Abstract
Protein tyrosine phosphatases (PTPs) are key regulators of different processes during development of the central nervous system. However, expression patterns and potential roles of PTPs in the developing superior colliculus remain poorly investigated. In this study, a degenerate primer-based reverse transcription-polymerase chain reaction (RT-PCR) approach was used to isolate seven different intracellular PTPs and nine different receptor-type PTPs (RPTPs) from embryonic E15 mouse superior colliculus. Subsequently, the expression patterns of 11 PTPs (TC-PTP, PTP1C, PTP1D, PTP-MEG2, PTP-PEST, RPTPJ, RPTPε, RPTPRR, RPTPσ, RPTPκ and RPTPγ) were further analyzed in detail in superior colliculus from embryonic E13 to postnatal P20 stages by quantitative real-time RT-PCR, Western blotting and immunohistochemistry. Each of the 11 PTPs exhibits distinct spatiotemporal regulation of mRNAs and proteins in the developing superior colliculus suggesting their versatile roles in genesis of neuronal and glial cells and retinocollicular topographic mapping. At E13, additional double-immunohistochemical analysis revealed the expression of PTPs in collicular nestin-positive neural progenitor cells and RC-2-immunoreactive radial glia cells, indicating the potential functional importance of PTPs in neurogenesis and gliogenesis.
Electronic supplementary material
The online version of this article (doi:10.1007/s00221-009-1963-6) contains supplementary material, which is available to authorized users.
Keywords: Superior colliculus, Protein tyrosine phosphatases, Real-time RT-PCR, Immunohistochemistry, Development
Introduction
The mouse retinocollicular system serves as an excellent model to explore the molecular and cellular mechanisms of axon growth and guidance as well as topographic map formation. Retinal ganglion cells (RGCs) differentiate within the retina at embryonic (E) day 11 (Cook 2003) and extend their axons toward the optic fissure. The first retinal axons travel out of the eye along the optic nerve and reach the optic chiasm at E14. After crossing, the retinal axons innervate their target, the superior colliculus, also known as the optic tectum in non-mammalian vertebrates, between E16 and postnatal (P) day 0 (Godement et al. 1984). Retinal inputs to the superior colliculus are organized in a precise topographic map, in which the temporal–nasal and dorsal–ventral axes of the retina correspond to the anterior–posterior and lateral–medial axes of the midbrain.
Extensive investigation of the developmental patterning of this topographic map has revealed gradients of transcription factors and cell surface receptors in the retina and tectum which guide its formation (Thanos and Mey 2001; O’Leary and McLaughlin 2005). Of interest here are the protein tyrosine phosphatases (PTPs), a large family of proteins that have been described to be implicated in axon growth and guidance (Garrity et al. 1999; Bixby 2000; Newsome et al. 2000; Stoker 2001) in different species. In RGC axons of Xenopus and chicken, RPTP-LAR, RPTPδ, RPTPμ and RPTPσ promote retinal neurite outgrowth (Burden-Gulley and Brady-Kalnay 1999; Ledig et al. 1999; Johnson et al. 2001), growth cone steering (Burden-Gulley et al. 2002) and targeting of retinal axons within the optic tectum (Rashid-Doubell et al. 2002). However, less is known regarding the potential role of PTPs in the development of the mammalian superior colliculus.
In order to identify PTPs that might contribute to signaling in the superior collicular development a degenerate primer-based reverse transcription polymerase chain reaction (PCR) approach was used to isolate cDNAs encoding PTPs from embryonic (E15) mouse superior colliculus. At this stage, neuronal and radial glial cells are generated (Gotz and Huttner 2005) as well as retinal ganglion axons first contact the superior colliculus. Using this approach seven different intracellular, non-transmembrane PTPs and nine different receptor-type PTPs (RPTPs) were identified. Subsequently, the expression pattern of 11 PTPs (TC-PTP, PTP1D, PTP1C, PTP-MEG2, PTP-PEST, RPTPJ, RPTPε, RPTPRR, RPTPκ, RPTPγ and RPTPσ) was analyzed in more detail in embryonic (E13, E15, E18) and postnatal (P0, P4, P12, and P20) superior colliculus by real-time RT-PCR, Western Blotting and immunohistochemistry. With ongoing maturation, all 11 PTPs displayed a distinct spatiotemporal regulation of mRNAs and proteins in the pre- and postnatal superior colliculus correlating with different processes such as proliferation, differentiation, axonal innervation and arborisation.
Methods
Animals
Adult NMRI mice were obtained from Charles River Laboratories (Sulzfeld, Germany) and mated over night. Females were checked for the presence of a vaginal plug, which corresponds to the gestational day 0.5 (E0.5). For all analyses, embryonic (E13, E15, E18) and postnatal (P0, P4, P12, P16, P20) stages were determined according to the staging criteria of Theiler (Bard et al. 1998).
RNA isolation and cDNA Synthesis
For RNA preparation, collicular tissue from each developmental stage was isolated, pooled, frozen in liquid nitrogen and stored at –70°C until RNA extraction. Total RNA was extracted according to the manufacturer’s instructions (RNeasy Mini or Midi kit, Qiagen, Hilden, Germany). Using a cDNA-synthesis kit (Fermentas GmbH, St. Leon-Rot, Germany) 1 μg of total RNA was used for reverse transcription.
PCR amplification of PTP sequences using degenerate primers
To produce PCR-generated DNA-fragments corresponding to PTP sequences in the conserved catalytic domain, cDNAs were reversely transcribed from E15 superior colliculus and were used as a template for the amplification with Taq polymerase (Eppendorf, Germany). Degenerate primers corresponding to amino acid sequences DFWQ(R/K/E/G)MI(M/V)WD(E/Q/H) (upstream) and HCSAGI(V/M)G (downstream) were synthesized by Invitrogen (Carlsbad, CA, USA). Low stringency PCR-reaction conditions were as follows: 5 min 94°C, followed by 36 cycles of 1 min at 94°C, 1 min at 50°C and 1 min at 72°C. The reaction products were run on 1.5% agarose gels, isolated, ligated into pCRII-TOPO Plasmids (Invitrogen) and used to transform competent E. coli TOP10 cells (Invitrogen, Carlsbad, CA, USA). Clones that contained inserts were sequenced using automated DNA sequencing (Department of Molecular Neurobiochemistry, Ruhr-University-Bochum). Obtained PCR fragment sequences were compared to sequences covered in the NCBI databases.
Quantitative real-time RT-PCR
Real-time-PCR using Syber Green I (Eurogentec) was performed on an Opticon-Cycler (MJ Research). Primer sequences of both housekeeping genes β-actin and cyclophilin and of the identified five intracellular PTPs and six RPTPs were designed (Horvat-Brocker et al. 2008). Their sequences, expected amplicon sizes and accession numbers are listed in Supplemental Table 1.
Primer concentration was optimized to a final concentration of 0.6 μM and combined with 20 ng RNA per well. Total three reactions per sample RNA (triplets) were set with a final volume of 20 μl per single reaction. Real-time PCR was performed as described previously (Ray et al. 2005; Horvat-Brocker et al. 2008). Briefly, each RT-PCR was performed from a pool of tissues originating from 15 different animals per each developmental stage. The average Ct values of three independent experiments (triplicates) were used to calculate the ratios for intracellular PTPs as described before (Pfaffl et al. 2002). In order to obtain amplification efficiencies of different primer sets, we generated standard curves by a twofold dilution series with template amounts ranging from 8 to 0.125 ng DNA per well. The efficiency of the PCR reaction was calculated for each primer pair according to the equation E = 10(−1/slope) (Pfaffl et al. 2002). The relative expression (R) of PTPs was calculated based on real-time PCR efficiency (E) and the threshold cycle (Ct) deviation of PTP at each developmental stage versus a control (embryonic stage 13, E13) according to the following equation: R = (Etarget)ΔCt target(MEAN control − MEAN sample)/(Ereference)ΔCt reference (MEAN control − MEAN sample). The Ct values for the reference genes (in our case β-actin and cyclophilin) were needed to take into account different mRNA levels of the sample. For statistical evaluation of Ct variations and calculated relative expression variations, data were analyzed for significant differences by a pair wise fixed reallocation and randomization test, as described before (Pfaffl et al. 2002).
Antibodies
The following primary antibodies were used for Western blot analyses and immunohistochemical stainings: rabbit anti-TC-PTP (Lammers et al. 1993), mouse anti-PTP1C (Santa Cruz Biotechnology, Santa Cruz, United States), rabbit anti-PTP1D (Stein-Gerlach et al. 1995; Tomic et al. 1995), rabbit anti-PTP-MEG2 (Sugen, Redwoodcity, CA, USA), rabbit anti-PTP-PEST (Eurogentec, Köln, Germany), rabbit anti-RPTPJ (Jallal et al. 1997) rabbit anti-RPTPκ (Fuchs et al. 1996; Anders et al. 2006), rabbit anti-RPTPγ (Horvat-Brocker et al. 2008), rabbit anti-RPTPε (Moller et al. 1995), rabbit anti-RPTPRR (Horvat-Brocker et al. 2008), rabbit anti-RPTPσ (Aicher et al. 1997), mouse anti-β-actin (Sigma, St. Louis, MO, USA), mouse anti-nestin (Chemicon, Temecula, CA, USA) and mouse anti-RC2 (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA, USA). The secondary antibodies used for Western blot analyses and immunohistochemical stainings were as follows: goat anti-mouse-CY3-coupled, goat anti-mouse-CY2-coupled, goat anti-rabbit-CY3-coupled, goat anti-rabbit-HRP-coupled and goat anti-mouse-HRP-coupled antibodies (all from Dianova, Hamburg, Germany).
Western blotting
For Western blotting, mouse collicular tissues from each developmental stage (E13, E15, E18, P0, P4, P12, P16 and P20) were isolated, pooled and immediately homogenized in ice-cold lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 5 mM EGTA, 10% v/v glycerin, 1% v/v Triton X-100, 0.1 mM Na3VO4, 10 μg/ml aprotinin, 10 μg/ml leupeptine, 1 mM phenylmethylsulfonyl fluoride). Lysates were clarified by centrifugation at 12,000×g for 20 min at 4°C. The protein concentration was determined using a BCA Protein Assay Kit (Pierce, Rockford, USA). 20 μg of each protein lysate was separated on 4–10% SDS-polyacrylamide gels (Roth, Karlsruhe, Germany). Immunoblotting and immunodetection were performed as described previously (Horvat-Brocker et al. 2008) using the following primary PTP-antibodies: rabbit anti-TC-PTP (1:2,000), mouse anti-PTP1C (1:2,000), rabbit anti-PTP1D (1:2,000), rabbit anti-PTP-MEG2 (1:1,000), rabbit anti-PTP-PEST (1:2,000), rabbit anti-RPTPJ (1:1,000), rabbit anti-RPTPκ (1:1,000), rabbit anti-RPTPγ (1:1,000), rabbit anti-RPTPε (1:1,000), rabbit anti-RPTPRR (1:1,000) and rabbit anti-RPTPσ (1:1,000). Immunoreactivities were detected using goat anti-rabbit-HRP-coupled or anti-mouse-HRP-coupled secondary antibodies (1:12000). Protein bands were detected using enhanced chemiluminescence ECL (Roth). After stripping with Restore Western blot Stripping Buffer (Pierce) all blots were reprobed using mouse anti-β-actin antibody (Sigma, 1:5,000), which indicates nearby equal amounts of proteins per each analysed developmental stage. Protein levels were quantified by using image J analysis software (Version 1.41, National Institutes of Health, USA). The relative expression values (normalized protein levels) of PTP-proteins were calculated based on β-actin signals and the expression level at E13 (set to 0).
Immunohistochemistry and confocal laser-scanning microscopy
For immunohistochemistry, whole embryo heads or postnatal brains were fixed in 4% w/v paraformaldehyde (PFA) in phosphate buffered saline (PBS) at 4°C and cryoprotected in 30% w/v sucrose/PBS. Tissue was sectioned in sagittal planes at 16 μm using a cryostat (Leica, Bensheim, Germany) and collected onto Superfrost plus object slides (Menzel-Glaeser, Braunschweig, Germany). Slides were stored at −80°C until required. Immunohistochemical labeling was carried out as described previously (Horvat-Brocker et al. 2008) by using the following primary antibodies: PTP-antibodies (all 1:500), anti-nestin (1:300) and anti-RC-2 (1:10). Adequate secondary antibodies were as follows: subclass-specific CY3-coupled anti-rabbit and anti-mouse antibodies (all 1:300, Dianova, Hamburg, Germany). Control slides without primary antibodies were immunonegative. All immunofluorescence stainings were analyzed by using a confocal laser-scanning microscope LSM 510 META (Zeiss, Göttingen, Germany). Laser lines and emission filters were optimized with the Zeiss LSM Image Browser software. Brightness and contrast of the images were adjusted using Adobe Photoshop software (version 8.0.1; Adobe Systems, Mountain View, CA, USA).
