Abstract
Eph receptors and their ephrin ligands are involved in normal hematopoietic development and tumorigenesis. Using methylated CpG island amplification/DNA promoter microarray, we identified several EPH receptor and EPHRIN genes as potential hypermethylation targets in acute lymphoblastic leukemia (ALL). We subsequently studied the DNA methylation status of the Eph/ephrin family by bisulfite pyrosequencing. Hypermethylation of EPHA2, -A4, -A5, -A6, -A7, -A10, EPHB1, -B2, -B3, -B4, EFNA1, -A3, -A5, and EFNB1 and -B2 genes was detected in leukemia cell lines and primary ALL bone marrow samples. Expression analysis of EPHB4, EFNB2, and EFNA5 genes demonstrated that DNA methylation was associated with gene silencing. We cloned the promoter region of EPHB4 and demonstrated that promoter hypermethylation can result in EPHB4 transcriptional silencing. Restoration of EPHB4 expression by lentiviral transduction resulted in reduced proliferation and apoptotic cell death in Raji cells in which EPHB4 is methylated and silenced. Finally, we demonstrated that phosphorylated Akt is down-regulated in Raji cells transduced with EPHB4. These results suggest that epigenetic silencing by hypermethylation of EPH/EPHRIN family genes contributes to ALL pathogenesis and that EPHB4 can function as a tumor suppressor in ALL.
Introduction
The erythroprotein-producing hepatoma amplified sequence (Eph) receptor tyrosine kinase family is the largest family of tyrosine kinases and includes at least 14 Eph receptors and 8 ligands.1 EphA1-A8 and -A10 are subclassified as EphA family members, whereas EphB1-B4 and B6 belong to the EphB family.2 EphA receptors generally bind the glycosylphosphatidylinositol (GPI)-linked ephrin-A ligand EFNA1-5, whereas EphB receptors bind the transmembrane ephrin-B ligand EFNB1-3.3 Both Eph receptors and ephrins localize to the cell surface. Upon ephrin binding, Eph receptor undergo clustering, phosphorylation, and kinase activation, followed by activation of downstream signaling cascades in both the receptor- and the ligand-expressing cells.4
The Eph/ephrin family is differentially expressed in various human tissues.5–7 It is involved in developmental processes, particularly in embryonic development, vasculature, and the nervous system.8,9 Recent evidence suggests that Eph receptors have both tumor-promoting and -suppressing activities, depending on their expression pattern in different tumors. Some of the EPH/EPHRIN genes are oncogenic and highly expressed in human cancers, including breast, colon, melanomas, lung, and prostate cancer.10–13 On the other hand, Eph receptors have also been proposed to act as tumor suppressors.14,15 Loss of expression of Eph receptors is evident in some tumors, for example, EphB2 and EphB4 in colorectal cancer and EphB6 in breast cancer.16–18 Knockout of EPHB2, EPHB3, or EPHA2 genes accelerates colorectal tumorigenesis and skin carcinogenesis in mouse models.14,19 In addition, somatic gene mutation of EPHB2 was identified in colorectal cancers and prostate tumors.20 In addition, ectopic expression of EPHB6 by transfection in neuroblastoma cells reduces cell growth in mouse xenografts.21
Aberrant CpG island (CGI) methylation can result in inactivation of tumor suppressor genes in cancer. Methylation of EPHA2, EPHA7, EPHB2, EPHB4, and EPHB6 has been reported in several human solid tumors, including colorectal, prostate, and breast cancer.16–18,22,23 However, hypermethylation and epigenetic regulation of EPH receptors and ephrins has not yet been investigated in hematologic malignancies in detail.
We have recently used a methylated CGI amplification (MCA)/DNA promoter microarray approach to a genome-wide screen for hypermethylated CGIs in acute lymphoblastic leukemia (ALL).24 Several EPH/EPHRIN genes were identified as potential targets of methylation by this screen. This prompted us to perform a comprehensive analysis of Eph/ephrin family members in ALL. Here, we demonstrated that 15 of the Eph/ephrin family genes are frequently hypermethylated in leukemia cell lines and primary ALL samples. In addition, we showed that forced restoration of EphB4 expression induces apoptosis and suppresses cell growth in EphB4 hypermethylated cells. These results suggest that epigenetic silencing by hypermethylation of Eph/ephrin family genes contributes to ALL pathogenesis.
Methods
Cell lines and ALL patient samples
The following human leukemia cell lines were studied: MOLT4, Jurkat, Peer, T-ALL1, CEM, J-TAG, B-JAB, RS4:11, ALL1, Raji, REH, and Ramos of lymphoid origin and K562, BV173, HL60, NB4, THP1, U937, OCI, HEL, MOLM13, and KBM5R of myeloid origin. T-ALL1 and Peer cell lines were obtained from the German Resource Center for Biological Material (DSMZ). K562, BV173, and KBM5R were from Dr Beran at M. D. Anderson Cancer Center (MDACC). The other cell lines were obtained from ATCC. All cell lines were cultured in RPMI 1640 (Invitrogen) supplemented with 10% fetal calf serum (FCS; Gemini Bio-Products) and penicillin-streptomycin (Invitrogen). Normal CD19+ cells were collected from 10 healthy persons (age, 32-70 years) in accordance with MDACC institutional guidelines. Cells were separated using the Human B Cell Isolation Kit (Miltenyi Biotec). Bone marrow (BM) aspiration specimens from patients with ALL were obtained before any therapy and were stored at established tissue banks at MDACC following institutional guidelines. All samples were collected using Ficoll-Paque density centrifugation. Patient characteristics are shown in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the page of the online article). DNA from leukemia cell lines, normal CD19+ cell controls, and BM samples from patients with ALL were extracted using standard phenol-chloroform methods. Consent for sample collection was used following institutional guidelines of MDACC and in accordance with the Declaration of Helsinki.
