Summary
The Salvador (Sav)/Warts (Wts)/Hippo (Hpo) (SWH) network controls tissue growth by inhibiting cell proliferation and promoting apoptosis. The core of the pathway consists of a MST and LATS family kinase cascade that ultimately phosphorylates and inactivates the YAP/Yorkie (Yki) transcription coactivator. The FERM domain proteins Merlin (Mer) and Expanded (Ex) represent one mode of upstream regulation controlling pathway activity. Here, we identify Kibra as a member of the SWH network. Kibra, which colocalizes and associates with Mer and Ex, also promotes the Mer/Ex association. Furthermore, the Kibra/Mer association is conserved in human cells. Finally, Kibra complexes with Wts and kibra depletion in tissue culture cells induces a marked reduction in Yki phosphorylation without affecting the Yki/Wts interaction. We suggest that Kibra is part of an apical scaffold that promotes SWH pathway activity.
Keywords: SIGNALING, CELLBIO, DEVBIO
Highlights
► The Salvador/Hippo/Warts network restricts tissue size ► We identify the scaffold protein Kibra as a SWH upstream regulator ► Kibra promotes SWH activity by complexing with multiple pathway members
Introduction
Growth regulation is a critical developmental process whose dysfunction can lead to many diseases, including cancer (Conlon and Raff, 1999). The Salvador (Sav)/Warts (Wts)/Hippo (Hpo) (SWH) network, identified in Drosophila and conserved in mammals, plays a major role in limiting growth by inhibiting cell proliferation and promoting apoptosis (Harvey and Tapon, 2007; Reddy and Irvine, 2008). Activation of the upstream kinase Hpo allows it to phosphorylate the downstream kinase Wts, which in turn phosphorylates and inhibits the transcription coactivator Yorkie (Yki). Scaffold proteins, such as Salvador (Sav) and Mats, potentiate the activity of the Hpo/Wts complex (Harvey and Tapon, 2007). One upstream input of the pathway is mediated via Merlin (Mer) and Expanded (Ex), two FERM (Four point one, Ezrin, Moesin, Radixin) domain proteins (Hamaratoglu et al., 2006). Recently, Ex was shown to bind Yki, and new experiments hint at the existence of an Ex/Hpo/Wts-containing apical complex anchoring Yki at the cortex (Badouel et al., 2009; Oh et al., 2009). These findings underline the importance of scaffold proteins in the regulation of the pathway. However, though some upstream members are known, how the SWH network is activated remains unclear.
Here, we identify the WW-domain-containing protein Kibra as a regulator of the SWH network. Human KIBRA (Kremerskothen et al., 2003) is known to be phosphorylated by Protein Kinase C ζ (PKCζ) (Buther et al., 2004) and has recently been reported to have a role in cell migration (Duning et al., 2008; Rosse et al., 2009). In Drosophila, Kibra had previously been recovered as a minor hit in several screens for growth regulators (Boutros et al., 2004; Muller et al., 2005; Ringrose et al., 2003; Tseng and Hariharan, 2002) but has not been further studied. Our experiments show that Kibra associates with Mer, Ex, and Wts and stabilizes the Mer/Ex interaction. This suggests that Kibra is a component of an apical scaffold that controls SWH pathway activation.
Results
kibra and wts RNAi Depletion Induce Similar Overgrowth Phenotypes
We performed an in vivo screen in the fly wing in order to identify genes implicated in growth control. Transgenic flies bearing RNA interference (RNAi) constructs generated by the Vienna Drosophila RNAi Centre (VDRC) (Dietzl et al., 2007) were crossed to the hedgehog-GAL4 (hh-GAL4) driver, leading to target gene silencing in the posterior compartment of the wing. We screened a collection of 12,000 lines targeting genes conserved between Drosophila and mammals. The results of this screen will be described elsewhere.
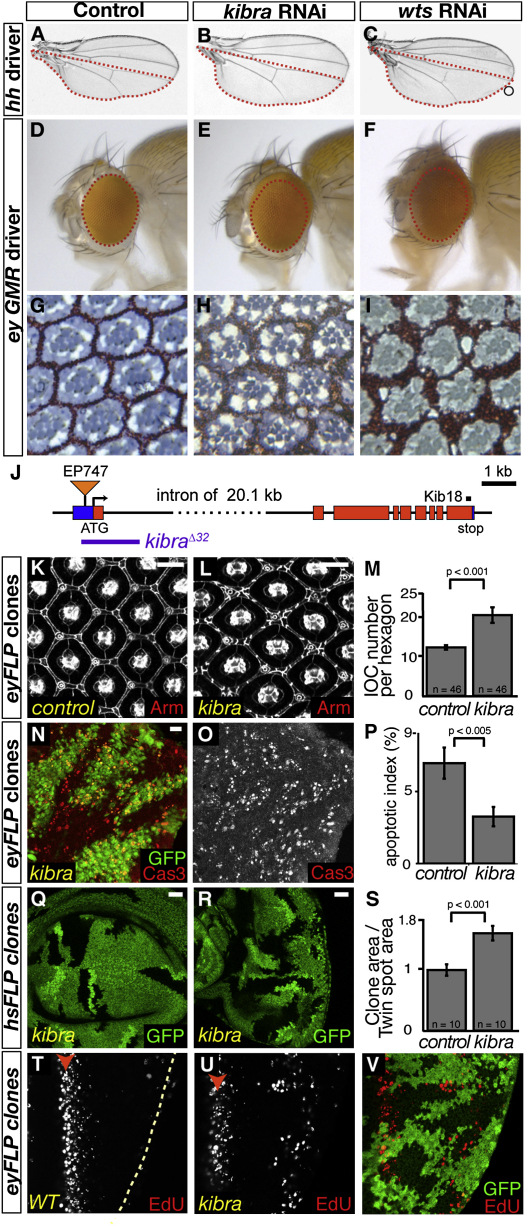
In this context, expressing an RNAi line directed against kibra induced overgrowth of the posterior wing compartment (Figure 1B) compared to control flies (Figure 1A). This phenotype was also observed upon wts depletion (Figure 1C). Driving the same kibra RNAi line in the eye also led to increased organ size (Figures 1D and 1E), similarly to a wts RNAi line (Figure 1F). To exclude off-target effects, we generated a transgenic line expressing a nonoverlapping RNAi construct and observed identical overgrowth phenotypes (data not shown). Furthermore, adult eye sections revealed that kibra knockdown retinas present an excess of interommatidial cells (IOCs) (Figures 1G and 1H). The IOCs, the last population of cells to differentiate in the eye primordium, give rise to the secondary and tertiary pigment cells that optically isolate the ommatidia in the compound eye from each other. Extra IOCs are produced during normal development but are then eliminated by apoptosis at the pupal stage to give rise to the adult lattice (Wolff and Ready, 1993). The presence of extra IOCs is a hallmark of SWH network loss of function (Kango-Singh et al., 2002; Tapon et al., 2002), which reduces retinal apoptosis, as seen in wts RNAi adult eye sections (Figure 1I). Thus, depletion of kibra elicits a similar phenotype to SWH network mutants, suggesting a potential role for Kibra in Hpo signaling.
