPYAAGYDL | H-2Ld |
| A5275–83 | VACV-A52 | KYGRLFNEI | H-2Kd |
| E3140–148 | VACV-E3 | VGPSNSPTF | H-2Dd |
| F-NP147–155 | Influenza A virus nucleoprotein | TYQRTRALV | H-2Kd |
| PB-CSP249–257 | P. Berghei circumsporozoite protein | SYIPSAEKI | H-2Kd |
| Pp89168–176 | Murine cytomegalovirus pp89 | YPHFMPTNL | H-2Ld |
| L-NP118–126 | Lymphocytic choriomeningitis virus nucleoprotein |
RPQASGVYM | H-2Ld |
| F-NP366–374 | Influenza A virus nucleoprotein | ASNENMDAM | H-2Db |
| E1A234–243 | Adenovirus 5 E1A | SGPSNTPPEI | H-2Db |
| Ova257–264 | Ovalbumin | SIINFEKL | H-2Kb |
| S-NP324–332 | Sendai virus nucleoprotein | FAPGNYPAL | H-2Kb |
Peptide immunizations
Groups of inbred H-2b mice or F1 hybrids were injected i.p. with 100 µg peptide mixed with 1 µg α-galactosylceramide (α-GalCer) (Alexis Biochemicals, Farmingdale, NY) in PBS/0.5% Tween 20 buffer. After 7 days, splenocytes were prepared and CD8+ T cell responses toward individual peptides were assessed by ICS as described above.
CD8+ T cell lines
Splenocytes were prepared from VACV-primed mice 3 weeks after infection. A portion of these splenocytes was pulsed with 10−8 M peptide in serum-free DMEM for 1 h at 37°C. Remaining splenocytes were resuspended in D10 containing 3 ng/ml rIL-2 (R&D Systems), 1:1000 β-mercaptoethanol (stock solution from Gibco, Cat No 21985-023) and 1:100 HEPES (stock solution from Gibco, Cat No 15630) (T cell medium). Unpulsed splenocytes (3×107) were mixed with 6×106 peptide-pulsed splenocytes and plated into a well of a 6-well plate in 10 ml of T cell medium. After 3 days, dead cells were removed by Ficoll gradient centrifugation. Cells were re-stimulated weekly by plating 1×107 cells from the T cell culture into a new well with 1/5 the number of peptide pulsed, mitomycin-treated splenocytes. After 3–4 rounds of re-stimulation, specificity of T cells was assessed by using a DimerX assay (see below). Once >80% of CD8+ T cells in the culture bound a DimerX reagent loaded with the appropriate peptide, Vβ usage on CD8+ cells was determined by staining with anti-CD8+-APC and the FITC-labelled anti-TCR Vβ panel.
DimerX assay to detect peptide-specific CD8+ T cells
Recombinant soluble dimeric mouse H-2Kb:Ig, H-2Ld:Ig and H-2Db:Ig fusion proteins were purchased from BD Biosciences and the DimerX assay was performed according to the manufacturer’s instructions. Briefly, 2 µg of H-2Kb:Ig or H-2Ld:Ig fusion proteins were incubated overnight at 37°C in PBS with a 40 molar excess of peptide. Four µg of H-2Db:Ig fusion protein were loaded with a 40 molar excess of peptide. Peptide-loaded dimers were then incubated for 1 h at room temperature with PE-coupled anti-mouse IgG1 (clone A85-1, BD Biosciences). Cells were labeled with DimerX and 1:200 anti-CD8-APC (clone 53-6.7, BD Biosciences) for 1 hr on ice and washed twice before acquisition using a FACS LSR II (BD Biosciences). Analysis was done using Flowjo software (Tree Star Inc.). Events were gated for live lymphocytes on FCS × SSC followed by CD8+ T cells × DimerX+ cells. Backgrounds as determined using irrelevant peptides were in the order of 0.5 to 0.8% and were subtracted from the values presented for test samples.
TCR Vβ profiles
For analysis of TCR Vβ expression by CD8+ T cells, a mouse Vβ TCR screening panel (BD Biosciences) was used according to the manufacturer’s instruction. This set of FITC-labeled antibodies detects 17 out of 24 known Vβ subfamilies in mice. To analyze TCR Vβ profiles ex vivo, splenocytes were prepared from mice infected i.p. for 7 days with 1×106 PFU of VACV. Splenocytes were labeled with peptide-loaded PE-labeled DimerX and Fc-block for 1 hr on ice. Cells were then washed, resuspended in PBS containing 2% FBS and plated into round-bottom microtitre plates. Anti-CD8-APC (clone 53-6.7; BD Biosciences) and FITC-labeled Vβ antibodies were added for 30 min on ice. Cells were washed twice before acquisition using a FACS LSR II and analysis was done with Flowjo software. Events were gated for live lymphocytes on FCS × SSC followed by CD8+ × DimerX+ × Vβ+ cells.
Enrichment of peptide-specific CD8+ T cells
Single-cell suspensions were prepared from spleens and lymph nodes (axillary, brachial, mesenteric, cervical and inguinal) from naïve mice. Cells were stained with peptide-loaded PE-labeled DimerX and Fc-block for 1 hr at room temperature. Cells were then washed, resuspended in 500 µl PBS containing 0.5% bovine serum albumin and 2 mM EDTA (MACS buffer) and labeled with 100 µl anti-PE microbeads (Miltenyi Biotech) for 30 min at 4°C. Cells were washed twice with MACS-buffer and passed over magnetized LS columns (Miltenyi Biotech). Columns were washed and bound cells were eluted. Cells were stained with anti-CD8 APC-Cy7, anti-CD3-PerCP-Cy5.5, anti-CD4-PE-Cy7, anti-CD62L-APC and anti-CD11b-, -CD11c-, -B220- and F4/80-FITC for 30 min on ice. Cells were washed twice before acquisition using a FACS LSR II and analysis with Flowjo software. Events were gated for live lymphocytes on FCS × SSC followed by FITC− (dump gate) × CD8+ cells. CD8+ cells were further gated on CD3+ × CD4− × CD62Lhigh × Dimer+ cells.
Statisitical analyses
Unless stated otherwise, statistical comparisons were done using an unpaired t-test. For the analysis of peptide-specific T cells in naive mice, Welch’s correction was used because population had unequal varience and the non-parametric Mann Whitney test was used for the Vβ analysis. All tests were done with the aid of GraphPad Prism software (GraphPad, La Jolla, CA).
Results
Some VACV epitopes are surprisingly poorly immunogenic in F1 mice
We have recently mapped and characterized VACV-derived CD8+ T cell determinants in H-2b- and H-2d-haplotype mice and established an immunodominance hierarchy (49–52). Of five epitopes identified in the first study of H-2b mice, B820 was identified as the most dominant epitope eliciting 6–10% of all splenic CD8+ T cells while the less dominant determinants K36 and A47138 induced responses of up to 2% of CD8+ T cells and together these three epitopes account for around 35% of the total response to VACV (51). In H-2d mice, the three epitopes F226, A5275 and E3140 made up approximately 40% of the total CD8+ T cell response to VACV, with F226 being as dominant in these mice as B820 was found to be on the H-2b haplotype (52). These six epitopes were used in the VACV model system to investigate CD8+ T cell immunodominance in F1 mice compared with the parental strains. Our expectation based on conclusions stated in the literature, was that responses to all epitopes would be reduced fairly equally and by around half in F1 compared with inbred mice. This reduction is required to accommodate responses to roughly twice the number of specificities in F1 mice when the total response to infection remains the same as in the inbred parents.
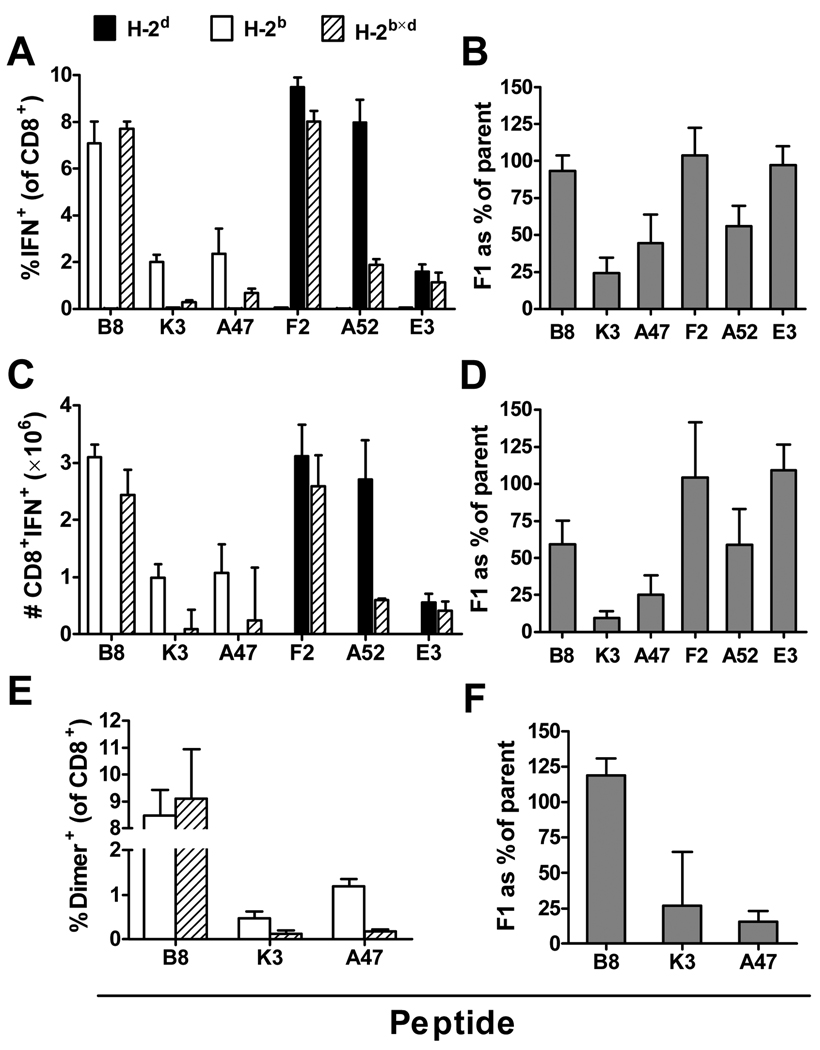
The first experiment examined the percentages and total numbers of CD8+ T cells elicited by the six epitopes in inbred H-2b and H-2d mice and in H-2b×d F1 mice at the peak of the acute response to VACV. Mice were infected i.p. with VACV and 7 days later, CD8+ T cell responses to peptides were measured by intracellular staining of IFN-γ after a 4 hour ex vivo stimulation (ICS). Percentages (Figure 1 A) and total numbers (Figure 1 C) of CD8+ T cells responding to three of the epitopes (H-2b-restricted B820 and H-2d-restricted F226 and E3140) were similar in parent and F1 mice. However, responses to the remaining three peptides were reduced in the F1, with H-2b–restricted K36 and A47138 being most affected. In the experiment shown the differences were statistically significant (p<0.05) for K36 and A5275. From this experiment (shown in figure 1A and 1C) the ratio of average CD8+ T cell responses in F1 mice compared to the relevant parent was calculated for each peptide. The experiment was then repeated three times providing four values of responses in F1 compared with inbred for each peptide and the means and SEM of these data plotted and shown in figure 1B and 1D. This set of experiments and analysis shows that while responses to some peptides are higher than expected, being equivalent in F1 and inbred mice, responses to other peptides are much weaker than expected. Of note, while the two dominant epitopes from each of the inbred strains remain dominant in F1 mice, the performance of the less dominant epitopes in the inbred strains does not predict the ratio of response in F1 compared to inbred mice. The best example here being E3140, the least dominant of the H-2d peptides that is as immunogenic in the F1 as it is in inbred H-2d mice. The peptides that were apparently most poorly immunogenic in F1 mice were K36 and A47138 and so to ensure there was no influence of the ICS assay used to measure these responses, we repeated experiments for just the H-2b epitopes and measured peptide-specific CD8+ T cells using DimerX reagents (a variant of peptide-MHC tetramer technology). Results are shown in figure 1E and 1F and these confirm those reported above with responses to K36 and A47138 being significantly lower (p<0.05) in F1 mice while responses to B8 are roughly equal in both strains.
