Summary
Synthetic sickness/lethality (SSL) can be exploited to develop therapeutic strategies for cancer. Deficiencies in the tumor suppressor proteins MLH1 and MSH2 have been implicated in cancer. Here we demonstrate that deficiency in MSH2 is SSL with inhibition of the DNA polymerase POLB, whereas deficiency in MLH1 is SSL with DNA polymerase POLG inhibition. Both SSLs led to the accumulation of 8-oxoG oxidative DNA lesions. MSH2/POLB SSL caused nuclear 8-oxoG accumulation, whereas MLH1/POLG SSL led to a rise in mitochondrial 8-oxoG levels. Both SSLs were rescued by silencing the adenine glycosylase MUTYH, suggesting that lethality could be caused by the formation of lethal DNA breaks upon 8-oxoG accumulation. These data suggest targeted, mechanism-based therapeutic approaches.
Keywords: CELLCYCLE
Graphical Abstract
Highlights
► A range of cancers are characterized by MLH1 or MSH2 deficiency ► Inhibition of DNA polymerase B selectively kills MSH2-deficient tumor cells ► Inhibition of DNA polymerase G selectively kills MLH1-deficient tumor cells ► These results suggest mechanism-based approaches to therapy
Significance
A range of tumor types, including colorectal cancers and in particular hereditary nonpolyposis colon cancer, are characterized by loss of either MLH1 or MSH2 tumor suppressor gene function. Where this occurs, MLH1 or MSH2 loss distinguishes tumor cells from normal cells. Here we exploit these differences to show that inhibition of particular DNA polymerases, when combined with either MLH1 or MSH2 loss, can lead to tumor cell death. These results suggest targeted therapeutic approaches to the treatment of MSH2- or MLH1-deficient cancers.
Introduction
One of the recurrent themes of modern cancer drug discovery has been the design of therapeutic approaches that exploit the underlying genetic makeup of tumor cells. For example, the efficacy of imatinib (Gleevec) in the treatment of leukemias carrying the BCR-ABL fusion (Druker et al., 2001) provides a clear demonstration of how a tumor-specific genetic change may be exploited clinically. Although targeting gain-of-function oncogenic events such as the BCR-ABL fusion is conceptually straightforward, it is not obvious how to pharmacologically target a tumor suppressor protein that is dysfunctional or even completely absent. To address this issue, the exploitation of synthetic sickness/lethality (SSL) relationships has been proposed (Hartwell et al., 1997; Kaelin, 2005). A synthetic lethal relationship between two genes or proteins exists when loss of function of either alone is compatible with viability but simultaneous loss of both causes death (Kaelin, 2005). This concept has been extended to the idea of synthetic sickness, where simultaneous mutation/loss of function of two genes impairs cellular fitness more than mutation of either gene alone (Kaelin, 2005). These concepts can be used to design therapeutic approaches that target the cancer cell-specific loss of tumor suppressor proteins (Luo et al., 2009). For example, if a tumor suppressor gene and a second gene are synthetically lethal or sick, inhibition of the second gene could selectively kill or impair the fitness of tumor cells.
SSL between DNA repair proteins and pathways has the potential to be exploited clinically. For example, inhibition of the base excision repair (BER) pathway protein PARP1 is SSL with deficiency of either the BRCA1 or BRCA2 tumor suppressor/DNA repair proteins (Bryant et al., 2005; Farmer et al., 2005; Fong et al., 2009). Similarly, Hartwell and colleagues (Hartwell et al., 1997) highlighted the potential for exploiting SSL interactions between mismatch repair (MMR) proteins and DNA polymerases, a hypothesis based primarily on studies in budding yeast (Morrison et al., 1993). For example, mutations in pol3-01 (the ortholog of the human POLD catalytic subunit) are SSL with loss of the orthologs of the MMR genes EXOI, MSH6, MSH2, MLH1, and PMS1 (Morrison et al., 1993; Argueso et al., 2002; Tran et al., 1997). Similarly, the yeast Msh6-PolH double mutant is not viable (Pavlov et al., 2001) and Msh2-pol2-4 (PolE) mutants are synthetically sick (Tran et al., 1997).
Germline mutations in the MMR genes MLH1 or MSH2 predispose to hereditary nonpolyposis colorectal cancer (HNPCC)/Lynch syndrome, which accounts for approximately 5% of all colorectal cancer cases (Jacob and Praz, 2002). Furthermore, aberrant MLH1 promoter methylation has been proposed as a cause of some sporadic colorectal cancers (Arnold et al., 2003; Bettstetter et al., 2007; Peltomaki and Vasen, 2004). Here, MSH2 and MLH1 are proposed to act as classical tumor suppressor genes, where tumor cells lose MSH2 or MLH1 function whereas normal cells retain at least one functional allele. Although other functions have been ascribed to both MSH2 and MLH1, their most well defined function is in DNA MMR (Jiricny, 2006). MMR is primarily concerned with repair of DNA lesions that occur during DNA replication, such as DNA polymerase errors that include base-base or insertion/deletion mismatches. Recent evidence suggests that MMR proteins are also involved in the repair of DNA lesions caused by oxidative damage (Colussi et al., 2002; Macpherson et al., 2005). In brief, DNA mismatches are recognized by the MutSα complex, an MSH2/MSH6 heterodimer. This interaction enables MutSα to recruit the MutLα complex, itself a heterodimer consisting of the MLH1 and PMS2 proteins. MutLα binding stabilizes the MutSα:DNA interaction and acts as an interface between mismatch recognition by MutSα and additional proteins that excise and repair the DNA mismatch (Jiricny, 2006).
Reactive oxygen species, generated by sources such as the normal metabolic processes of cells as well as environmental agents, are genotoxic, producing DNA base lesions such as 8-oxoguanine (8-oxoG) (Kamiya, 2003). At replication, 8-oxoG residues can direct the incorporation of either C or A bases, which if left unrepaired can be mutagenic, primarily by causing GC→TA transversions. Mammalian cells have a number of mechanisms for dealing with 8-oxoG lesions, including BER and also processes involving MLH1 and MSH2. Repair of 8-oxoG lesions by BER is initiated by removal of the oxidized base by the DNA glycosylase OGG1. The apurinic (AP) site that results is further processed by the endonuclease APE1. APE1 hydrolyzes the 5′ phosphodiester bond of the sugar backbone at the AP site, giving rise to a single-strand DNA break (SSB) flanked by 3′ hydroxyl and 5′ deoxyribose-5-phosphate (5′dRp) termini. 5′dRp termini are then processed via the lyase activity of DNA polymerase β (POLB). POLB also incorporates the correct base at the SSB, after which the final break in the DNA sugar backbone is sealed by DNA ligase III to complete the repair process (Klungland and Bjelland, 2007). The direct involvement of POLB in this pathway is indicated by the inability of lysates from Polb−/− cells to repair synthetic 8-oxoG lesions (Horton et al., 2002). DNA polymerase γ (POLG) has been implicated in the repair of 8-oxoG lesions in mitochondria (Lewis et al., 2007), and like POLB, POLG also possesses a lyase activity that allows the removal of 5′dRp termini. Both MLH1 and MSH2 have also been implicated in 8-oxoG repair, and deficiency in either gene causes an accumulation of 8-oxoG lesions (Colussi et al., 2002; Macpherson et al., 2005).
During replication, if adenine is inserted opposite 8-oxoG, the enzyme MUTYH excises the adenine opposite 8-oxoG, causing the formation of an abasic site (Oka et al., 2008). This activity is thought to limit the number of GC→TA transversions that are otherwise caused by persistent 8-oxoG lesions. Further processing of the abasic sites by AP endonuclease results in the formation of SSBs (Oka et al., 2008). Accumulation of these SSBs can present a significant threat to the integrity of the genome and the viability of cells. For example, SSBs can stall replication forks and cause the formation of potentially lethal double-strand DNA breaks (DSBs) (Saffhill and Ockey, 1985). It has also been shown that cell death caused by an accumulation of 8-oxoG lesions is dependent upon MUTYH activity and characterized by an increase in SSB formation (Oka et al., 2008).
Given the demonstration of SSL relationships involving MLH1 and MSH2 orthologs and DNA polymerases in lower organisms, we investigated whether similar SSL relationships existed in human tumor cells and explored their therapeutic potential.