Results
Identification of PTPs expressed in the mouse superior colliculus at embryonic stage 15 (E15) by degenerate primer RT-PCR
In order to identify PTPs that might contribute to the signaling in the embryonic (E15) mouse superior colliculus, a degenerate primer-based RT-PCR approach was used. At E15, retinal ganglion axons start to innervate the superior colliculus (Edwards et al. 1986; Herrera et al. 2003) and intensive genesis processes (differentiation, proliferation, migration) of neuronal and radial glial cells occur (Altman and Bayer 1981; Edwards et al. 1986; Galileo et al. 1990). The primers used in this study were synthesized based on the catalytic domains of PTPs (DFWQ(R/K/E/G)MI(M/V)WD(E/Q/H) (upstream) and HCSAGIG (downstream)). Single PCR products were isolated and ligated into a pCRII-TOPO Vector (Invitrogen). Three hundred and ten clones were sequenced, and fragments containing PTP-like domains were identified. These 310 cDNA-fragments encoded 16 different PTPs in the mouse superior colliculus at E15. Novel PTP sequences were not found. From a total of 310 clones 50 clones (16%) coded for seven different intracellular PTPs, whereas 260 clones (84%) coded for nine different RPTPs. The relative abundance of different intracellular PTPs and RPTPs among the RT-PCR-generated cDNAs is shown in Table 1. The four most abundant cDNAs were RPTPσ (158 clones), RPTP-LAR (62 clones), TC-PTP (23 clones) and PTP1B (17 clones). The fifth most abundant cDNAs (12 clones) encoded RPTPJ. Nine cDNAs encoded RPTPRR and RPTPε, while five cDNAs encoded RPTPδ. RPTPα and PTP-MEG2 were represented by three clones each. For the intracellular protein tyrosine phosphatases PTP36, PTP1C and PTP1D each two clones were identified. RPTPκ, RPTPγ and PTP-PEST were represented by only one single clone each.
Table 1.
Cloning and identification of intracellular and receptor-type protein tyrosine phosphatases (PTPs) from embryonic E15 mouse superior colliculus using a degenerate primer-based RT-PCR approach
| PTPs | No. of PTP clones = 310 (%) |
|---|---|
| RPTPσ | 158 (51) |
| RPTP-LAR | 62 (20) |
| TC-PTP | 23 (7.4) |
| PTP1B | 17 (5.5) |
| RPTPJ | 12 (4) |
| RPTPRR | 9 (3) |
| RPTPε | 9 (3) |
| RPTPδ | 5 (1.6) |
| RPTPα | 3 (0.9) |
| PTP-MEG2 | 3 (0.9) |
| PTP36 | 2 (0.6) |
| PTP1C | 2 (0.6) |
| PTP1D | 2 (0.6) |
| RPTPκ | 1 (0.3) |
| RPTPγ | 1 (0.3) |
| PTP-PEST | 1 (0.3) |
The reamplified intracellular PTPs and receptor-type protein tyrosine phosphatases (RPTPs) cloned by RT-PCR were ranked by the relative abundance of the PCR products. The seven different intracellular PTPs (TC-PTP, PTP1B, PTP-MEG2, PTP36, PTP1C, PTP1D and PTP-PEST) recovered in the E15 mouse superior colliculus screen represented 50 (16%) from a total of 310 clones. The nine different RPTPs (RPTPσ, RPTP-LAR, RPTPJ, RPTPRR, RPTPε, RPTPδ, RPTPα, RPTPκ and RPTPβ/ζ) were represented by 260 clones (84%)
Quantitative real-time RT-PCR and Western blotting revealed differential regulation of PTP mRNAs and proteins during development of mouse superior colliculus
Since antibodies for all PTPs, which were identified with degenerate primers in the E15 mouse superior colliculus, were not available in this study, we focused on the 11 PTPs: five intracellular PTPs (TC-PTP, PTP1C, PTP1D, PTP-MEG2 and PTP-PEST) and six receptor-type PTPs (RPTPJ, RPTPκ, RPTPγ, RPTPε, RPTPRR and RPTPσ). To study the role of these PTPs in the developing mouse superior colliculus in detail, the regulation of the corresponding mRNAs and proteins was first investigated by quantitative real-time RT-PCR and Western blotting at different developmental time points (E13, E15, E18, P0, P4, P8, P12, P16 and P20). The embryonic and postnatal stages from E13 to P12 cover different developmental processes including genesis of neuronal and glial cells, formation of synapses and retinocollicular topographic mapping. Sequences of primer pairs used in real-time RT-PCR experiments are shown in Table 1 (Supplemental data).
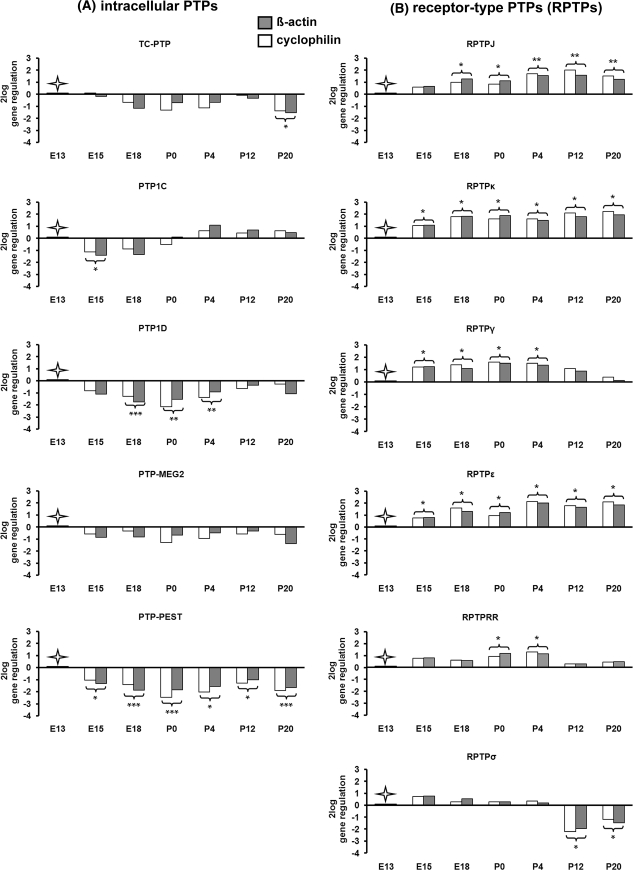
The results of the quantitative real-time RT-PCRs revealed equal amounts of mRNAs (Ct values = mean threshold cycle) for the reference genes ß-actin and cyclophilin during collicular development (Supplemental data, Table 2). At E13, the Ct values for the investigated PTPs varied between 22 and 29, indicating different expression levels of the PTPs at this early developmental stage (Supplemental data, Table 2). With ongoing development of the superior colliculus, the PTPs showed differential mRNA patterns when compared to the expression level at E13 (set to 0) (Fig. 1a, b, Supplemental data, Table 2).
Fig. 1.
Temporal regulation of intracellular and receptor-type PTP (RPTP) mRNAs as revealed by quantitative real time RT-PCR in the developing mouse superior colliculus. The relative gene regulation of TC-PTP, PTP1C, PTP1D, PTP-MEG2, PTP-PEST, RPTPJ, RPTPκ, RPTPγ, RPTPε, RPTPRR and RPTPσ (R = here displayed as 2log) was calculated based on real-time PCR efficiency (E) and the threshold cycle (Ct) deviation of PTPs at each developmental stage (E15, E18, P0, P4, P12 and P20) versus a control (embryonic stage 13, E13) according to the following equation: R = (Etarget)ΔCt target(MEAN control − MEAN sample)/(Ereference)ΔCt reference (MEAN control − MEAN sample). All values represent the relative expression ratio of PTPs in relation to each house keeping gene (reference) (β-actin and cyclophilin) and were normalized to the expression level at E13 ( control, set to 0). *P < 0.05; **P < 0.01; ***P < 0.001
control, set to 0). *P < 0.05; **P < 0.01; ***P < 0.001
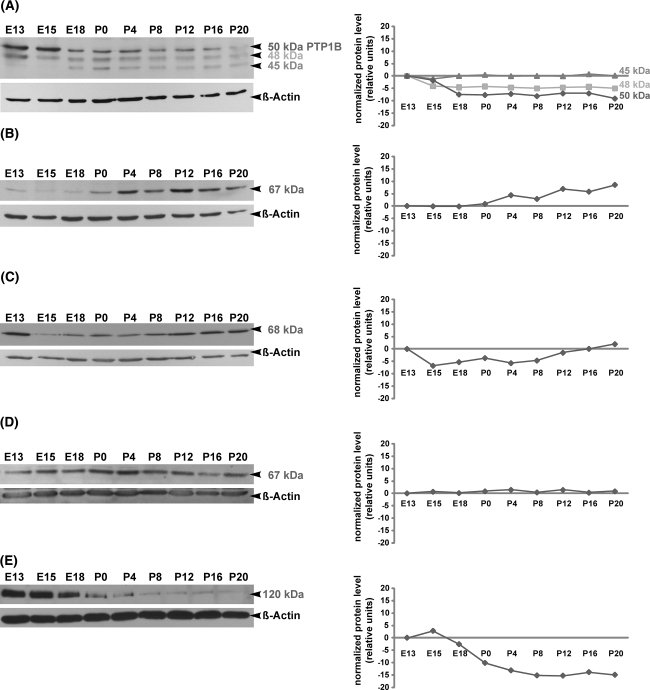
In comparison to E13, the expression of TC-PTP mRNA decreased slightly during collicular development and reached a significant (P < 0.05) 3(21.5)-fold downregulation at P20 (Fig. 1a). In Western Blot analysis, TC-PTP antibody recognized three protein bands at 45-, 48- and ~50 kDa (Fig. 2a). The 48 and 45 kDa bands correspond to the previously described two TC-PTP human isoforms (Tiganis et al. 1998, 1999; Lam et al. 2001). The 50 kDa band corresponds probably to the intracellular protein tyrosine phosphatase PTP1B, which shows more than 70% homology to TC-PTP (Ibarra-Sanchez et al. 2000; Bourdeau et al. 2005). During development of the superior colliculus only the 48 kDa TC-PTP isoform revealed almost the same regulation pattern as TC-PTP mRNA, whereas the 45 kDa isoform was uniformly expressed. Strong signals of the 50 kDa protein, corresponding probably to PTP1B, were detected at E13 and E15 before dropping between E18 and P16. At P20 a faint 50 kDa band was detected. However, it has to be mentioned that primer pairs for TC-PTP amplification in real-time RT-PCR experiments did not amplify PTP1B.
Fig. 2.
Temporal regulation of intracellular PTP proteins as revealed by Western blot analysis in the developing mouse superior colliculus. Protein lysates of superior colliculus tissues from E13, E15, E18, P0, P4, P8, P12, P16 and P20 were separated by SDS-PAGE, and immunoblotted with TC-PTP (a), PTP1C (b), PTP1D (c), PTP-MEG2 (d) and PTP-PEST (e) antibodies, respectively. Molecular weights of intracellular PTP proteins are indicated on the right. All blots were reprobed for β-actin to demonstrate the equal protein loading. Changes in the relative protein expression (normalized protein level) of each PTP protein are shown in the graphical representations. In the case of PTP1C, PTP1D, PTP-MEG2 and PTP-PEST each PTP antibody recognized only one band, which corresponded to the predicted molecular weights of these PTPs. However, the TC-PTP antibody recognized three protein bands at 45, 48 and ~50 kDa (Fig. 2a). The 48 kDa and 45 kDa bands correspond to the previously described two TC-PTP human isoforms, whereas the 50 kDa band corresponds most probably to the intracellular protein tyrosine phosphatase PTP1B. Note that proteins of the investigated intracellular PTPs are differentially regulated during superior collicular development
When compared with E13, the quantification of PTP1C-mRNA using real-time RT-PCR revealed a slight downregulation at E15 (P < 0.05) and E18 (Fig. 1a). At P0 the PTP1C mRNA expression returned to the level found at E13. From P4 to P20, a slight upregulation of the gene was observed (statistically not significant). A similar expression pattern was found at protein level, as revealed in the immunoblot with specific anti-PTP1C antibody, which recognized a single protein band at 67 kDa (Fig. 2b). This band represents the full PTP1C protein as calculated from the amino acid sequence.