Figure 1. Peptide-specific CD8+ T cell responses to VACV at the peak of the acute response in inbred and F1 mice.
Groups of C57Bl/6 (H-2b), DBA/2 (H-2d) or BDF1 (H-2b×d) mice were infected i.p. with 106 pfu VACV strain WR. Seven days later, peptide-specific CD8+ T cell responses in the spleen were measured by ICS (A–D) or DimerX assays (E,F). Percentages (A) and total numbers (C) of CD8+ T cells that produce IFN-γ in ex vivo stimulations with the indicated individual peptides; data are shown from one representative experiment and are means and SEM of groups of 4 mice. Ratios of responses in F1 mice compared with relevant parent strain are shown based on percentages (B) and total numbers (D); data represent means and SEM of 3 or 4 experiments (B and D respectively). Detection of epitope-specific CD8+ T cells with peptide-loaded recombinant DimerX reagents (E) and ratios of responses in F1 compared with H-2b mice based on E (F); data are means and SEM of 6 mice (B820, A47138) or 7 mice (K36) from two experiments.
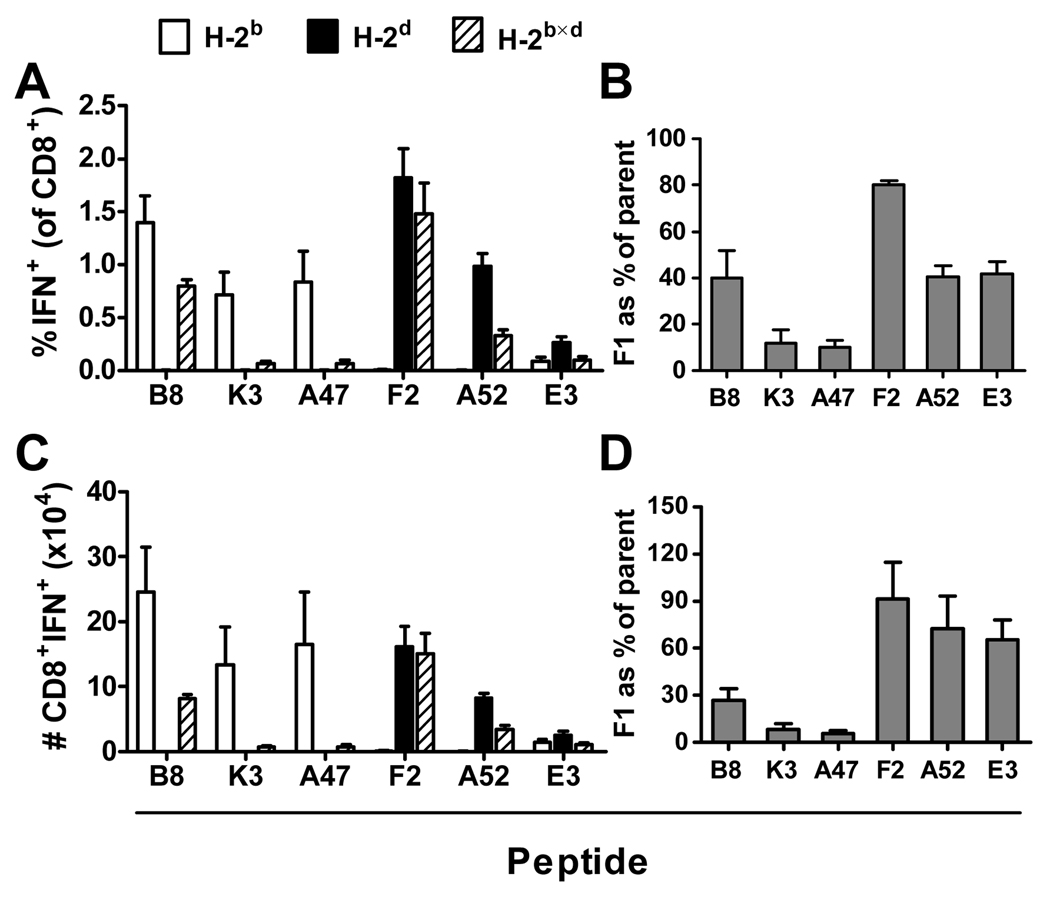
Next, we ask whether the same changes in immunodominance patterns were maintained in memory. H-2b, H-2d and H-2b×d mice were infected i.p. with VACV and CD8+ T cells responses were measured by ICS after 3 months. Figure 2 shows that if anything, differences in immunogenicity for individual peptides differed more between inbred and F1 mice in a memory response and again the size of CD8+ T cell responses differed significantly (p<0.05) between inbred and F1 mice for peptides K36 and A47138 and A5275. As in figure 1, data shown in panels A and C are from a single experiment and the ratios shown in panels B and D are from 3 experiments. Of note here, the immunodominant peptides from each parent which were co-dominant in F1 mice at acute times now show a clear hierarchy with the H-2d-restricted F226 peptide being superior.
Figure 2. Peptide-specific memory CD8+ T cell responses to VACV in inbred and F1 mice.
Groups of C57Bl/6 (H-2b), DBA/2 (H-2d) and BDF1 (H-2b×d) were infected i.p. with 106 pfu VACV strain WR. Three months later CD8+ T cell responses in the spleen were measured by ICS. Percentages (A) and total numbers (C) of CD8+ T cells that produce IFN-γ in ex vivo stimulations with the indicated individual peptides from a representative experiment; data are means and SEM from groups of 4 mice. Ratios of responses in F1 mice compared with parent strains are shown based on percentages (B) and total numbers (D); data are means and SEM of 3 experiments.
Together these data do not support the hypothesis that CD8+ T cell immunodominance in F1 mice is predictable from the hierarchies established using their inbred parents.
Variable immunogenicity of VACV epitopes in F1 mice is linked to MHC
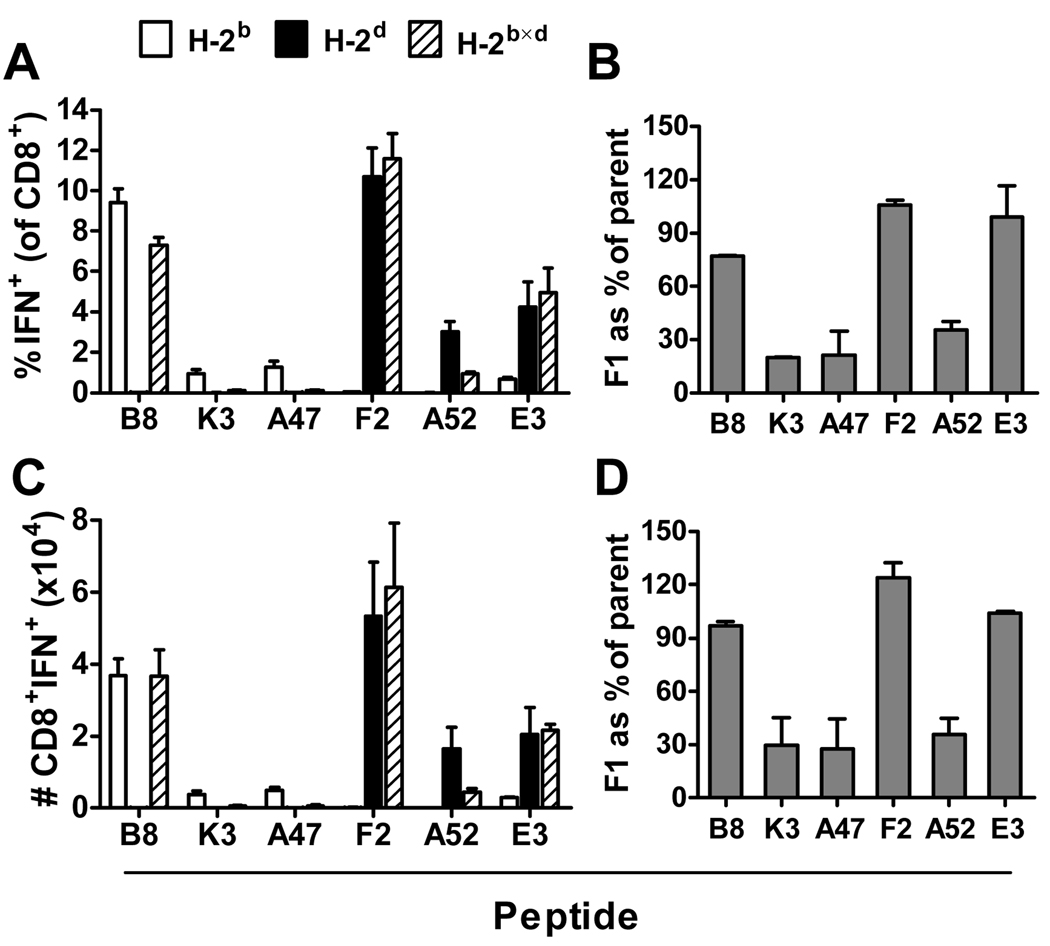
The inbred strains used thus far differ at many loci across their genomes and so it is possible that changes in immunogenicity of various epitopes in F1 mice were a result of these differences and not the addition of new MHC restriction elements. To test whether genes outside the MHC region have an effect on the immunogenicity of some epitopes, we repeated experiments described above with MHC class I congenic mice: C57Bl/10 (H-2b), B10.D2 (H-2d) and B10×D2 F1 (H-2b×d). Before doing experiments with these mice and their F1 progeny we directly confirmed that the CD8+ T cell hierarchy in the relevant congenic strains was similar to that found in C57Bl/6 and DBA/2 mice (not shown). In figure 3 the percentages and total numbers of epitope-specific CD8+ T cells in each of these strains are shown at the peak of the response, seven days after infection. Panels A and C show results of a single experiment with groups of three mice, while panels B and D show ratios from 2 experiments. As was seen in previous experiments, CD8+ T cell responses in parent mice and F1 hybrids were similar for peptides B820, F226 and E3140 but responses in F1 mice were reduced compared to the relevant parent for K36, A47138 and A5275 (p<0.05 for all in the experiment shown in figure 3A). An additional experiment using these congenic mice was done to look at CD8+ T cell responses six weeks after infection with VACV and again results paralleled findings made using standard inbred strains mice (not shown). The similarity of results obtained using standard inbred strains and these congenic mice led us to conclude that the variable immunogenicity of VACV CD8+ T cell epitopes seen between inbred and F1 mice was due to the additional MHC antigens and not heterozygosity at other loci. Having established this, all remaining experiments were done using standard inbred strains C57Bl/6, DBA/2 and their F1 progeny.