Figure 1.
Kibra Loss of Function Induces a Phenotype Similar to SWH Network Loss of Function
(A–C) Control wings (A) and wings where the kibra (B) or wts (C) genes were silenced by RNAi in the posterior compartment (red dotted line).
(D–I) Control fly eyes (D and G) and eyes expressing the same RNAi lines against kibra (E and H) or wts (F and I). In adult eye sections (G–I), interommatidial cells (IOCs) are in red and photoreceptors in blue.
(J) Schematic of the kibra locus showing the localization of the kibraΔ32 allele, generated by excision of the EP747 P element (triangle). The coding sequence is in red; 5′- and 3′-UTRs are in blue. In black is the peptide recognized by the Kib18 antibody.
(K–M) 40 hr APF retinas containing kibra mutant clones. (K and L) Cell outlines are visualized by anti-Arm staining (scale bar = 10 μm). (M) Quantification of the IOCs. The p value from a Mann-Whitney test is shown.
(N–P) Apoptotic index in 28 hr APF retinas containing kibra mutant clones (absence of GFP). A retina stained for activated Caspase-3 (Cas3) merged with GFP is shown in (N). The Cas3 staining for the whole retina is shown in (O). (P) Quantification of the apoptotic index. The p value from a Mann-Whitney test is shown.
(Q–S) Wing disc (Q) and eye disc (R) containing kibra clones (lack of GFP). Scale bars = 20 μm. (S) Quantification of the proliferation advantage of WT or kibra mutant cells by calculating the ratio between total clonal area (no GFP) and total twin spot area (two copies of GFP) for discs containing either WT or kibra clones. The p value from a Mann-Whitney test is shown.
(T–V) EdU labeling (shown in gray or red) of WT discs (T) and of discs containing kibra clones (U and V). Posterior is to the right, and the SMW is indicated by a red arrowhead. See also Figure S1.
Error bars represent standard deviations.
kibra Loss of Function Causes Excess Proliferation and a Reduced Apoptotic Rate, Similarly to SWH Network Loss of Function
To study kibra loss of function, we generated the kibraΔ32 allele by imprecise excision of the EP747 transposon (see Figure 1J and Experimental Procedures). This deletion allele, which removes the translation initiation site, is homozygous lethal and may be a null allele for kibra. kibraΔ32 FLP/FRT mutant clones in 40 hr after-puparium-formation (APF) retinas present extra IOCs (Figure 1K–1M), similarly to what was observed in adult eyes with kibra knockdown (Figures 1G–1I). Duplication of bristles or missing bristles can also be observed. We determined apoptotic indexes (Colombani et al., 2006) during the retinal apoptosis wave (28 hr APF) in pupal retinas containing kibra mutant clones stained with an anti-active Caspase-3 antibody. kibra mutant tissue presents a reduced apoptotic index compared with wild-type (WT) areas in the same retinas (Figures 1N–1P; see Figures S1A–S1A″′ available online). Thus, extra IOCs persist in kibra mutant clones as a result of decreased developmental apoptosis.
We assessed the proliferation rate of kibra mutant cells in imaginal discs, the larval precursors to the adult appendages. By using the FLP/FRT system under the control of the heat-shock promoter, kibra mutant cells and their WT sister clones were generated through single recombination events from heterozygous mother cells (Brumby and Richardson, 2005). After several rounds of divisions, the sizes of mutant clones (no GFP) and WT twin spots (two copies of GFP) were compared, allowing us to estimate the relative proliferation rates of mutant versus WT cells. The total kibra clone area is 1.57 (±0.12)-fold larger than the control twin spot area, compared to a ratio of 0.98 (±0.09) when both clones and twin spots are WT (Figures 1Q–1S), indicating that kibraΔ32 mutant cells grow 1.6 times faster than WT cells.
In addition to cell cycle rates, the timing of cell cycle exit can readily be measured in the eye disc, where cell divisions follow a spatially determined pattern (Wolff and Ready, 1993). During the third larval instar, the morphogenetic furrow, a wave of differentiation, sweeps the eye disc from posterior to anterior. Anterior to the furrow cells still proliferate asynchronously, while in the furrow cells synchronize in G1. Immediately posterior to the furrow, cells enter a final round of synchronous S phases, the second mitotic wave (SMW). Posterior to the SMW, most cells permanently exit the cell cycle. Thus, in WT discs, no S phases (marked by EdU incorporation) can be observed posterior to the SMW (Figure 1T). As expected, hpo mutant cells fail to exit from the cell cycle in a timely manner and present ectopic EdU-positive staining posterior to the SWM (Figures S1B and S1B′). kibra mutant cells exhibit a less pronounced but similar phenotype (Figures 1U and 1V). Thus, kibra mutant tissues have a proliferative advantage and an apoptosis defect, consistent with an involvement in the SWH network. The overgrowth defect appears more subtle than that of core pathway members such as wts and is more akin to upstream regulators (e.g., ex and mer).