The quantification of PTP1D mRNA using real-time RT-PCR revealed a 2(21)-4(22)-fold decrease in the expression at the late embryonic stage E18 (P < 0.001) and the early postnatal stages P0 and P4 (P < 0.01) when compared to E13 (Fig. 1a). From P12 to P20, PTP1D mRNA expression returned to the level found at E13. Western Blot analysis revealed a similar expression pattern (Fig. 2c). Using a specific PTP1D-antibody, a single 68 kDa band corresponding to the predicted molecular weight of PTP1D was detected. A large amount of 68 kDa protein was found at E13 before dropping between E15 and P8. The signal intensity of 68 kDa protein increased in the later postnatal stages (from P12 to P20).
The MEG2 mRNA and protein revealed constant expression levels from E13 to P20 (Fig. 1a). In Western Blot analysis, a specific PTP-MEG2 antibody recognized a single protein band at 67 kDa (Fig. 2d), corresponding to the predicted size.
In contrast to other intracellular PTPs, the PTP-PEST mRNA expression was characterized through a continuous 2(21)-5(22.5)-fold decrease in superior colliculus development (P < 0.05 at E15, P4 and P12; P < 0.001 at E18, P0 and P20, Fig. 1a). Note that the regulation of the PTP-PEST protein, which was detected at ~120 kDa using Western blot analyses (Fig. 2e), correlates with the mRNA expression as detected by quantitative real-time RT-PCR. The predicted molecular weight of the murine PTP-PEST is 86 kDa. As revealed by other groups (Davidson et al. 1997; Sirois et al. 2006; Halle et al. 2007) and as shown by our analyses, PTP-PEST migrates in the range of 120 kDa.
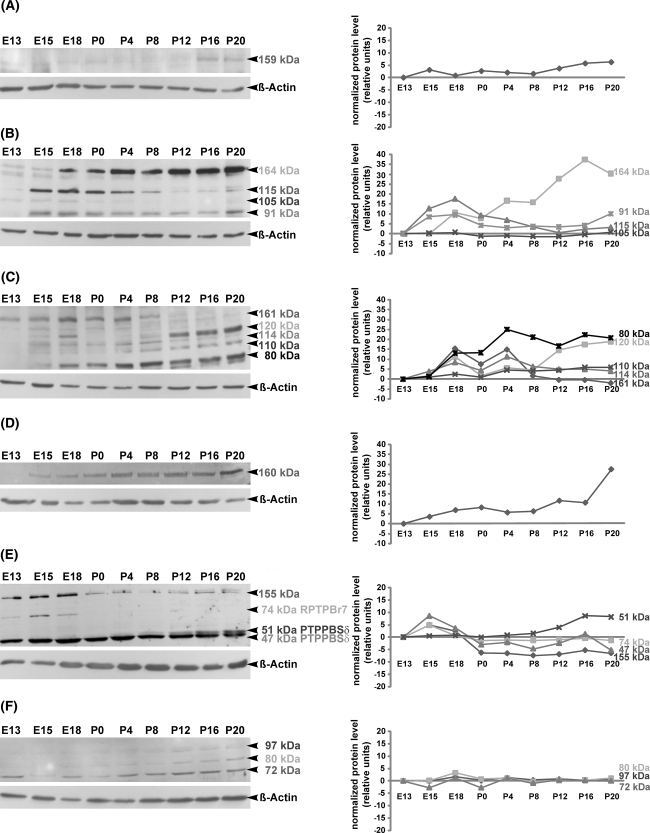
When compared with E13, the mRNA regulation of the receptor protein tyrosine phosphatase RPTPJ was characterized through a 2(21)-4(22)-fold increase in the expression from late embryonic stage E18 until P20 (at E18 and P0, P < 0.05; at P4, P12 and P20, P < 0.01) (Fig. 1b). Nearby the same expression pattern was found at the protein level, as revealed in the immunoblot with specific anti-RPTPJ antibody, which recognized a 159 kDa band (Fig. 3a). Although the calculated molecular weight of RPTPJ is 134 kDa (Kuramochi et al. 1996), our data indicate that during collicular development RPTPJ might be glycosylated.
Fig. 3.
Temporal regulation of RPTP proteins as revealed by Western blot analysis in the developing mouse superior colliculus. Protein lysates of superior colliculus tissue from E13, E15, E18, P0, P4, P8, P12, P16 and P20 were separated by SDS-PAGE and immunoblotted with RPTPJ (a), RPTPκ (b), RPTPγ (c), RPTPε (d), RPTPRR (e) and RPTPσ (f) antibodies, respectively. Molecular weights of RPTP proteins are indicated on the right. All blots were reprobed for β-actin to demonstrate the equal protein loading. Changes in the relative protein expression (normalized protein level) of each RPTP protein are shown in the graphical representations. RPTPJ (a) and RPTPε (d) proteins are detected with appropriate antibodies each as single bands at 159 and 160 kDa, respectively. Immunoblots of RPTPκ (b), RPTPγ (c) and RPTPσ (f) reveal several bands, which correspond to the integral proteins and their derived cleavage products. The anti-RPTPκ antibody detects four bands: 164 kDa (full protein) and its 115, 105 and 91 kDa cleavage products (b). The anti-RPTPγ antibody recognizes five bands (c): 161 kDa (full protein) and its 120, 114, 110 and 80 kDa cleavage products. As shown in (f), the antibody against RPTPσ recognizes only the cleavage products at 97, 80 and 72 kDa, whereas the full protein RPTPσ (168 kDa) could not be detected. RPTPRR antibody detects three bands, which correspond to the alternatively spliced receptor-like isoforms of the RPTPRR gene (74 kDa RPTPBr7, 51 kDa PTPPBSδ (+) and 47 kDa PTPPBSδ (−)). An additional 155 kDa band detected by anti-RPTPRR might represent the glycosylated form of one of the receptor-like isoforms (e). Note that the investigated RPTPs and their products (either cleavage products or alternatively spliced isoforms) are differentially regulated during collicular development, providing an evidence for differential regulation at the level of posttranslational modification through proteolytic cleavage or alternative splicing
The quantification of RPTPκ mRNA using real-time RT-PCR revealed a 2(21)- to 4(22)-fold increase of the expression level from E15 to P20 (P < 0.05) in comparison with E13 (Fig. 1b). Western blot analysis using specific anti-RPTPκ antibody (Fig. 3b) resulted in the detection of four bands of 164, 115, 105 and 91 kDa. The 164 kDa band represents the full RPTPκ protein (Jiang et al. 1993), while the 115-, 105- and 91 kDa bands correspond to the proteolytic cleavage products of RPTPκ (Anders et al. 2006). Weak signals of the full RPTPκ protein (164 kDa) were detected at E13 and E15 before being gradually upregulated between E18 and P20. At E13, the 115-, 105- and 91 kDa proteins were weakly expressed. A strong upregulation of the 115 kDa protein was detected between E15 and P8 prior to its gradual downregulation between P12 and P20. The 105 kDa protein band was weakly but nearby constantly present during collicular development. In contrast, the 91 kDa protein was prominently detectable at E15 and E18 and remained constantly present during postnatal collicular development until P20. Taken together, only the complete RPTPκ protein (164 kDa) and the 91 kDa protein revealed the same regulation pattern as RPTPκ mRNA.
When compared with E13, the RPTPγ mRNA revealed a significant increase 2(21)-fold to 3(21.5) of the expression level from E15 to P4 (P < 0.05). At P20 the expression returned to the level found at E13 (Fig. 1b). In Western blot analysis, the anti-RPTPγ antibody recognized five bands of 161, 120, 114, 110 and 80 kDa (Fig. 3c). The 161 kDa band corresponds to the predicted size deduced from the amino acid sequences (Barnea et al. 1993; Shintani and Marunaka 1996). The 120, 114, 110 and 80 kDa bands might represent the cleaved products of RPTPγ (van Niekerk and Poels 1999). The full RPTPγ protein (161 kDa) showed the same regulation pattern as RPTPγ mRNA. The large amount of 120 kDa protein was first detected in later postnatal stages (from P12 onward). A constantly weak signal from E15 onward was detected for the 114 kDa band. The small amount of the 110 kDa protein was detected between E15 and P0 before increasing between P4 and P20. The amount of the 80 kDa protein was almost undetectable until E15 and started to increase from E18 to P20.
The quantification of RPTPε mRNA revealed a 2(21)-fold to 4(22) constant upregulation from E15 onward (P < 0.05) when compared with E13 (Fig. 1b). A similar regulation pattern of RPTPε was detected at the protein level (Fig. 3d). The RPTPε antibody (Moller et al. 1995) recognized a 160 kDa band that became gradually upregulated with the maturation of the superior colliculus. The observed 160 kDa band corresponds to the highly glycosylated form of RPTPε, which has been reported in the mouse brain (Elson and Leder 1995).
Since several transmembrane and cytoplasmatic isoforms of the RPTPRR gene are known (Augustine et al. 2000a; Chirivi et al. 2004), primer pairs which span the sequence between the extracellular and intracellular part of the transmembrane RPTPBr7 isoform were used in the quantitative real-time-PCR (Horvat-Brocker et al. 2008). When compared with E13, a significant 2-(21)-fold upregulation of RPTPRR/RPTPBr7 mRNA was found at P0 and P4 (P < 0.05), while in other embryonic and postnatal stages the expression level was almost the same as at E13 (Fig. 1b). The anti-RPTPRR antibody, which was raised against the intracellular domain (Horvat-Brocker et al. 2008), recognized in Western blot four bands: 155-, 74-, 51- and 47 kDa (Fig. 3e). The 74-, 51- and 47 kDa proteins correspond to the receptor like-RPTPRR isoforms previously identified as RPTPBr7, RPTPPBSδ (+) and RPTPPBSδ (−) (Ogata et al. 1995; Sharma and Lombroso 1995; Augustine et al. 2000b), which are the products of alternative splicing of the RPTPRR gene (Augustine et al. 2000b; Chirivi et al. 2004). The 155 kDa band might represent a glycosylated form of one of the receptor-like isoforms. The strong signals of the 155 kDa protein, which was detected in all embryonic stages (from E13 to E18), decreased sharply at the day of birth (P0) and remained weakly expressed during the whole postnatal period. The RPTPBr7 protein (74 kDa) was detected only from E13 to E18, whereas no band could be detected from P0 onward. During collicular development, the RPTPPBSδ (−) protein appeared slightly downregulated, whereas the RPTPPBSδ (+) isoform was first identified at P4 and reached the maximum level at late postnatal stages (P16 and P20).
In contrast to the other investigated RPTPs, the RPTPσ mRNA showed a slight but significant 4-(22)- and 2-(21)-fold downregulation at P12 (P < 0.05) and P20 (P < 0.05), respectively (Fig. 1b). As shown in Fig. 3f, an antibody against RPTPσ (Aicher et al. 1997) recognized three proteins of 97-, 80- and 72 kDa. These bands represent three proteolytically processed products of RPTPσ as described before (Aicher et al. 1997). The 97- and 80 kDa bands were almost undetectable in the embryonic (E13 to E18) and early postnatal stages. In contrast, the 72 kDa protein band was firstly detected at E13. Taken together, all three proteins showed an overlapping and nearby constant pattern of regulation.
Spatiotemporal expression pattern of PTPs in the embryonic E13 and postnatal P20 mouse superior colliculus as revealed by immunohistochemistry
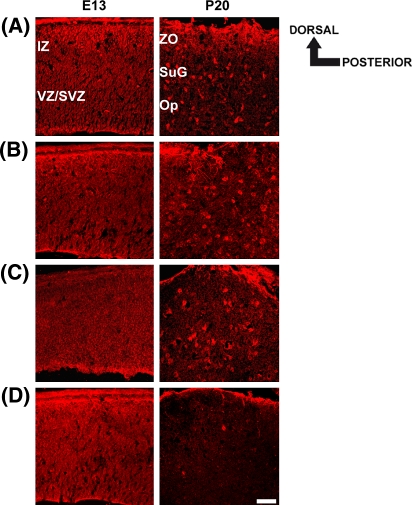
To determine the spatial and temporal localization of PTP1C, PTP1D, PTP-MEG2, PTP-PEST, RPTPJ, RPTPκ, RPTPγ, RPTPε, RPTPRR and RPTPσ during development of the mouse superior colliculus, immunohistochemical analyses were performed at the early embryonic stage E13 and the late postnatal stage P20 (Figs. 4a–d, 5a–f). Due to the cross-reactivity of the anti-TC-PTP antibody, which also recognized PTP1B protein (Fig. 2a), no immunhistochemical analysis was performed for this PTP. For the other PTPs, appropriate antibodies already used in Western blot analyses (Figs. 2b–e, 3a–f) were applied to sagittal sections of the mouse superior colliculus. The specificity of the immunhistochemical signals was determined in control experiments using species-specific secondary CY3-labeled antibodies. In these control experiments, no signals were detected (data not shown). With the exception of RPTPJ, that displayed a decreasing anterior–posterior protein gradient (insert, Fig. 5a), the examination of the superior colliculus in both anteroposterior and dorsoventral orientation revealed no significant protein expression gradients of the investigated PTPs (Figs. 4a–d, 5b–f).