Figure 3. Peptide-specific CD8+ T cell responses to VACV at the peak of the acute response in MHC congenic mice and their F1 progeny.
Groups of C57Bl/10 (H-2b), C57Bl/10.D2 (H-2d) or B10×D2F1 (H-2b×d) mice were infected i.p. with 106 pfu VACV strain WR. Seven days later CD8+ T cell responses in the spleen were measured by ICS. Percentages (A) and total numbers (C) of CD8+ T cells that produce IFN-γ in ex vivo stimulations with the indicated peptides from one representative experiment; data are means and SEM of groups of 4 mice. The ratios of responses in F1 mice compared with parent strains are shown based on percentages (B) and total numbers (D); data are means and SEM of 2 experiments.
Variable immunogenicity of VACV epitopes in F1 mice affects an experimental recombinant vaccine
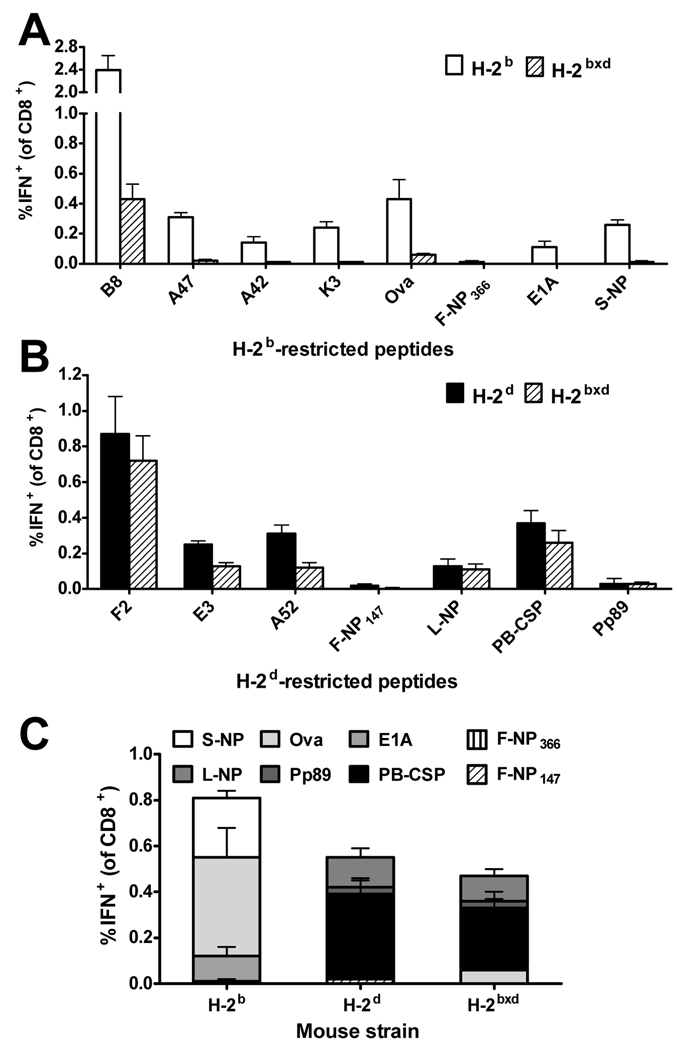
VACV is one of the more frequently used vectors for recombinant vaccines in current clinical trials. For this reason we investigated the effects of additional MHC alleles on a memory CD8+ T cell response to a recombinant VACV vaccine called murine-pt-rVV (57). The recombinant antigen in murine-pt-rVV is based on the ‘string of beads’ or ‘polytope’ concept with 10 CD8+ T cell epitopes, including eight restricted by H-2b or H-2d (listed in Table I). This virus was made using VACV strain WR with the polytope inserted into the thymidine kinase gene and would be expected to be attenuated compared with the wild type virus used for other experiments. H-2b, H-2d and H-2b×d were infected i.p. with murine-pt-rVV and splenic CD8+ T cell responses to VACV and polytope epitopes measured by ICS after 3 months. Comparison of memory CD8+ T cell responses to the two dominant epitopes from each parent using this virus confirms the finding shown in figure 2 using wild type VACV, with B820 becoming subdominant to F226 in F1 mice. Indeed responses to all of the H-2b-restricted peptides, including those encoded by the polytope, were very poor in F1 compared to H-2b mice (significantly lower, p<0.05 for all) and responses to three of four relevant polytope peptides were reduced to the limit of detection (Figure 4A). In comparison, the H-2d-restricted epitopes were in general not highly compromised in F1 mice, with responses to two polytope epitopes being no different in parent and F1 mice. Notably, one of these (MCMV Pp89168) is one of the weakest H-2d-restricted epitopes and so the rank order of a peptide in the inbred parent clearly does not predict whether or not responses will be compromised in an F1 hybrid. These data also show that the hierarchy in F1 mice was not simply derived by merging those of the parents: in H-2b×d F1 hybrids, LCMV NP118 and MCMV pp89168, which are the second and third from the bottom-ranked epitopes in H-2d mice, became equivalent or more dominant than the second and third highest ranked peptides from H-2b mice. Finally we added up responses to all polytope epitopes in each strain of mice (figure 4C). This analysis shows that the sum of responses to all peptides in the polytope construct were highest in H-2b mice that could only present four peptides and lowest in the H-2b×d mice where eight peptides could be presented. From this we conclude that testing the performance of such vaccines in inbred mouse strains might lead to an overestimation of their immunogenicity in an outbred population.
Figure 4. Memory CD8+ T cell responses to epitopes contained in a multi-epitope construct expressed from VACV.
Groups of C57BL/6 (H-2b), DBA/2 (H-2d) and BDF1 (H-2b×d) mice were infected i.p. with 106 pfu of murine-pt-rVV. Three months later, CD8+ T cell responses in the spleen were measured by ICS. Percentages of CD8+ T cells that produce IFN-γ in ex vivo stimulations with H-2b-(A) or H-2d-(B) restricted peptides are shown. (C) Comparison of CD8+ T cell responses to all polytope peptides in parent strains and F1 mice. Data represent means and SEM of 4 mice per group. The experiment was repeated with similar results.
Poor immunogenicity in F1 mice is a function of epitopes and not presenting MHC haplotype
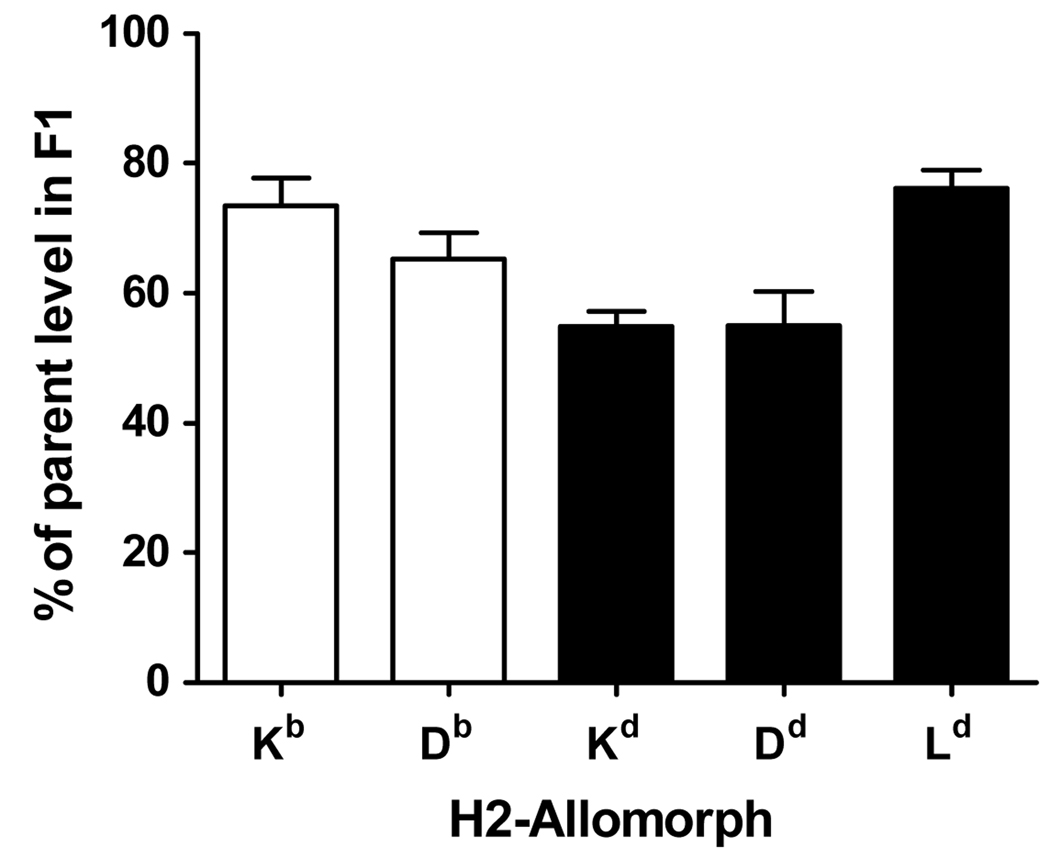
In the preceding experiments and especially those shown in figure 4, it would appear that as a general rule, responses to H-2b-restricted epitopes are compromised in H-2b×d F1 mice. This possibility was examined from two perspectives. The first was to rule out the possibility that surface expression of H-2b alleles was reduced as a result of co-expression of H-2d alleles in F1 mice. This question has been examined previously and competition between MHC class I proteins observed, but not with the same mouse strains used here (58). Using the same set of antibodies as this previous study, expression of individual H-2 allomorphs on splenocytes from C57Bl/6, DBA/2 and BDF1 mice was measured by flow cytometry. Levels of H-2b and H-2d alleles on splenocytes from F1 mice ranged from 60–80% of those seen in inbred mice (figure 5). Notably expression of H-2Kb and H-2Db were not substantially lower than the H-2d class I alleles. This result is in agreement with the previous work and suggests that poor immunogenicity of the H-2b-restricted epitopes in F1 mice is not linked to poor surface expression of H-2b alleles in these mice.