Bisulfate pyrosequencing and bisulfite sequencing
Bisulfite treatment of genomic DNA was performed as described.24 Bisulfite pyrosequencing and bisulfite sequencing was performed as described.24,25 Primer sequences and conditions for bisulfite polymerase chain reaction (PCR) and pyrosequencing are shown in supplemental Table 2.
RNA extraction and real-time PCR
RNA extraction, reverse transcription reactions, and real-time PCR analysis were performed as described.24,26 Whole BM cDNA from normal control was purchased from Cell Systems (All Cells LLC). TaqMan probes were purchased from Applied Biosystems.
5-Aza-2′-deoxycytidine and/or trichostatin A treatment
Leukemia cells were cultured in media supplemented with 2μM 5-Aza-2′-deoxycytidine (DAC; Sigma-Aldrich) daily for 4 days, 2μM DAC for 4 days, followed by 500nM trichostatin A (TSA; ICN Biomedicals) for the last 24 hours, or 500nM TSA for 24 hours alone as described.24
Luciferase assay
The EPHB4 regulatory sequence from 512 bp upstream to 243 bp downstream of the EPHB4 transcription start site (TSS) was cloned by genomic PCR using human genomic DNA as a template. Various size deletions of the promoter were generated, also by PCR, with various 5′ primers and a fixed 3′ primer (supplemental Table 2). The expected sizes of PCR fragments were 753, 201, 582, 422, and 245 bp. All the amplified EPHB4 promoter fragments were cloned into luciferase reporter vector pGL3 (Promega). The resultant constructs were designated as pGL3-p EphB4-1, -2, -3, -4, and -5, respectively. For in vitro methylation, the EPHB4 promoter was released from the construct pGL3-pEphB4-1 and -4 with KpnII and NheI digestion and then treated with SssI methylase (New England BioLabs), according to the manufacturer's instructions. Methylated promoter was purified and ligated back into the pGL3-basic luciferase vector. Human renal epithelial cell line 293T cells were transiently transfected with the methylated and unmethylated EphB4-pGL3 constructs or pGL3-basic vector, together with 0.1 ng of pRL-TK control vector, which encodes Renilla luciferase (Promega) using the FuGENE 6 Transfection Reagent (Roche Applied Science). Luciferase activity was detected using a Dual Luciferase Assay System (Promega). The ability to stimulate transcription was defined as the ratio of the luciferase activity in cells transfected with pGL3-1, -2, -3, -4, or -5 relative to the activity in cells transfected with empty vector (pGL3-basic). All experiments were repeated at least 3 times.
Lentivirus constructs and gene transduction
Human EPHB4 lentiviral construct was generated by inserting human full-length EphB4 cDNA into a lentiviral-cytomegalovirus long terminal repeat (LTR)–ubiquitin-internal ribosome entry site-green fluorescent protein transfer vector (FUGW), as described previously.26,27 In this vector, green fluorescent protein (GFP) is coexpressed with EPHB4 in an internal ribosome entry site (IRES) expression cassette, and both are placed under the control of a cytomegalovirus enhancer and ubiquitin promoter. Lentivirus production was conducted by cotransfection of 293T cells with lentiviral gene transfer vector carrying hEphB4 and lentivirus packaging plasmids, as described.24 For transduction of leukemia cells, cells were seeded at 1 × 105 cells/well in 6-well plates and infected by adding 2 mL of viral supernatant supplemented with 4 μg/mL polybrene, as described.26
Cell growth and colony formation in soft agarose
Cell growth and colony formation assays were performed as previously described.24
Cell-cycle and apoptosis analysis
Cell-cycle analysis was performed using propidium iodide staining.24 Induction of apoptosis was quantified by measuring annexin V-PE–positive cells with flow cytometry using the Annexin V PE Apoptosis Detection Kit (BioVision).
Immunoprecipitation and Western blot analysis
For Ephrin-B2 Fc stimulation, Raji/EphB4 and Raji/vector cells were serum starved overnight and then stimulated with 3 μg/mL Ephrin-B2 Fc (R&D Systems) at the indicated time points. Cells were collected and lysed in RIPA buffer. Samples were analyzed by SDS-PAGE followed by Western blot analysis, as previously described.24 Monoclonal EphB4 was purchased from Invitrogen; phosphotyrosine and Crk antibodies were from BD Biosciences; and Akt, Phospho-Akt, Src, Phospho-Src, ERK1/2, pERK1/2, MEK1, and p-MEK1 were from Cell Signaling (Santa Cruz Biotechnology).
Immunofluorescence microscopy
EphB4 immunostaining was performed essentially as described with minor modifications.24 Briefly, GFP+ Raji cells were fixed in 10% formalin for 1 hour and processed for 5-μm paraffin sectioning. Slides were incubated with an anti-EphB4 primary antibody (1:50 dilution) and then with the fluorescence-conjugated secondary antibody Alexa Fluor 594 (1:100 dilution; Invitrogen). Nuclei were counterstained and mounted with Vectashield mounting medium with 4′,6-diamidino-2-phenylindole (Vector Laboratories). Images were obtained using an Olympus AX70 fluorescence confocal laser microscope.
cDNA microarray
cDNA microarray analysis was performed at the MDACC Genomic Core Laboratory. Total RNA was isolated from Raji/EphB4 and Raji/empty vector cells. RNA amplification and labeling was performed as described.28,29 We hybridized Cy3-labeled RNA from Raji/vector and Cy5-labeled RNA from Raji/EphB4 on a cDNA microarray (manufactured by the Cancer Genomic Core Laboratory, MDACC) that contains 4704 genes. cDNA array results were filtered using the filter on fold change option to identify genes up-regulated or down-regulated 3-fold in tested samples.
Statistical analysis
Statistical analysis was performed using Prism 4 (GraphPad Software) or Statistica 6 software (Statistica for Windows version 6.0, StatSoft). The Fisher exact test and t tests were used to compare gene methylation frequencies in leukemia cell lines or ALL patients and normal control groups. The Spearman nonparametric test was used to analyze correlations. All reported P values were 2-sided, and a P value less than .05 was considered statistically significant. Survival was calculated from initial presentation at MDACC using the standard method.