Results
Synthetic Sickness/Lethality Relationships between DNA Polymerases and MMR Genes
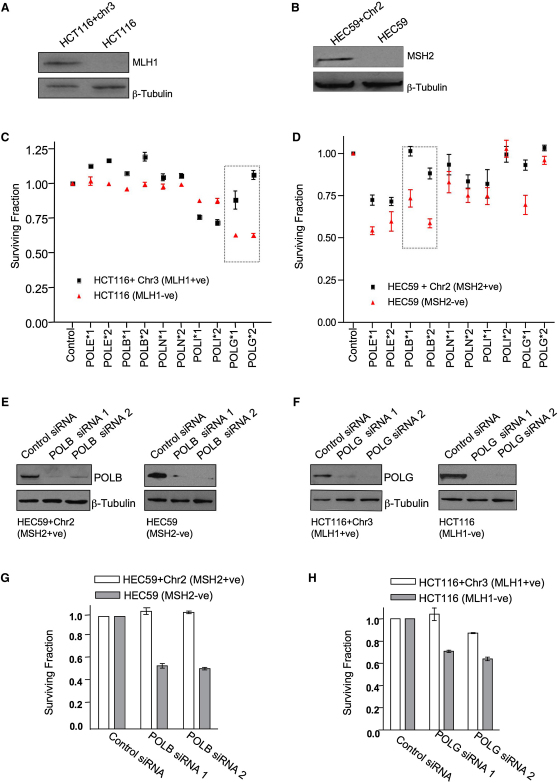
To assess SSL interactions in human cells, we used isogenic models of MLH1 or MSH2 deficiency. To model MLH1 deficiency, we used the previously characterized human colon adenocarcinoma cancer cell line HCT116, which carries a homozygous mutation of the MLH1 gene (Umar et al., 1997), and its MLH1-proficient derivative HCT116+Chr3 cell line, which carries an extra human chromosome 3 expressing a functional MLH1 allele (Koi et al., 1994) (Figure 1A). To model MSH2 deficiency, we used the previously characterized human endometrial cell line HEC59, which harbors compound heterozygous MSH2 nonsense mutations (Boyer et al., 1995), and its MSH2-proficient derivative HEC59+Chr2 cell line, which carries an extra human chromosome 2 expressing a functional MSH2 allele (Umar et al., 1997) (Figure 1B). We screened each of these four cell lines with a panel of short interfering RNAs (siRNAs) targeting five DNA polymerases (POL ɛ, β, η, ι, and γ: two siRNAs per gene; Echeverri et al., 2006) and measured the effect of each siRNA on cell viability. Most strikingly, targeting of POLG was selective for MLH1 deficiency (Figures 1C and 1F; see Tables S1 and S2 available online) and MSH2 selectivity could be achieved by targeting POLB (Figures 1D and 1E; Tables S1 and S2). To more accurately assess the scale of these effects, we used longer-term clonogenic assays, similar to those used to assess SSL between PARP1 silencing and loss of BRCA1 or BRCA2 function (Farmer et al., 2005). In clonogenic assays, the scale of the MSH2/POLB and MLH1/POLG effects were similar to that observed using RNA interference-mediated silencing of PARP1 in BRCA1- or BRCA2-deficient cells (Farmer et al., 2005) (Figures 1G and 1H). To assess the generality of these observations, we used additional isogenic cell models. POLG silencing was also selective in A2780cp70 cells, where MLH1 deficiency occurs by epigenetic silencing of the MLH1 gene (Strathdee et al., 1999) (Figure S1A; Table S2), as well as in HeLa and MCF7 cells, where MLH1 and POLG were silenced by using combinations of siRNAs (Figure S1B). Similarly, the MSH2/POLB SSL was observed in HeLa cells with stable silencing of MSH2 deficiency (Figure S1C; Table S2) as well as in HeLa and MCF7 cells transfected with siRNA combinations (Figure S1D). Both MSH2/POLB and MLH1/POLG SSLs were also apparent in non-tumor cells such as the human breast epithelial line MCF10A (Figure S1E) and also in mouse embryonic fibroblasts (MEFs) (Figures S1F–S1H). To further validate our observations, we exploited small-molecule inhibitors of POLB and POLG. Although potent or highly specific inhibitors of POLG and POLB do not currently exist, menadione is known to inhibit POLG (Sasaki et al., 2008), and masticadienonic acid inhibits POLB (Boudsocq et al., 2005). These compounds were able to elicit either MLH1 or MSH2 selectivity (Figures S1I and S1J), supporting the observations made using RNA interference.
Figure 1.
MSH2 and MLH1 Deficiencies Are Synthetically Sick/Lethal with Silencing of DNA Polymerases
(A) Cell lysates from HCT116 and HCT116+Chr3 cells were analyzed by western blotting using MLH1- and β-tubulin-specific antibodies.
(B) Cell lysates from HEC59 and HEC59+Chr2 cells were analyzed by western blotting using MSH2- and β-tubulin-specific antibodies.
(C) HCT116 (MLH1-deficient) and HCT116+Chr3 (MLH1-proficient) cells were transfected with siRNAs targeting DNA polymerases, and cell viability was estimated 5 days later using CellTiter-Glo reagent.
(D) HEC59 and HEC59+Chr2 cells were transfected with siRNAs targeting DNA polymerases, and cell viability was estimated 5 days later using CellTiter-Glo reagent.
(E) HEC59 cells were transfected with siRNA, and cell lysates were analyzed by western blotting 72 hr later. POLB- and β-tubulin-specific antibodies were used as shown.
(F) HCT116 cells were transfected with siRNA, and cell lysates were analyzed by western blotting 72 hr later. POLG and β-tubulin antibodies were used as shown.
(G) HEC59 and HEC59+Chr2 cells were transfected with either control siRNA or siRNA targeting POLB, and clonogenic assays were performed.
(H) HCT116 and HCT116+Chr3 cells were transfected with either control siRNA or siRNA targeting POLG, and clonogenic assays were performed.
Error bars for each individual experiment represent standard errors of the mean. See also Figure S1.
MLH1 Deficiency Is Associated with Elevated POLG Expression Levels and MSH2 Deficiency Is Associated with Elevated POLB Expression
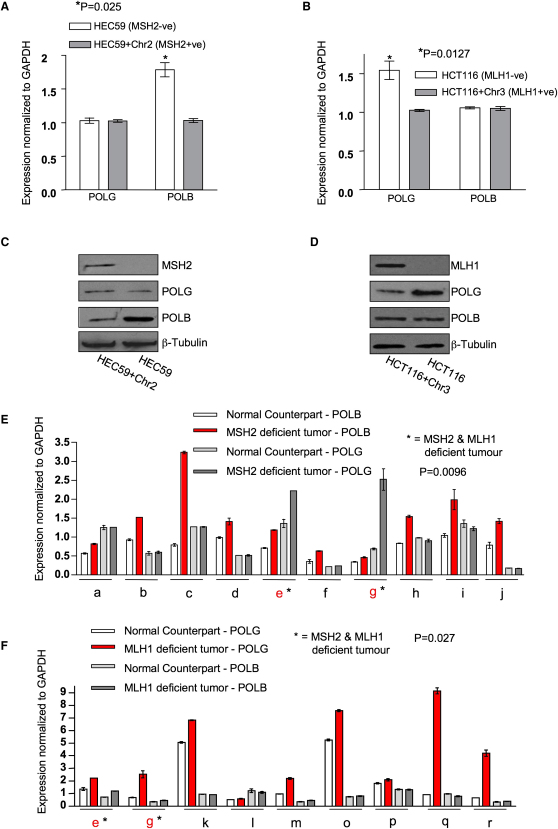
To further investigate the MSH2/POLB and MLH1/POLG SSLs, we measured POLB and POLG mRNA levels in cells with either MSH2 or MLH1 deficiencies. POLB mRNA levels were significantly higher (p = 0.025) in the MSH2-deficient HEC59 cell line than in the MSH2-proficient HEC59+Chr2 cell line (Figure 2A). Similarly, POLG mRNA levels in the MLH1-deficient HCT116 cells were significantly higher (p = 0.0127) than in HCT116+Chr3 cells (Figure 2B). The MSH2 or MLH1 specificity of these effects was suggested by the absence of POLG mRNA upregulation in MSH2-deficient cells and an absence of POLB upregulation in MLH1-deficient cells (Figures 2A and 2B). Corresponding changes in POLB and POLG protein levels were observed in HEC59 and HCT116 cells, respectively (Figures 2C and 2D), as well as in additional isogenic models of MSH2 or MLH1 deficiency (Figures S2A–S2C; Table S3). An increase in Msh2 expression was also observed in Polb−/− MEFs when compared with their wild-type counterparts (Figure S2C).
Figure 2.
MSH2 and MLH1 Deficiencies Are Associated with Particular Increases in DNA Polymerase Expression
(A) POLB mRNA levels were analyzed by qRT-PCR using GAPDH expression as a control. ∗p = 0.0254 compared to the MSH2-proficient HEC59+Chr2 cells (Student's t test).
(B) POLG mRNA levels were analyzed by qRT-PCR using GAPDH expression as a control. ∗p = 0.0127 compared to the MLH1-proficient HCT116+Chr3 cells (Student's t test).
(C) Cell lysates from HEC59 and HEC59+Chr2 cells were analyzed by western blotting using MSH2-, POLB-, POLG-, and β-tubulin-specific antibodies as shown.