Figure 5. Relative levels of MHC class I allomorphs on splenocytes from F1 mice compared with parent strains.
Splenocytes were prepared from C57Bl/6 (H-2b), DBA/2 (H-2d) and BDF1 (H-2b×d) mice and stained with PE-labeled anti-H-2Kb, anti-H2-Db, anti-H-2Kd, anti-H-2Dd or anti-H-2Ld antibodies. Cells were gated on lymphocytes in the forward/side scatter and the mean fluorescence intensity of PE-labeled cells was assessed. Ratios of MHC allomorph expression in F1 mice versus parent strains are shown. Data represent means and SEM of 4 mice from at least two experiments.
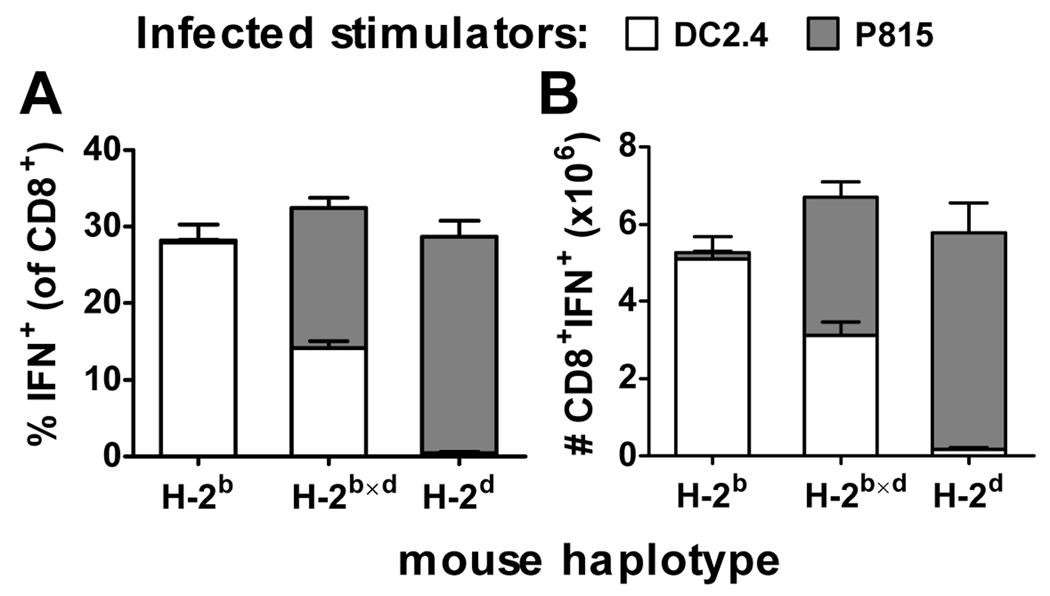
To look more directly at whether responses to H-2b-restricted epitopes are generally compromised in H-2b×d mice, the contribution of each parental haplotype to the anti-VACV CD8+ T cell response was determined. To do this, instead of using peptides to stimulate splenocytes from VACV-infected mice in ICS assays, we used VACV-infected cell lines (DC2.4 (H-2b) and P815 (H-2d)) to capture a picture of the total VACV-specific response. Mice were infected with VACV and after 7 days, the percent and number of CD8+ splenocytes that could recognize DC2.4 or P815 infected with VACV were measured. Figure 6 shows that as expected, the total anti-VACV CD8+ T cell response across inbred and F1 mice was similar. However surprisingly given the consistently very poor responses seen for most H-2b-restricted peptides, MHC alleles from each parent restricted between 40–50% of the total anti-VACV response in F1 mice. This demonstrates that H-2b×d mice are not generally compromised in their ability to mount primary H-2b-restricted CD8+ T cell responses.
Figure 6. Contribution of H-2b and H-2d haplotypes to anti-VACV CD8+ T cell responses in H-2b×d mice.
Groups of C57Bl/6 (H-2b), DBA/2 (H-2d) and BDF1 (H-2b×d) mice were infected i.p. with 106 pfu VACV strain WR. After seven days, the number of CD8+ T cells from the spleen that could respond to VACV-infected DC2.4 (H-2b) or P815 (H-2d) cells by making IFN-γ in an ICS assay was determined. Percentages (A) and total numbers (B) of responding CD8+ T cells are shown. Data are means and SEM of 8 mice collected over two experiments.
Poor immunogenicity of epitopes in F1 mice is reflected by reduced ability to respond to peptide alone
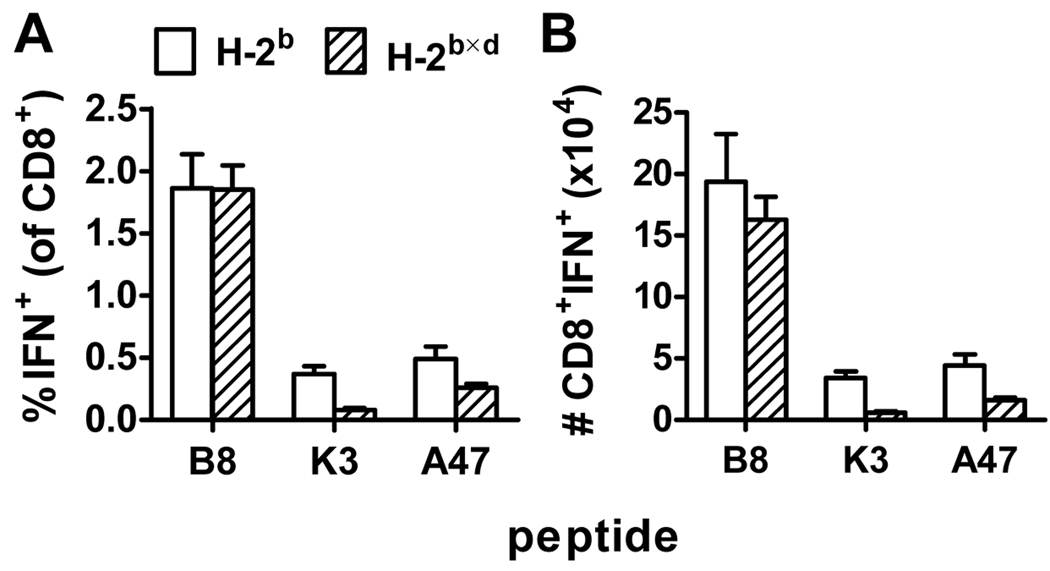
Having shown that poor immunogenicity in the context of additional restriction elements is an inherent property of individual epitopes, we wanted to probe the mechanism further. It has been suggested that CD8+ T cell responses to a given peptide can depend on the presence or absence of other specificities through the phenomenon of immunodomination. If this is the case for VACV it is conceivable that the additional epitope diversity in F1 hybrids contributes to the poor responses to some peptides in these mice. To examine this, we focused on the three VACV-derived H-2b-restricted peptides and used peptide immunization so that responses to each could be measured in the absence of any competing specificities. H-2b and H-2b×d mice were immunized with B820, K36 or A47138 adjuvanted with αGalCer (59) and seven days later CD8+ T cell responses towards these peptides were assessed by ICS. As shown in Figure 7, percentage and total number of CD8+ T cells responding to B820 were similar in parent H-2b and hybrid F1 mice. However, in F1 mice the responses to A47138 and especially K36 were reduced (p<0.05). For each of the three peptides, the data here echo those obtained seven days after VACV infection (compare with figure 1) and suggest that responses to A47138 and K36 are not reduced because of increased immunodomination in F1 mice. The finding that B820 responses are the same in both strains of mice demonstrates that F1 mice have no general defect in mounting immune responses to peptide/α-GalCer. Given this, the most likely reason for the relatively poor response to K36 and A47138 seen in F1 mice is a change in the naïve repertoire of CD8+ T cells.
Figure 7. CD8+ T cell responses to H-2b-restricted VACV epitopes in peptide-immunized inbred and F1 mice.
Groups of C57Bl/6 (H-2b) and BDF1 (H-2b×d) mice were immunized with 100 µg B820, K36 or A47138 in 1 µg αGalCer. Seven days later CD8+ T cell responses in the spleen were measured by ICS. Percentages (A) and total numbers (B) of CD8+ T cells that produce IFN-γ in ex vivo stimulations with the indicated peptides are shown. Data are means and SEM from 8 mice (B820, A47138) or 6 mice (K36) from two experiments.