Fig. 4.
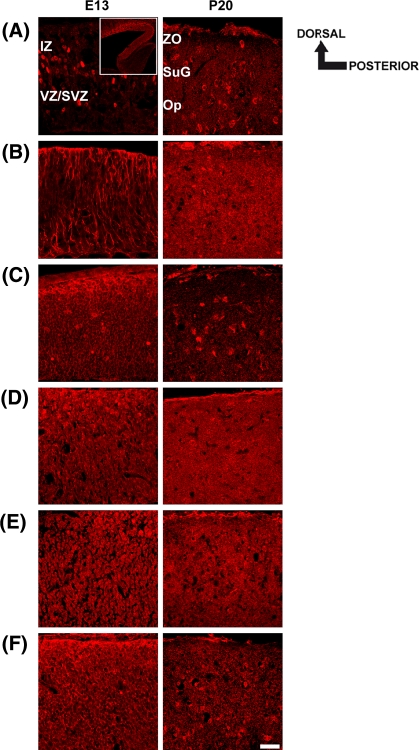
Spatiotemporal expression pattern of PTP1C (a), PTP1D (b), PTP-MEG2 (c) and PTP-PEST (d) in the embryonic (E13) and postnatal (P20) mouse superior colliculus. Sagittal sections through the embryonic (E13) and late postnatal (P20) superior colliculus were labeled with appropriate intracellular PTP antibodies. Dorsal and posterior orientation is schematically presented. a–d At E13, immunofluorescent staining using antibodies against PTP1C, PTP1D, PTP-MEG2 and PTP-PEST reveals a more uniform distribution throughout the whole superior colliculus. (b–d) The strongest signals for PTP1C, PTP-1D and PTP-PEST were obtained in the outer dorsal edge of the intermediate zone (IZ). a–d In addition, a prominent immunoreactivity for all intracellular PTPs was found in the ventricular zone (VZ) of the embryonic superior colliculus. a–c At P20, PTP1C-, PTP1D- and PTP-MEG2-immunoreactivities were found in single cells scattered throughout the stratum griseum superficiale (SuG) and the stratum opticum (Op) of the superior colliculus. d In contrast, no immunoreactivity was found for PTP-PEST at P20. Here, rabbit non-immune serum labels only the pial surface non-specifically. IZ intermediate zone, VZ/SVZ ventricular zone/subventricular zone, ZO stratum zonale, SuG stratum griseum superficiale, Op stratum opticum. Scale bar 50 μm
Fig. 5.
Spatiotemporal expression pattern of RPTPJ (a), RPTPκ (b), RPTPγ (c), RPTPε (d), RPTPRR (e) and RPTPσ (f) in the embryonic (E13) and mature (P20) mouse superior colliculus. Sagittal sections through the embryonic (E13) and late postnatal superior colliculus (P20) were labeled with appropriate RPTP-antibodies. Dorsal and posterior orientation is schematically presented. a At E13, a polyclonal antibody against RPTPJ detects single immunoreactive cells in the subventricular zone (SVZ) of the embryonic superior colliculus. In addition we found RPTPJ expressing cells in a decreasing anterior–posterior gradient (insert, Fig. 5a). Insert: lower magnification of a RPTPκ stained sagittal section of the E13 superior colliculus (scale bar = 200 μm). In the P20 superior colliculus, single cells, scattered throughout the stratum griseum superficiale (SuG) and stratum opticum (SO), were labeled. b The RPTPκ epitope is located on radial processes in the embryonic superior colliculus, whereas in the P20 superior colliculus a broad RPTPκ-immunoreactivity was found within all visual layers. (c, d and f) The strongest RPTPγ-, RPTPε- and RPTPσ-immunoreactivities were found in the dorsal part of the E13 superior colliculus (IZ intermediate zone). At P20, RPTPγ and RPTPσ immunoreactive cells were restricted to the stratum griseum superficiale (SuG) and stratum opticum (SO), whereas RPTPε immunoreactivity displays a more dispersed distribution pattern. e Immunolabeling using a polyclonal RPTPRR antibody reveals a weak and uniform staining pattern in the embryonic as well as in the P20 superior colliculus. IZ intermediate zone, VZ/SVZ ventricular zone/subventricular zone, ZO stratum zonale, SuG stratum griseum superficiale, Op stratum opticum. Scale bar 50 μm
At E13, immunolabeling using antibodies for the intracellular PTPs PTP1C, PTP1D, PTP-MEG2 and PTP-PEST revealed a more uniform distribution throughout the whole embryonic superior colliculus. Nevertheless, prominent immunoreactivities for PTP1C, PTP1D and PTP-PEST were found in the outer dorsal edge of the intermediate zone (IZ) of the superior colliculus (Fig. 4a, b and d). In addition, all intracellular PTPs displayed stronger immunoreactivities in the ventricular zone (VZ) (Fig. 4a–d). At P20, PTP1C-, PTP1D- and PTP-MEG2-immunoreactive cells were found dispersed throughout the stratum griseum superficiale (SuG) and the stratum opticum (Op) of the superior colliculus (Fig. 4a, b and c). In contrast, no PTP-PEST immunoreactivity was detected at P20 (Fig. 4d). Here, rabbit non-immune serum labels only the pial surface non-specifically.
In comparison to the intracellular PTPs, the receptor-type PTPs RPTPJ and RPTPκ revealed unique immunostainings at E13. The RPTPJ-antibody strongly labeled cells in the ventricular zone (VZ) and subventricular zone (SVZ) of the embryonic superior colliculus (Fig. 5a), whereas the RPTPκ epitope was located on radial processes that span the whole tissue (Fig. 5b). The most prominent RPTPγ- (Fig. 5c), RPTPε- (Fig. 5d) and RPTPσ-immunoreactivities (Fig. 5f) were found in the intermediate zone (IZ) of the E13 superior colliculus. In comparison to the other RPTPs, RPTPRR immunoreactivity was found uniformly distributed throughout the E13 superior colliculus (Fig. 5e). At P20, single RPTPJ- (Fig. 5a), RPTPγ- (Fig. 5c) and RPTPσ-immunoreactive cells (Fig. 5f) seemed to be restricted to the stratum griseum superficiale (SuG) and stratum opticum (Op), whereas immunofluorescence signals of RPTPκ (Fig. 5b), RPTPε (Fig. 5d) and RPTPRR (Fig. 5e) revealed a more dispersed staining pattern within all visual layers of the P20 superior colliculus.
Cell-type specific expression of PTPs in collicular neural progenitors and radial glia cells at E13
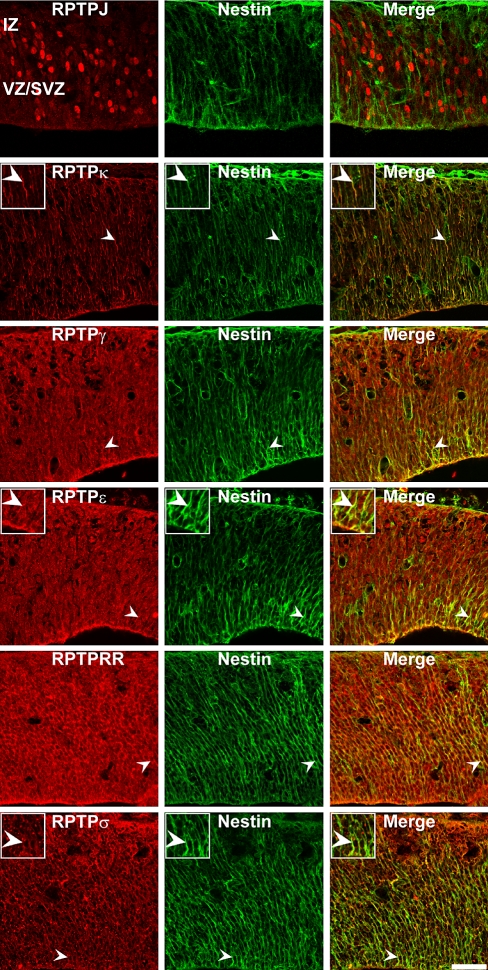
Based on our observations of a prominent PTP expression in the early embryonic superior colliculus at E13, a period of intense neurogenesis and gliogenesis, we hypothesized a possible involvement of PTP proteins in the regulation of early collicular neural progenitors and radial glia cells. To determine whether PTPs are expressed by these specific cell types, we performed double-immunohistochemistry on E13 superior colliculus slides using antibodies against PTPs and two different markers of early neuronal precursor cells, nestin, and a subtype of radial glia cells, RC-2 (Figs. 6–9). Within the superior colliculus, both nestin- and RC-2 antibodies strongly labeled radially oriented processes, which span the whole tissue. The most prominent immunoreactivities for nestin as well as for RC-2 positive cells/processes were found in close association with the subventricular and ventricular zones. In comparison to the observed nestin-immunoreactivity, decreased label intensity from the subventricular/ventricular zone (SVZ/VZ) to the intermediate zone (IZ) was found for RC-2.
Fig. 6.
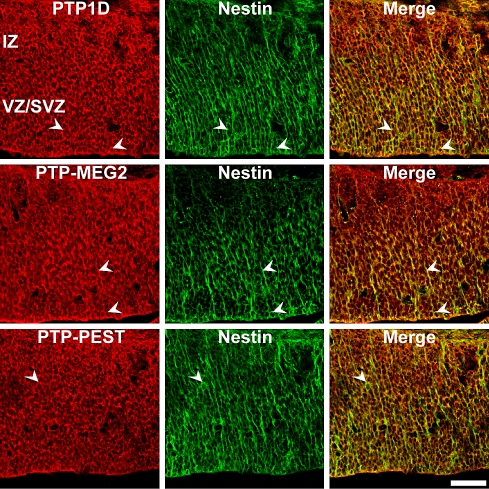
Co-localization of PTP1D, PTP-MEG2 and PTP-PEST proteins in nestin-positive neural progenitor cells in the E13 mouse superior colliculus. Sagittal sections through the embryonic E13 superior colliculus were double-labeled with antibodies against PTPs and the neural progenitor marker nestin. Arrowheads indicate examples of double-labeled cells. PTP1D, PTP-MEG2 and PTP-PEST epitopes partially co-localize with nestin-positive neural progenitor cells. Because the nestin- and PTP1C antibodies originated from the same species (both mouse, IgG), it was not possible to analyse the co-localization of the PTP1C protein with nestin-positive precursor cells. Note that the antibodies directed at intracellular PTPs label mainly nuclei/perikarya of collicular cells; both markers reveal an incomplete overlapping expression pattern (merge). IZ intermediate zone, VZ/SVZ ventricular zone/subventricular zone. Scale bar 50 μm
Fig. 9.
Co-localization of RPTPJ, RPTPκ, RPTPγ, RPTPε, RPTPRR and RPTPσ proteins in RC-2-positive radial glia cells in the E13 mouse superior colliculus. Sagittal sections through the embryonic E13 superior colliculus were double-labeled with antibodies against RPTPs and the radial glia marker RC-2. Arrowheads indicate examples of double-labeled cells. RC-2 is restricted to radial glia processes in the superior colliculus, and only RPTPκ immunoreactivity completely overlaps with radial fibers (merge). RPTPJ, RPTPγ, RPTPε, RPTPRR, RPTPσ immunoreactivities were mainly restricted to the nuclei/perikarya of collicular radial glia. Nevertheless, a partial overlap was observed for RPTPγ, RPTPε, RPTPRR and RPTPσ, whereas RPTPJ positive nuclei showed no association with RC-2 positive fibers. IZ intermediate zone, VZ/SVZ ventricular zone/subventricular zone. Inserts: Higher magnification of immunopositive cells (merge figures represent co-immunoreactive cells). Scale bar 50 μm
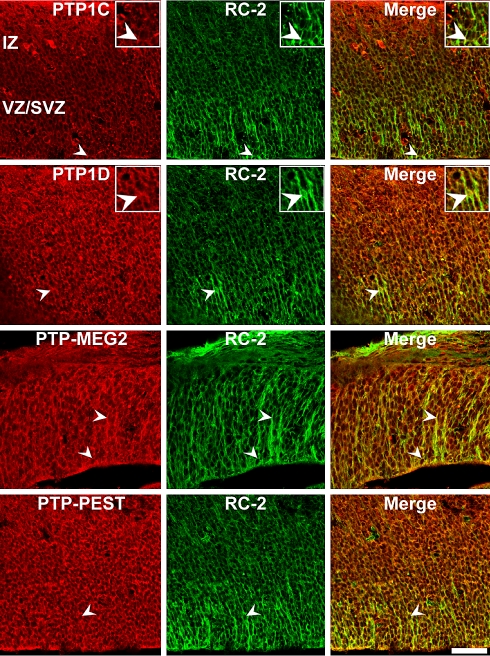
Double-immunofluorescence analysis revealed that the immunoreactivity for the intracellular PTPs PTP1D, PTP-MEG2 and PTP-PEST colocalized partially with the precursor cell marker nestin (Fig. 6) and the radial glia cell marker RC-2 (Fig. 8). The most prominent signals were observed in the VZ and SVZ. PTP1C immunoreactivity, which was found most prominently in the outer edge of the intermediate zone and in single cells of the ventricular zone, showed reduced co-localization with RC-2 positive cells in the ventricular zone and did not co-localize with minor labeled cells in the intermediate zone. Considering the species of the nestin and PTP1C antibodies (both mouse, IgG) it was not possible to determine the co-localization of the PTP1C protein with nestin-positive precursor cells. We speculate that the overlap should be minor, if any. This interpretation is in accordance with the PTP1C-immunoreactivity, which was mostly found in more mature cells of the IZ.