(D) Cell lysates from HCT116 and HCT116+Chr3 cells were analyzed by western blotting using MLH1-, POLG-, POLB-, and β-tubulin-specific antibodies as shown.
(E and F) Polymerase mRNA expression in MSH2- and MLH1-deficient tumors and adjacent nontumor material from the same patient was determined by qRT-PCR. MSH2-deficient tumors are shown in (E) and MLH1-deficient tumors are shown in (F). Patient samples are labeled alphabetically, with patients e and g having tumors with both MSH2 and MLH1 deficiency (labeled by ∗). MSH2 and MLH1 protein expression was determined by immunohistochemical analysis. The relative polymerase mRNA levels were measured using qRT-PCR and normalized to a housekeeping gene, GAPDH.
(E) POLB expression was elevated in MSH2-deficient tumors, compared to patient-matched nontumor material; p = 0.0096 (pairwise Student's t test).
(F) POLG expression was elevated in MLH1-deficient tumors, compared to patient-matched nontumor material; p = 0.027 (pairwise Student's t test).
Error bars for each individual experiment represent standard errors of the mean. See also Figure S2.
To assess whether these alterations in POLB and POLG expression occurred in human tumors, we measured POLB and POLG mRNA levels in MSH2- or MLH1-deficient (as defined by immunohistochemistry; IHC) colorectal tumor biopsies and corresponding adjacent normal tissues. POLB mRNA levels were consistently elevated in MSH2-negative tumors compared with those in patient-matched MSH2-positive normal tissue (p = 0.0096) (Figure 2E). The specificity of this observation was suggested by an absence of POLG mRNA elevation in the MSH2-negative tumors (Figure 2E). Similarly, MLH1-negative tumors exhibited elevated levels of POLG expression (p = 0.027), whereas POLB mRNA was not consistently altered (Figure 2F). In two tumor/normal matched biopsy pairs where the tumors were deficient of both MSH2 and MLH1, both POLB and POLG mRNAs were elevated in the tumors (Figures 2E and 2F). These results support the conclusion that POLB expression is elevated in MSH2-deficient tumors and POLG expression is increased in MLH1-deficient tumors.
It has previously been suggested that SSL interactions between genes, proteins, and indeed pathways may be as a consequence of functional compensation or “buffering” (Kaelin, 2005). In an SSL interaction, one gene may buffer the effect of changes in a second gene but this buffering is lost when the function of both genes is absent (Kaelin, 2005 and references therein). It is possible, therefore, that the increases in POLB or POLG expression in MSH2- or MLH1-deficient cells and tumors reflect such a buffering process.
Increased 8-OxoG Accumulation Correlates with SSL in Both MLH1- and MSH2-Deficient Cells
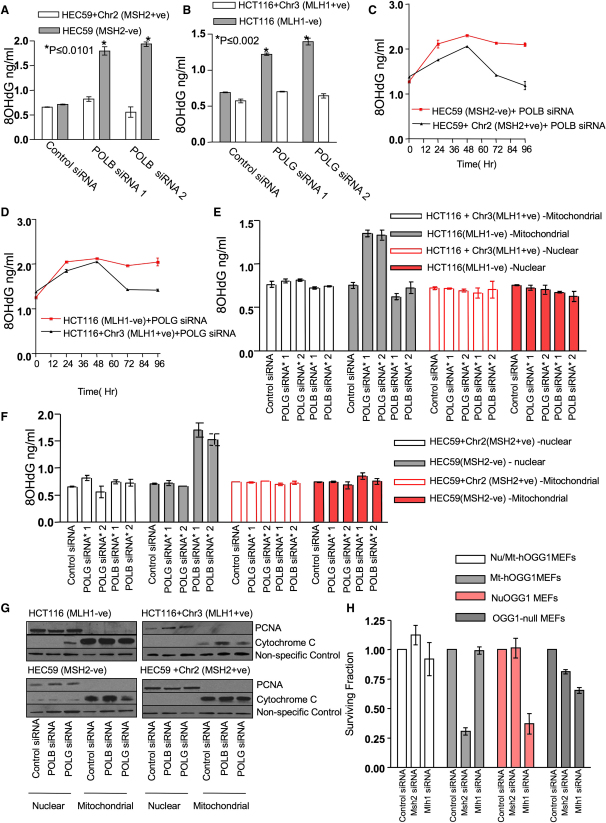
Given the selectivity of POLB or POLG inhibition for either MSH2 or MLH1 deficiencies and the changes in POLB or POLG expression in MSH2- and MLH1-deficient cells, we considered whether these observations could be explained by failure to repair a particular DNA lesion. We investigated whether the accumulation of 8-oxoG correlated with the MSH2/POLB and MLH1/POLG SSL effects. Using an ELISA method, we assessed 8-oxoG levels from DNA extracted from total cell lysates. Seventy-two hours after siRNA transfection, POLB silencing caused a significant increase in 8-oxoG levels in MSH2-deficient HEC59 cells when compared with MSH2-proficient cells (Figure 3A). POLG silencing in MLH1-deficient HCT116 cells also resulted in an increase in 8-oxoG lesions (Figure 3B). These effects were reminiscent of 8-oxoG increases in Msh2−/−;Ogg1−/− MEFs (Colussi et al., 2002), and we were able to demonstrate that silencing of OGG1 by siRNA caused elevation of 8-oxoG lesions in cells deficient in MSH2 or MLH1 as well as synthetic lethality between OGG1 and MSH2 and also OGG1 and MLH1 (Figures S3A–S3C). Silencing of other DNA polymerases in MLH1- or MSH2-deficient cells did not elicit the same increases in 8-oxoG levels (Figure S3D), suggesting that these effects were specific to POLB or POLG.
Figure 3.
Increased 8-OxoG Accumulation Correlates with Polymerase Inhibition and MSH2 or MLH1 Deficiency
(A) HEC59 and HEC59+Chr2 cells were transfected with siRNA. Seventy-two hours after transfection, DNA was isolated from cells and analyzed for 8-oxoG accumulation using an ELISA assay. Oxidized lesions were quantified according to a standard curve generated using known amounts of 8-oxoG. Assays were performed in triplicate. ∗p ≤ 0.0101 compared to the similarly transfected MSH2-proficient HEC59+Chr2 cells (Student's t test).
(B) HCT116 and HCT116+Chr3 cells were transfected with siRNA and analyzed as in (A). ∗p ≤ 0.002 compared to the similarly transfected MLH1-proficient HCT116+Chr3 cells (Student's t test).
(C) HEC59 and HEC59+Chr2 cells were transfected with siRNA and DNA and analyzed as in (A) over a 96 hr time course. Assays were performed in triplicate.
(D) HCT116 and HCT116+Chr3 cells were transfected with siRNA and DNA and analyzed as in (B) over a 96 hr time course. Assays were performed in triplicate.
(E) HCT116 and HCT116+Chr3 cells were transfected with siRNA. Nuclear and mitochondrial DNA isolated from transfected cells were analyzed for 8-oxoG accumulation as in (B). Assays were performed in triplicate.
(F) HEC59 and HEC59+Chr2 cells were transfected with siRNA. Nuclear and mitochondrial DNA isolated from transfected cells were analyzed for 8-oxoG accumulation as in (A). Assays were performed in triplicate.
(G) Validation of nuclear and mitochondrial fractionation. Cells were transfected with siRNA, and nuclear and mitochondrial lysates were analyzed by western blotting. PCNA- and cytochrome c-specific antibodies were used to determine nuclear and mitochondrial fractionations, respectively.
(H) Silencing of Mlh1 is synthetically lethal with loss of a mitochondrial isoform of Ogg1, whereas silencing of Msh2 is synthetically lethal with loss of a nuclear isoform of Ogg1. Fibroblasts expressing either a nuclear isoform of Ogg1, a mitochondrial isoform of Ogg1, or both Ogg1 isoforms were transfected with either nontargeting control siRNA, Msh2 siRNA, or Mlh1 siRNA. After 5 days, cell survival was estimated using CellTiter-Glo reagent.
Error bars for each individual experiment represent standard errors of the mean. See also Figure S3.
We extended our observations by examining the time course of 8-oxoG formation. Silencing of POLB caused an initial increase in 8-oxoG levels in both MSH2-deficient and -proficient cells (Figure 3C). However, 48 hr after POLB siRNA transfection, 8-oxoG levels started to decrease in MSH2-proficient cells but continued to rise in MSH2-deficient cells (Figure 3C). POLG inhibition caused a very similar effect in MLH1-deficient cells (Figure 3D). These observations suggested that inhibition of specific DNA polymerases initially leads to the formation of 8-oxoG lesions that are eventually repaired in cells proficient in MLH1 or MSH2, whereas these lesions accumulate in MLH1- or MSH2-deficient cells. In these latter cells, it seems possible that the level of 8-oxoG eventually reaches a threshold that is incompatible with either cell viability or the ability to divide/proliferate.
