Abstract
Genetic screens in Drosophila have identified many genes involved in neural development and function. However, until recently, it has been impossible to monitor neural signals in Drosophila central neurons, and it has been difficult to make specific perturbations to central neural circuits. This has changed in the past few years with the development of new tools for measuring and manipulating neural activity in the fly. Here we review how these new tools enable novel conceptual approaches to “cracking circuits” in this important model organism. We discuss recent studies aimed at defining the cognitive demands on the fly brain, identifying the cellular components of specific neural circuits, mapping functional connectivity in those circuits, and defining causal relationships between neural activity and behavior.
What is circuit-cracking?
The Oxford English Dictionary defines to crack as “to puzzle out, make out, solve, discuss”. To completely solve a neural circuit would require
describing a behavior whose neural circuit mechanisms we seek to understand,
identifying which neurons are involved,
determining what drives activity in each type of neuron and how these signals are transformed through the circuit,
discovering the cellular, synaptic, and circuit mechanisms underlying these neural transformations, and
understanding why these neural transformations are useful intermediates in producing this behavior.
Why the fly?
Cracking every neural circuit in every species would be impossible and pointless. But a detailed comparision of several circuits in different species should help reveal what features of neural circuits are fundamental and which are specializations. Many (though not all) neuroscientists believe that some neural circuits in Drosophila are worth including in this research program.
The most obvious reason is the power of the Drosophila genetic toolbox. Mouse neurogenetic tools are beginning to rival those of the fly, however, and so this reason is no longer preeminent. A more basic reason is numerical simplicity: there are about 1,000-fold fewer neurons in the brain of the fruit fly. Another advantage is the identified neuron—a stereotyped neuron that can be located (in theory) in every fly. Many Drosophila neurons are identifiable in this way. C. elegans is even simpler, and each of its 302 neurons is uniquely identifiable, but the behavioral repertoire of the worm is less extensive. Thus, the fly is a useful compromise between tractability and richness.
In this review, we have generally restricted our focus to studies published within the last four years. Several excellent reviews have summarized technical innovations in the genetic control of Drosophila neurons during this time period [1, 2]. Here, we instead examine how these tools succeed (and occasionally fail) in advancing the broad goal of understanding neural circuits. We also look critically at whether the anticipated power of this model system—to rapidly achieve complete understanding of a neural circuit—is currently realistic.
Defining the behavioral task
Circuit-cracking generally begins with an observable behavior that we seek to understand. Many classical Drosophila behavioral paradigms were designed to screen many flies simultaneously for profound defects. But neuroscientists are increasingly interested in fly behavior for its own sake, rather than simply viewing it as a tool for isolating genetic mutations [3].
A number of recent behavioral studies have expanded our notion of the fly's cognitive ability. For example, Drosophila can discriminate between subtly different abstract visual symbols, and can recognize a familiar object regardless of where in the visual field that object previously appeared [4-6]. Flies can also remember the spatial position of an object that has been removed from their environment [7]. They can detect tiny changes in odor concentration [8, 9], and can integrate olfactory information with visual signals [10-13] or other olfactory cues [9, 14]. A male can tell the difference between a virgin and non-virgin, and can learn to modify his courtship strategy after sexual rejection [15, 16]. Females can discriminate between the courtship songs of males of different species [17, 18]. Flies will fight over food or mates, and can learn to modify their fighting strategy based on past encounters [19]. None of the neural circuits mediating these behaviors are currently understood.
Behavioral observations can also reveal what algorithms might be used to accomplish a particular cognitive task. This is useful because it constrains the underlying neural computations. For example, a recent study asked how flying flies discriminate between a dangerous object (like a predator or colliding fly) and an object that would make a good landing site [20]. Flies were tethered inside a controlled visual environment where they could beat their wings freely. When any small visual object was presented to a fly, it attempted to turn away by adjusting the relative movements of its two wings. However, a visual edge elicited the opposite reaction. These results suggest that a fly may avoid collisions simply by avoiding any small object. This avoidance response is evidently suppressed by edges, which more likely correspond to safe landing sites. This simple algorithm suggests the existence of two opposing visuomotor circuits.
The ultimate goal of many behavioral experiments is to establish a baseline for genetic manipulations. Since some neural mechanisms are only required within a limited performance regime, a systematic approach is more likely to reveal a phenotype. For example, one recent study used a novel visual motion stimulus to systematically probe motor responses to patterns of varying velocity, contrast, luminance, spatial density, and coherence [21]. Different velocities evoked walking either toward or against the motion of the stimulus. Genetic manipulations revealed that some neurons in the optic lobe are selectively involved in one of these behaviors. Another recent study systematically explored how flies navigate along temperature gradients [22]. Flies were found to avoid both warm and cold extremes, preferring a narrow range of temperatures. Mutations in a Trp channel were found to selectively affect warmth avoidance, without affecting responses to cold. These sorts of studies are beginning to reveal that many behaviors are mediated by multiple neural circuits acting in parallel, some with opposing effects. This is perhaps not surprising, but it implies that it may be unrealistic to expect to find a unitary neural circuit underlying any particular behavior.
What neurons are involved?