Kibra Regulates SWH Network Targets in Ovarian Posterior Follicle Cells
Several transcriptional targets of the SWH network have been identified, such as the Drosophila Inhibitor of Apoptosis 1 (DIAP1) gene, the cell cycle regulator cycE, the miRNA bantam, as well as ex (Harvey and Tapon, 2007; Saucedo and Edgar, 2007). In kibra mutant wing or eye discs, we could not detect a strong change in DIAP1, CycE, or ex-lacZ reporter levels (data not shown). Since overgrowth of kibra mutant cells in the wing is subtle compared to wts mutants, it is possible that Kibra plays a relatively minor role in SWH signaling in the wing. Accordingly, using an anti-Kibra antibody we generated (Figures S2A–S2C), we noted that Kibra staining in the wing disc is weak and consists of a punctate apical staining which can clearly be observed when kibra is overexpressed in a stripe of cells. Thus, the extent to which Kibra is required may vary in different tissues.
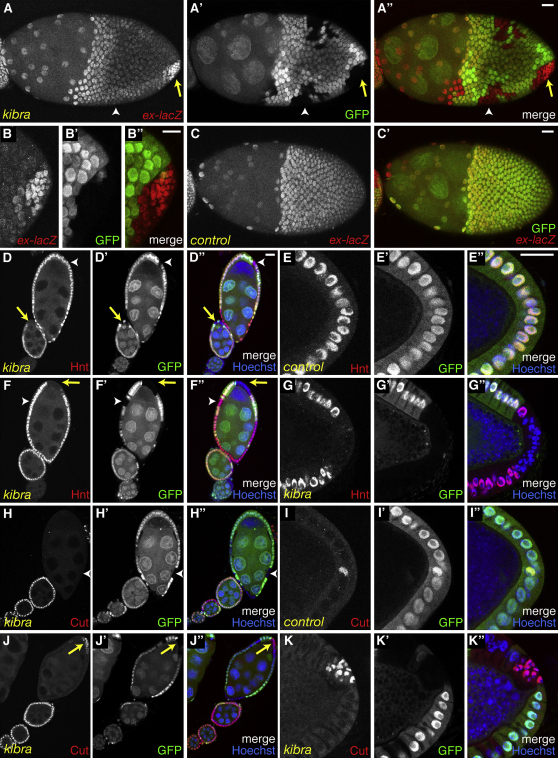
We and others have previously reported that ovarian posterior follicle cells (PFCs) are particularly sensitive to SWH loss of function (Meignin et al., 2007; Polesello and Tapon, 2007; Yu et al., 2008), leading us to study the kibraΔ32 phenotype in the ovary. First, we noted that Kibra protein levels are higher in follicle cells than in the wing discs (Figure S2D–S2E′). Kibra staining is mainly apical and is severely reduced in kibraΔ32 clones. Similarly to hpo or wts loss of function, kibra loss of function in the PFCs induces an upregulation of the ex-lacZ reporter (Figures 2A–2B″, compare with Figures 2C–2C″). hpo or wts mutant PFCs also show a misregulation of the Notch (N) pathway and ectopic cell divisions (Meignin et al., 2007; Polesello and Tapon, 2007; Yu et al., 2008). The N target Hindsight (Hnt) is normally repressed in all follicle cells up to stage 6 and switched on from stage 7 to stage 10B (Figures 2D–2E″) (Poulton and Deng, 2007). Cut, which is repressed by Hnt, presents an opposite pattern of expression (Figures 2H–2I″). In kibra mutant PFCs from stage 7–10B egg chambers, Hnt expression is lost (Figures 2D–2D″ and 2F–2G″), while Cut is ectopically expressed (Figures 2J–2K″). This indicates that N signaling is downregulated in kibra mutant PFCs. Loss of kibra also leads to perturbation of epithelial integrity, as mutant PFCs show an accumulation of the apical polarity protein aPKC and the N receptor (Figures S2F–S2G″) as well as multilayering of the follicular epithelium (Figures 2I–2I″ and 2K–2K″). Ectopic mitotic divisions are also observed in PFCs clones after stage 6, as detected by phospho-histone H3 (PH3) staining (Figures S2H–S2H″). Together, these phenotypes are identical to those observed in hpo or wts loss of function, suggesting that Kibra is indeed a member of the SWH network.
Figure 2.
Kibra Regulates SWH Pathway Targets in Ovarian Posterior Follicle Cells
Egg chambers containing heat-shock-induced kibra mutant clones (absence of GFP). Yellow arrows point to kibra mutant cells in the PFC region while white arrowheads point to kibra cells in lateral follicle cells. Scale bars = 20 μm. (A–C′) Stage 10B egg chambers stained for β-galactosidase (gray or red), which monitors the activity of the ex-lacZ reporter. (B–B″) Close-up of the PFC region in (A)–(A″). (D–K″) Egg chambers stained for the N target Hindsight (Hnt) (D–G″) or for Cut (H–K″). Nuclei are shown in blue (Hoechst). (E)–(E″), (G)–(G″), (I)–(I″), and (K)–(K″) are close-ups of the PFC region of stage 10B egg chambers. (D–D″) The arrow points to a stage 8 clone; the arrowhead points to a stage 9 clone. (F–F″ and J–J″) Arrows and arrowheads point to clones in stage 9 egg chambers. See also Figure S2.