POLB has been implicated as an important nuclear DNA polymerase, whereas POLG is believed to be the only DNA polymerase active in mitochondria (Loeb and Monnat, 2008). In light of this, we hypothesized that the difference in MSH2/POLB and MLH1/POLG SSLs might be explained by 8-oxoG accumulation in either the nucleus or mitochondria. To address this, we transfected MSH2- or MLH1-proficient and -deficient cells with control, POLB, or POLG siRNA as detailed above. However, instead of isolating and analyzing total DNA, we fractionated cells into mitochondrial and nuclear compartments, and then extracted DNA and measured 8-oxoG levels by ELISA (Figures 3E–3G). Increased 8-oxoG accumulation was observed in the mitochondrial DNA fraction from the MLH1-deficient cells transfected with POLG siRNA, whereas no significant accumulation of this lesion was observed in the nuclear DNA fraction (Figure 3E). POLB silencing in MLH1-deficient cells did not, however, increase nuclear or mitochondrial 8-oxoG levels (Figure 3E). In contrast, mitochondrial and nuclear DNA extracts isolated from MSH2-deficient cells transfected with POLG siRNA showed no increase in 8-oxoG accumulation (Figure 3F). However, nuclear DNA from MSH2-deficient cells transfected with POLB siRNA demonstrated a significant increase in 8-oxoG, whereas no increase was observed in the DNA isolated from mitochondria (Figure 3F). These observations were further validated in additional cellular models (Figures S3E and S3F).
Repair of 8-oxoG lesions by BER is initiated by removal of the oxidized base by the DNA glycosylase OGG1. As shown above (Figures S3A–S3C), silencing of OGG1 caused elevation of 8-oxoG lesions in cells deficient in MSH2 or MLH1. We therefore examined the nuclear/mitochondrial dichotomy in MEFs expressing (1) only a nuclear isoform of human OGG1 (Nu-hOGG1), (2) a mitochondrial isoform of hOGG1 (Mt-OGG1), or (3) no OGG1 at all (OGG1 null) (Oka et al., 2008). Silencing of MLH1 caused a loss of viability in OGG1 null cells that was rescued by OGG1 expression in the mitochondria but not by OGG1 expression in the nucleus (Figure 3H). Conversely, silencing of MSH2 caused loss of viability in OGG1 null cells that was rescued in cells expressing the nuclear isoform of OGG1 but not by cells expressing only a mitochondrial isoform of OGG1 (Figure 3H). Taken together, these observations suggested that in MEFs, the SSL interaction between MSH2 and OGG1 was most likely mediated by loss of a nuclear OGG1 activity and that the MLH1/OGG1 SSL effect was mediated by loss of a mitochondrial OGG1 activity. It is possible that the MSH2/OGG1 and MLH1/OGG1 SSLs observed in human tumor cells (Figure S3A) could also be explained by a similar mechanism.
Supporting the hypothesis that the MLH1/POLG SSL effect could be due to a mitochondrial dysfunction, we also demonstrated that MLH1 expression was present in both the mitochondria and the nucleus of human tumor cells, whereas MSH2 expression was restricted to the nucleus (Figure S3G). This latter observation was also supported by a previous analysis of the mitochondrial proteome, which identified MLH1, but not MSH2, as a mitochondrial protein (Mootha et al., 2003). Similarly, analysis of the amino acid sequences of MSH2 and MLH1 using the MitoProt algorithm that predicts the presence or absence of mitochondrial target sequences (Claros and Vincens, 1996) also suggested that MLH1 was localized to the mitochondria (MLH1 P[0lg] score = 0.9188, MSH2 P[0lg] = 0.0166, where P[0lg] > 0.5 indicates a predicted mitochondrial localization; the known mitochondrial protein, POLG, has a P[0lg] = 0.8941).
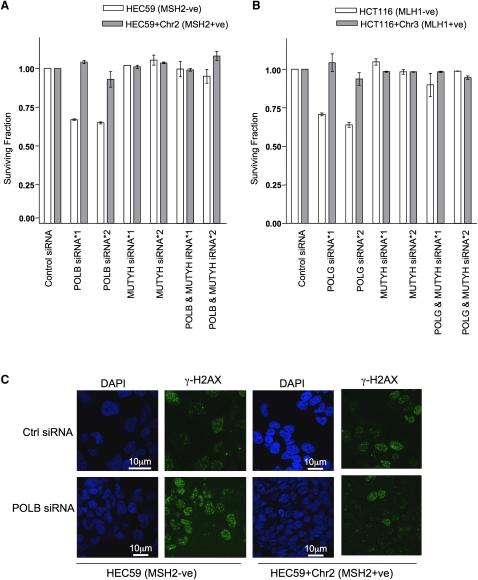
MLH1/POLG and MSH2/POLB SSLs Are Rescued by MUTYH Silencing
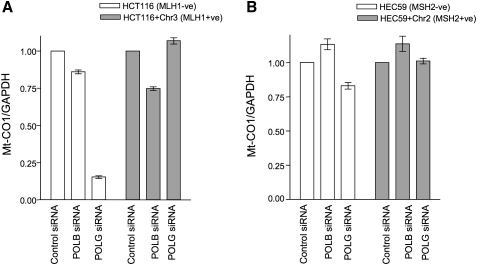
As cell death caused by an accumulation of 8-oxoG lesions requires MUTYH activity (Oka et al., 2008), we investigated whether the MLH1/POLG and MSH2/POLB SSLs were also MUTYH dependent. Silencing of MUTYH rescued both MSH2/POLB and MLH1/POLG SSLs (Figures 4A and 4B; Figure S4A), suggesting dependency upon MUTYH and the possibility that the formation of DSBs could explain the MSH2/POLB and MLH1/POLG lethalities. The formation of nuclear DSBs can be efficiently monitored by the detection of nuclear γ-H2AX foci (Bonner et al., 2008). Silencing of POLB elicited an elevation in γ-H2AX foci in MSH2-deficient cells, suggesting an increase in DSB formation (Figure 4C; Figure S4B). To assess the integrity of mitochondrial DNA, we used quantitative PCR to measure relative levels of a gene located in mitochondrial DNA, COX1 (aka MT-CO1) (Oka et al., 2008). Silencing of POLG caused a significant depletion of COX1 DNA in MLH1-deficient cells when compared with MLH1-proficient cells (Figures 5A and 5B), suggesting that the integrity of mitochondrial DNA was compromised in MLH1/POLG-deficient cells, perhaps contributing to the MLH1/POLG SSL.
Figure 4.
MLH1/POLG and MSH2/POLB SSL Phenotypes Are Rescued by MUTYH Silencing
(A) HEC59 and HEC59+Chr2 cells were transfected with either control, POLB, MUTYH siRNA, or with siRNAs in combination, and clonogenic assays were performed.
(B) HCT116 and HCT116+Chr3 cells were transfected with either control, POLB, MUTYH siRNA, or with siRNAs in combination, and clonogenic assays were performed.
(C) HEC59 and HEC59+Chr2 cells were transfected with either control or POLB siRNA. Seventy-two hours after transfection, γ-H2AX foci were quantified by immunofluorescence. Nuclei are shown in blue and γ-H2AX foci are in green. The scale bars represent 10 μm.
Error bars for each individual experiment represent standard errors of the mean. See also Figure S4.
Figure 5.
Depletion of Mt-CO1 DNA upon Inhibition of POLG in MLH1-Deficient Cells
(A) HCT116 and HCT116+Chr3 cells were transfected with either control, POLB, or POLG siRNA. Mt-CO1 DNA levels were analyzed by qPCR with GAPDH used as a control.
(B) HEC59 and HEC59+Chr2 cells were transfected with either control, POLB, or POLG siRNA. Mt-CO1 DNA levels were analyzed by qPCR with GAPDH used as a control.
Error bars for each individual experiment represent standard errors of the mean.
OGG1 Expression Is Decreased in the Absence of POLB Expression
We demonstrated that POLB silencing caused an increase in 8-oxoG levels in MSH2-deficient cells (Figure 3A). The role of POLB in the repair of 8-oxoG lesions is well established and is considered to act downstream of OGG1 (Boiteux and Radicella, 2000). Therefore, it was initially not clear why POLB silencing would cause an increase in 8-oxoG lesions.
Like many other protein networks, components of the BER pathway are coregulated, most likely as a means to closely coordinate pathway activity. For example, expression of the BER scaffold protein XRCC1 determines the stability of other BER proteins including POLB (Parsons et al., 2008). Similarly, POL κ deficiency causes an increase in the persistence of UV photodimers in DNA, presumably by modulating the activity and/or stability of upstream DNA repair proteins (Ogi and Lehmann, 2006). On this basis, we reasoned that a decrease in POLB expression could modulate OGG1 expression and by this mechanism cause an increase in nuclear 8-oxoG levels in MSH2-deficient cells.