Results
Identification of hypermethylated Eph receptor and Ephrin family genes in leukemia cell lines
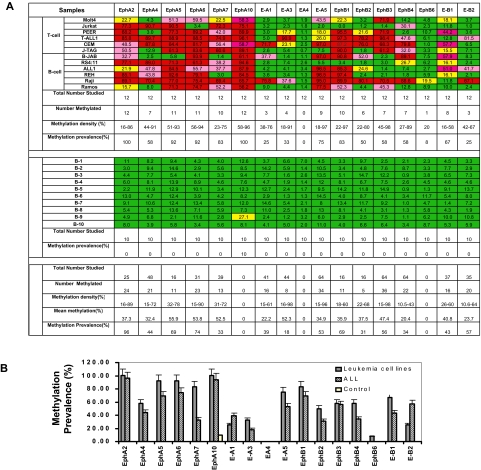
Using MCA/microarray, we identified 6 Eph receptor and ephrin genes (EPHA5, EFNA4, EFNA5, EPHB2, EPHB4, and EFNB3) as potential targets of aberrant DNA methylation in ALL. We then analyzed the dendogram of Eph/ephrin family by aligning the entire Eph/ephrin family members by their cDNA sequence homology (data not shown). This diagram revealed that the Eph/ephrin gene family is highly homologous. Most Eph/ephrin genes harbor a CGI in the proximity of the promoter. We further investigated the methylation profile of all Eph/ephrin family members in leukemia cell lines and normal peripheral blood CD19+ controls. Because EPHA1, EPHA3, and EFNA2 do not contain a CGI in their promoter region, they were not included in this study, as they cannot be physically methylated. The methylation profile of the other 17 members of the Eph/ephrin family in leukemia cell lines and normal controls is shown in Figure 1A. An example of pyrograms and results on myeloid cell lines is shown on the supplemental figure. EFNA4 was not methylated in any of the leukemia cell lines and normal controls. EPHB6, which was identified as being hypermethylated in breast cancer,18 only had low-level methylation (20%) in Raji cells but was unmethylated in other leukemia cell lines. In contrast, frequent hypermethylation of EPHA2, -A4, -A5, -A6, -A7, -A10, EPHB1, -B2, -B3, -B4, EFNA1, -A3, -A5, and EFNB1 and -B2 genes was detected in various leukemia cell lines but not (or rarely) in normal control samples (Figure 1A). The methylation prevalence (number of cases methylated) and density (percentage of CpG sites methylated for a particular promoter) of each gene is shown in Figures 1B and 2.
Figure 1.
Validation of methylation status of Eph/ephrin family genes in leukemia cell lines, primary ALL samples, and normal controls. (A) Methylation profile of Eph receptor/ephrin genes in leukemia cell lines and normal controls. Bisulfite pyrosequencing was performed to determine methylation status. Green indicates methylation density < 15%; yellow, methylation density between 15% and 29.99%; pink, methylation density between 30% and 59.99%; and red, methylation density > 60%. Methylation density > 15% was used to define a sample as methylated. (B) Methylation prevalence of 17 Eph/ephrin family genes in leukemia cell lines, ALL patients, and normal CD19+ controls. Methylation density > 15% was used as the cutoff for hypermethylation. Methylation prevalence was calculated as the percentage of positive methylated samples vs the total numbers studied for each gene. Error bars indicate range.
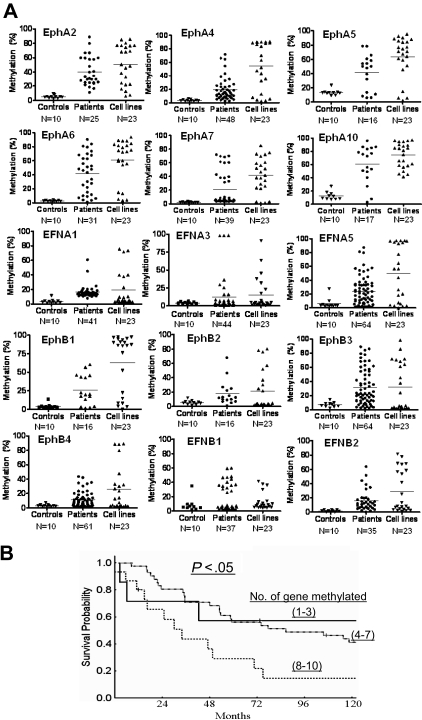
Figure 2.
Methylation characteristics of 15 Eph/ephrin family genes in normal controls, ALL patients, and leukemia cell lines. (A) Bisulfite pyrosequencing was performed to determine methylation density. N represents the number of cases in each group. Methylation (%) indicates methylation density levels. Each symbol represents a data point. (B) Survival analysis of patients with ALL based on number of methylated genes. Patients were grouped in 3 different sets: methylation of 1 to 3 genes, 4 to 7 genes, and 8 to 10 genes.
Prevalence and clinical implications of Eph receptor and Ephrin gene methylation in primary ALL
We subsequently evaluated the methylation status of EPHA2, -A4, -A5, -A6, -A7, -A10, EPHB1, -B2, -B3, -B4, EFNA1, -A3, -A5, and EFNB1 and -B2 genes in pretreatment bone marrow samples obtained before any therapy from 64 patients with ALL. Patient characteristics are shown in supplemental Table 1. All patients had been homogenously treated with hyper cyclophosphamide, vincristine, adriamycin, and dexamethasone (hyper-CVAD)–based chemotherapy at MDACC.
Methylation density for each gene is shown in Figure 2. By using a methylation density cutoff of 15%, methylation frequencies (percentage of patients in whom a gene is methylated) in ALL ranged from 18% to 96% (Figure 1A-B). Most genes were found frequently methylated, and in most cases, 2 or more EphA and EphB receptors were found concomitantly methylated in patient samples (data not shown).