Genetic Experiments Place kibra Upstream of the Core SWH Kinase Cassette
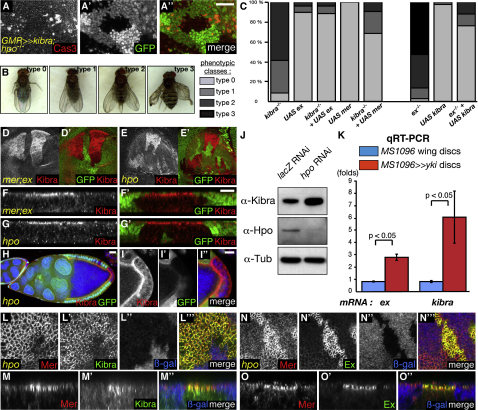
To further explore the role of Kibra in the SWH network, genetic interaction and epistasis experiments were performed. Overexpressing kibra in the eye under the GMR (Glass Multimer Reporter) promoter elicits the formation of a small rough eye with frequent ommatidial fusions (Figures S3A–S3B′). This phenotype can be partially rescued by removing one copy of the hpo gene (Figures S3C and S3C′). In contrast, overexpressing kibra could not rescue the hpo-like overgrowth phenotype induced by yki overexpression (Figures S3D and S3E), suggesting that Kibra may be an upstream regulator of the pathway.
To conduct epistasis experiments between kibra and yki, we used the MARCM system to generate clones of mutant cells while simultaneously overexpressing or depleting other pathway components (Lee and Luo, 1999). MARCM clones expressing yki RNAi generated with eyFLP lead to the formation of a normal eye, because yki-depleted cells are eliminated by apoptosis (data not shown) and replaced by WT cells (Figure S3F). As expected, eyFLP kibra MARCM clones cause eye overgrowth (Figure S3G). This overgrowth is rescued by yki depletion in the mutant cells (Figure S3H), indicating that the kibra overgrowth phenotype is yki dependent. Furthermore, overexpressing kibra in the eye under the GMR promoter induces apoptosis in third instar eye discs, which is suppressed by loss of hpo (Figures 3A–3A″). Together, these epistasis experiments are consistent with Kibra being a member of the SWH network acting upstream of Yki and Hpo.
Figure 3.
kibra Is Epistatic to hpo and Is a Transcriptional Target of the SWH Network
(A–A″) Third instar eye imaginal disc expressing kibra under the GMR promoter and containing hpo mutant cells (absence of GFP). Apoptosis is visualized by an anti-activated Caspase-3 staining. Posterior is to the bottom.
(B and C) Epistatic relationships between Kibra, Mer and Ex. Examples of the 4 phenotypic classes used to score are shown in (B). The percentages of each class for each genotype are summarized in (C).
(D–G′) Third instar imaginal discs containing clones (marked by absence of GFP) of cells mutant for mer;ex (D, D′, F, and F′) or hpo (E, E′, G, and G′). (D)–(E′) are XY sections while (F)–(G′) are XZ transverse sections (apical is to the top).
(H–I″) Stage 10A egg chamber containing a hpo clone (marked by absence of GFP) in the PFCs and stained for Kibra (red). (I)–(I″) show a higher magnification of the PFC region in (H). Nuclei are stained with Hoechst (blue). Scale bars = 20 μm (A–A″, D–E′, and H), 10 μm (F–G′ and I–I″).
(J) Lysates from S2R+ cells treated with lacZ RNAi or hpo RNAi and probed for Kibra and Tubulin.
(K) A graph presenting a comparison of kibra and ex mRNA levels between yki overexpressing and WT wing discs, as measured by qRT-PCR, is shown. p values from Mann-Whitney tests are shown. Error bars represent standard deviations.
(L–O″) XY and transverse sections of third instar imaginal wing discs containing hpo mutant clones (labeled by absence of βgal) and stained for Kibra (L–L″ and N–N″), Ex (M–M″ and O–O″), and Mer (L–O″). See also Figure S3.
Genetic interactions between kibra, mer, and ex, upstream members of the SWH network, were then investigated. Expressing a kibra, an ex, or a mer RNAi line in the eye under the GMR promoter induces eye overgrowth (Figures S3I–S3L). Combined depletion of either Ex/Kibra or Mer/Kibra shows stronger phenotypes than individual depletion of these proteins (Figures S3M and S3N). We used the MARCM technique to evaluate epistatic relationships between those three genes. hsFLP MARCM clones of various genotypes were generated and scored according to the severity of the wing overgrowth phenotypes, with type 0 representing normal wings and type 4 the strongest overgrowth (Figures 3B and 3C). Overexpressing ex or mer in kibra mutant clones significantly rescues the overgrowth of kibra mutant clones (p < 0.0001 for both genotypes). Reciprocally, kibra overexpression was also able to suppress the ex overgrowth phenotype (p < 0.0001). Thus, we could not determine a strict epistatic relationship between kibra, ex, and mer, consistent with a model whereby kibra, ex, and mer cooperate to control SWH pathway activity.
Kibra Is a Transcriptional Target of the SWH Network and Colocalizes with Mer and Ex
As well as being an upstream regulator of the SWH network, ex is also one of its transcriptional targets (Hamaratoglu et al., 2006), as are other upstream regulators (e.g., mer, four-jointed, dachsous). Since epistasis experiments place Kibra at the level of Mer and Ex, we wished to test whether this is also the case for kibra. Kibra levels were highly upregulated in mer;ex or hpo clones (Figures 3D–3E′), showing an apical localization (Figures 3F–3G′). The same is true in hpo clones in follicle cells (Figures 3H–3I″). Similarly, hpo-depleted cultured Drosophila S2R+ cells have increased Kibra levels (Figure 3J). To determine whether kibra is a transcriptional SWH network target, quantitative RT-PCR experiments were performed on yki-overexpressing and control wing imaginal discs (Figure 3K). As expected, ex mRNA levels were increased (2.97 ± 0.25-fold) in yki-expressing discs compared to control discs. Interestingly, kibra mRNA levels were also upregulated in yki-expressing discs (6.24 ± 2.12-fold), confirming that kibra is a Yki transcriptional target and suggesting the existence of a possible negative feedback loop regulating Kibra expression.