Changes in the Vβ usage of peptide-specific CD8+ T cells between inbred and F1 mice
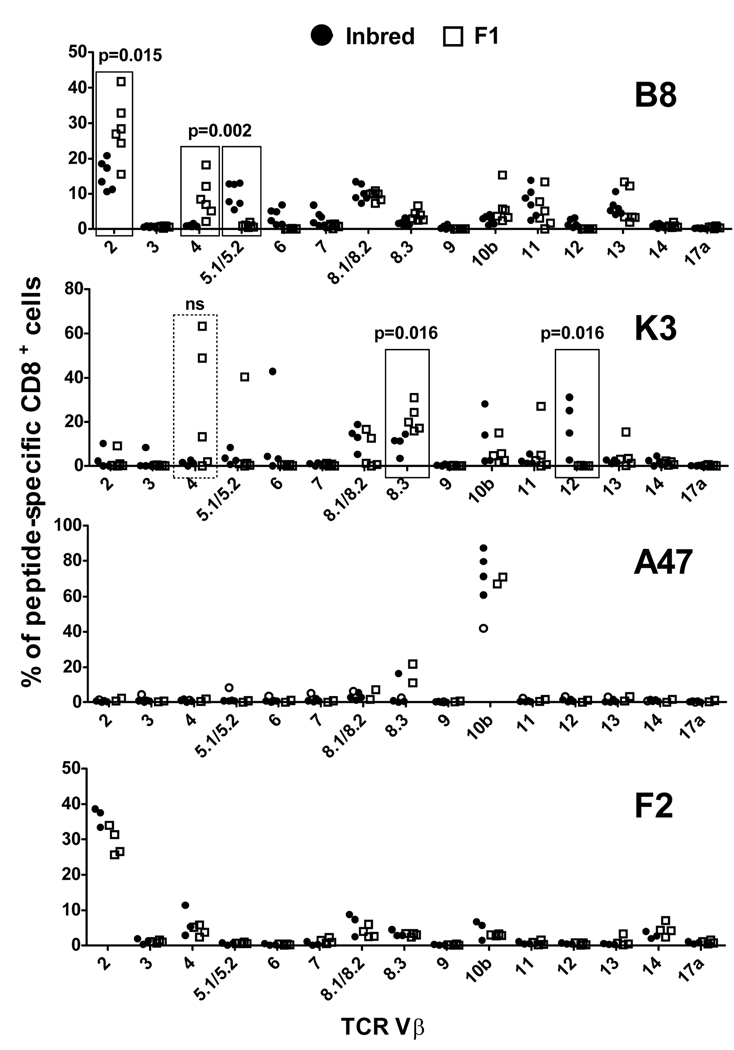
In an influenza virus infection model, the poor immunogenicity of a peptide in F1 mice was found to be associated with an altered TCR repertoire (44). To find evidence of differences in TCR repertoires between inbred and F1 mice, a broad analysis of TCR Vβ usage was done for B820-, K36-, A47138- and F226-specific CD8+ T cells in the relevant mice after infection with VACV (figure 8). Where possible this analysis was done directly ex vivo, however K36- and A47138-specific cells in F1 mice were too rare for ex vivo analysis and so we obtained data from multiple T cell lines.
Figure 8. TCR Vβ usage in VACV-specific CD8+ T cells from inbred and F1 mice.
TCR Vβ usage in CD8+ T cells with peptide-specificity as shown in the top right of each graph was analysed in inbred (circles) and F1 (squares) mice. C57Bl/6 (H-2b) or DBA/2 (H-2d) and BDF1 (H-2b×d) mice were infected i.p. with 106 pfu VACV strain WR. Seven days later, splenocytes were prepared from individual mice and analyzed for Vβ expression on CD8+, DimerX+ cells (B820, all; A47138, filled circles for inbred only; F226, all). Alternatively, peptide-specific CD8+ T cell lines were prepared from splenocytes from individual mice and Vβ expression on CD8+ cells was assessed after 3–4 rounds of restimulation with peptide (K36, all; A47138 open circles inbred and all F1). Notable differences between inbred and F1 mice are boxed and p-values determined using a Mann-Whitney (nonparametic) test.
Vβ usage for B820-specific CD8+ T cells both from inbred and F1 mice was highly diverse, however there were some significant differences in the frequency of usage for some Vβ segments. These included more prevalent use of Vβ 5.1/5.2 in H-2b (p=0.002) and of Vβ 2 (p=0.015) and Vβ 4 (p=0.002) in H-2b×d mice. To determine Vβ profiles for K36-specific CD8+ T cells, multiple T cell lines were made from VACV-infected H-2b and H-2b×d mice. While this is not as direct as ex vivo measurement, some clear differences were apparent: Use of Vβ 12 and Vβ 8.3 were significantly (p=0.016 for both) greater for H-2b and H-2b×d mice, respectively. A closer look at Vβ 12 usage showed that this segment was used by lines from three of four H-2b mice, but no lines from H-2b×d mice. Conversely, Vβ 4 was used only by H-2b×d lines (three of five), dominating two of them, but because of the variance, this was not statistically significant. Unlike the broad Vβ usage seen thus far, A47138-specific CD8+ T cells had a very strong bias with up to 80% cells from some mice measured directly ex vivo using Vβ 10b with very little contribution from other segments. This result was essentially the same when experiments were done ex vivo (four mice) and using a T cell line for H-2b mice and in lines from two H-2b×d mice and all have been plotted together. Finally, F226-specific CD8+ T cells (all analysed ex vivo) also showed biased Vβ usage, this time with Vβ 2 being most preferred both in H-2d and H-2b×d mice. Minor usage of other Vβ segments was also similar for F226-specific T cells from inbred and F1 mice. Taken together, CD8+ T cells with four specificities were examined and while two of these showed significant differences in Vβ usage, this did not predict the epitopes that had worse than expected immunogenicity in F1 mice.
Poor immunogenicity of epitopes in F1 mice is predicted by reduced numbers of epitope-specific CD8+ T cells in the naïve repertoire
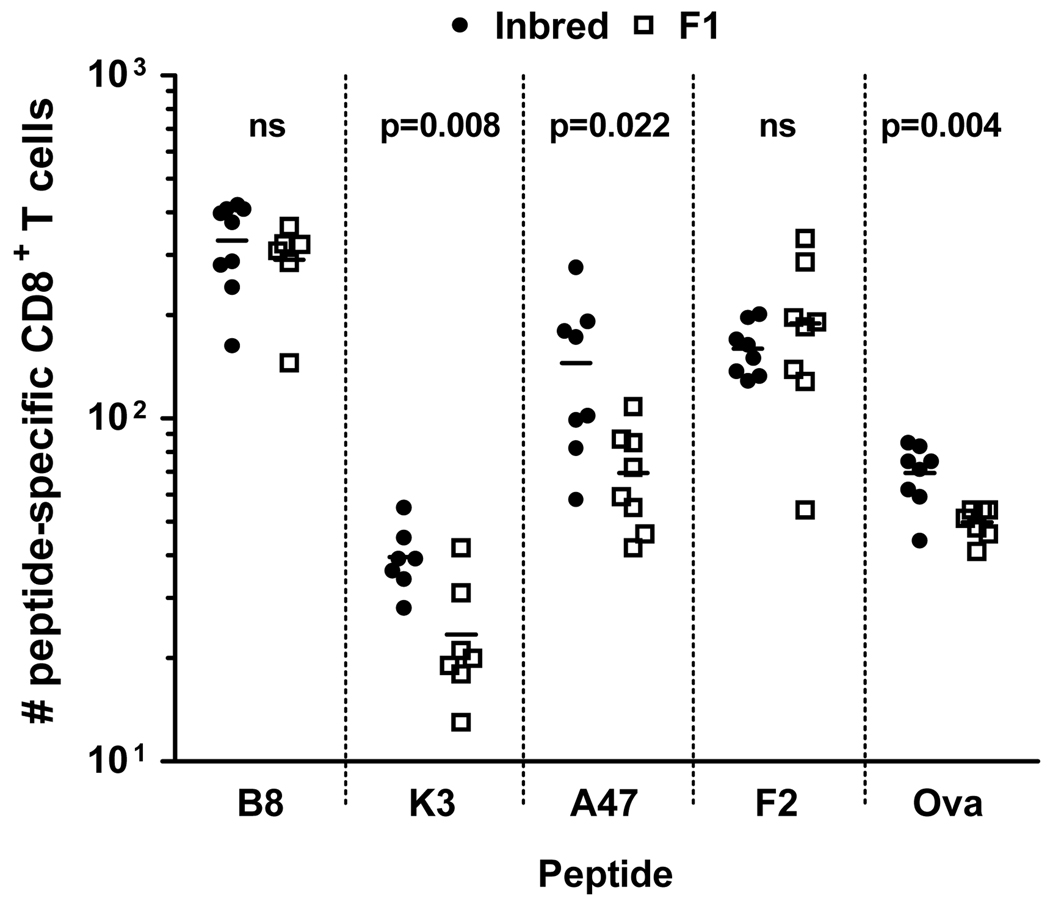
The experiments above suggest that while the repertoire of TCRs interacting with viral peptides differs between inbred and F1 mice, these changes are not necessarily associated with poor immunogenicity in the latter. Therefore the next step was to directly assess the numbers of peptide-specific CD8+ T cells in the naïve repertoire for a number of relevant epitopes using the recently developed methodology for magnetic bead-based enrichment of these rare cells (figure 9) (13, 18). For this analysis we chose the four peptides used in the Vβ experiments as well as ova257 (SIINFEKL) so that we could benchmark our estimates against the literature (18). We used Dimer-X reagents rather than traditional tetramers and were satisfied that these had the required specificity through the use of infected mice as positive controls (not shown). In C57Bl/6 mice, the number of CD8+ T cells in naïve mice that recognized these peptides ranged from around 450 for B820 in some mice down to less than 30 for K36. In our hands, the number that recognized SIINFEKL was a little lower than previosuly published (70–90 compared with around 100) but was close enough for us to continue with confidence. In DBA/2 mice the number of CD8+ T cells recognising the dominant F226 peptide was relatively high (upto 200), but somewhat surprisingly lower than for the dominant peptide in C57Bl/6 mice B820 (p<0.001) and not higher than the less dominant A47138, (p=0.6). Finally, when the number of naïve T cells recognising these peptides were compared in F1 mice versus the relevant inbred parent, these were significantly lower for K36 (p=0.008), A47138 (p=0.022) and SIINFEKL (p=0.004), but not B820 or F226. The peptide-specific differences are not the result of the total numbers of CD8+ T cells in the various mice, which were not significantly different between H-2b and H-2b×d mice (not shown). Thus a reduced number of cells in the naïve repertoire in F1 compared with the inbred predicts poor immunogenicity in F1 mice.
Figure 9. Numbers of peptide-specific CD8+ T cell precursors in naïve inred and F1 mice.
Single cells suspensions were prepared from spleens and lymph nodes of individual naïve C57Bl/6 (H-2b) or DBA/2 (H-2d) (inbred, filled circles) and BDF1 (H-2b×d) (F1, open squares) mice. Cells were stained with peptide-loaded PE-labeled DimerX and labelled cells were enriched by magnetic sorting using anti-PE magnetic beads. Further stains were applied as stated in the methods section and cells analysed by flow cytometry. Each point represents an individual mouse and the horizontal lines are means, data were obtained from more than one experiment for each peptide/mouse. Differences between means were tested using a t-test with Welch’s correction for unequal varience and p-values are noted for each peptide.