We further analyzed associations between gene methylation status and overall survival in ALL patients. To perform this exploratory analysis, patients were divided into 3 groups: those with methylation (≥ 15%) of 1 to 3 genes, 4 to 7 genes, and 8 to 10 genes. The median survival of patients in the first group of patients had not been reached, whereas it was 120 weeks for the second group and 48 weeks for the third group (P = .048; Figure 2B). This analysis cannot exclude that the survival effect observed is due to the presence of a hypermethylator phenotype instead of epigenetic inactivation of the genes analyzed here.
Expression of EPHB4, EFNB2, and EFNA5 in leukemia cells and their responses to 5-aza-2′-deoxycitidine treatment
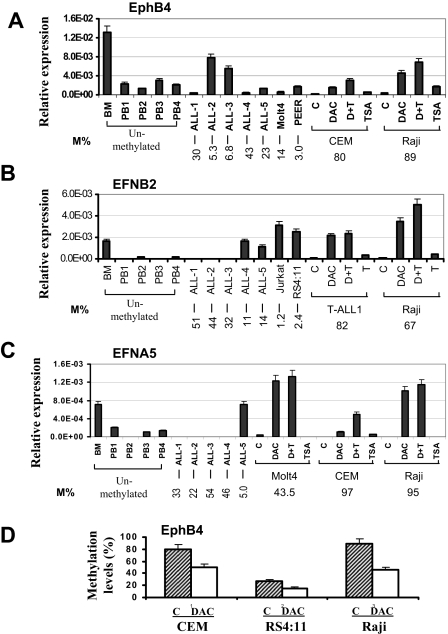
To examine the relationship between DNA methylation and gene expression, mRNA levels of EPHB4, EFNB2, and EFNA5 were analyzed by quantitative real-time PCR in normal bone marrow, normal peripheral blood cells, leukemia cell lines, and ALL bone marrow samples. EPHB4, EFNB2, and EFNA5 genes were expressed in normal bone marrow but were expressed at relatively low levels in normal peripheral blood cells (Figure 3A-C). These genes were either not expressed or very weakly expressed in hypermethylated primary ALL bone marrow cells and cell lines but were expressed in unmethylated or partially methylated ALL bone marrow cells and ALL cell lines (Figure 3A-C). In general, expression of EPHB4, EFNB2, and EFNA5 was restored in methylated leukemia cell lines treated with DAC with or without TSA (Figure 3A-C). Finally, we analyzed EPHB4 methylation status in CEM, RS4:11, and the Raji cell line before and after therapy with DAC. This resulted in a significant decrease in the methylation density of EPHB4 in these cell lines as measured by bisulfite pyrosequencing (Figure 3D). This suggests that the restoration of EPHB4 gene expression occurs via hypomethylation of the EPHB4 DNA sequence. These data also indicate that the DNA methylation of these genes is associated with suppressed gene expression.
Figure 3.
Gene expression and effect of demethylating treatment. Expression of EPHB4 (A), EFNB2 (B), and EFNA5 (C) genes was determined in normal BM, normal peripheral blood cells CD19+ (PB), primary ALL bone marrow, and leukemia cell lines. Leukemia cells were either untreated (C) or treated with DAC only, TSA only, or both (D + T) as described in “RNA extraction and real-time PCR” and “5-Aza-2′-deoxycytidine and/or trichostatin A treatment.” The relative gene expression was determined by real-time PCR assays and normalized to that of GAPDH. Methylation density (M%) for each samples and pretreatment cell lines are shown on the bottom. (D) Methylation levels of EphB4 before and after DAC treatment. Pyrosequencing was performed to determine methylation density. Error bars indicate range.
Role of DNA methylation on the promoter activity of EPHB4
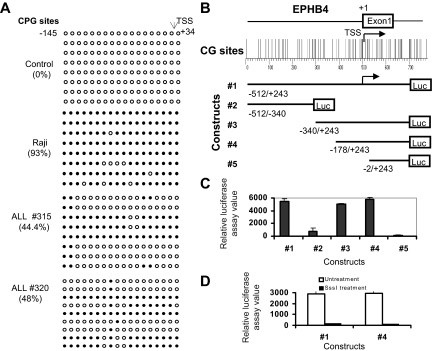
Because it is difficult to study all members of the Eph receptor/ephrin family in detail, we decided to focus on EPHB4 because this gene was originally identified and abundantly expressed in human BM CD34+ cells.27 To explore the potential role of EPHB4 promoter methylation in silencing gene transcription, we designed 5 pairs of pyrosequencing assays, each covering several CpG sites across the entire EPHB4 promoter CGI. We also performed bisulfite sequencing of region from −145 to +34 of the EPHB4 promoter encompassing 18 CpG sites. Methylation mapping by pyrosequencing and bisulfite sequencing revealed that EPHB4 was methylated over its 1.2-kb entire CGI in EPHB4-negative Raji cells and in primary ALL samples but not in normal controls (Figure 4A and data not shown). To determine whether hypermethylation of the CGI is associated with transcriptional silencing of the EPHB4 gene, we tested the promoter activity of the EPHB4 CGI in 293 T cells using 5 pGL3 luciferase reporter constructs containing serial deletion fragments of the EPHB4 promoter (Figure 4B). We observed a remarkable increase in luciferase activity of constructs 1, 3, and 4 and greatly reduced luciferase activity of constructs 2 and 5 (Figure 4B). These results suggest that the CGI around EPHB4 promoter, especially the region near EPHB4 TSS, contain promoter activity. To investigate the role of DNA methylation in regulating EPHB4 expression, we excised the promoter sequence from constructs 1 and 4, treated DNA fragments with SssI methylase, and then religated methylated DNA fragments into the pGL3 vector. Promoter activity of the methylated pGL3-EphB4 1 and 4 constructs were 45 and 31 times lower (and virtually silenced) than those of the unmethylated constructs 1 and 4 (Figure 4C). Taken together, these results indicate that promoter hypermethylation influences EPHB4 gene transcription.