Because hpo clones present increased levels of Kibra as well as Mer and Ex, these constitute a good system to evaluate the colocalization of those proteins. Indeed, Kibra colocalizes with Mer in the wing disc (Figures 3L–3M″). As expected, Mer and Ex also colocalize (Figures 3N–3O″). Thus, Kibra, Mer, and Ex colocalize apically in imaginal disc cells, but are dispensable for each other's apical sorting, because Kibra is still apical in mer;ex clones and Mer/Ex are normally localized in kibra clones (Figures 3D, 3D′, 3F, and 3F′ and data not shown).
Kibra Associates with Ex and Mer, and the Kibra/Mer Complex Is Conserved in Human Cells
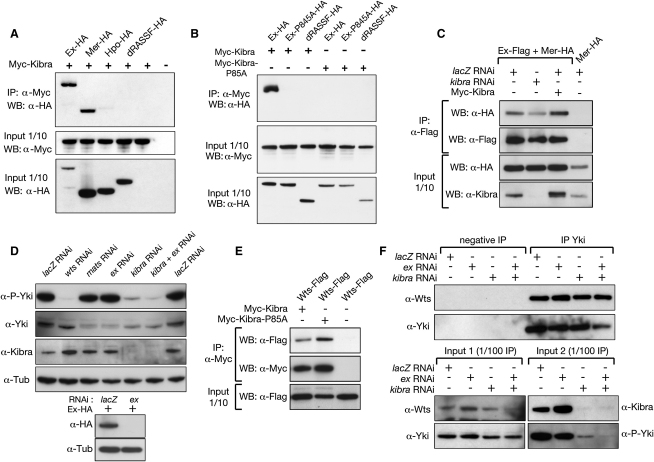
Because Kibra colocalizes with Mer/Ex, a possible association between those proteins was examined by conducting coimmunoprecipitation (co-IP) assays in S2R+ cells. Kibra was found to co-IP with Ex and Mer, but not with Hpo or with the negative regulator of Hpo, dRASSF (Polesello et al., 2006) (Figure 4A). Interestingly, Kibra was reported to interact with Mer in a large-scale yeast two-hybrid screening study (Formstecher et al., 2005). Kibra possesses two WW domains, which are predicted to mediate protein-protein interactions by binding to PPXY motifs. Furthermore, the first WW domain of human KIBRA was shown to recognize the consensus motif RXPPXY in vitro (Kremerskothen et al., 2003). In flies, Mer does not contain any PPXY sites, while Ex has two PPXY sites (P786PPY and P1203PPY) and an RXPPXY site (R842DPPPY). We therefore further investigated the association between Kibra and Ex by mutating amino acids that are known to be required for WW domains and PPXY sites to interact (Kremerskothen et al., 2003; Otte et al., 2003). A Kibra protein mutant for its first WW domain (P85A) could no longer co-IP WT Ex. Reciprocally, WT Kibra could not co-IP an Ex protein deficient for its RXPPXY site (P845A) (Figure 4B). Thus, Kibra associates with Ex through its first WW domain and the Ex RXPPXY motif.
Figure 4.
Kibra Associates with Mer and Ex, and kibra Depletion Strongly Reduces Yki Phosphorylation without Interfering with the Yki/Wts Interaction
(A and B) Western blots of coimmunoprecipitation assays between Myc-Kibra and HA-tagged members of the SWH network are shown.
(C) Western blots of co-IP assays between Mer-HA and Ex-Flag in presence or absence of Kibra are shown.
(D) Lysates of S2R+ cells treated with RNAi against different members of the SWH network and probed for P-Ser168 Yki (P-Yki), pan-Yki, Kibra, and Tubulin are shown (the anti-Kibra blot was performed on a parallel run of the same samples). The bottom panel shows efficiency of the ex dsRNAi treatment.
(E) Western blots of co-IP assays between Wts-Flag and different versions of Myc-tagged Kibra are shown.
(F) Endogenous co-IP assays between Yki and Wts in control S2 cells and in cells treated with ex and/or kibra RNAi are shown. The anti-Wts input blot was performed on a parallel run of the same samples. See also Figure S4.
In contrast, mutating either one or both Kibra WW domains does not affect Kibra/Mer association (Figure S4A). Further assays reveal that both Kibra N- and C-terminal fragments are sufficient for the association with Mer, but a central stretch (aa 484–857) is not (Figure S4A). Because the WW motifs, which are required for the Ex/Kibra association, are located in the N-terminal fragment, this suggests that Mer can complex with Kibra both through Ex and independently of Ex. To further test this possibility, coimmunoprecipitation assays between Mer and Kibra were performed in ex-depleted cells (Figure S4B). The Kibra/Mer immunoprecipitation is not affected by ex depletion, suggesting that Ex is not required for the Kibra/Mer association.
Because the Hpo pathway is highly conserved from Drosophila to humans, we tested for potential interactions between human KIBRA and the human orthologs of Ex (FRMD6), Mer (NF2/MER), Hpo (MST2), and dRASSF (RASSF6). We used split-TEV as readout, which is based on TEV protease complementation and represents a sensitive method for detecting interactions between membrane-associated proteins (Wehr et al., 2006, 2008). We found that KIBRA associates with NF2/MER but did not interact with FRMD6, MST2, or RASSF6 (Figure S4C). Interestingly, FRMD6 contains only an N-terminally conserved sequence of Ex but lacks the entire C-terminal part, which in Ex harbors the PPXY motifs. Thus, the missing PPXY motifs and the generally limited level of sequence conservation in FRMD6 likely explain the absence of interaction between KIBRA and FRMD6. These results imply that the ability to associate with Kibra evolved in an ancestral Ex/Mer-like FERM domain protein and was later lost in FRMD6 but retained in NF2/MER. Alternatively, an Ex functional homolog other than FRMD6 may exist.