Discussion
Several aspects of this study of CD8+ T cell immunodominance in response to virus infection of F1 and inbred mice distinguish it from previously published work. First, and most importantly, previous work has tended to focus on the similar overall appearance of hierarchies rather than variation in immunogenicity of individual epitopes between inbred and F1 mice. This has led to a general view that changes in immunodominance in F1 compared with inbred animals are unlikely to be substantial or of relevance to vaccine development or our understanding of immunity to infectious disease. In contrast, we show that responses to some epitopes can be very severely compromised in F1 mice, that a surprisingly large number of epitopes are likely to be affected and that this can directly affect assessment of vaccination strategies in an animal model. This is perhaps best illustrated by the polytope experiment where five epitopes out of 14 had responses in the F1 reduced by more than 90% compared with the inbred controls when measured as a percent of CD8+ T cells. If total numbers of epitope-specific cells are calculated, responses to six of the 14 are reduced by more than 90% in the F1 mice (not shown). In addition, the data show clearly that the assessment of immunogenicity of epitopes in the polytope is very different if F1 rather than inbred mice are used. Second, this is the first study of immunodominance in F1 mice that has confirmed that the phenomeneon is linked to MHC. This was done using MHC-congenic mice, so it remains possible that genes within the MHC region other than the classical class I genes are involved, but the relative lack of polymorphism in these genes suggest that this is unlikely. Third we are using VACV, attenuated strains of which are a potential vaccine vector in current clinical trials and which also has relevance in the context of a renewed desire to have vaccines available that protect against smallpox (60–65). The large genome size and therefore greater diversity of antigens presented to the immune system distinguishes this virus from those more commonly used to study immunodominance and the acute nature of infections (53). This logic holds even though we have not exploited the large number of epitopes now mapped for H-2b and H-2d mice (49, 50). While similar studies have been done with MCMV another large DNA virus, in this model altered responses linked to MHC were not nearly as dramatic as those related to genetic background, which had a strong influence on the total anti-viral CD8+ T cell response as well as immunodominance (45). Finally, this is the first study to directly link poor immunogenicity of certain peptides in F1 mice to a reduction in the number of peptide-specific CD8+ T cells in the naïve repertoire compared with an inbred parent.
The findings here also provide a cautionary tale about assuming that results obtained with a limited set of epitopes are relevant to all epitopes. The data shown in the first four figures led us to hypothesize that epitopes restricted by H-2b allomorphs were at a disadvantage compared with H-2d-restricted epitopes in H-2b×d F1 mice. This conclusion could be supported statistically when data for individual epitopes were collapsed and analyzed by presenting haplotype (not shown). It was also consistent with the earliest publications looking at the effect of non-restricting MHC on cytotoxic T cell responses (41, 42). These considerations led us to confirm published accounts showing that surface expression of H-2b alleles are not unduly compromised by co-expression of H-2d alleles in H-2b×d F1 mice (58 and figure 5). We then went on to examine the contribution of each parental haplotype to anti-VACV responses in H-2b×d F1 mice. These experiments showed unambiguously first, that roughly the same number of CD8+ T cells respond to VACV in the parental and F1 mice and second, that MHC alleles from each parent restrict roughly half of the total anti-VACV CD8+ T cells in the F1 mice. Together these data suggest that our selection of epitopes was simply unlucky, such that by chance the H-2b-restricted epitopes chosen for this study were poorly immunogenic in the F1 mice. In this context it is worth remembering the broad specificity of anti-VACV CD8+ T cells, with more than 100 epitopes mapped across the two haplotypes and more than half of the H-2d-restricted response still unaccounted for (50, 66). So while we have used 14 different peptides here (in the polytope experiments), this may be tracking only 10% or so of the possible specificities. This issue remains a challenge in models based on large viruses.
In pursuing the mechanistic basis of our findings we focused on the T cell side of immunodominance because it seemed less likely that antigen presentation for some, but not all peptides presented by a single MHC allomorph would differ between inbred strains and their F1 progeny. First we examined functional avidity by measuring responsiveness to limiting amounts of peptide in ICS assays both for IFN-γ and TNF-α and found no substantial differences for B820-, K36- and A47138-specific CD8+ T cells in H-2b and H-2b×d F1 mice (data not shown). Next we asked whether responses to these peptides were always poor, or were poor only in the context of VACV where immunodomination might be occurring. We found that when used as sole immunogens, A47138 and especially K36, but not B820 were poorly immunogenic in H-2b×d compared with H-2b mice. From this it was concluded that poor CD8+ T cell responses to some epitopes in F1 mice was most likely due to changes in the TCR repertoire.
To further support changes in the naïve repertoire as the cause of poor responses to some epitopes in F1 mice an analysis of Vβ segment usage in peptide-specific T cells and a direct estimation of the number of CD8+ T cells with the relevant specificities in naïve mice were done. The Vβ analysis was in part a response to previous work that showed changes in Vβ bias for influenza virus PA224–specific CD8+ T cells in F1 mice (44) where there is an apparent loss of a prominent clone or set of clones in the presence of additional MHC alleles. We examined T cells that recognised two peptides that were poorly immunogenic, and two that maintained immunogenicity in F1 mice (figure 8). While differences in Vβ usage were found, this occurred for B820- and K36-specific populations which are well and poorly represented in F1 mice respectively. In addition, the changes were largely in alignment with differences in Vβ preference seen in the wider CD8+ compartment in naïve mice (not shown). These results demonstrate that changes in TCR usage can be tolerated without loss of immunogenicity and where such a loss occurs this is not limited to peptides which are recognised by a narrow spectrum of receptors. The remaining two peptide-specific T cell population demonstrated substantial bias in Vβ preference, but no difference between inbred and F1 mice. It remains possible that many of the Vβ 10b+ A47138-specific cells in H-2b mice are from a dominant clone that is missing in F1 mice, but unlike PA224 from influenza, there is no evidence of Vβ diversification in these mice. Finally, this study also allows a first comparison of Vβ usage for T cells recognizing two dominant epitopes from the same virus, but in the context of different MHC haplotypes. We found that B820- and F226-specific T cells had broad and narrow Vβ usage respectively, which shows that the breadth of TCRs used per se is not an important factor in immunodominance.
In contrast to the Vβ analysis, a reduction in number of naïve precursors in F1 compared with inbred was able to predict the peptides that would be poorly immunogenic in F1 mice. We were somewhat surprised that the differences were relatively modest, in the order of two-fold, given the substantial differences seen after VACV or peptide immunization. Indeed differences in numbers of naïve precursors across the peptides in the different mice ranged over 10-fold. The reasons why a relatively small difference in precursors is exaggerated during expansion in response to immunization, even in the absence of competing specificities as in the peptide experiments, will be of ongoing interest. A second look at the data from mice examined 6 weeks after immunization shows that differences can also be increased further in the retraction phase VACV infection (compare figure 1 and figure 2) and this also requires further exploration. These observations suggest that while changes in the naïve repertoire of T cells contribute substantially to this phenomenon, deletion of clones in the thymus due to cross-reactivity with the additional MHC molecules may not be complete explanation. We speculate the the quality of the clones remaining may also play a role, though as noted above in this case we ruled out the simplest explanation, which is poor avidity.
This is also the initial estimation of the frequency of VACV paptide-specific T cells in the naïve repertoire so it is the first time that the relationship between this frequency and immunodominance can be examined in this model. Looking at our precursor data alone suggests that in H-2b×d mice, responses to B820 and F226 should be substantially different and not similar as observed. Perhaps in this case a lack of precursors for F226 is compensated by a higher affinity for MHC (49, 52) or processing differences. However in another case where precursor numbers fail to predict position in hierarchy, a resolution is less easy to find: after VACV immunization, A47138 and K36 induced similar numbers of CD8+ T cells in H-2b mice, yet more T cells recognized A47138 in naïve mice and this peptide has a 10-fold advantage in affinity for MHC compared with K36 (49) Moreover, in peptide vaccinations (figure 7) processing issues are avoided. These data suggest that immunodominance is not as well explained by number of precursors and affinity for MHC as recent papers have suggested (13, 18), with the caveat that poor affinity of peptide for MHC in the DimerX reagents may compromises the efficiency of naïve CD8+ T cell enrichment.
Overall the data presented here suggest that the presence of non-presenting MHC may affect more epitopes more profoundly than previously appreciated and indeed can affect immunodominance hierarchies. This has implications for the preclinical assessment of experimental vaccines in inbred mice. For example, proof of concept testing of multi-epitope constructs aiming to cover several haplotypes, such as the one used here (56), is most appropriately done in F1 hybrids rather than a series of inbred mice. This phenomenon may also help to explain why immunodominance hierarchies are relatively difficult to define in outbred human populations. Epitopes to which responses are found in the majority of individuals sharing a particular HLA molecule are likely to be those whose immunogenicity is less frequently affected by other restriction elements, rather than those that elicit the largest responses in any given individual. In the context of testing new smallpox vaccines based on VACV, this presents some challenges as to how all the new human CD8+ T cell epitope information might best be used (60, 67, 68).
Acknowledgements
We wish to thank Leanne Morrison, Mick Devoy and Stewart Smith for technical assistance and Jack Bennink and Weisan Chen for helpful discussions.
Footnotes
This work was supported by an Australian NHMRC project grant #389819 and Career Development Award #418108 and NIH grant RO1AI067401 to DCT. YW was supported by an NHMRC Training Fellowship #316978 and TC by an NHMRC Biomedical Postgraduate Scholarship.