Figure 4.
Promoter hypermethylation silences expression of EPHB4. (A) Methylation status of EPHB4 gene was analyzed by bisulfite sequencing of the promoter region. Each row of circles represents the sequence of an individual clone; ○, unmethylated CpG sites; and ●, methylated CpG sites. (B) Diagram of the human EphB4 promoter region studied. CpG sites are indicated by short vertical bars. Arrows point to TSS. Below are serial deletion constructs of the EphB4 regulatory sequence. (C) Relative luciferase activity of different portions of the unmethylated EphB4 promoter constructs pGL3-EphB4 1, 2 3, 4, and 5 in 293T cells. Error bars indicate range. (D) Relative luciferase activities of unmethylated (□) and methylated (■) pGL3-EphB4 1 and 4 vectors in 293T cells. Promoter methylation of the EphB4 5′ region inhibits luciferase activity.
Ectopic expression of EphB4 in Raji cell lines induces apoptosis and suppresses cell growth and colony formation
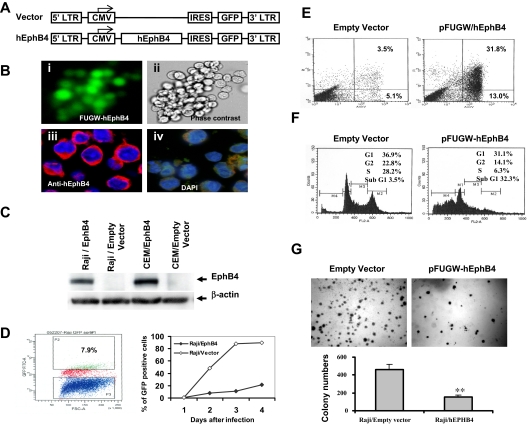
To investigate the relevance of EPHB4 function in leukemia cells, we expressed human EPHB4 with a lentiviral vector (FUGW-hEphB4) in Raji and CEM cell lines, in which the endogenous EPHB4 gene is hypermethylated and silenced. FUGW-hEphB4 contains an IRES that permits the expression of hEphB4 and GFP from a single bicistronic mRNA. Therefore, the expression of GFP was used to identify successfully transduced cells. EPHB4 transgene in Raji and CEM cells was confirmed by flow cytometry, immunofluorescence analysis, real-time PCR, and Western blot (Figure 5A-C). The expression of exogenous EPHB4 in Raji cells was detected in the cytoplasm and on the cell surface, especially at leading membrane edges (Figure 5B). Overexpression of EPHB4 in Raji cells dramatically suppressed cell growth rate, as determined by GFP sorting. Conversely, no significant effects were observed in cells infected with FUGW empty vector (Figure 5D). Annexin V–PE staining of FUGW-hEphB4 transduced Raji cells demonstrated approximately 45% of apoptotic cells 2 days after transduction, whereas only 9% of empty vector transduced GFP+ cells stained positively for annexin V–PE (Figure 5E). Moreover, by comparison with vector transduced cells, FUGW-hEphB4 lentivirus infected Raji cells displayed a significant appearance of a sub-G1 fraction by flow cytometric analysis 3 days after transduction. No significant alteration in the cell-cycle profile was observed in Raji cells infected with FUGW empty vector (Figure 5F). The effect of FUGW-hEphB4 overexpression on Raji cell growth was further examined by colony formation assays. The colony numbers of FUGW-hEphB4-lentivirus transduced cells significantly decreased compared with that of empty vector-transduced cells (P < .001; Figure 5G). Similar results were also observed in CEM cell line (data not shown). Taken together, these data suggest that epigenetic silencing of EPHB4 in Raji and CEM cells may be critical for their survival, supporting the notion that EphB4 functions as a tumor suppressor in cells where the EPHB4 gene is hypermethylated and transcriptionally silenced.
Figure 5.
Functional consequences of reintroducing EPHB4 expression into Raji cells. (A) Lentiviral constructs for transducing EPHB4 and controls. (B) Ectopic expression and membrane localization of lentivirus-delivered EPHB4 in Raji cells. Raji cells were infected with FUGW-hEphB4 or FUGW-lentivirus. Cells were examined by fluorescence microscopy (i, ×40) or phase contrast (ii, ×40). Raji cells transduced with FUGW- hEphB4 resulted in the expression of EPHB4 as shown by immunofluorescence staining with Alexa 594 (red) on the cell membrane (iii, ×60), whereas the empty vector had no effect and had only shown 4′,6-diamidino-2-phenylindole nuclear staining (iv, ×60). For more information see “Immunofluorescence microscopy.” (C) Western blot analysis of EPHB4 expression in Raji and CEM cells transduced with EphB4 lentivirus or empty vector. (D) Assessment of GFP positive Raji cell proliferation after infected with FUGW-EphB4 lentivirus or control using flow cytometric GFP sorting. Growth curves for GFP-positive Raji cells were determined for each of 3 replicates averaged at the indicated time points. (E) Analysis of apoptosis in Raji cells 2 days after lentivirus infection using flow cytometry and annexin V–PE staining. (F) FACS analysis of cell-cycle distributions measured 3 days after lentivirus infection. The percentage of cells in sub-G1 is presented. (G) For colony formation assay, Raji cells infected with FUGW-EphB4 lentivirus or controls were grown in soft agarose. Bar graph (bottom) represents experiments shown on top. **P < .01.
Molecular mechanisms of EphB4 tumor suppression effect in Raji cells
To analyze global molecular changes after transduction of EPHB4 in Raji cells, we performed a cDNA microarray analysis. compared with Raji/empty vector cells, the Raji/EphB4 cells showed a different gene expression pattern. In fact, 292 genes were found up-regulated (supplemental Table 3), whereas 170 were down-regulated after EPHB4 transduction (supplemental Table 4). Further details on this analysis are shown in the supplemental data.