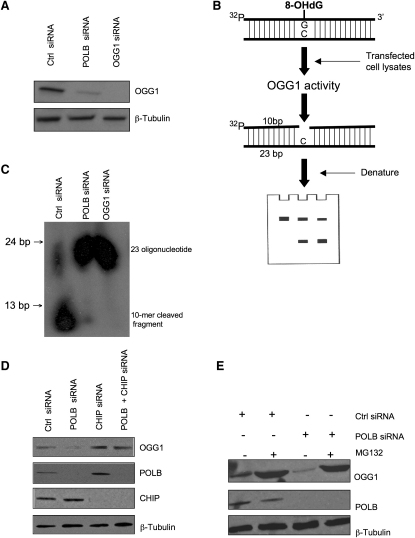
POLB silencing did indeed cause a reduction in OGG1 protein levels (Figure 6A; Figure S5A), an effect that appeared specific to OGG1; the levels of the other BER proteins that we examined were not ostensibly changed (Figure S5B). Furthermore, with the exception of APE1 silencing, the inhibition of other BER proteins did not elicit SSL with either MSH2 or MLH1 deficiency (Figures S5C and S5D). We also observed a decrease in Ogg1 expression in Polb−/− MEFs when compared with wild-type MEFs (Figure S5E). We next investigated whether the reduction in OGG1 protein levels caused by POLB silencing could actually elicit a reduction in 8-oxoG removal, commensurate with the effects observed in the MSH2/POLB SSL. To assess this, we used an in vitro OGG1 activity assay. HeLa cells were transfected with either a nontargeting control siRNA, OGG1 siRNA, or siRNA targeting POLB. Protein extracts were prepared from transfected cells and incubated with a 23 bp oligonucleotide duplex containing one 32P-labeled strand with an 8-oxoG lesion at the 11th base annealed to an unlabeled complementary strand containing dC at the opposite base position to the 8-oxoG (Figure 6B). The oligonucleotide strands were then electrophoresed on a denaturing gel and cleaved product was detected by autoradiography (Figure 6B). As expected, extracts from HeLa cells transfected with control siRNA caused cleavage of the 8-oxoG-radiolabeled strand, resulting in a labeled 10 base cleavage product (Figure 6C) suggestive of normal OGG1 activity. However, cells transfected with POLB siRNA exhibited a significant decrease in OGG1-mediated cleavage of 8-oxoG similar to that observed upon silencing of OGG1 itself (Figure 6C). This suggested that OGG1-mediated cleavage of 8-oxoG was most likely dependent on POLB expression.
Figure 6.
OGG1 Cleavage Activity Is Decreased in the Absence of POLB Expression
(A) HeLa cells were transfected with siRNA, and lysates were analyzed for OGG1 expression 72 hr later.
(B) Schematic model for in vitro OGG1 assay. Briefly, protein was isolated from transfected cells. The DNA substrate is a 23 base oligonucleotide containing 8-oxoG at its 11th base, labeled with 32P at its 5′ end, and annealed to its complementary strand (containing dC at the opposite base position to the 8-oxoG). Upon cleavage of the substrate by the OGG1 enzyme, the oligonucleotides were electrophoresed on a denaturing PAGE gel, followed by autoradiography.
(C) HeLa cells were transfected with either control, POLB, or OGG1 siRNA and incubated with an oligonucleotide substrate containing 8-oxoG, as described above. The oligonucleotides were electrophoresed and a 10 base fragment (labeled cleavage product) was revealed in addition to the original 23 base oligonucleotide. Autoradiography revealed that in the absence of POLB expression, cleavage of the 8-OHdG lesion was significantly decreased.
(D) HeLa cells were transfected with siRNA, and cell lysates were analyzed 72 hr later. OGG1 expression was suppressed by POLB siRNA but rescued by combined silencing of POLB and CHIP.
(E) Cell lysates from HeLa cells were transfected with siRNA and, after 48 hr, cells were treated with MG132 (50 μM). Lysates were analyzed 18 hr later by western blotting. Antibodies directed against OGG1, POLB, and β-tubulin were used to demonstrate reduction in expression of OGG1 after transfection with POLB siRNA, which was rescued by treatment with the proteasomal inhibitor MG132. See also Figure S5.
Recent work has shown that BER proteins such as POLB, ligase III, and XRCC1, when not involved in repair complexes, are ubiquitylated by the carboxyl terminus of Hsc70-interacting protein (CHIP) and then degraded by the proteasome (Parsons et al., 2008). It seemed possible, therefore, that the reduction in OGG1 levels caused by POLB silencing could also be mediated by a similar process. To assess this, HeLa cells were transfected with POLB siRNA alone or POLB and CHIP siRNA together (Figure 6D). As before, POLB silencing mediated a decrease in OGG1 levels. However, OGG1 levels in POLB-silenced cells could be restored by silencing of CHIP (Figure 6D). Furthermore, treatment of POLB-silenced cells with the proteasomal inhibitor MG132 restored the expression of OGG1 (Figure 6E; Figure S5F). Taken together, these results suggested that the reduction in OGG1 levels caused by a reduction in POLB expression were mediated by CHIP ubiquitination and proteasomal degradation of OGG1.
Discussion
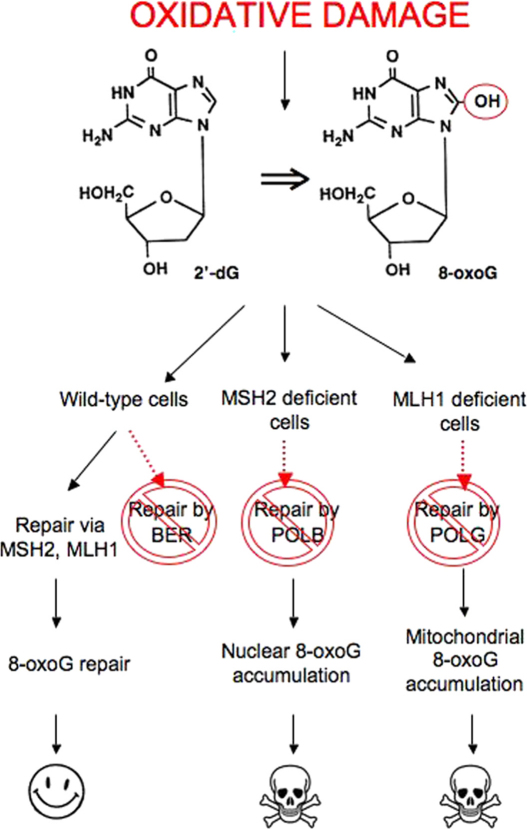
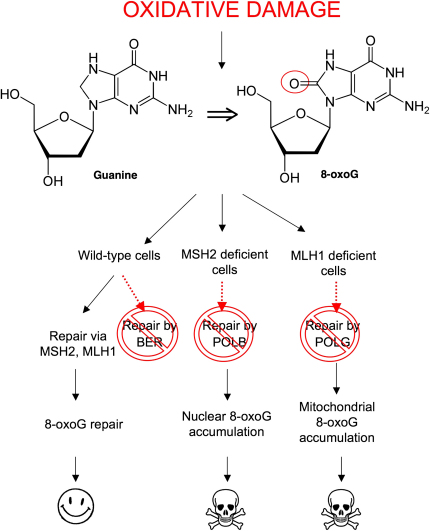
In their 1997 review describing how genetic approaches may be used to identify therapeutic approaches for the treatment of cancer, Hartwell and colleagues used data from studies in budding yeast to highlight the potential of exploiting SSL in the treatment of cancers characterized by specific MMR gene defects (Hartwell et al., 1997). Here we show that SSLs involving MLH1 or MSH2 and DNA polymerases also exist in human tumor cells and can be exploited to specifically target cells carrying these underlying gene defects. We also investigate the characteristics of these SSLs and suggest that increases in oxidized DNA lesions may, in part at least, explain how combining such DNA repair defects limits the viability of tumor cells. We propose a model (Figure 7) where MSH2 gene defects, when combined with inhibition of POLB, lead to the accumulation of oxidized DNA base damage. Likewise, POLG inhibition, when imposed upon cells with MLH1 deficiency, also leads to the accumulation of oxidized DNA damage. In both scenarios, the persistent DNA lesions that occur either cause a loss of fitness by interfering with the normal operation of the cell or eventually cause the formation of more lethal DNA lesions such as double-strand DNA breaks.
Figure 7.
A Model for the Selective Effects of DNA Polymerase Inhibition in MLH1- or MSH2-Deficient Cells
Oxidized DNA lesions, including 8-oxoG, can be repaired by either MSH2/MLH1-dependent processes or BER. In wild-type cells, inhibition of POLB or POLG leads to repair of these lesions by MSH2/MLH1. In the absence of MSH2, POLB is essential for 8-oxoG repair. Inhibition of POLB in MSH2-deficient cells leads to the accumulation of 8-oxoG in nuclear DNA. Cells harboring these unrepaired lesions may permanently arrest or die. These effects may be mediated by MUTYH, which in attempting to reverse C→A transversions opposite oxidized bases ultimately causes the formation of single-strand DNA breaks which in turn cause the formation of lethal double-strand DNA breaks. In cells with MLH1 deficiency, POLG inhibition leads to the accumulation of 8-oxoG in mitochondrial DNA. Again, this accumulation either becomes incompatible with viability or limits the cell's replicative potential.