Fig. 8.
Co-localization of PTP1C, PTP1D, PTP-MEG2 and PTP-PEST proteins in RC-2-positive radial glia cells in the E13 mouse superior colliculus. Sagittal sections through the embryonic E13 superior colliculus were double-labeled with antibodies against intracellular PTPs and the radial glia marker RC-2. Arrowheads indicate examples of double-labeled cells. PTP1D, PTP-MEG2 and PTP-PEST epitopes partially co-localize with RC-2-positive radial glia cells. In contrast, PTP1C immunoreactivity shows a less pronounced co-localization with RC-2 positive cells in the ventricular zone and does not co-localize with minor RC-2-positive cells in the intermediate zone. Note that the antibodies directed at intracellular PTPs label mainly nuclei/perikarya of collicular cells; both markers reveal an incomplete overlapping expression pattern (merge). IZ intermediate zone, VZ/SVZ ventricular zone/subventricular zone. Inserts: Higher magnification of immunopositive cells (merge figures represent co-immunoreactive cells). Scale bar 50 μm
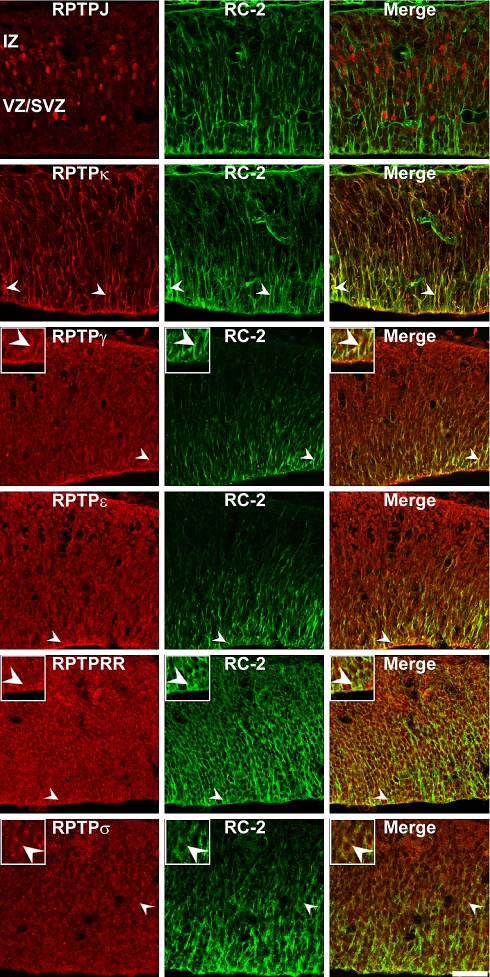
In addition, no co-localization with nestin-positive progenitor cells (Fig. 7) and RC-2 positive radial glia cells (Fig. 9) was observed for the receptor-type PTP RPTPJ. In contrast, the RPTPκ epitope was strongly co-expressed by nestin- and by RC-2-positive cells, as revealed by a nearby completely overlapping expression pattern (Figs. 7, 9). RPTPγ, RPTPε, RPTPRR and RPTPσ were partially expressed in nestin- and RC-2- double-positive cells/processes (Figs. 7, 9).
Fig. 7.
Co-localization of RPTPJ, RPTPκ, RPTPγ, RPTPε, RPTPRR and RPTPσ proteins in nestin-positive neural progenitor cells in the E13 mouse superiors colliculus. Sagittal sections through the embryonic E13 superior colliculus were double-labeled with antibodies against RPTPs and the neural progenitor marker nestin. Arrowheads indicate examples of double-labeled cells. Note that RPTPγ, RPTPε, RPTPRR and RPTPσ epitopes partially co-localize with nestin-positive progenitors. In contrast, RPTPJ was not co-localized with nestin-positive cells. Since nestin produces strictly radial staining whereas RPTPJ, RPTPγ, RPTPε, RPTPRR and RPTPσ antibodies label mainly nuclei/perikarya of collicular cells, both markers reveal a faint overlapping expression pattern (merge). As shown for RC-2 immunostainings, RPTPκ immunoreactivity directly overlaps with nestin-intermediate filament staining (merge). In comparison to the RC-2/RPTPκ overlap, the correspondence of nestin and RPTPκ appears less extensive. IZ intermediate zone, VZ/SVZ ventricular zone/subventricular zone. Inserts: Higher magnification of immunopositive cells (merge figures represent co-immunoreactive cells). Scale bar 50 μm
Discussion
Our study of the PTP family highlights the identification and differential expression pattern of eleven PTPs during critical periods of mouse superior colliculus histogenesis. Quantitative real-time PCR and Western blotting of TC-PTP, PTP1C, PTP1D, PTP-MEG2, PTP-PEST, RPTPJ, RPTPκ, RPTPγ, RPTPε, RPTPRR and RPTPσ revealed distinct temporal changes of gene and protein expression during collicular development. Moreover, double-immunohistochemical analysis demonstrated for the first time that the intracellular PTPs PTP1D, PTP-MEG2 and PTP-PEST and the receptor-type PTPs RPTPκ, RPTPγ, RPTPε, RPTPRR and RPTPσ are co-expressed in a diverse pattern by collicular neural progenitors and radial glia cells at E13. In the light of these particular regulatory patterns and drawing on reports relating to other systems, we propose that the tyrosine phosphatases play an important regulatory role for the neural stem/progenitor cells in the superior colliculus.
Using a degenerate primer-based RT-PCR approach we identified TC-PTP and PTP1B as the most abundant intracellular PTPs. As reported by previous studies, TC-PTP shares 71% amino-acid identity with PTP1B (Bourdeau et al. 2005; Ibarra-Sanchez et al. 2000). Based on the cross-reactivity of the TC-PTP antibody used in Western blotting, PTP1B was identified as a 50 kDa protein band. PTP1B has been detected in RGCs where it may function as positive modulator of nerve fiber outgrowth (Pathre et al. 2001). Therefore, it is conceivable that PTP1B could play a crucial role for the establishment of topographic retino-collicular projections. Consistent with this view, PTP1B protein exhibited the most prominent expression at early stages (E13 and E15), when the first RGC fibers begin to grow out and innervate the superior colliculus. Different from the case of PTP1B, the expression of TC-PTP is described here for the first time for the murine CNS, where two alternatively spliced isoforms were visualized by Western blot. The constitutively expressed 48 kDa TC-PTP isoform was gradually downregulated during development, whereas the 45 kDa isoform remained constant. Previously, two alternatively spliced variants have been described in the human: a 48 kDa form, which is localized to the endoplasmatic reticulum by a hydrophobic C-terminus, and a 45 kDa form which lacks the hydrophobic C-terminus and is located in the nucleus (Tiganis et al. 1998, 1999; Lam et al. 2001). Earlier investigations reported a regulatory involvement of TC-PTP in EGF- and PDGF-signal transduction pathways (Tiganis et al. 1997; Persson et al. 2004; Mattila et al. 2005), which are implicated in cell proliferation and cell adhesion processes. Furthermore, TC-PTP has been described as a negative regulator of colony-stimulating factor 1 signaling and macrophage differentiation (Simoncic et al. 2006). Consistent with these reports, our data suggest different potential roles of TC-PTP during collicular development, especially in early developmental stages.
PTP1C mRNAs and proteins were weakly expressed during the embryonic stages and at the day of birth, while a strong expression was detected from P4 onward. In addition, immunohistochemistry revealed a prominent expression of PTP1C in the intermediate zone (IZ) of the superior colliculus, which consists of more mature cells at E13. Furthermore, PTP1C was detected in cells scattered throughout the SuG and Op of the superior colliculus at P20. As shown by Jena et al. (1997), PTP1C is associated with synaptic vesicles and interacts directly with the vesicular protein synaptophysin, which suggests an involvement of PTP1C in synaptic transmission. Indeed, the expression profile of PTP1C in the superior colliculus correlates well with synaptogenesis, which starts approximately at P1 (Lo and Mize 1999). Furthermore, synaptophysin is up-regulated during neuronal development and maturation (Becher et al. 1999a, b). In addition, previous studies reported that PTP1C is involved in proliferation and differentiation of astrocytes, microglia (Horvat et al. 2001; Wishcamper et al. 2001) and oligodendrocytes (Massa et al. 2004). These cell types are generated at late embryonic and early postnatal stages.
The peak amount of PTP1D mRNA and protein was detected at E13 and at late postnatal stages. The strong expression of PTP1D at E13, the sharp drop at late embryonic and early postnatal stages and the subsequent increased expression at late developmental stages are in agreement with the assumption that PTP1D could adopt different functions during the early and late development of the superior colliculus. PTP1D may affect cell growth, cell adhesion and cell motility either as a positive or a negative regulator of distinct signal transduction pathways (Li et al. 1994; Matozaki et al. 1994; Noguchi et al. 1994; Miao et al. 2000; Poliakov et al. 2004; Zhang et al. 2004). In addition, PTP1D directly interacts with EphA2 and influences directional cell migration. In this context, the impact of the Eph family in patterning and establishing the topographic mapping within the visual system is well described (Flanagan and Vanderhaeghen 1998; Feldheim et al. 2000, 2004). Furthermore, we could show that PTP1D is expressed in RC-2 positive radial glia cells during early collicular development, which could hint at a potential functional role of this PTP in neuronal migration processes.
PTP-MEG2 mRNA and protein were continuously expressed from E13 to P20, indicating its important role during development as well as in the maintenance of a mature superior colliculus. A strong PTP-MEG2 immunoreactivity was detected throughout all layers of E13 superior colliculus, whereas at P20 the protein was restricted to single cells of the SuG and Op. Interestingly, PTP-MEG2 deficient mice display several severe neurodevelopmental deficits such as neural tube defects associated with craniofacial abnormalities, exencephaly, encephalocoeles and meningomyelocoeles. In addition, the brains of neonatal PTP-MEG2 null-mice are grossly smaller in comparison to wild type animals (Wang et al. 2005). Extrapolating from the severe developmental defects that were observed in the null-mice, PTP-MEG2 might be involved in a diversity of signal transduction pathways that regulate neurulation processes. Consistent with this view, PTP-MEG2 controls the development and growth of erythrocyte precursors cells (Xu et al. 2003). A potential neuro-developmental role of PTP-MEG2 is supported by its expression in nestin-positive radial glia cells and neuronal collicular progenitors. Therefore, we believe that this PTP might act as a molecule which impacts on the differentiation pathway of these specialized cell types.
In contrast to other intracellular PTPs, PTP-PEST mRNA and protein levels were dramatically downregulated from P0 onward. Immunohistochemical analysis revealed a prominent expression of this PTP in early collicular progenitor- and radial glia cells, while it was absent at P20. These findings suggest that PTP-PEST might be involved in the regulation of early developmental processes, such as cell proliferation, differentiation and migration. Indeed, similar to PTP1D, PTP-PEST acts as a negative regulator of cell migration processes (Davidson et al. 1997; Garton et al. 1997; Shen et al. 1998; Garton and Tonks 1999; Davidson and Veillette 2001).
The receptor PTPs RPTPRR and RPTPσ, RPTPJ RPTPκ, RPTPγ and RPTPε displayed a constant up-regulation of mRNAs and proteins during collicular development, suggesting an important role both in developmental processes and in homeostasis of the mature superior colliculus.