EphB4 activation inhibits Akt phosphorylation after Ephrin B2 stimulation
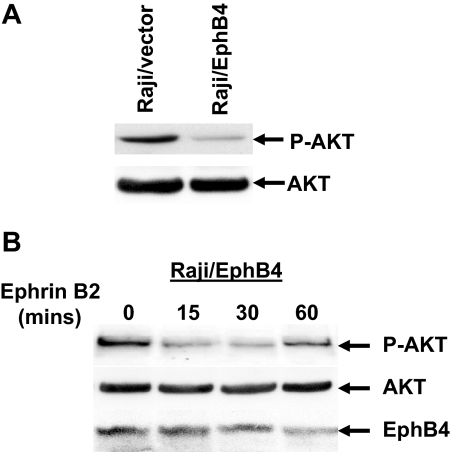
To investigate in part the molecular mechanisms of EphB4 mediated leukemia cell growth inhibition and apoptosis, we analyzed the potential involvement of signaling pathways in EphB4 action by analyzing the phosphorylation status of several proteins commonly associated with receptor tyrosine kinase signaling transduction pathways. To do so, we investigated the effects of ligand treatment with ephrinB2 in EphB4-expressing Raji cells. By comparing Raji cells transduced with empty vector, the phosphorylation of AKT at ser473 was significantly decreased in Raji/EphB4 cells. In contrast, the level of total AKT protein was not affected in Raji/EphB4 or Raji/empty vector cells (Figure 6A). We further stimulated Raji/EphB4 cells with clustered human FC-Ephrin B2 at different time points. Phosphorylated AKT was significantly decreased in Raji/EphB4 cells after 15 to 30 minutes of EphrinB2 stimulation (Figure 6B). No apparent effect of EphB4 on CRK, SRC, and ERK, MEK1 phosphorylation was noted in Raji/EphB4 cells (data not shown). These data suggest that down-regulation of the AKT pathway is involved in EphB4-mediated growth inhibition in ALL.
Figure 6.
Ephrin-B2 stimulation of Raji cells lead to down-regulation of AKT phosphorylation. (A) Raji cells transduced with empty vector (Raji/vector) or Ephb4 lentivirus construct (Raji/EphB4) were serum-starved for 24 hours and stimulated with 3 μg/mL clustered human FC-Ephrin B2 for 15 minutes. Cell lysates were analyzed by Western blotting to determine the phosphorylation status of AKT and total AKT levels. (B) Raji/EphB4 cells were serum-starved for 24 hours and stimulated with 3 μg/mL clustered human FC-Ephrin B2 for the indicated time points. Lysates were then probed with anti–phospho-AKT, total AKT, and EphB4.
Discussion
Using MCA/microarray, we identified several Eph receptor and ephrin genes as potential hypermethylation targets in ALL patients. We further investigated the methylation profile of the entire Eph/ephrin family members in leukemia cell lines and primary ALL bone marrows by pyrosequencing. Fifteen Eph/ephrin family genes, including EPHA2, -A4, -A5, -A6, -A7, -A10, EPHB1, -B2, -B3, -B4, EFNA1, -A3, -A5, and EFNB1 and -B2, were frequently hypermethylated in leukemia cells. The methylation characteristics of specific Eph/ephrin gene are different in different tumor types. EPHA2, EPHA4, EPHA5, EPHA6, EPHA7, EPHA10, EPHB2, and EPHB3 are found commonly methylated in various leukemia cell lines and ALL samples. EPHB6, although frequently methylated in breast cancer,18 was found unmethylated in the leukemia cell lines studied. EPHB4 was found more frequently methylated in T- and B-lineage lymphocytic cell lines compared with myeloid cell lines. In primary ALL samples, 15 EPH/EPHRIN family members were found frequently methylated, and in most cases, 2 or more EPHA and EPHB receptors were found hypermethylated in a concomitant fashion. Furthermore, methylation of multiple Eph/ephrin family genes was associated with a worse outcome. Those patients with methylation of more than 4 genes appeared to have a significant worse outcome. That said, these results should be considered as exploratory, as it is possible that the survival effect observed is due to the presence of a hypermethylator phenotpye and not merely to Ephrin family inactivation. Although these results need to be validated in larger clinical samples, they suggest that Eph/ephrin methylation have an important role in ALL pathogenesis. This survival analysis should be considered as exploratory, as we cannot conclude that the survival effect observed is not due to a hypermethylation phenomenon instead of individual Eph/ephrine methylation. Larger, more detailed analysis will be needed to define these results.
We here examined the expression profile of EPHB4, EFNB2, and EFNA5 in leukemia cell lines and ALL samples. We confirmed methylation-associated transcription silencing of these genes based on their gene expression profile and demethylation treatment analysis. We further confirmed that EPHB4 was frequently silenced in T- and B-lineage lymphocytic leukemia cell lines and ALL bone marrows and that its promoter hypermethylation is a mechanism for regulating EPHB4 transcription.
We further characterized the promoter methylation, transcriptional silencing, and the putative tumor suppressor function of EPHB4 in human leukemia cell lines. We cloned full-length human EphB4 cDNA into lentivirus vector and transduced them into Raji cells. Forced restoration of EPHB4 in EPHB4-silenced Raji cells resulted in apoptotic cell death and decreased cell growth and colony formation. These results indicate that EphB4 functions as a tumor suppressor in ALL. Cancer cells may elude the tumor suppressor activities of EPHB4 or other EPH receptors by hypermethylation and epigenetic down-regulation of the EPH receptor or ephrin expression in leukemia.
We also studied the molecular effect of EPHB4 in Raji cells using a cDNA microarray. We identified a significant variation in approximately 462 transcripts (292 overexpression and 170 down-regulation) associated with the exogenous restoration of EPHB4 expression. Interestingly, 64 genes whose putative function is that of a tumor suppressor were found as being up-regulated, and 54 genes whose function related to oncogenes were down-regulated. All these data together suggest that various pathways involved in cell survival, proliferation, signaling, and development mediate the effects of EphB4 in ALL.