Although the clinical assessment of SSLs in cancer therapy is still at an early stage, the preliminary data are encouraging, with a number of patients with BRCA mutation-associated cancer showing favorable and sustained responses to high-potency PARP inhibitors (Fong et al., 2009). Although highly potent and selective POLB or POLG inhibitors are not yet available for use in the context of MSH2 or MLH1 deficiency, our work suggests that targeting these proteins and the pathways they control is worthy of further investigation. How might such inhibitors be used clinically? First, we propose that mechanism-based biomarkers be used to identify patients likely to respond favorably to therapy. In this case, DNA sequencing-based detection of MSH2 or MLH1 mutations in blood and tumor biopsies would be used (Hegde and Roa, 2009), alongside IHC detection of MSH2 and MLH1 proteins in tumor samples to confirm loss of heterozygosity (Shia, 2008). The work presented here suggests that MSH2 and MLH1 deficiencies are associated with increased expression of POLB and POLG, respectively, and therefore IHC detection of these latter proteins in tumor biopsies may also serve as a predictive biomarker. Our work also suggests that combined MSH2/POLB or MLH1/POLG inhibition is characterized by a sustained increase in the levels of 8-oxoG. Measuring 8-oxoG using an ELISA assay may therefore provide a mechanism-based marker of drug efficacy to be used alongside more standard measures of tumor response. Finally, we propose that POLG or POLB inhibitors be used for a limited time course rather than extended periods. Such a regime would ideally maximize the antitumor response while limiting the potential for either deleterious long-term effects of DNA repair pathway inhibition or the development of drug resistance, including the potential reversion of MSH2 or MLH1 gene defects, as has been documented for BRCA gene defects (Edwards et al., 2008).
To realize the potential of our proposed synthetic lethality approach, the development of highly potent and selective POLB and POLG inhibitors is required. Although somewhat specific POLB inhibitors have already been identified, these are of low potency. For example, koetjapic acid (Sun et al., 1999), pamoic acid (Hu et al., 2004), prunasin (Mizushina et al., 1999), solanapyrone A (Mizushina et al., 2002), trans-communic acid, mahureone A, and masticadienonic acid (Boudsocq et al., 2005) all inhibit POLB. Of these, masticadienonic acid (IC50 of 8 μM) is the most potent POLB inhibitor identified to date. The development of POLG inhibitors is also at a relatively early stage of development; vitamin K3 (VK3; menadione) is known to inhibit POLG with an IC50 in the micromolar range. Interestingly, VK3 causes impairment of mitochondrial DNA replication and repair, and induces a significant increase in reactive oxygen species, leading to cellular apoptosis (Sasaki et al., 2008). Lamivudine-TP (LVD-TP), adefovir-diphosphate (ADV-DP), and tenofovir-DP (TFV-DP), as well as zalcitabine-TP (ddCTP) and zidovudine-TP (AZT-TP), all inhibit POLG activity in vitro, although the extent of inhibition varies widely (Lewis et al., 1994; Mazzucco et al., 2008). It is also worth considering whether similar synthetic lethalities could be achieved by targeting proteins other than POLB or POLG. Silencing of the DNA glycosylase OGG1 was also able to elicit selectivity in cells with either MLH1 or MSH2 deficiency (Figure S3A), and it is possible that the catalytic activity of this protein may be more amenable to pharmacological inhibition. It is also possible that other SSL effects could be exploited in the treatment of MMR-defective cancers. MMR-defective mouse and human cell lines have been reported to be sensitive to drugs inducing interstrand crosslinks (ICLs) including CCNU (N-[2-chloroethyl]-N′-cyclohexyl-N-nitrosourea) and MMC (mitomycin C) (Aquilina et al., 1998; Wu et al., 2005). The tolerance and repair of ICLs is extremely complex and most likely involves a number of pathways including MMR, translesion synthesis, and homologous recombination (Zheng et al., 2006). This interplay of these repair pathways may suggest that MMR-deficient tumor cell sensitivity to ICL-causing agents could be increased in a synergistic fashion, by inhibiting additional DNA repair pathways.
We do note that the SSL involving POLB is specific to MSH2 deficiency whereas the POLG SSL is specific to cells with MLH1 defects, suggesting that the SSLs described here are genotype specific rather than being associated with MMR deficiency per se. Interestingly, there is also a growing body of evidence to suggest that MLH1 and MSH2 not only have distinct roles outside of the canonical MMR pathway but their deficiencies also lead to distinct phenotypic and clinical manifestations. For example, MLH1 mutations are particularly associated with an increased risk of colonic cancers, whereas MSH2 mutations have a higher incidence of extracolonic tumors (Kastrinos et al., 2008; Palomaki et al., 2009). Similarly, differences in MLH1 and MSH2 function have been identified that suggest that the canonical MMR pathway is perhaps but one mechanism by which these proteins maintain the integrity of the genome. Notably, the response to certain chemotherapies differs between MLH1- and MSH2-deficient tumor cells (Martin et al., 2009; Wu and Vasquez, 2008; Zhang et al., 2002), and MLH1 has been shown to regulate homologous recombination independently of MSH2 (Siehler et al., 2009). Intriguingly, de Souza-Pinto and colleagues report that mitochondria possess a distinct version of MMR that is MSH2 independent (de Souza-Pinto et al., 2009), suggesting that noncanonical forms of MMR exist that are perhaps worthy of additional study.
In summary, we provide the proof of principle that SSLs could be exploited to design targeted therapeutic approaches to the treatment of cancers characterized by specific MMR gene defects. Clinical trials of these approaches will require the development of suitable drug-like inhibitors.
Experimental Procedures
Cell Culture
The human endometrial cell lines HEC59 and HEC59+Chr2 were the kind gift of Dr. T. Kunkel (National Institute of Environmental Health Sciences). The human colon cancer cell lines HCT116 and HCT116+Chr3 were the kind gift of Dr. A. Clark (NIEHS). The human ovarian tumor cell lines A2780cp70+chr3/A2 and A2780cp70+chr3/E1 were a kind gift from Dr. R. Brown (ICR). Wild-type and Polb−/− mouse embryonic fibroblasts were obtained from the American Type Culture Collection. Masticadienonic acid was a kind gift of Dr. C. Cazaux (Institut de Pharmacologie et de Biologie Structurale) and Dr. G. Massiot (Institut de Recherche, Pierre Fabre). Detailed methods for cell culture are described in Supplemental Experimental Procedures.
Protein Analysis
Cell pellets were lysed in 20 mmol/L Tris (pH 8), 200 mmol/L NaCl, 1 mmol/L EDTA, 0.5% (v/v) NP40, 10% (v/v) glycerol, and protease inhibitors. Mitochondrial and nuclear protein were extracted using a mitochondrial isolation kit (ab65321; Abcam). For western blotting, lysates were electrophoresed on Novex precast gels (Invitrogen) and immunoblotted. Antibody details are described in Supplemental Experimental Procedures.
Use of RNA Interference to Assess Synthetic Lethality
Short-term and clonogenic assays were performed as described in Farmer et al. (2005) and Martin et al. (2009). For 96-well plate-based cell viability assays, HeLa, HCT116, HCT116+Chr3, HEC59, and HEC59+Chr2 cells were transfected with individual siRNAs using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. A2780cp70+chr3/A2 and A2780cp70+chr3/E1 cells were transfected with individual siRNAs using Lipofectamine RNAi Max (Invitrogen) according to the manufacturer's instructions. For clonogenic assays, exponentially growing cells were seeded at various densities in six-well plates. Cells were transfected with siRNA as before. Further experimental details and siRNA target sequences are detailed in Supplemental Experimental Procedures.
Detection of γ-H2AX Foci by Immunofluorescence
Transfected cells were seeded onto coverslips. Seventy-two hours posttransfection, cells were fixed, permeabilized, and then incubated with γ-H2AX antibody (Millipore) for 24 hr at 4°C. Coverslips were stained with DAPI, mounted, and viewed using a Leica TCS-SP2 confocal microscope. The experimental procedure is described in full in Supplemental Experimental Procedures.
Immunohistochemical Staining
The use of human tissue for this study was reviewed and approved by the Beaumont Hospital Ethics Committee and samples were obtained after informed consent. Four micrometer sections were cut from formalin-fixed paraffin-embedded samples for the purpose of immunohistochemistry. A section of normal colon tissue was used as a positive control, and negative controls were performed by replacing the antibody with Tris-buffered saline. Further experimental details are described in Supplemental Experimental Procedures.