As Kibra complexes with both Ex and Mer and Ex/Mer have been reported to directly interact (McCartney et al., 2000), we tested the possibility that Kibra could affect the Mer/Ex interaction. We performed co-IP assays between Mer and Ex in cells expressing different levels of Kibra protein (Figure 4C). The Mer/Ex interaction is reduced in kibra-depleted cells compared to WT cells, whereas the interaction is strengthened in cells that express a Myc-Kibra construct. Thus, the presence of Kibra is required to fine-tune the stability of the Mer/Ex interaction.
Kibra Associates with Wts and Is Required for Yki Phosphorylation, but Not for Yki/Wts Binding
Because Kibra complexes with Ex and a Yki/Ex interaction has recently been described (Badouel et al., 2009), we sought to determine whether Kibra can affect Yki activity. S2R+ cells were treated with RNAi against several SWH pathway components, and Yki phophorylation on Ser168 was monitored by western blotting (Figure 4D). The phosphorylation of Yki by Wts at Ser168 leads to Yki inactivation and sequestration in the cytoplasm, where it has been reported to bind Ex, Wts, Hpo, and 14.3.3 (Badouel et al., 2009; Huang et al., 2005; Oh and Irvine, 2008; Oh et al., 2009). lacZ RNAi-treated cells show a high basal level of phospho-Yki (P-Yki). As expected, Yki phosphorylation is abolished when Wts is depleted, and mildly reduced when the Wts cofactor Mats (Lai et al., 2005) is depleted. In wts treated RNAi cells, a Yki downward shift can also be observed using a pan-Yki antibody (Figure 4D, second row). ex RNAi treatment has only a mild effect on P-Yki levels. Interestingly, kibra depletion leads to a marked reduction in P-Yki. When depleted in conjunction with ex, the P-Yki signal becomes even further reduced.
This suggests that Kibra and Ex are required for Wts activity on Yki, which prompted us to investigate whether Kibra could associate with Wts. Co-IP assays reveal that Kibra interacts with Wts (Figure 4E). Wts does not seem to compete with Ex for Kibra association, because it could still complex with a form of Kibra mutant for its first WW domain. Because Kibra associates with Wts and Ex interacts with Yki, we investigated whether Wts requires Kibra/Ex to bind Yki. Endogenous IPs between Yki and Wts were performed in S2 cells treated with various dsRNAs (Figure 4F). In these conditions, the effect of kibra and ex depletion on Yki phosphorylation can also be observed (see input). In control cells, Wts binding to Yki is detected after immunoprecipitating Yki. This endogenous interaction is unaffected by the individual or combined depletion of ex and kibra. These results suggest that Ex and Kibra are required to activate the SWH pathway by nucleating an active Hpo/Wts kinase cassette, rather than promoting the Wts/Yki interaction.
Discussion
Our data identify Kibra as a regulator of the SWH network that associates with Ex and Mer, with which it is colocalized apically and transcriptionally coregulated. Given that the apical surface of epithelial cells is instrumental in both cell-cell signaling and tissue morphogenesis, we speculate that Kibra may cooperate with Ex and Mer to transduce an extracellular signal, or relay information about epithelial architecture, via the SWH network, to control tissue growth and morphogenesis.
Recent data have suggested that an apical scaffold machinery containing Hpo, Wts, and Ex recruits Yki to the apical membrane, facilitating its inhibitory phosphorylation by Wts (Badouel et al., 2009; Oh et al., 2009). Since Kibra associates with Ex and is also apically localized, we can hypothesize that Kibra is also part of this scaffold and participates in nucleating an active Hpo/Wts complex and recruiting Yki for inactivation. This view is supported by our finding that Kibra complexes with Wts and that combined depletion of Kibra and Ex leads to a strong decrease in Yki phosphorylation, but does not disrupt the Wts/Yki interaction. Our data also suggest that the importance of Kibra may be tissue-specific since we observe robust phenotypes in ovaries and hemocyte-derived S2R+ cells, but weaker effects in imaginal discs. Thus, considering the relative levels of expression of Ex, Mer, and Kibra may be important in determining pathway activation. Finally, since mammalian KIBRA complexes with the NF2/MER tumor suppressor, our findings raise the possibility that human KIBRA may contribute to tumor suppression in human neurofibromas and potentially other tumors.
Experimental Procedures
For Drosophila genotypes, primer sequences, and further experimental details, see Supplemental Experimental Procedures.
kibra Mutant
The P element of EP747 (Bloomington stock center) was mobilized using standard genetic techniques, and excisions were screened by PCR (see Supplemental Experimental Procedures for details).
Immunostainings
Immunostainings and confocal image acquisitions were performed as in Genevet et al. (2009). Mouse β-galactosidase (Promega), rabbit anti-Cleaved Caspase-3 (Asp175) (Cell Signaling Technology), rabbit anti-aPKC (Santa Cruz), and mouse NICD (C17.9C6, Development Studies Hybridoma Bank) antibodies were used at 1/500. Mouse anti-Arm, anti-Cut, and anti-Hnt (N2 7A1, 2B10 and 1G9, DSHB) were used at 1/10 and 1/20. The EdU staining was performed as described in the Click-iT EdU Alexa Fluor Imaging Kit (Invitrogen). Guinea pig anti-Mer, a gift from R. Fehon, was used at 1/7500, and rabbit anti-Ex, a gift from A. Laughon, at 1/400. Rabbit anti-Kibra antibody (Kib18, 1/100) was generated by Eurogentec SA (Seraing, Belgium) against a peptide corresponding to the last 15 amino acids of Kibra.