Recent studies of Eph receptors and ephrin signaling have indicated the involvement of several different of pathways, including activation or inhibition of MAPK/ERK pathways by EphB2 receptor,20 inhibition of Ras/ERK1/2 signaling cascade by EphA2,19 and phosphorylation of Src kinases and Akt by EphA2 and EphA4. However, signaling pathways involved in EphB4 tyrosine kinase in hematopoiesis have not been fully determined. In this study, we examined the phosphorylation status of the major components of the MAPK pathway, including ERK, MAPK, AKT, CRK, and ABL tyrosine kinase. We observed that ectopic expression of EphB4 inhibited AKT phosphorylation in Raji cells. These results suggested that EphB4-mediated growth inhibition occurs via modulation of AKT signaling. In addition, a recent report describing that activation of EphB4 signaling inhibits breast cancer cell proliferation provides further evidence linking EphB4 with mitogen-induced signaling pathways.15 Thus, investigation of the signal transduction pathways activated by the Eph tyrosine kinases in leukemia will help understand additional aberrant molecular mechanisms in ALL and to define the biologic function of the Eph/ephrin system in normal hematopoiesis and leukemogenesis. That said, a limitation of this study is that we have limited the functional analysis just to one member of the family (EphB4). Analysis of more members and interactions would help in our understanding of Eph/Ephrin dysregulation in ALL.
In summary, we identified that 15 of Eph/ephrin family genes are frequently hypermethylated and down-regulated in human leukemia cell lines and ALL samples. We showed that EPHB4 expression is frequently reduced or lost in ALL and that this loss is potentially associated with poor outcome in ALL. Reintroduction of EPHB4 into EphB4-deficient cells significantly induced apoptosis and reduced cell growth, suggesting that EPHB4 has tumor suppressor activities in ALL. Because several Eph/ephrin family members exhibits tumor suppressor functions,14,14 epigenetic dysregulation of their signaling pathways may selectively lead to the aberrant methylation part of the downstream genes and provide a growth advantage to tumor cells. Further in-depth investigation of these transcriptional regulators or signal pathways is required to define the role of Eph/ephrins in carcinogenesis.
Acknowledgments
We thank Dr Scadden at Harvard Medical School for scientific support.
This work was supported by a Leukemia SPORE (P50 CA100632) Career Development Award to S.-Q.K. and by National Cancer Institute grants CA100067 and CA105771, the Leukemia & Lymphoma Society of America, and an M. D. Anderson Cancer Center Physician-Scientist Award from the Commonwealth Cancer Foundation for Research (all to G.G.-M.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: S.-Q.K. performed all experiments, wrote the manuscript, analyzed the data, and partially funded the study; H.B., Z.-H.F., G.L., H.Y., W.T., and Z.Z.W. contributed to the experimental procedures of the manuscript and data analysis; and G.G.-M. designed the study, wrote the manuscript, analyzed the data, and funded the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for S.-Q.K. is Medical Genetics, University of Texas Health Science Center at Houston.
Correspondence: Guillermo Garcia-Manero, Department of Leukemia, University of Texas M. D. Anderson Cancer Center, Box 428, 1515 Holcombe Blvd, Houston, TX 77030; e-mail: ggarciam@mdanderson.org.
References
- 1.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298(5600):1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 2.Unified nomenclature for Eph family receptors and their ligands, the ephrins. Eph Nomenclature Committee. Cell. 1997;90(3):403–404. doi: 10.1016/s0092-8674(00)80500-0. [DOI] [PubMed] [Google Scholar]
- 3.Pasquale EB. Eph-ephrin promiscuity is now crystal clear. Nat Neurosci. 2004;7(5):417–418. doi: 10.1038/nn0504-417. [DOI] [PubMed] [Google Scholar]
- 4.Bruckner K, Klein R. Signaling by Eph receptors and their ephrin ligands. Curr Opin Neurobiol. 1998;8(3):375–382. doi: 10.1016/s0959-4388(98)80064-0. [DOI] [PubMed] [Google Scholar]
- 5.Hafner C, Schmitz G, Meyer S, et al. Differential gene expression of Eph receptors and ephrins in benign human tissues and cancers. Clin Chem. 2004;50(3):490–499. doi: 10.1373/clinchem.2003.026849. [DOI] [PubMed] [Google Scholar]
- 6.Fox BP, Tabone CJ, Kandpal RP. Potential clinical relevance of Eph receptors and ephrin ligands expressed in prostate carcinoma cell lines. Biochem Biophys Res Commun. 2006;342(4):1263–1272. doi: 10.1016/j.bbrc.2006.02.099. [DOI] [PubMed] [Google Scholar]
- 7.Hafner C, Becker B, Landthaler M, Vogt T. Expression profile of Eph receptors and ephrin ligands in human skin and downregulation of EphA1 in nonmelanoma skin cancer. Mod Pathol. 2006;19(10):1369–1377. doi: 10.1038/modpathol.3800660. [DOI] [PubMed] [Google Scholar]
- 8.Himanen JP, Nikolov DB. Eph receptors and ephrins. Int J Biochem Cell Biol. 2003;35(2):130–134. doi: 10.1016/s1357-2725(02)00096-1. [DOI] [PubMed] [Google Scholar]