A strong expression of the RPTPJ-epitope was restricted to single cells within the subventricular and ventricular zones (SZV/VZ) of the E13 superior colliculus. These areas contain predominantly immature proliferative and early postmitotic cells. Interestingly, in C. elegans RPTPJ regulates signaling of the EGF-receptor and enables Notch-dependent determination and differentiation of certain cell types (Berset et al. 2005); in the human and mouse, RPTPJ exhibits tumor-suppressor activity (Keane et al. 1996; Trapasso et al. 2000; Ruivenkamp et al. 2002). Thus, we assume that the expression of RPTPJ in the embryonic superior colliculus might be necessary to promote neurogenic/gliogenic determination and differentiation of specific collicular progenitors. In fact, we found no co-localization of RPTPJ with early immature neural progenitors or radial glia cells, which supports the notion of a potential role in cell fate determination and specification. In addition, RPTPJ was expressed in a decreasing anterior-posterior gradient-like pattern, which suggests that the determination of specific cell types starts in the most anterior part during superior colliculus development. This gradient like pattern was restricted to the early E13 superior colliculus and was not observed in other CNS structures.
In contrast to other PTPs, RPTPκ displayed a completely overlapping expression with nestin-positive progenitor cells and RC-2-positive radial glial cell processes in the E13 superior colliculus. Recently, we showed that RPTPκ is expressed on radial processes in the embryonic retina (Horvat-Brocker et al. 2008) and co-localizes with nestin-positive retinal progenitor cells. Because radial glia processes serve as cellular scaffold during CNS development and function as guide for neuronal migration (Newman and Reichenbach 1996; Bauch et al. 1998), the expression on radial processes might implicate RPTPκ in the process of neuronal migration within the developing superior colliculus. In support of this view, it has been shown that RPTPκ stimulates the migration of cancer cells (Kim et al. 2006) and acts as cell adhesion molecule with homophilic adhesion properties (Jiang et al. 1993; Sap et al. 1994). Western blot analysis for RPTPκ demonstrated a complex temporal regulation of the full RPTPκ protein (164 kDa) and the proteolytic cleavage products (115-, 105- and 91 kDa). Thus, we surmise that the integral RPTPκ protein and its isoforms are functionally relevant during mouse collicular development.
As shown for RPTPε and RPTPσ, RPTPγ exhibits a predominant expression in the intermediate zone (IZ) of the embryonic mouse superior colliculus. At E13, this layer consists of more mature post-mitotic cells, which are generated in an inside-out sequence (Altman and Bayer 1981; Edwards et al. 1986). Indeed, as revealed by previous studies in the rat, RPTPγ mRNA is most prominently expressed by post-mitotic neurons in the superficial layers of the postnatal cortex (Sahin et al. 1995). However, our data additionally indicate that the RPTPγ protein is expressed by a subset of neural progenitor and radial glia cells within the ventricular zone. With regard to the described prominent expression of RPTPγ in post-mitotic cells, real-time PCR and Western Blot analysis revealed an up-regulation of the RPTPγ mRNA and RPTPγ proteolytic cleavage products (120-, 114-, 110- and 80 kDa) during collicular development. Nevertheless, considering our observation that RPTPγ is subject to marked proteolytic cleavage, we suppose a diversity of functions of RPTPγ during collicular development. For example, RPTPγ is involved in the differentiation of oligodendrocytes (Ranjan and Hudson 1996; Fraser et al. 2006) and inhibits NGF-induced neurite outgrowth of PC12D cells (Shintani et al. 2001).
Our analyses of RPTPε revealed a minor mRNA- and protein-expression within the embryonic superior colliculus, which was followed by a gradual up-regulation of mRNA and protein during collicular maturation. In some systems, RPTPε can downregulate mitogenic signaling by inhibiting MAP-kinase activity (Wabakken et al. 2002; Toledano-Katchalski et al. 2003) and JAK-STAT-signaling in M1 leukemia cells (Tanuma et al. 2000, 2001, 2003). Furthermore, RPTPε negatively regulates proliferation of endothelial cells (Thompson et al. 2001). Based on these studies, we assume that the faint expression of RPTPε is required to permit proliferation of early collicular cells. RPTPε was widely expressed with ongoing maturation, while at early embryonic stages it was mainly confined to the superficial layer of the superior colliculus. In fact, studies of the physiological roles of RPTPε in the brain discuss the biological significance of this PTP in neuronal differentiation processes (Mukouyama et al. 1997). This assumption is in accordance with the observed minor co-expression of RPTPε in immature, partially proliferative, nestin- and RC-2-positive precursor/radial glia cells within the ventricular zone.
Previous studies pointed out that several transmembrane and cytoplasmatic isoforms of the RPTPRR gene are known (Augustine et al. 2000a; Chirivi et al. 2004). In the developing superior colliculus, our Western blot results indicate the differential expression of three alternative splicing variants (RPTPBr7 (74 kDa), RPTPPBSδ (+) (51 kDa) and RPTPPBSδ (−) (47 kDa)) of the RPTPRR gene. In addition, we identified a 155 kDa protein band, which might represent a glycosylated form of one of the described receptor-like isoforms. With the exception of RPTPPBSδ (+), all RPTPRR proteins were found prominently expressed in early embryonic collicular stages. A similar developmental regulation of the RPTPRR gene and the corresponding alternatively spliced variants has been reported in the retina (Horvat-Brocker et al. 2008), in Purkinje cells (van den Maagdenberg et al. 1999) and during chondrogenesis (Augustine et al. 2000a). Considering the generation and developmentally distinct regulation of multiple isoforms from a single gene through alternative splicing, it seems plausible that the identified RPTPRR proteins regulate various biological and physiological processes at different developmental time points.
In contrast to the other RPTPs, RPTPσ mRNA was downregulated in the late postnatal stages (at P12 and at P20), correlating well with the completion of the retino-collicular map. On protein level, we observed a nearby constant expression during development. In RGC axons of Xenopus and chicken, RPTPσ promotes retinal neurite outgrowth (Burden-Gulley and Brady-Kalnay 1999; Ledig et al. 1999; Johnson et al. 2001), growth cone guidance (Burden-Gulley et al. 2002) and targeting of retinal axons within the optic tectum (Rashid-Doubell et al. 2002). We assume an analogous functional role of RPTPσ in the mouse superior colliculus. Furthermore, our results parallel the previously described expression of RPTPσ in the ventricular and subventricular zones of the developing CNS (Walton et al. 1993; Meathrel et al. 2002), where RPTPσ plays an important role in neural stem cell differentiation. Accordingly, the expression of RPTPε in early collicular progenitors and radial glia cells is suggestive of a functional relevance of this PTP in stem cell development.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We gratefully acknowledge the excellent technical assistance of Stephanie Chun, Marion Voelzkow and Sabine Kindermann. This work was supported by the German Research Council (DFG, SFB 509: neuronal mechanisms of vision), the Research School of the Ruhr-University Bochum (DFG GSC98/1) and the grant "Regulation of retinal stem cells by neural extracellular matrix and tyrosinephosphatases" (DFG, FA 159/14-1).
Footnotes
J. Reinhard and A. Horvat-Bröcker contributed equally to this work.
References
- Aicher B, Lerch MM, Muller T, Schilling J, Ullrich A. Cellular redistribution of protein tyrosine phosphatases LAR and PTPsigma by inducible proteolytic processing. J Cell Biol. 1997;138:681–696. doi: 10.1083/jcb.138.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman J, Bayer SA. Time of origin of neurons of the rat superior colliculus in relation to other components of the visual and visuomotor pathways. Exp Brain Res. 1981;42:424–434. doi: 10.1007/BF00237507. [DOI] [PubMed] [Google Scholar]
- Anders L, Mertins P, Lammich S, Murgia M, Hartmann D, Saftig P, Haass C, Ullrich A. Furin-, ADAM 10-, and gamma-secretase-mediated cleavage of a receptor tyrosine phosphatase and regulation of beta-catenin’s transcriptional activity. Mol Cell Biol. 2006;26:3917–3934. doi: 10.1128/MCB.26.10.3917-3934.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustine KA, Rossi RM, Silbiger SM, Bucay N, Duryea D, Marshall WS, Medlock ES. Evidence that the protein tyrosine phosphatase (PC12, Br7, Sl) gamma (−) isoform modulates chondrogenic patterning and growth. Int J Dev Biol. 2000;44:361–371. [PubMed] [Google Scholar]
- Augustine KA, Silbiger SM, Bucay N, Ulias L, Boynton A, Trebasky LD, Medlock ES. Protein tyrosine phosphatase (PC12, Br7, S1) family: expression characterization in the adult human and mouse. Anat Rec. 2000;258:221–234. doi: 10.1002/(SICI)1097-0185(20000301)258:3<221::AID-AR1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Bard JL, Kaufman MH, Dubreuil C, Brune RM, Burger A, Baldock RA, Davidson DR. An internet-accessible database of mouse developmental anatomy based on a systematic nomenclature. Mech Dev. 1998;74:111–120. doi: 10.1016/S0925-4773(98)00069-0. [DOI] [PubMed] [Google Scholar]
- Barnea G, Silvennoinen O, Shaanan B, Honegger AM, Canoll PD, D’Eustachio P, Morse B, Levy JB, Laforgia S, Huebner K, et al. Identification of a carbonic anhydrase-like domain in the extracellular region of RPTP gamma defines a new subfamily of receptor tyrosine phosphatases. Mol Cell Biol. 1993;13:1497–1506. doi: 10.1128/mcb.13.3.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauch H, Stier H, Schlosshauer B. Axonal versus dendritic outgrowth is differentially affected by radial glia in discrete layers of the retina. J Neurosci. 1998;18:1774–1785. doi: 10.1523/JNEUROSCI.18-05-01774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher A, Drenckhahn A, Pahner I, Ahnert-Hilger G. The synaptophysin–synaptobrevin complex is developmentally upregulated in cultivated neurons but is absent in neuroendocrine cells. Eur J Cell Biol. 1999;78:650–656. doi: 10.1016/S0171-9335(99)80050-8. [DOI] [PubMed] [Google Scholar]
- Becher A, Drenckhahn A, Pahner I, Margittai M, Jahn R, Ahnert-Hilger G. The synaptophysin–synaptobrevin complex: a hallmark of synaptic vesicle maturation. J Neurosci. 1999;19:1922–1931. doi: 10.1523/JNEUROSCI.19-06-01922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berset TA, Hoier EF, Hajnal A. The C. elegans homolog of the mammalian tumor suppressor Dep-1/Scc1 inhibits EGFR signaling to regulate binary cell fate decisions. Genes Dev. 2005;19:1328–1340. doi: 10.1101/gad.333505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bixby JL. Receptor tyrosine phosphatases in axon growth and guidance. NeuroReport. 2000;11:R5–R10. doi: 10.1097/00001756-200007140-00001. [DOI] [PubMed] [Google Scholar]