Measurement of 8-OxoG
8-oxoG levels in DNA were measured using an 8-oxoG-specific ELISA method (Cell Biolabs) described in full in Supplemental Experimental Procedures.
In Vitro OGG1 Assay
OGG1 glycosylase activity was analyzed using the OGG1 assay kit (Sigma-Aldrich). Detailed methods are described in Supplemental Experimental Procedures.
Quantitative RT-PCR
Total RNA from cell lines was extracted using Trizol (Invitrogen) according to the manufacturer's instructions. Total RNA from patient biopsies was purified from 10 μm sections using the High Pure RNA Paraffin Kit (Roche Diagnostics). Detailed methods for cDNA synthesis and quantitative RT-PCR are described in Supplemental Experimental Procedures.
Acknowledgments
We thank Dr. T. Kunkel, Dr. A. Clark, and Dr. R. Brown for the provision of cell lines. We thank Dr. C. Cazaux and Dr. G. Massiot for providing masticadienonic acid. We also thank Dr. M. Hewish for helpful discussions. C.J.L., A.A., and S.A.M. are inventors on a patent pertaining to this work and may financially gain under the ICR rewards to investors scheme. This work was supported by grants from Cancer Research UK and Breakthrough Breast Cancer. We acknowledge NHS funding to the NIHR Biomedical Research Centre.
Published: March 15, 2010
Footnotes
Supplemental Information includes five figures, three tables, and Supplemental Experimental Procedures and can be found with this article online at doi:10.1016/j.ccr.2009.12.046.
Contributor Information
Christopher J. Lord, Email: chris.lord@icr.ac.uk.
Alan Ashworth, Email: alan.ashworth@icr.ac.uk.
Supplemental Information
References
- Aquilina G., Ceccotti S., Martinelli S., Hampson R., Bignami M. N-(2-chloroethyl)-N′-cyclohexyl-N-nitrosourea sensitivity in mismatch repair-defective human cells. Cancer Res. 1998;58:135–141. [PubMed] [Google Scholar]
- Argueso J.L., Smith D., Yi J., Waase M., Sarin S., Alani E. Analysis of conditional mutations in the Saccharomyces cerevisiae MLH1 gene in mismatch repair and in meiotic crossing over. Genetics. 2002;160:909–921. doi: 10.1093/genetics/160.3.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold C.N., Goel A., Boland C.R. Role of hMLH1 promoter hypermethylation in drug resistance to 5-fluorouracil in colorectal cancer cell lines. Int. J. Cancer. 2003;106:66–73. doi: 10.1002/ijc.11176. [DOI] [PubMed] [Google Scholar]
- Bettstetter M., Dechant S., Ruemmele P., Grabowski M., Keller G., Holinski-Feder E., Hartmann A., Hofstaedter F., Dietmaier W. Distinction of hereditary nonpolyposis colorectal cancer and sporadic microsatellite-unstable colorectal cancer through quantification of MLH1 methylation by real-time PCR. Clin. Cancer Res. 2007;13:3221–3228. doi: 10.1158/1078-0432.CCR-06-3064. [DOI] [PubMed] [Google Scholar]
- Boiteux S., Radicella J.P. The human OGG1 gene: structure, functions, and its implication in the process of carcinogenesis. Arch. Biochem. Biophys. 2000;377:1–8. doi: 10.1006/abbi.2000.1773. [DOI] [PubMed] [Google Scholar]
- Bonner W.M., Redon C.E., Dickey J.S., Nakamura A.J., Sedelnikova O.A., Solier S., Pommier Y. γH2AX and cancer. Nat. Rev. Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq F., Benaim P., Canitrot Y., Knibiehler M., Ausseil F., Capp J.P., Bieth A., Long C., David B., Shevelev I. Modulation of cellular response to cisplatin by a novel inhibitor of DNA polymerase β. Mol. Pharmacol. 2005;67:1485–1492. doi: 10.1124/mol.104.001776. [DOI] [PubMed] [Google Scholar]
- Boyer J.C., Umar A., Risinger J.I., Lipford J.R., Kane M., Yin S., Barrett J.C., Kolodner R.D., Kunkel T.A. Microsatellite instability, mismatch repair deficiency, and genetic defects in human cancer cell lines. Cancer Res. 1995;55:6063–6070. [PubMed] [Google Scholar]
- Bryant H.E., Schultz N., Thomas H.D., Parker K.M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N.J., Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- Claros M.G., Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Colussi C., Parlanti E., Degan P., Aquilina G., Barnes D., Macpherson P., Karran P., Crescenzi M., Dogliotti E., Bignami M. The mammalian mismatch repair pathway removes DNA 8-oxodGMP incorporated from the oxidized dNTP pool. Curr. Biol. 2002;12:912–918. doi: 10.1016/s0960-9822(02)00863-1. [DOI] [PubMed] [Google Scholar]
- de Souza-Pinto N.C., Mason P.A., Hashiguchi K., Weissman L., Tian J., Guay D., Lebel M., Stevnsner T.V., Rasmussen L.J., Bohr V.A. Novel DNA mismatch-repair activity involving YB-1 in human mitochondria. DNA Repair (Amst.) 2009;8:704–719. doi: 10.1016/j.dnarep.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druker B.J., Talpaz M., Resta D.J., Peng B., Buchdunger E., Ford J.M., Lydon N.B., Kantarjian H., Capdeville R., Ohno-Jones S., Sawyers C.L. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N. Engl. J. Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- Echeverri C.J., Beachy P.A., Baum B., Boutros M., Buchholz F., Chanda S.K., Downward J., Ellenberg J., Fraser A.G., Hacohen N. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat. Methods. 2006;3:777–779. doi: 10.1038/nmeth1006-777. [DOI] [PubMed] [Google Scholar]
- Edwards S.L., Brough R., Lord C.J., Natrajan R., Vatcheva R., Levine D.A., Boyd J., Reis-Filho J.S., Ashworth A. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–1115. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- Farmer H., McCabe N., Lord C.J., Tutt A.N., Johnson D.A., Richardson T.B., Santarosa M., Dillon K.J., Hickson I., Knights C. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Fong P.C., Boss D.S., Yap T.A., Tutt A., Wu P., Mergui-Roelvink M., Mortimer P., Swaisland H., Lau A., O'Connor M.J. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- Hartwell L.H., Szankasi P., Roberts C.J., Murray A.W., Friend S.H. Integrating genetic approaches into the discovery of anticancer drugs. Science. 1997;278:1064–1068. doi: 10.1126/science.278.5340.1064. [DOI] [PubMed] [Google Scholar]
- Hegde M.R., Roa B.B. Genetic testing for hereditary nonpolyposis colorectal cancer (HNPCC) Curr. Protoc. Hum. Genet. 2009 doi: 10.1002/0471142905.hg1012s61. Chapter 10, Unit 10.12. [DOI] [PubMed] [Google Scholar]
- Horton J.K., Baker A., Berg B.J., Sobol R.W., Wilson S.H. Involvement of DNA polymerase β in protection against the cytotoxicity of oxidative DNA damage. DNA Repair (Amst.) 2002;1:317–333. doi: 10.1016/s1568-7864(02)00008-3. [DOI] [PubMed] [Google Scholar]
- Hu H.Y., Horton J.K., Gryk M.R., Prasad R., Naron J.M., Sun D.A., Hecht S.M., Wilson S.H., Mullen G.P. Identification of small molecule synthetic inhibitors of DNA polymerase β by NMR chemical shift mapping. J. Biol. Chem. 2004;279:39736–39744. doi: 10.1074/jbc.M402842200. [DOI] [PubMed] [Google Scholar]
- Jacob S., Praz F. DNA mismatch repair defects: role in colorectal carcinogenesis. Biochimie. 2002;84:27–47. doi: 10.1016/s0300-9084(01)01362-1. [DOI] [PubMed] [Google Scholar]
- Jiricny J. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- Kaelin W.G., Jr. The concept of synthetic lethality in the context of anticancer therapy. Nat. Rev. Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- Kamiya H. Mutagenic potentials of damaged nucleic acids produced by reactive oxygen/nitrogen species: approaches using synthetic oligonucleotides and nucleotides: survey and summary. Nucleic Acids Res. 2003;31:517–531. doi: 10.1093/nar/gkg137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastrinos F., Stoffel E.M., Balmana J., Steyerberg E.W., Mercado R., Syngal S. Phenotype comparison of MLH1 and MSH2 mutation carriers in a cohort of 1,914 individuals undergoing clinical genetic testing in the United States. Cancer Epidemiol. Biomarkers Prev. 2008;17:2044–2051. doi: 10.1158/1055-9965.EPI-08-0301. [DOI] [PubMed] [Google Scholar]
- Klungland A., Bjelland S. Oxidative damage to purines in DNA: role of mammalian Ogg1. DNA Repair (Amst.) 2007;6:481–488. doi: 10.1016/j.dnarep.2006.10.012. [DOI] [PubMed] [Google Scholar]