Abbreviations: αGalCer, α-galactosylceramide; DMEM, Dulbecco’s modified Eagle’s medium; D10; DMEM with glutamine and 10% FBS; ICS, intracellular cytokine staining; LCMV, choriomeningitis virus; VACV, vaccinia virus; WR, Western reserve
References
- 1.Yewdell JW, Bennink JR. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu. Rev. Immunol. 1999;17:51–88. doi: 10.1146/annurev.immunol.17.1.51. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Anton LC, Bennink JR, Yewdell JW. Dissecting the multifactorial causes of immunodominance in class I- restricted T cell responses to viruses. Immunity. 2000;12:83–93. doi: 10.1016/s1074-7613(00)80161-2. [DOI] [PubMed] [Google Scholar]
- 3.La Gruta NL, Kedzierska K, Pang K, Webby R, Davenport M, Chen W, Turner SJ, Doherty PC. A virus-specific CD8+ T cell immunodominance hierarchy determined by antigen dose and precursor frequencies. PNAS. 2006;103:994–999. doi: 10.1073/pnas.0510429103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weidt G, Utermohlen O, Heukeshoven J, Lehmann-Grube F, Deppert W. Relationship among immunodominance of single CD8+ T cell epitopes, virus load, and kinetics of primary antiviral CTL response. J. Immunol. 1998;160:2923–2931. [PubMed] [Google Scholar]
- 5.van der Most RG, Murali-Krishna K, Lanier JG, Wherry EJ, Puglielli MT, Blattman JN, Sette A, Ahmed R. Changing immunodominance patterns in antiviral CD8 T-cell responses after loss of epitope presentation or chronic antigenic stimulation. Virol. 2003;315:93–102. doi: 10.1016/j.virol.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Probst HC, Tschannen K, Gallimore A, Martinic M, Basler M, Dumrese T, Jones E, van den Broek MF. Immunodominance of an antiviral cytotoxic T cell response is shaped by the kinetics of viral protein expression. J. Immunol. 2003;171:5415–5422. doi: 10.4049/jimmunol.171.10.5415. [DOI] [PubMed] [Google Scholar]
- 7.Bousso P, Levraud JP, Kourilsky P, Abastado JP. The Composition of a Primary T Cell Response Is Largely Determined by the Timing of Recruitment of Individual T Cell Clones. J. Exp. Med. 1999;189:1591–1600. doi: 10.1084/jem.189.10.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen W, Norbury CC, Cho Y, Yewdell JW, Bennink JR. Immunoproteasomes shape immunodominance hierarchies of antiviral CD8(+) T cells at the levels of T cell repertoire and presentation of viral antigens. J. Exp. Med. 2001;193:1319–1326. doi: 10.1084/jem.193.11.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisenlohr LC, Yewdell JW, Bennink JR. Flanking sequences influence the presentation of an endogenously synthesized peptide to cytotoxic T lymphocytes. J. Exp. Med. 1992;175:481–487. doi: 10.1084/jem.175.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Del Val M, Schlicht HJ, Ruppert T, Reddehase MJ, Koszinowski UH. Efficient processing of an antigenic sequence for presentation by MHC class I molecules depends on its neighboring residues in the protein. Cell. 1991;66:1145–1153. doi: 10.1016/0092-8674(91)90037-y. [DOI] [PubMed] [Google Scholar]
- 11.Pang KC, Sanders MT, Monaco JJ, Doherty PC, Turner SJ, Chen W. Immunoproteasome subunit deficiencies impact differentially on two immunodominant influenza virus-specific CD8+ T cell responses. J. Immunol. 2006;177:7680–7688. doi: 10.4049/jimmunol.177.11.7680. [DOI] [PubMed] [Google Scholar]
- 12.Deng Y, Yewdell JW, Eisenlohr LC, Bennink JR. MHC affinity, peptide liberation, T cell repertoire, and immunodominance all contribute to the paucity of MHC class I-restricted peptides recognized by antiviral CTL. J. Immunol. 1997;158:1507–1515. [PubMed] [Google Scholar]
- 13.Kotturi MF, Scott I, Wolfe T, Peters B, Sidney J, Cheroutre H, von Herrath MG, Buchmeier MJ, Grey H, Sette A. Naive precursor frequencies and MHC binding rather than the degree of epitope diversity shape CD8+ T cell immunodominance. J. Immunol. 2008;181:2124–2133. doi: 10.4049/jimmunol.181.3.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sette A, Vitiello A, Reherman B, Fowler P, Nayersina R, Kast WM, Melief CJ, Oseroff C, Yuan L, Ruppert J, Sidney J, del Guercio MF, Southwood S, Kubo RT, Chesnut RW, Grey HM, Chisari FV. The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J. Immunol. 1994;153:5586–5592. [PubMed] [Google Scholar]
- 15.van der Most RG, Sette A, Oseroff C, Alexander J, Murali-Krishna K, Lau LL, Southwood S, Sidney J, Chesnut RW, Matloubian M, Ahmed R. Analysis of cytotoxic T cell responses to dominant and subdominant epitopes during acute and chronic lymphocytic choriomeningitis virus infection. J. Immunol. 1996;157:5543–5554. [PubMed] [Google Scholar]
- 16.Busch DH, Pamer EG. MHC class I/peptide stability: implications for immunodominance, in vitro proliferation, and diversity of responding CTL. J. Immunol. 1998;160:4441–4448. [PubMed] [Google Scholar]
- 17.Daly K, Nguyen P, Woodland DL, Blackman MA. Immunodominance of major histocompatibility complex class I-restricted influenza virus epitopes can be influenced by the T-cell receptor repertoire. J. Virol. 1995;69:7416–7422. doi: 10.1128/jvi.69.12.7416-7422.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Obar JJ, Khanna KM, Lefrançois L. Endogenous naive CD8+ T cell crecursor frequency regulates primary and memory responses to infection. Immunity. 2008;28:859–869. doi: 10.1016/j.immuni.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perkins DL, Wang YS, Fruman D, Seidman JG, Rimm IJ. Immunodominance is altered in T cell receptor (beta-chain) transgenic mice without the generation of a hole in the repertoire. J. Immunol. 1991;146:2960–2964. [PubMed] [Google Scholar]
- 20.Choi EY, Christianson GJ, Yoshimura Y, Sproule TJ, Jung NJ, Joyce S, Roopenian DC. Immunodominance of H60 is caused by an abnormally high precursor T cell pool directed against its unique minor histocompatibility antigen peptide. Immunity. 2002;17:593–603. doi: 10.1016/s1074-7613(02)00428-4. [DOI] [PubMed] [Google Scholar]
- 21.Haeryfar SMM, Hickman HD, Irvine KR, Tscharke DC, Bennink JR, Yewdell JW. Terminal deoxynucleotidyl transferase establishes and broadens anti-viral CD8+ T cell immunodominance hierarchies. J. Immunol. 2008;181:649–659. doi: 10.4049/jimmunol.181.1.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wallace ME, Bryden M, Cose SC, Coles RM, Schumacher TN, Brooks A, Carbone FR. Junctional biases in the naive TCR repertoire control the CTL response to an immunodominant determinant of HSV-1. Immunity. 2000;12:547–556. doi: 10.1016/s1074-7613(00)80206-x. [DOI] [PubMed] [Google Scholar]
- 23.Trautmann L, Rimbert M, Echasserieau K, Saulquin X, Neveu B, Dechanet J, Cerundolo V, Bonneville M. Selection of T cell clones expressing high-affinity public TCRs within human cytomegalovirus-specific CD8 T cell responses. J. Immunol. 2005;175:6123–6132. doi: 10.4049/jimmunol.175.9.6123. [DOI] [PubMed] [Google Scholar]
- 24.Dzutsev AH, Belyakov IM, Isakov DV, Margulies DH, Berzofsky JA. Avidity of CD8 T cells sharpens immunodominance. Int. Immunol. 2007;19:497–507. doi: 10.1093/intimm/dxm016. [DOI] [PubMed] [Google Scholar]
- 25.Liu F, Whitton JL, Slifka MK. The rapidity with which virus-specific CD8+ T cells initiate IFN-γ synthesis increases markedly over the course of infection and correlates with immunodominance. J. Immunol. 2004;173:456–462. doi: 10.4049/jimmunol.173.1.456. [DOI] [PubMed] [Google Scholar]
- 26.Badovinac VP, Tvinnereim AR, Harty JT. Regulation of antigen-specific CD8+ T Cell homeostasis by perforin and interferon-gamma. Science. 2000;290:1354. doi: 10.1126/science.290.5495.1354. [DOI] [PubMed] [Google Scholar]
- 27.Haeryfar SMM, DiPaolo RJ, Tscharke DC, Bennink JR, Yewdell JW. Regulatory T cells suppress CD8+ T cell responses induced by direct priming and cross-priming and moderate immunodominance disparities. J. Immunol. 2005;174:3344–3351. doi: 10.4049/jimmunol.174.6.3344. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez F, Harkins S, Slifka MK, Whitton JL. Immunodominance in virus-induced CD8+ T-cell responses is dramatically modified by DNA immunization and is regulated by gamma interferon. J. Virol. 2002;76:4251–4259. doi: 10.1128/JVI.76.9.4251-4259.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andreansky SS, Stambas J, Thomas PG, Xie W, Webby RJ, Doherty PC. Consequences of immunodominant epitope deletion for minor influenza virus-specific CD8+ T cell responses. J. Virol. 2005;79:4329–4339. doi: 10.1128/JVI.79.7.4329-4339.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jenkins MR, Webby R, Doherty PC, Turner SJ. Addition of a prominent epitope affects influenza A virus-specific CD8+ T cell immunodominance hierarchies when antigen is limiting. J. Immunol. 2006;177:2917–2925. doi: 10.4049/jimmunol.177.5.2917. [DOI] [PubMed] [Google Scholar]
- 31.Roy-Proulx G, Baron C, Perreault C. CD8 T-cell ability to exert immunodomination correlates with T-cell receptor: epitope association rate. Biol Blood Marrow Transplant. 2005;11:260–271. doi: 10.1016/j.bbmt.2004.12.334. [DOI] [PubMed] [Google Scholar]
- 32.Thomas PG, Brown SA, Keating R, Yue W, Morris MY, So J, Webby RJ, Doherty PC. Hidden epitopes emerge in secondary influenza virus-specific CD8+ T cell reponses. J. Immunol. 2007;178:3091–3098. doi: 10.4049/jimmunol.178.5.3091. [DOI] [PubMed] [Google Scholar]
- 33.Lewicki HA, Von Herrath MG, Evans CF, Whitton JL, Oldstone MB. CTL escape viral variants. II. Biologic activity in vivo. Virol. 1995;211:443–450. doi: 10.1006/viro.1995.1426. [DOI] [PubMed] [Google Scholar]
- 34.Fischer MA, Tscharke DC, Donohue KB, Truckenmiller ME, Norbury CC. Reduction of vector gene expression increases foreign antigen-specific CD8+ T-cell priming. J. Gen. Virol. 2007;88:2378–2386. doi: 10.1099/vir.0.83107-0. [DOI] [PubMed] [Google Scholar]