- Bourdeau A, Dube N, Tremblay ML. Cytoplasmic protein tyrosine phosphatases, regulation and function: the roles of PTP1B and TC-PTP. Curr Opin Cell Biol. 2005;17:203–209. doi: 10.1016/j.ceb.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Burden-Gulley SM, Brady-Kalnay SM. PTPmu regulates N-cadherin-dependent neurite outgrowth. J Cell Biol. 1999;144:1323–1336. doi: 10.1083/jcb.144.6.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burden-Gulley SM, Ensslen SE, Brady-Kalnay SM. Protein tyrosine phosphatase-mu differentially regulates neurite outgrowth of nasal and temporal neurons in the retina. J Neurosci. 2002;22:3615–3627. doi: 10.1523/JNEUROSCI.22-09-03615.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirivi RG, Dilaver G, Vorstenbosch R, Wanschers B, Schepens J, Croes H, Fransen J, Hendriks W. Characterization of multiple transcripts and isoforms derived from the mouse protein tyrosine phosphatase gene Ptprr. Genes Cells. 2004;9:919–933. doi: 10.1111/j.1365-2443.2004.00773.x. [DOI] [PubMed] [Google Scholar]
- Cook T. Cell diversity in the retina: more than meets the eye. Bioessays. 2003;25:921–925. doi: 10.1002/bies.10356. [DOI] [PubMed] [Google Scholar]
- Davidson D, Veillette A. PTP-PEST, a scaffold protein tyrosine phosphatase, negatively regulates lymphocyte activation by targeting a unique set of substrates. EMBO J. 2001;20:3414–3426. doi: 10.1093/emboj/20.13.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson D, Cloutier JF, Gregorieff A, Veillette A. Inhibitory tyrosine protein kinase p50csk is associated with protein–tyrosine phosphatase PTP-PEST in hemopoietic and non-hemopoietic cells. J Biol Chem. 1997;272:23455–23462. doi: 10.1074/jbc.272.37.23455. [DOI] [PubMed] [Google Scholar]
- Edwards MA, Schneider GE, Caviness VS., Jr Development of the crossed retinocollicular projection in the mouse. J Comp Neurol. 1986;248:410–421. doi: 10.1002/cne.902480309. [DOI] [PubMed] [Google Scholar]
- Elson A, Leder P. Protein–tyrosine phosphatase epsilon. An isoform specifically expressed in mouse mammary tumors initiated by v-Ha-ras OR neu. J Biol Chem. 1995;270:26116–26122. doi: 10.1074/jbc.270.44.26116. [DOI] [PubMed] [Google Scholar]
- Feldheim DA, Kim YI, Bergemann AD, Frisen J, Barbacid M, Flanagan JG. Genetic analysis of ephrin-A2 and ephrin-A5 shows their requirement in multiple aspects of retinocollicular mapping. Neuron. 2000;25:563–574. doi: 10.1016/S0896-6273(00)81060-0. [DOI] [PubMed] [Google Scholar]
- Feldheim DA, Nakamoto M, Osterfield M, Gale NW, DeChiara TM, Rohatgi R, Yancopoulos GD, Flanagan JG. Loss-of-function analysis of EphA receptors in retinotectal mapping. J Neurosci. 2004;24:2542–2550. doi: 10.1523/JNEUROSCI.0239-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan JG, Vanderhaeghen P. The ephrins and Eph receptors in neural development. Annu Rev Neurosci. 1998;21:309–345. doi: 10.1146/annurev.neuro.21.1.309. [DOI] [PubMed] [Google Scholar]
- Fraser L, Wysocki P, Ciereszko A, Plucienniczak G, Kotlowska M, Kordan W, Wojtczak M, Dietrich G, Strzezek J. Application of biochemical markers for identification of biological properties of animal semen. Reprod Biol. 2006;6(Suppl 1):5–20. [PubMed] [Google Scholar]
- Fuchs M, Muller T, Lerch MM, Ullrich A. Association of human protein–tyrosine phosphatase kappa with members of the armadillo family. J Biol Chem. 1996;271:16712–16719. doi: 10.1074/jbc.271.18.10588. [DOI] [PubMed] [Google Scholar]
- Galileo DS, Gray GE, Owens GC, Majors J, Sanes JR. Neurons and glia arise from a common progenitor in chicken optic tectum: demonstration with two retroviruses and cell type-specific antibodies. Proc Natl Acad Sci USA. 1990;87:458–462. doi: 10.1073/pnas.87.1.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity PA, Lee CH, Salecker I, Robertson HC, Desai CJ, Zinn K, Zipursky SL. Retinal axon target selection in Drosophila is regulated by a receptor protein tyrosine phosphatase. Neuron. 1999;22:707–717. doi: 10.1016/S0896-6273(00)80730-8. [DOI] [PubMed] [Google Scholar]
- Garton AJ, Tonks NK. Regulation of fibroblast motility by the protein tyrosine phosphatase PTP-PEST. J Biol Chem. 1999;274:3811–3818. doi: 10.1074/jbc.274.6.3811. [DOI] [PubMed] [Google Scholar]
- Garton AJ, Burnham MR, Bouton AH, Tonks NK. Association of PTP-PEST with the SH3 domain of p130cas; a novel mechanism of protein tyrosine phosphatase substrate recognition. Oncogene. 1997;15:877–885. doi: 10.1038/sj.onc.1201279. [DOI] [PubMed] [Google Scholar]
- Godement P, Salaun J, Imbert M. Prenatal and postnatal development of retinogeniculate and retinocollicular projections in the mouse. J Comp Neurol. 1984;230:552–575. doi: 10.1002/cne.902300406. [DOI] [PubMed] [Google Scholar]
- Gotz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Halle M, Liu YC, Hardy S, Theberge JF, Blanchetot C, Bourdeau A, Meng TC, Tremblay ML. Caspase-3 regulates catalytic activity and scaffolding functions of the protein tyrosine phosphatase PEST, a novel modulator of the apoptotic response. Mol Cell Biol. 2007;27:1172–1190. doi: 10.1128/MCB.02462-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera E, Brown L, Aruga J, Rachel RA, Dolen G, Mikoshiba K, Brown S, Mason CA. Zic2 patterns binocular vision by specifying the uncrossed retinal projection. Cell. 2003;114:545–557. doi: 10.1016/S0092-8674(03)00684-6. [DOI] [PubMed] [Google Scholar]
- Horvat A, Schwaiger F, Hager G, Brocker F, Streif R, Knyazev P, Ullrich A, Kreutzberg GW. A novel role for protein tyrosine phosphatase shp1 in controlling glial activation in the normal and injured nervous system. J Neurosci. 2001;21:865–874. doi: 10.1523/JNEUROSCI.21-03-00865.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvat-Brocker A, Reinhard J, Illes S, Paech T, Zoidl G, Harroch S, Distler C, Knyazev P, Ullrich A, Faissner A. Receptor protein tyrosine phosphatases are expressed by cycling retinal progenitor cells and involved in neuronal development of mouse retina. Neuroscience. 2008;152:618–645. doi: 10.1016/j.neuroscience.2008.01.016. [DOI] [PubMed] [Google Scholar]
- Ibarra-Sanchez MJ, Simoncic PD, Nestel FR, Duplay P, Lapp WS, Tremblay ML. The T-cell protein tyrosine phosphatase. Semin Immunol. 2000;12:379–386. doi: 10.1006/smim.2000.0220. [DOI] [PubMed] [Google Scholar]
- Jallal B, Mossie K, Vasiloudis G, Knyazev P, Zachwieja J, Clairvoyant F, Schilling J, Ullrich A. The receptor-like protein–tyrosine phosphatase DEP-1 is constitutively associated with a 64-kDa protein serine/threonine kinase. J Biol Chem. 1997;272:12158–12163. doi: 10.1074/jbc.272.18.12158. [DOI] [PubMed] [Google Scholar]
- Jena B, Webster P, Geibel JP, Pol AN, Sritharan KC. Localization of SH-PTP1 to synaptic vesicles: a possible role in neurotransmission. Cell Biol Int. 1997;21:469–476. doi: 10.1006/cbir.1997.0185. [DOI] [PubMed] [Google Scholar]
- Jiang YP, Wang H, D’Eustachio P, Musacchio JM, Schlessinger J, Sap J. Cloning and characterization of R-PTP-kappa, a new member of the receptor protein tyrosine phosphatase family with a proteolytically cleaved cellular adhesion molecule-like extracellular region. Mol Cell Biol. 1993;13:2942–2951. doi: 10.1128/mcb.13.5.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KG, McKinnell IW, Stoker AW, Holt CE. Receptor protein tyrosine phosphatases regulate retinal ganglion cell axon outgrowth in the developing Xenopus visual system. J Neurobiol. 2001;49:99–117. doi: 10.1002/neu.1068. [DOI] [PubMed] [Google Scholar]
- Keane MM, Lowrey GA, Ettenberg SA, Dayton MA, Lipkowitz S. The protein tyrosine phosphatase DEP-1 is induced during differentiation and inhibits growth of breast cancer cells. Cancer Res. 1996;56:4236–4243. [PubMed] [Google Scholar]
- Kim YS, Kang HY, Kim JY, Oh S, Kim CH, Ryu CJ, Miyoshi E, Taniguchi N, Ko JH. Identification of target proteins of N-acetylglucosaminyl transferase V in human colon cancer and implications of protein tyrosine phosphatase kappa in enhanced cancer cell migration. Proteomics. 2006;6:1187–1191. doi: 10.1002/pmic.200500400. [DOI] [PubMed] [Google Scholar]
- Kuramochi S, Matsuda S, Matsuda Y, Saitoh T, Ohsugi M, Yamamoto T. Molecular cloning and characterization of Byp, a murine receptor-type tyrosine phosphatase similar to human DEP-1. FEBS Lett. 1996;378:7–14. doi: 10.1016/0014-5793(95)01415-2. [DOI] [PubMed] [Google Scholar]
- Lam MH, Michell BJ, Fodero-Tavoletti MT, Kemp BE, Tonks NK, Tiganis T. Cellular stress regulates the nucleocytoplasmic distribution of the protein–tyrosine phosphatase TCPTP. J Biol Chem. 2001;276:37700–37707. doi: 10.1074/jbc.M105128200. [DOI] [PubMed] [Google Scholar]
- Lammers R, Bossenmaier B, Cool DE, Tonks NK, Schlessinger J, Fischer EH, Ullrich A. Differential activities of protein tyrosine phosphatases in intact cells. J Biol Chem. 1993;268:22456–22462. [PubMed] [Google Scholar]
- Ledig MM, Haj F, Bixby JL, Stoker AW, Mueller BK. The receptor tyrosine phosphatase CRYPalpha promotes intraretinal axon growth. J Cell Biol. 1999;147:375–388. doi: 10.1083/jcb.147.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Gaits F, Ragab A, Ragab-Thomas JM, Chap H. Translocation of an SH2-containing protein tyrosine phosphatase (SH-PTP1) to the cytoskeleton of thrombin-activated platelets. FEBS Lett. 1994;343(1):89–93. doi: 10.1016/0014-5793(94)80613-6. [DOI] [PubMed] [Google Scholar]
- Lo FS, Mize RR. Retinal input induces three firing patterns in neurons of the superficial superior colliculus of neonatal rats. J Neurophysiol. 1999;81:954–958. doi: 10.1152/jn.1999.81.2.954. [DOI] [PubMed] [Google Scholar]
- Massa PT, Wu C, Fecenko-Tacka K. Dysmyelination and reduced myelin basic protein gene expression by oligodendrocytes of SHP-1-deficient mice. J Neurosci Res. 2004;77:15–25. doi: 10.1002/jnr.20155. [DOI] [PubMed] [Google Scholar]
- Matozaki T, Suzuki T, Uchida T, Inazawa J, Ariyama T, Matsuda K, Horita K, Noguchi H, Mizuno H, Sakamoto C, et al. Molecular cloning of a human transmembrane-type protein tyrosine phosphatase and its expression in gastrointestinal cancers. J Biol Chem. 1994;269(3):2075–2081. [PubMed] [Google Scholar]
- Mattila E, Pellinen T, Nevo J, Vuoriluoto K, Arjonen A, Ivaska J. Negative regulation of EGFR signalling through integrin-alpha1beta1-mediated activation of protein tyrosine phosphatase TCPTP. Nat Cell Biol. 2005;7:78–85. doi: 10.1038/ncb1209. [DOI] [PubMed] [Google Scholar]
- Meathrel K, Adamek T, Batt J, Rotin D, Doering LC. Protein tyrosine phosphatase sigma-deficient mice show aberrant cytoarchitecture and structural abnormalities in the central nervous system. J Neurosci Res. 2002;70:24–35. doi: 10.1002/jnr.10382. [DOI] [PubMed] [Google Scholar]
- Miao W, Luo Z, Kitsis RN, Walsh K. Intracoronary, adenovirus-mediated Akt gene transfer in heart limits infarct size following ischemia-reperfusion injury in vivo. J Mol Cell Cardiol. 2000;32(12):2397–2402. doi: 10.1006/jmcc.2000.1283. [DOI] [PubMed] [Google Scholar]
- Moller NP, Moller KB, Lammers R, Kharitonenkov A, Hoppe E, Wiberg FC, Sures I, Ullrich A. Selective down-regulation of the insulin receptor signal by protein-tyrosine phosphatases alpha and epsilon. J Biol Chem. 1995;270:23126–23131. doi: 10.1074/jbc.270.39.23126. [DOI] [PubMed] [Google Scholar]
- Mukouyama Y, Kuroyanagi H, Shirasawa T, Tomoda T, Saffen D, Oishi M, Watanabe T. Induction of protein tyrosine phosphatase epsilon transcripts during NGF-induced neuronal differentiation of PC12D cells and during the development of the cerebellum. Brain Res Mol Brain Res. 1997;50:230–236. doi: 10.1016/S0169-328X(97)00193-9. [DOI] [PubMed] [Google Scholar]