- Koi M., Umar A., Chauhan D.P., Cherian S.P., Carethers J.M., Kunkel T.A., Boland C.R. Human chromosome 3 corrects mismatch repair deficiency and microsatellite instability and reduces N-methyl-N′-nitro-N-nitrosoguanidine tolerance in colon tumor cells with homozygous hMLH1 mutation. Cancer Res. 1994;54:4308–4312. [PubMed] [Google Scholar]
- Lewis W., Simpson J.F., Meyer R.R. Cardiac mitochondrial DNA polymerase-γ is inhibited competitively and noncompetitively by phosphorylated zidovudine. Circ. Res. 1994;74:344–348. doi: 10.1161/01.res.74.2.344. [DOI] [PubMed] [Google Scholar]
- Lewis W., Day B.J., Kohler J.J., Hosseini S.H., Chan S.S., Green E.C., Haase C.P., Keebaugh E.S., Long R., Ludaway T. Decreased mtDNA, oxidative stress, cardiomyopathy, and death from transgenic cardiac targeted human mutant polymerase γ. Lab. Invest. 2007;87:326–335. doi: 10.1038/labinvest.3700523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb L.A., Monnat R.J., Jr. DNA polymerases and human disease. Nat. Rev. Genet. 2008;9:594–604. doi: 10.1038/nrg2345. [DOI] [PubMed] [Google Scholar]
- Luo J., Solimini N.L., Elledge S.J. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson P., Barone F., Maga G., Mazzei F., Karran P., Bignami M. 8-oxoguanine incorporation into DNA repeats in vitro and mismatch recognition by MutSα. Nucleic Acids Res. 2005;33:5094–5105. doi: 10.1093/nar/gki813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S.A., McCarthy A., Barber L.J., Burgess D.J., Parry S., Lord C.J., Ashworth A. Methotrexate induces oxidative DNA damage and is selectively lethal to tumour cells with defects in the DNA mismatch repair gene MSH2. EMBO Mol. Med. 2009;1:323–337. doi: 10.1002/emmm.200900040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzucco C.E., Hamatake R.K., Colonno R.J., Tenney D.J. Entecavir for treatment of hepatitis B virus displays no in vitro mitochondrial toxicity or DNA polymerase γ inhibition. Antimicrob. Agents Chemother. 2008;52:598–605. doi: 10.1128/AAC.01122-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushina Y., Takahashi N., Ogawa A., Tsurugaya K., Koshino H., Takemura M., Yoshida S., Matsukage A., Sugawara F., Sakaguchi K. The cyanogenic glucoside, prunasin (D-mandelonitrile-β-D-glucoside), is a novel inhibitor of DNA polymerase β. J. Biochem. 1999;126:430–436. doi: 10.1093/oxfordjournals.jbchem.a022468. [DOI] [PubMed] [Google Scholar]
- Mizushina Y., Kamisuki S., Kasai N., Shimazaki N., Takemura M., Asahara H., Linn S., Yoshida S., Matsukage A., Koiwai O. A plant phytotoxin, solanapyrone A, is an inhibitor of DNA polymerase β and λ. J. Biol. Chem. 2002;277:630–638. doi: 10.1074/jbc.M105144200. [DOI] [PubMed] [Google Scholar]
- Mootha V.K., Bunkenborg J., Olsen J.V., Hjerrild M., Wisniewski J.R., Stahl E., Bolouri M.S., Ray H.N., Sihag S., Kamal M. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115:629–640. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- Morrison A., Johnson A.L., Johnston L.H., Sugino A. Pathway correcting DNA replication errors in Saccharomyces cerevisiae. EMBO J. 1993;12:1467–1473. doi: 10.1002/j.1460-2075.1993.tb05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogi T., Lehmann A.R. The Y-family DNA polymerase κ (pol κ) functions in mammalian nucleotide-excision repair. Nat. Cell Biol. 2006;8:640–642. doi: 10.1038/ncb1417. [DOI] [PubMed] [Google Scholar]
- Oka S., Ohno M., Tsuchimoto D., Sakumi K., Furuichi M., Nakabeppu Y. Two distinct pathways of cell death triggered by oxidative damage to nuclear and mitochondrial DNAs. EMBO J. 2008;27:421–432. doi: 10.1038/sj.emboj.7601975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomaki G.E., McClain M.R., Melillo S., Hampel H.L., Thibodeau S.N. EGAPP supplementary evidence review: DNA testing strategies aimed at reducing morbidity and mortality from Lynch syndrome. Genet. Med. 2009;11:42–65. doi: 10.1097/GIM.0b013e31818fa2db. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons J.L., Tait P.S., Finch D., Dianova I.I., Allinson S.L., Dianov G.L. CHIP-mediated degradation and DNA damage-dependent stabilization regulate base excision repair proteins. Mol. Cell. 2008;29:477–487. doi: 10.1016/j.molcel.2007.12.027. [DOI] [PubMed] [Google Scholar]
- Pavlov Y.I., Nguyen D., Kunkel T.A. Mutator effects of overproducing DNA polymerase η (Rad30) and its catalytically inactive variant in yeast. Mutat. Res. 2001;478:129–139. doi: 10.1016/s0027-5107(01)00131-2. [DOI] [PubMed] [Google Scholar]
- Peltomaki P., Vasen H. Mutations associated with HNPCC predisposition—update of ICG-HNPCC/INSiGHT mutation database. Dis. Markers. 2004;20:269–276. doi: 10.1155/2004/305058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffhill R., Ockey C.H. Strand breaks arising from the repair of the 5-bromodeoxyuridine-substituted template and methyl methanesulphonate-induced lesions can explain the formation of sister chromatid exchanges. Chromosoma. 1985;92:218–224. doi: 10.1007/BF00348697. [DOI] [PubMed] [Google Scholar]
- Sasaki R., Suzuki Y., Yonezawa Y., Ota Y., Okamoto Y., Demizu Y., Huang P., Yoshida H., Sugimura K., Mizushina Y. DNA polymerase γ inhibition by vitamin K3 induces mitochondria-mediated cytotoxicity in human cancer cells. Cancer Sci. 2008;99:1040–1048. doi: 10.1111/j.1349-7006.2008.00771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shia J. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome. Part I. The utility of immunohistochemistry. J. Mol. Diagn. 2008;10:293–300. doi: 10.2353/jmoldx.2008.080031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siehler S.Y., Schrauder M., Gerischer U., Cantor S., Marra G., Wiesmuller L. Human MutL-complexes monitor homologous recombination independently of mismatch repair. DNA Repair (Amst.) 2009;8:242–252. doi: 10.1016/j.dnarep.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathdee G., MacKean M.J., Illand M., Brown R. A role for methylation of the hMLH1 promoter in loss of hMLH1 expression and drug resistance in ovarian cancer. Oncogene. 1999;18:2335–2341. doi: 10.1038/sj.onc.1202540. [DOI] [PubMed] [Google Scholar]
- Sun D.A., Starck S.R., Locke E.P., Hecht S.M. DNA polymerase β inhibitors from Sandoricum koetjape. J. Nat. Prod. 1999;62:1110–1113. doi: 10.1021/np990104r. [DOI] [PubMed] [Google Scholar]
- Tran H.T., Keen J.D., Kricker M., Resnick M.A., Gordenin D.A. Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol. Cell. Biol. 1997;17:2859–2865. doi: 10.1128/mcb.17.5.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umar A., Koi M., Risinger J.I., Glaab W.E., Tindall K.R., Kolodner R.D., Boland C.R., Barrett J.C., Kunkel T.A. Correction of hypermutability, N-methyl-N′-nitro-N-nitrosoguanidine resistance, and defective DNA mismatch repair by introducing chromosome 2 into human tumor cells with mutations in MSH2 and MSH6. Cancer Res. 1997;57:3949–3955. [PubMed] [Google Scholar]
- Wu Q., Vasquez K.M. Human MLH1 protein participates in genomic damage checkpoint signaling in response to DNA interstrand crosslinks, while MSH2 functions in DNA repair. PLoS Genet. 2008;4:e1000189. doi: 10.1371/journal.pgen.1000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Christensen L.A., Legerski R.J., Vasquez K.M. Mismatch repair participates in error-free processing of DNA interstrand crosslinks in human cells. EMBO Rep. 2005;6:551–557. doi: 10.1038/sj.embor.7400418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Lu X., Zhang X., Peterson C.A., Legerski R.J. hMutSβ is required for the recognition and uncoupling of psoralen interstrand cross-links in vitro. Mol. Cell. Biol. 2002;22:2388–2397. doi: 10.1128/MCB.22.7.2388-2397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Wang X., Legerski R.J., Glazer P.M., Li L. Repair of DNA interstrand cross-links: interactions between homology-dependent and homology-independent pathways. DNA Repair (Amst.) 2006;5:566–574. doi: 10.1016/j.dnarep.2006.01.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.