- 35.Wallace ME, Keating R, Heath WR, Carbone FR. The cytotoxic T-cell response to herpes simplex virus type 1 infection of C57BL/6 mice is almost entirely directed against a single immunodominant determinant. J. Virol. 1999;73:7619–7626. doi: 10.1128/jvi.73.9.7619-7626.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moss DJ, Burrows SR, Silins SL, Misko I, Khanna R. The immunology of Epstein-Barr virus infection. Philosophical Transactions of the Royal Society of London Series B-Biological Sciences. 2001;356:475–488. doi: 10.1098/rstb.2000.0784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hislop AD, Annels NE, Gudgeon NH, Leese AM, Rickinson AB. Epitope-specific evolution of human CD8+ T cell responses from primary to persistent phases of Epstein-Barr virus infection. J. Exp. Med. 2002;195:893–905. doi: 10.1084/jem.20011692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khan N, Hislop A, Gudgeon N, Cobbold M, Khanna R, Nayak L, Rickinson AB, Moss PAH. Herpesvirus-specific CD8 T cell immunity in old age: cytomegalovirus impairs the response to a coresident EBV infection. J. Immunol. 2004;173:7481–7489. doi: 10.4049/jimmunol.173.12.7481. [DOI] [PubMed] [Google Scholar]
- 39.Pudney VA, Leese AM, Rickinson AB, Hislop AD. CD8+ immunodominance among Epstein-Barr virus lytic cycle antigens directly reflects the efficiency of antigen presentation in lytically infected cells. J. Exp. Med. 2005;201:349–360. doi: 10.1084/jem.20041542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koelle DM, Liu Z, McClurkan CL, Cevallos RC, Vieira J, Hosken NA, Meseda CA, Snow DC, Wald A, Corey L. Immunodominance among herpes simplex virus-specific CD8 T cells expressing a tissue-specific homing receptor. PNAS. 2003;100:12899–12904. doi: 10.1073/pnas.2131705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doherty PC, Biddison WE, Bennink JR, Knowles BB. Cytotoxic T-cell responses in mice infected with influenza and vaccinia viruses vary in magnitude with H-2 genotype. J. Exp. Med. 1978;148:534–543. doi: 10.1084/jem.148.2.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zinkernagel RM, Althage A, Cooper S, Kreeb G, Klein PA, Sefton B, Flaherty L, Stimpfling J, Shreffler D, Klein J. Ir-genes in H-2 regulate generation of anti-viral cytotoxic T cells. Mapping to K or D and dominance of unresponsiveness. J. Exp. Med. 1978;148:592–606. doi: 10.1084/jem.148.2.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mullbacher A, Blanden RV, Brenan M. Neonatal tolerance of major histocompatibility complex antigens alters Ir gene control of the cytotoxic T cell response to vaccinia virus. J. Exp. Med. 1983;157:1324–1338. doi: 10.1084/jem.157.4.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Belz GT, Stevenson PG, Doherty PC. Contemporary analysis of MHC-related immunodominance hierarchies in the CD8+ T cell response to influenza A viruses. J. Immunol. 2000;165:2404–2409. doi: 10.4049/jimmunol.165.5.2404. [DOI] [PubMed] [Google Scholar]
- 45.Munks MW, Gold MC, Zajac AL, Doom CM, Morello CS, Spector DH, Hill AB. Genome-wide analysis reveals a highly diverse CD8 T cell response to murine cytomegalovirus. J. Immunol. 2006;176:3760–3766. doi: 10.4049/jimmunol.176.6.3760. [DOI] [PubMed] [Google Scholar]
- 46.Chen W, Bennink JR, Morton PA, Yewdell JW. Mice deficient in perforin, CD4+ T Cells, or CD28-mediated signaling maintain the typical immunodominance hierarchies of CD8+ T-cell responses to influenza virus. J. Virol. 2002;76:10332–10337. doi: 10.1128/JVI.76.20.10332-10337.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rutigliano JA, Ruckwardt TJ, Martin JE, Graham BS. Relative dominance of epitope-specific CD8+ T cell responses in an F1 hybrid mouse model of respiratory syncytial virus infection. Virol. 2007;362:314–319. doi: 10.1016/j.virol.2006.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burrows SR, Silins SL, Moss DJ, Khanna R, Misko IS, Argaet VP. T cell receptor repertoire for a viral epitope in humans is diversified by tolerance to a background major histocompatibility complex antigen. J. Exp. Med. 1995;182:1703–1715. doi: 10.1084/jem.182.6.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moutaftsi M, Peters B, Pasquetto V, Tscharke DC, Sidney J, Bui HH, Grey H, Sette A. A consensus epitope prediction approach identifies the breadth of murine TCD8+-cell responses to vaccinia virus. Nat Biotechnol. 2006;24:817–819. doi: 10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]
- 50.Oseroff C, Peters B, Pasquetto V, Moutaftsi M, Sidney J, Panchanathan V, Tscharke DC, Maillere B, Grey H, Sette A. Dissociation between epitope hierarchy and immunoprevalence in CD8 responses to vaccinia virus Western Reserve. J. Immunol. 2008;180:7193–7202. doi: 10.4049/jimmunol.180.11.7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tscharke DC, Karupiah G, Zhou J, Palmore T, Irvine KR, Haeryfar SMM, Williams S, Sidney J, Sette A, Bennink JR, Yewdell JW. Identification of poxvirus CD8+ T cell determinants to enable rational design and characterization of smallpox vaccines. J. Exp. Med. 2005;201:95–104. doi: 10.1084/jem.20041912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tscharke DC, Woo W-P, Sakala IG, Sidney J, Sette A, Moss DJ, Bennink JR, Karupiah G, Yewdell JW. Poxvirus CD8+ T-cell determinants and cross-reactivity in BALB/c mice. J. Virol. 2006;80:6318–6323. doi: 10.1128/JVI.00427-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y, Flesch IEA, Tscharke DC. Vaccinia virus CD8+ T cell dominance hierarchies cannot be altered by prior immunization with individual peptides. J. Virol. 2009 doi: 10.1128/JVI.00410-09. JVI.00410-00409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harrington LE, van der Most R, Whitton JL, Ahmed R. Recombinant vaccinia virus-induced T-cell immunity: quantitation of the response to the virus vector and the foreign epitope. J. Virol. 2002;76:3329–3337. doi: 10.1128/JVI.76.7.3329-3337.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith CL, Mirza F, Pasquetto V, Tscharke DC, Palmowski MJ, Dunbar PR, Sette A, Harris AL, Cerundolo V. Immunodominance of poxviral-specific CTL in a human trial of recombinant-modified vaccinia Ankara. J. Immunol. 2005;175:8431–8437. doi: 10.4049/jimmunol.175.12.8431. [DOI] [PubMed] [Google Scholar]
- 56.Thomson SA, Elliott SL, Sherritt MA, Sproat KW, Coupar BE, Scalzo AA, Forbes CA, Ladhams AM, Mo XY, Tripp RA, Doherty PC, Moss DJ, Suhrbier A. Recombinant polyepitope vaccines for the delivery of multiple CD8 cytotoxic T cell epitopes. J. Immunol. 1996;157:822–826. [PubMed] [Google Scholar]
- 57.Thomson SA, Sherritt MA, Medveczky J, Elliott SL, Moss DJ, Fernando GJP, Brown LE, Suhrbier A. Delivery of Multiple CD8 Cytotoxic T Cell Epitopes by DNA Vaccination. J. Immunol. 1998;160:1717–1723. [PubMed] [Google Scholar]
- 58.Tourdot S, Gould KG. Competition between MHC class I alleles for cell surface expression alters CTL responses to influenza A virus. J. Immunol. 2002;169:5615–5621. doi: 10.4049/jimmunol.169.10.5615. [DOI] [PubMed] [Google Scholar]
- 59.Silk JD, Hermans IF, Gileadi U, Chong TW, Shepherd D, Salio M, Mathew B, Schmidt RR, Lunt SJ, Williams KJ, Stratford IJ, Harris AL, Cerundolo V. Utilizing the adjuvant properties of CD1d-dependent NK T cells in T cell-mediated immunotherapy. J. Clin. Invest. 2004;114:1800–1811. doi: 10.1172/JCI22046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tscharke DC. Adaptive immunity to vaccinia virus; revisiting an old friend. Future Virol. 2007;2:163–172. [Google Scholar]
- 61.Greenberg RN, Kennedy JS, Clanton DJ, Plummer EA, Hague L, Cruz J, Ennis FA, Blackwelder WC, Hopkins RJ. Safety and immunogenicity of new cell-cultured smallpox vaccine compared with calf-lymph derived vaccine: a blind, single-centre, randomised controlled trial. Lancet. 2005;365:398–409. doi: 10.1016/S0140-6736(05)17827-1. [DOI] [PubMed] [Google Scholar]
- 62.Schraeder TL, Campion EW. Smallpox Vaccination -- The Call to Arms. The New England Journal of Medicine. 2003;348:381–382. doi: 10.1056/NEJMp020177. [DOI] [PubMed] [Google Scholar]
- 63.Bozzette SA, Boer R, Bhatnagar V, Brower JL, Keeler EB, Morton SC, Stoto MA. A Model for a Smallpox-Vaccination Policy. The New England Journal of Medicine. 2003;348:416–425. doi: 10.1056/NEJMsa025075. [DOI] [PubMed] [Google Scholar]
- 64.Gomez CE, Najera JL, Krupa M, Esteban M. The poxvirus vectors MVA and NYVAC as gene delivery systems for vaccination against infectious diseases and cancer. Curr Gene Ther. 2008;8:97–120. doi: 10.2174/156652308784049363. [DOI] [PubMed] [Google Scholar]
- 65.Drexler I, Staib C, Sutter G. Modified vaccinia virus Ankara as antigen delivery system: how can we best use its potential? Curr. Opin. Biotechnol. 2004;15:506–512. doi: 10.1016/j.copbio.2004.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moutaftsi M, Peters B, Pasquetto V, Sidney J, Bui HH, Grey H, Tscharke D, Yewdell J, Sette A. Towards the complete account of T cell specificities directed against vaccinia virus. J. Immunol. 2006;176:S116–S116. [Google Scholar]
- 67.Jing L, Chong TM, McClurkan CL, Huang J, Story BT, Koelle DM. Diversity in the acute CD8 T cell response to vaccinia virus in humans. J. Immunol. 2005;175:7550–7559. doi: 10.4049/jimmunol.175.11.7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oseroff C, Kos F, Bui H-H, Peters B, Pasquetto V, Glenn J, Palmore T, Sidney J, Tscharke DC, Bennink JR, Southwood S, Grey HM, Yewdell JW, Sette A. HLA class I-restricted responses to vaccinia recognize a broad array of proteins mainly involved in virulence and viral gene regulation. Proceedings of the National Academy of Sciences, USA. 2005;102:13980–13985. doi: 10.1073/pnas.0506768102. [DOI] [PMC free article] [PubMed] [Google Scholar]