- 9.Kullander K, Klein R. Mechanisms and functions of Eph and ephrin signalling. Nat Rev Mol Cell Biol. 2002;3(7):475–486. doi: 10.1038/nrm856. [DOI] [PubMed] [Google Scholar]
- 10.Surawska H, Ma PC, Salgia R. The role of ephrins and Eph receptors in cancer. Cytokine Growth Factor Rev. 2004;15(6):419–433. doi: 10.1016/j.cytogfr.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 11.Zelinski DP, Zantek ND, Stewart JC, Irizarry AR, Kinch MS. EphA2 overexpression causes tumorigenesis of mammary epithelial cells. Cancer Res. 2001;61(5):2301–2306. [PubMed] [Google Scholar]
- 12.Nakada M, Drake KL, Nakada S, Niska JA, Berens ME. Ephrin-B3 ligand promotes glioma invasion through activation of Rac1. Cancer Res. 2006;66(17):8492–8500. doi: 10.1158/0008-5472.CAN-05-4211. [DOI] [PubMed] [Google Scholar]
- 13.Foubert P, Silvestre JS, Souttou B, et al. PSGL-1-mediated activation of EphB4 increases the proangiogenic potential of endothelial progenitor cells. J Clin Invest. 2007;117(6):1527–1537. doi: 10.1172/JCI28338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Batlle E, Bacani J, Begthel H, et al. EphB receptor activity suppresses colorectal cancer progression. Nature. 2005;435(7045):1126–1130. doi: 10.1038/nature03626. [DOI] [PubMed] [Google Scholar]
- 15.Noren NK, Foos G, Hauser CA, Pasquale EB. The EphB4 receptor suppresses breast cancer cell tumorigenicity through an Abl-Crk pathway. Nat Cell Biol. 2006;8(8):815–825. doi: 10.1038/ncb1438. [DOI] [PubMed] [Google Scholar]
- 16.Alazzouzi H, Davalos V, Kokko A, et al. Mechanisms of inactivation of the receptor tyrosine kinase EPHB2 in colorectal tumors. Cancer Res. 2005;65(22):10170–10173. doi: 10.1158/0008-5472.CAN-05-2580. [DOI] [PubMed] [Google Scholar]
- 17.Davalos V, Dopeso H, Castano J, et al. EPHB4 and survival of colorectal cancer patients. Cancer Res. 2006;66(18):8943–8948. doi: 10.1158/0008-5472.CAN-05-4640. [DOI] [PubMed] [Google Scholar]
- 18.Fox BP, Kandpal RP. Transcriptional silencing of EphB6 receptor tyrosine kinase in invasive breast carcinoma cells and detection of methylated promoter by methylation specific PCR. Biochem Biophys Res Commun. 2006;340(1):268–276. doi: 10.1016/j.bbrc.2005.11.174. [DOI] [PubMed] [Google Scholar]
- 19.Guo H, Miao H, Gerber L, et al. Disruption of EphA2 receptor tyrosine kinase leads to increased susceptibility to carcinogenesis in mouse skin. Cancer Res. 2006;66(14):7050–7058. doi: 10.1158/0008-5472.CAN-06-0004. [DOI] [PubMed] [Google Scholar]
- 20.Huusko P, Ponciano-Jackson D, Wolf M, et al. Nonsense-mediated decay microarray analysis identifies mutations of EPHB2 in human prostate cancer. Nat Genet. 2004;36(9):979–983. doi: 10.1038/ng1408. [DOI] [PubMed] [Google Scholar]
- 21.Tang XX, Zhao H, Robinson ME, et al. Implications of EPHB6, EFNB2, and EFNB3 expressions in human neuroblastoma. Proc Natl Acad Sci U S A. 2000;97(20):10936–10941. doi: 10.1073/pnas.190123297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nosho K, Yamamoto H, Takahashi T, et al. Genetic and epigenetic profiling in early colorectal tumors and prediction of invasive potential in pT1 (early invasive) colorectal cancers. Carcinogenesis. 2007;28(6):1364–1370. doi: 10.1093/carcin/bgl246. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, Kataoka H, Suzuki M, Sato N, Nakamura R, Tao H, et al. Downregulation of EphA7 by hypermethylation in colorectal cancer. Oncogene. 2005;24(36):5637–5647. doi: 10.1038/sj.onc.1208720. [DOI] [PubMed] [Google Scholar]
- 24.Kuang SQ, Tong WG, Yang H, et al. Genome-wide identification of aberrantly methylated promoter associated CpG islands in acute lymphocytic leukemia. Leukemia. 2008;22(8):1529–1538. doi: 10.1038/leu.2008.130. [DOI] [PubMed] [Google Scholar]
- 25.Shu J, Jelinek J, Chang H, et al. Silencing of bidirectional promoters by DNA methylation in tumorigenesis. Cancer Res. 2006;66(10):5077–5084. doi: 10.1158/0008-5472.CAN-05-2629. [DOI] [PubMed] [Google Scholar]
- 26.Kuang SQ, Ling X, Sanchez-Gonzalez B, Yang H, Andreeff M, Garcia-Manero G. Differential tumor suppressor properties and transforming growth factor-beta responsiveness of p57KIP2 in leukemia cells with aberrant p57KIP2 promoter DNA methylation. Oncogene. 2007;26(10):1439–1448. doi: 10.1038/sj.onc.1209907. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z, Miura N, Bonelli A, et al. Receptor tyrosine kinase, EphB4 (HTK), accelerates differentiation of select human hematopoietic cells. Blood. 2002;99(8):2740–2477. doi: 10.1182/blood.v99.8.2740. [DOI] [PubMed] [Google Scholar]
- 28.Sun X, Zhang W, Ramdas L, et al. Comparative analysis of genes regulated in acute myelomonocytic leukemia with and without inv(16)(p13q22) using microarray techniques, real-time PCR, immunohistochemistry, and flow cytometry immunophenotyping. Mod Pathol. 2007;20(8):811–820. doi: 10.1038/modpathol.3800829. [DOI] [PubMed] [Google Scholar]
- 29.Yendamuri S, Trapasso F, Ferracin M, et al. Tumor suppressor functions of ARLTS1 in lung cancers. Cancer Res. 2007;67(16):7738–7745. doi: 10.1158/0008-5472.CAN-07-1481. [DOI] [PubMed] [Google Scholar]