- Newman E, Reichenbach A. The Muller cell: a functional element of the retina. Trends Neurosci. 1996;19:307–312. doi: 10.1016/0166-2236(96)10040-0. [DOI] [PubMed] [Google Scholar]
- Newsome TP, Asling B, Dickson BJ. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development. 2000;127:851–860. doi: 10.1242/dev.127.4.851. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Matozaki T, Horita K, Fujioka Y, Kasuga M. Role of SH-PTP2, a protein-tyrosine phosphatase with Src homology 2 domains, in insulin-stimulated Ras activation. Mol Cell Biol. 1994;14:6674–6682. doi: 10.1128/mcb.14.10.6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary DD, McLaughlin T. Mechanisms of retinotopic map development: Ephs, ephrins, and spontaneous correlated retinal activity. Prog Brain Res. 2005;147:43–65. doi: 10.1016/S0079-6123(04)47005-8. [DOI] [PubMed] [Google Scholar]
- Ogata M, Sawada M, Fujino Y, Hamaoka T. cDNA cloning and characterization of a novel receptor-type protein tyrosine phosphatase expressed predominantly in the brain. J Biol Chem. 1995;270:2337–2343. doi: 10.1074/jbc.270.5.2337. [DOI] [PubMed] [Google Scholar]
- Pathre P, Arregui C, Wampler T, Kue I, Leung TC, Lilien J, Balsamo J. PTP1B regulates neurite extension mediated by cell–cell and cell–matrix adhesion molecules. J Neurosci Res. 2001;63:143–150. doi: 10.1002/1097-4547(20010115)63:2<143::AID-JNR1006>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Persson C, Savenhed C, Bourdeau A, Tremblay ML, Markova B, Bohmer FD, Haj FG, Neel BG, Elson A, Heldin CH, Ronnstrand L, Ostman A, Hellberg C. Site-selective regulation of platelet-derived growth factor beta receptor tyrosine phosphorylation by T-cell protein tyrosine phosphatase. Mol Cell Biol. 2004;24:2190–2201. doi: 10.1128/MCB.24.5.2190-2201.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucl Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliakov A, Cotrina M, Wilkinson DG. Diverse roles of eph receptors and ephrins in the regulation of cell migration and tissue assembly. Dev Cell. 2004;7:465–480. doi: 10.1016/j.devcel.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Ranjan M, Hudson LD. Regulation of tyrosine phosphorylation and protein tyrosine phosphatases during oligodendrocyte differentiation. Mol Cell Neurosci. 1996;7:404–418. doi: 10.1006/mcne.1996.0029. [DOI] [PubMed] [Google Scholar]
- Rashid-Doubell F, McKinnell I, Aricescu AR, Sajnani G, Stoker A. Chick PTPsigma regulates the targeting of retinal axons within the optic tectum. J Neurosci. 2002;22:5024–5033. doi: 10.1523/JNEUROSCI.22-12-05024.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Zoidl G, Weickert S, Wahle P, Dermietzel R. Site-specific and developmental expression of pannexin1 in the mouse nervous system. Eur J Neurosci. 2005;21:3277–3290. doi: 10.1111/j.1460-9568.2005.04139.x. [DOI] [PubMed] [Google Scholar]
- Ruivenkamp CA, Wezel T, Zanon C, Stassen AP, Vlcek C, Csikos T, Klous AM, Tripodis N, Perrakis A, Boerrigter L, Groot PC, Lindeman J, Mooi WJ, Meijjer GA, Scholten G, Dauwerse H, Paces V, Zandwijk N, Ommen GJ, Demant P. Ptprj is a candidate for the mouse colon-cancer susceptibility locus Scc1 and is frequently deleted in human cancers. Nat Genet. 2002;31:295–300. doi: 10.1038/ng903. [DOI] [PubMed] [Google Scholar]
- Sahin M, Dowling JJ, Hockfield S. Seven protein tyrosine phosphatases are differentially expressed in the developing rat brain. J Comp Neurol. 1995;351:617–631. doi: 10.1002/cne.903510410. [DOI] [PubMed] [Google Scholar]
- Sap J, Jiang YP, Friedlander D, Grumet M, Schlessinger J. Receptor tyrosine phosphatase R-PTP-kappa mediates homophilic binding. Mol Cell Biol. 1994;14:1–9. doi: 10.1128/mcb.14.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma E, Lombroso PJ. A neuronal protein tyrosine phosphatase induced by nerve growth factor. J Biol Chem. 1995;270:49–53. doi: 10.1074/jbc.270.49.29578. [DOI] [PubMed] [Google Scholar]
- Shen Y, Schneider G, Cloutier JF, Veillette A, Schaller MD. Direct association of protein–tyrosine phosphatase PTP-PEST with paxillin. J Biol Chem. 1998;273:6474–6481. doi: 10.1074/jbc.273.11.6474. [DOI] [PubMed] [Google Scholar]
- Shintani Y, Marunaka Y. Regulation of single Cl- channel conductance by insulin and tyrosine phosphatase. Biochem Biophys Res Commun. 1996;218:142–147. doi: 10.1006/bbrc.1996.0025. [DOI] [PubMed] [Google Scholar]
- Shintani T, Maeda N, Noda M. Receptor-like protein tyrosine phosphatase gamma (RPTPgamma), but not PTPzeta/RPTPbeta, inhibits nerve-growth-factor-induced neurite outgrowth in PC12D cells. Dev Neurosci. 2001;23:55–69. doi: 10.1159/000048696. [DOI] [PubMed] [Google Scholar]
- Simoncic PD, Bourdeau A, Lee-Loy A, Rohrschneider LR, Tremblay ML, Stanley ER, McGlade CJ. T-cell protein tyrosine phosphatase (Tcptp) is a negative regulator of colony-stimulating factor 1 signaling and macrophage differentiation. Mol Cell Biol. 2006;26:4149–4160. doi: 10.1128/MCB.01932-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirois J, Cote JF, Charest A, Uetani N, Bourdeau A, Duncan SA, Daniels E, Tremblay ML. Essential function of PTP-PEST during mouse embryonic vascularization, mesenchyme formation, neurogenesis and early liver development. Mech Dev. 2006;123:869–880. doi: 10.1016/j.mod.2006.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein-Gerlach M, Kharitonenkov A, Vogel W, Ali S, Ullrich A. Protein-tyrosine phosphatase 1D modulates its own state of tyrosine phosphorylation. J Biol Chem. 1995;270:24635–24637. doi: 10.1074/jbc.270.42.24635. [DOI] [PubMed] [Google Scholar]
- Stoker AW. Receptor tyrosine phosphatases in axon growth and guidance. Curr Opin Neurobiol. 2001;11:95–102. doi: 10.1016/S0959-4388(00)00179-3. [DOI] [PubMed] [Google Scholar]
- Tanuma N, Nakamura K, Shima H, Kikuchi K. Protein-tyrosine phosphatase PTPepsilon C inhibits Jak-STAT signaling and differentiation induced by interleukin-6 and leukemia inhibitory factor in M1 leukemia cells. J Biol Chem. 2000;275:28216–28221. doi: 10.1074/jbc.M003661200. [DOI] [PubMed] [Google Scholar]
- Tanuma N, Shima H, Nakamura K, Kikuchi K. Protein tyrosine phosphatase epsilonC selectively inhibits interleukin-6- and interleukin- 10-induced JAK-STAT signaling. Blood. 2001;98:3030–3034. doi: 10.1182/blood.V98.10.3030. [DOI] [PubMed] [Google Scholar]
- Tanuma N, Shima H, Shimada S, Kikuchi K. Reduced tumorigenicity of murine leukemia cells expressing protein-tyrosine phosphatase, PTPepsilon C. Oncogene. 2003;22:1758–1762. doi: 10.1038/sj.onc.1206267. [DOI] [PubMed] [Google Scholar]
- Thanos S, Mey J. Development of the visual system of the chick II. Mechanisms of axonal guidance. Brain Res Brain Res Rev. 2001;35:205–245. doi: 10.1016/S0165-0173(01)00049-2. [DOI] [PubMed] [Google Scholar]
- Thompson LJ, Jiang J, Madamanchi N, Runge MS, Patterson C. PTP-epsilon, a tyrosine phosphatase expressed in endothelium, negatively regulates endothelial cell proliferation. Am J Physiol Heart Circ Physiol. 2001;281:H396–H403. doi: 10.1152/ajpheart.2001.281.1.H396. [DOI] [PubMed] [Google Scholar]
- Tiganis T, Flint AJ, Adam SA, Tonks NK. Association of the T-cell protein tyrosine phosphatase with nuclear import factor p97. J Biol Chem. 1997;272:21548–21557. doi: 10.1074/jbc.272.34.21548. [DOI] [PubMed] [Google Scholar]
- Tiganis T, Bennett AM, Ravichandran KS, Tonks NK. Epidermal growth factor receptor and the adaptor protein p52Shc are specific substrates of T-cell protein tyrosine phosphatase. Mol Cell Biol. 1998;18:1622–1634. doi: 10.1128/mcb.18.3.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiganis T, Kemp BE, Tonks NK. The protein–tyrosine phosphatase TCPTP regulates epidermal growth factor receptor-mediated and phosphatidylinositol 3-kinase-dependent signaling. J Biol Chem. 1999;274:27768–27775. doi: 10.1074/jbc.274.39.27768. [DOI] [PubMed] [Google Scholar]
- Toledano-Katchalski H, Kraut J, Sines T, Granot-Attas S, Shohat G, Gil-Henn H, Yung Y, Elson A. Protein tyrosine phosphatase epsilon inhibits signaling by mitogen-activated protein kinases. Mol Cancer Res. 2003;1:541–550. [PubMed] [Google Scholar]
- Tomic S, Greiser U, Lammers R, Kharitonenkov A, Imyanitov E, Ullrich A, Bohmer FD. Association of SH2 domain protein tyrosine phosphatases with the epidermal growth factor receptor in human tumor cells. Phosphatidic acid activates receptor dephosphorylation by PTP1C. J Biol Chem. 1995;270:21277–21284. doi: 10.1074/jbc.270.36.21277. [DOI] [PubMed] [Google Scholar]
- Trapasso F, Iuliano R, Boccia A, Stella A, Visconti R, Bruni P, Baldassarre G, Santoro M, Viglietto G, Fusco A. Rat protein tyrosine phosphatase eta suppresses the neoplastic phenotype of retrovirally transformed thyroid cells through the stabilization of p27(Kip1) Mol Cell Biol. 2000;20:9236–9246. doi: 10.1128/MCB.20.24.9236-9246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Maagdenberg AM, Schepens JT, Schepens MT, Merkx GF, Darroudi F, Wieringa B, Geurts van Kessel A, Hendriks WJ. Assignment1 of the PTP-SL/PTPBR7 gene (Ptprr/PTPRR) to mouse chromosome region 8A2 by in situ hybridization. Cytogenet Cell Genet. 1999;84:243–244. doi: 10.1159/000015268. [DOI] [PubMed] [Google Scholar]
- Niekerk CC, Poels LG. Reduced expression of protein tyrosine phosphatase gamma in lung and ovarian tumors. Cancer Lett. 1999;137:61–73. doi: 10.1016/S0304-3835(98)00344-9. [DOI] [PubMed] [Google Scholar]
- Wabakken T, Hauge H, Finne EF, Wiedlocha A, Aasheim H. Expression of human protein tyrosine phosphatase epsilon in leucocytes: a potential ERK pathway-regulating phosphatase. Scand J Immunol. 2002;56:195–203. doi: 10.1046/j.1365-3083.2002.01126.x. [DOI] [PubMed] [Google Scholar]
- Walton KM, Martell KJ, Kwak SP, Dixon JE, Largent BL. A novel receptor-type protein tyrosine phosphatase is expressed during neurogenesis in the olfactory neuroepithelium. Neuron. 1993;11:387–400. doi: 10.1016/0896-6273(93)90193-U. [DOI] [PubMed] [Google Scholar]
- Wang Y, Vachon E, Zhang J, Cherepanov V, Kruger J, Li J, Saito K, Shannon P, Bottini N, Huynh H, Ni H, Yang H, McKerlie C, Quaggin S, Zhao ZJ, Marsden PA, Mustelin T, Siminovitch KA, Downey GP. Tyrosine phosphatase MEG2 modulates murine development and platelet and lymphocyte activation through secretory vesicle function. J Exp Med. 2005;202:1587–1597. doi: 10.1084/jem.20051108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishcamper C, Coffin JD, Lurie DI. Lack of the protein tyrosine phosphatase SHP-1 results in decreased numbers of glia within the motheaten (me/me) mouse brain. J Comp Neurol. 2001;441:118–133. doi: 10.1002/cne.1401. [DOI] [PubMed] [Google Scholar]
- Xu MJ, Sui X, Zhao R, Dai C, Krantz SB, Zhao ZJ. PTP-MEG2 is activated in polycythemia vera erythroid progenitor cells and is required for growth and expansion of erythroid cells. Blood. 2003;102:4354–4360. doi: 10.1182/blood-2003-04-1308. [DOI] [PubMed] [Google Scholar]
- Zhang SQ, Yang W, Kontaridis MI, Bivona TG, Wen G, Araki T, Luo J, Thompson JA, Schraven BL, Philips MR, Neel BG. Shp2 regulates SRC family kinase activity and Ras/Erk activation by controlling Csk recruitment. Mol Cell. 2004;13:341–355. doi: 10.1016/S1097-2765(04)00050-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.