Quantification of Apoptotic Indexes in Mutant Clones versus WT Tissue
Apoptotic indexes were quantified on 6 retinas as described in Colombani et al. (2006).
Standard Growth Conditions and Size Measurements
The experiment was performed as described in Genevet et al. (2009). Ten wing discs for each genotype were analyzed.
Fly Eye Scanning Electron Microscopy
Scanning microscopy of adult flies was performed as described in Polesello et al. (2006).
RNAi Treatment and Coimmunoprecipitation of Proteins
For RNAi treatment, S2 or S2R+ cells were treated with dsRNAs as indicated for 4 days. For coimmunoprecipitations, S2R+ or S2 cells transfected with the indicated plasmids were used. See Supplemental Experimental Procedures for details.
Quantitative RT-PCR
MS1096 > > (control) and MS1096 > > yki wing discs were dissected in PBS and snap-frozen in liquid nitrogen. RNA isolation and subsequent qRT-PCR reactions were performed as described in Genevet et al. (2009). See Supplemental Experimental Procedures for primer sequences.
Statistical Analysis
All error bars displayed represent standard deviations. All statistical analyses performed (except epistasis analysis) were assessed by Mann-Whitney nonparametric tests using the website http://elegans.swmed.edu/∼leon/stats/utest.html.
The epistasis analysis was made by pairwise comparison after correction for the batch effect on 42 to 310 flies of each genotype, divided in 4 to 6 cohorts. The approach used was a three-way log-linear model, against a null-hypothesis of no interaction between phenotype and population. The p value indicates whether the pair of populations differ in their phenotype profiles: p value(kibra−/−; kibra−/− + UAS ex) = 2.69 × 10−63, p value(kibra−/−; kibra−/− +UAS mer) = 6.07 × 10−35, p value(ex−/−; ex−/− + UAS kibra) = 1.76 × 10−20.
Acknowledgments
We thank D. Pan, R. Fehon, G. Halder, A. Laughon, K. Irvine, J. Kremerskothen, G. Struhl, T. Igaki, H. McNeill, the Bloomington and VDRC stock centres, and the DGRC for fly stocks and reagents. We are very grateful to G. Dietzl, B. Dickson, and S. Cohen for their support in carrying out the RNAi screen. We are grateful to K. Blight, A. Weston, and L. Collinson from the LRI Electron Microscopy Facility and to P. Jordan from the Light Microscopy Facility for technical help, to G. Kelly for help with statistics, to F. Josué for help with apoptotic index analysis, as well as to T. Gilbank, S. Murray, and S. Maloney from the Fly Facility for technical support. We thank S. Cohen and C. Polesello for discussions and comments on the manuscript. We thank H. Stocker and E. Hafen for discussing data prior to publication. M.C.W. is supported by an EMBO long-term fellowship. The Tapon and Thompson laboratories are supported by Cancer Research UK.
Published: February 15, 2010
Footnotes
Supplemental Information includes four figures, Supplemental Experimental Procedures, and Supplemental References and can be found with this article online at doi:10.1016/j.devcel.2009.12.011.
Contributor Information
Barry J. Thompson, Email: barry.thompson@cancer.org.uk.
Nicolas Tapon, Email: nicolas.tapon@cancer.org.uk.
Supplemental Information
References
- Badouel C., Gardano L., Amin N., Garg A., Rosenfeld R., Le Bihan T., McNeill H. The FERM-domain protein expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Dev. Cell. 2009;16:411–420. doi: 10.1016/j.devcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- Boutros M., Kiger A.A., Armknecht S., Kerr K., Hild M., Koch B., Haas S.A., Paro R., Perrimon N. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- Brumby A.M., Richardson H.E. Using Drosophila melanogaster to map human cancer pathways. Nat. Rev. Cancer. 2005;5:626–639. doi: 10.1038/nrc1671. [DOI] [PubMed] [Google Scholar]
- Buther K., Plaas C., Barnekow A., Kremerskothen J. KIBRA is a novel substrate for protein kinase Czeta. Biochem. Biophys. Res. Commun. 2004;317:703–707. doi: 10.1016/j.bbrc.2004.03.107. [DOI] [PubMed] [Google Scholar]
- Colombani J., Polesello C., Josue F., Tapon N. Dmp53 activates the Hippo pathway to promote cell death in response to DNA damage. Curr. Biol. 2006;16:1453–1458. doi: 10.1016/j.cub.2006.05.059. [DOI] [PubMed] [Google Scholar]
- Conlon I., Raff M. Size control in animal development. Cell. 1999;96:235–244. doi: 10.1016/s0092-8674(00)80563-2. [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K.C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Duning K., Schurek E.M., Schluter M., Bayer M., Reinhardt H.C., Schwab A., Schaefer L., Benzing T., Schermer B., Saleem M.A. KIBRA modulates directional migration of podocytes. J. Am. Soc. Nephrol. 2008;19:1891–1903. doi: 10.1681/ASN.2007080916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formstecher E., Aresta S., Collura V., Hamburger A., Meil A., Trehin A., Reverdy C., Betin V., Maire S., Brun C. Protein interaction mapping: a Drosophila case study. Genome Res. 2005;15:376–384. doi: 10.1101/gr.2659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genevet A., Polesello C., Blight K., Robertson F., Collinson L., Pichaud F., Tapon N. The Hippo pathway regulates apical domain size independently of its growth control function. J. Cell Sci. 2009;122:2360–2370. doi: 10.1242/jcs.041806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaratoglu F., Willecke M., Kango-Singh M., Nolo R., Hyun E., Tao C., Jafar-Nejad H., Halder G. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- Harvey K., Tapon N. The Salvador-Warts-Hippo pathway—an emerging tumour-suppressor network. Nat. Rev. Cancer. 2007;7:182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- Huang J., Wu S., Barrera J., Matthews K., Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Kango-Singh M., Nolo R., Tao C., Verstreken P., Hiesinger P.R., Bellen H.J., Halder G. Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development. 2002;129:5719–5730. doi: 10.1242/dev.00168. [DOI] [PubMed] [Google Scholar]
- Kremerskothen J., Plaas C., Buther K., Finger I., Veltel S., Matanis T., Liedtke T., Barnekow A. Characterization of KIBRA, a novel WW domain-containing protein. Biochem. Biophys. Res. Commun. 2003;300:862–867. doi: 10.1016/s0006-291x(02)02945-5. [DOI] [PubMed] [Google Scholar]
- Lai Z.C., Wei X., Shimizu T., Ramos E., Rohrbaugh M., Nikolaidis N., Ho L.L., Li Y. Control of cell proliferation and apoptosis by Mob as tumor suppressor, Mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Lee T., Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- McCartney B.M., Kulikauskas R.M., LaJeunesse D.R., Fehon R.G. The neurofibromatosis-2 homologue, Merlin, and the tumor suppressor expanded function together in Drosophila to regulate cell proliferation and differentiation. Development. 2000;127:1315–1324. doi: 10.1242/dev.127.6.1315. [DOI] [PubMed] [Google Scholar]
- Meignin C., Alvarez-Garcia I., Davis I., Palacios I.M. The Salvador-Warts-Hippo pathway is required for epithelial proliferation and axis specification in Drosophila. Curr. Biol. 2007;17:1871–1878. doi: 10.1016/j.cub.2007.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller D., Kugler S.J., Preiss A., Maier D., Nagel A.C. Genetic modifier screens on Hairless gain-of-function phenotypes reveal genes involved in cell differentiation, cell growth and apoptosis in Drosophila melanogaster. Genetics. 2005;171:1137–1152. doi: 10.1534/genetics.105.044453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H., Irvine K.D. In vivo regulation of Yorkie phosphorylation and localization. Development. 2008;135:1081–1088. doi: 10.1242/dev.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H., Reddy B.V., Irvine K.D. Phosphorylation-independent repression of Yorkie in Fat-Hippo signaling. Dev. Biol. 2009;335:188–197. doi: 10.1016/j.ydbio.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otte L., Wiedemann U., Schlegel B., Pires J.R., Beyermann M., Schmieder P., Krause G., Volkmer-Engert R., Schneider-Mergener J., Oschkinat H. WW domain sequence activity relationships identified using ligand recognition propensities of 42 WW domains. Protein Sci. 2003;12:491–500. doi: 10.1110/ps.0233203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesello C., Tapon N. Salvador-warts-hippo signaling promotes Drosophila posterior follicle cell maturation downstream of Notch. Curr. Biol. 2007;17:1864–1870. doi: 10.1016/j.cub.2007.09.049. [DOI] [PubMed] [Google Scholar]
- Polesello C., Huelsmann S., Brown N.H., Tapon N. The Drosophila RASSF homolog antagonizes the Hippo pathway. Curr. Biol. 2006;16:2459–2465. doi: 10.1016/j.cub.2006.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulton J.S., Deng W.M. Cell-cell communication and axis specification in the Drosophila oocyte. Dev. Biol. 2007;311:1–10. doi: 10.1016/j.ydbio.2007.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy B.V., Irvine K.D. The Fat and Warts signaling pathways: new insights into their regulation, mechanism and conservation. Development. 2008;135:2827–2838. doi: 10.1242/dev.020974. [DOI] [PubMed] [Google Scholar]
- Ringrose L., Rehmsmeier M., Dura J.M., Paro R. Genome-wide prediction of Polycomb/Trithorax response elements in Drosophila melanogaster. Dev. Cell. 2003;5:759–771. doi: 10.1016/s1534-5807(03)00337-x. [DOI] [PubMed] [Google Scholar]
- Rosse C., Formstecher E., Boeckeler K., Zhao Y., Kremerskothen J., White M.D., Camonis J.H., Parker P.J. An aPKC-exocyst complex controls paxillin phosphorylation and migration through localised JNK1 activation. PLoS Biol. 2009;7:e1000235. doi: 10.1371/journal.pbio.1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucedo L.J., Edgar B.A. Filling out the Hippo pathway. Nat. Rev. Mol. Cell Biol. 2007;8:613–621. doi: 10.1038/nrm2221. [DOI] [PubMed] [Google Scholar]
- Tapon N., Harvey K.F., Bell D.W., Wahrer D.C., Schiripo T.A., Haber D.A., Hariharan I.K. salvador promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Tseng A.S., Hariharan I.K. An overexpression screen in Drosophila for genes that restrict growth or cell-cycle progression in the developing eye. Genetics. 2002;162:229–243. doi: 10.1093/genetics/162.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehr M.C., Laage R., Bolz U., Fischer T.M., Grunewald S., Scheek S., Bach A., Nave K.A., Rossner M.J. Monitoring regulated protein-protein interactions using split TEV. Nat. Methods. 2006;3:985–993. doi: 10.1038/nmeth967. [DOI] [PubMed] [Google Scholar]
- Wehr M.C., Reinecke L., Botvinnik A., Rossner M.J. Analysis of transient phosphorylation-dependent protein-protein interactions in living mammalian cells using split-TEV. BMC Biotechnol. 2008;8:55. doi: 10.1186/1472-6750-8-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff T., Ready D.F. Pattern formation in the Drosophila retina. In: Bate M., Arias A.M., editors. Volume 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1993. (The Development of Drosophila melanogaster). [Google Scholar]
- Yu J., Poulton J., Huang Y.C., Deng W.M. The hippo pathway promotes Notch signaling in regulation of cell differentiation, proliferation, and oocyte polarity. PLoS ONE. 2008;3:e1761. doi: 10.1371/journal.pone.0001761. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.