We have little notion of what neurons are involved in many fly behaviors. For example, we have no idea which neurons mediate somatosensory, gustatory, or auditory behavior (except primary sensory cells and motorneurons). Although we know where the axons of the relevant primary sensory cells project, this doesn't make it trivial to find their postsynaptic targets. This is because the fly brain and thoracic ganglion are small (∼200 microns) relative to the size of an individual fly neuron (which can have a dendritic arbor >50 microns wide, and an axon >100 microns long). This means that one neuron can reach across multiple brain regions (Fig. 1). Additionally, neural somata are anatomically segregated from axons and dendrites: somata are restricted to the rind of the brain and thoracic ganglion, with axons and dendrites localized to the neuropil core of these structures. Thus, a neuron might have a dendrite or axon in one part of the brain, and a soma lying some distance away. Despite these difficulties several complementary approaches hold some promise for identifying candidate cellular components of neural circuits in the fly.
Figure 1. Drosophila neurons can span large distances in the brain.
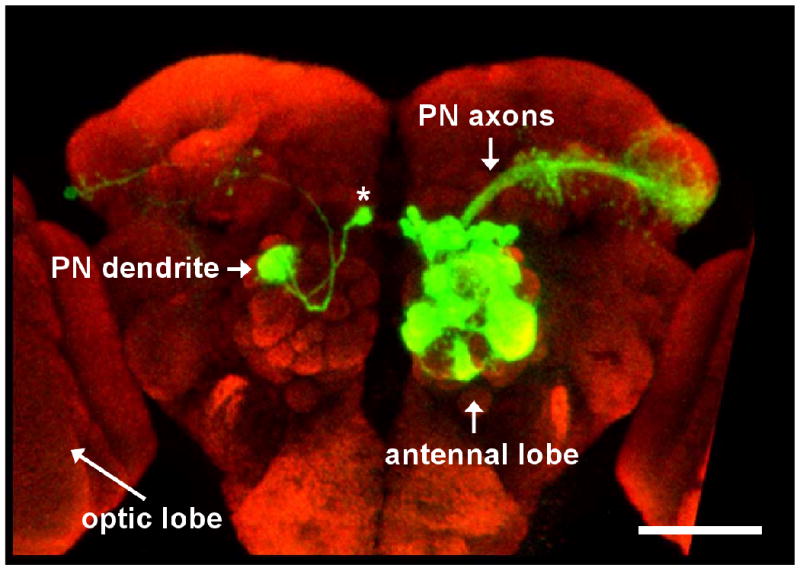
(a) On the right hemisphere of the brain a large number of PNs are labeled with GFP. On the left hemisphere a single PN is labeled revealing both its dendritic and axonal morphology (the cell body is indicated by the asterisk). Note the large distance the axon traverses. The optic lobes from both hemispheres of the brain are cropped in this image. The scale bar corresponds to approximately 50 μm. Modified with permission from Ref. 100.
Anatomical methods
In lucky cases, it can be possible to identify candidate cellular components of a circuit simply by visually screening Gal4 lines. The Gal4/UAS technique provides a way to drive gene ectopic expression in a genetically-defined subpopulation of cells (see [2, 23] for reviews). When crossed to a green fluorescent protein (GFP) reporter line, a Gal4 line will sometimes label only a small number of neurons. Gal4 labeling can also be further restricted according to a neuron's lineage [24]. If the GFP-labeled neurons reside in a highly-ordered brain region, their participation in a circuit can sometimes be obvious. This approach has been used to describe cellular components of three orderly circuits—the antennal lobe [25], mushroom body [25], and optic lobe [26, 27].
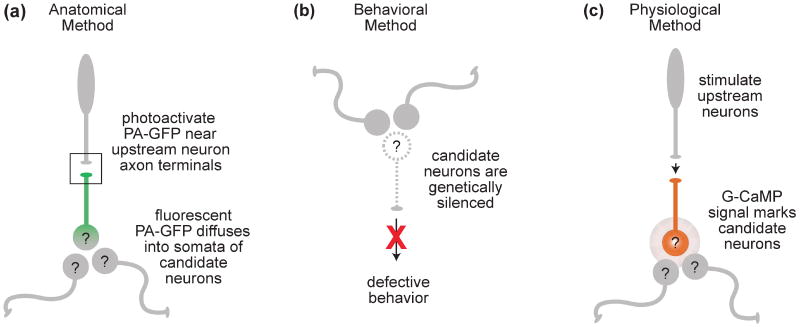
However, if GFP labeling is widespread, or if the brain region of interest is disorderly, this approach is less useful. A new approach to this problem uses a genetically-restricted form of targeted anatomical tracing [28]. Photoactivatible GFP is expressed under the control of either a pan-neuronal Gal4 line or a more restricted promoter. Photoactivation in a small region of neuropil can label all the neurons intersecting that region that express Gal4. For example, photoactivating a region where an axon tract terminates should label the dendrites of postsynaptic partners. Diffusion of photoactivated GFP out of these dendrites then labels the somata of the candidate cells (Fig. 2a).
Figure 2. Strategies for identifying neurons in sensory and behavioral circuits.
(a) A broadly expressed promoter is used to express PA-GFP in a large population of candidate neurons (labeled “?”). Photostimulation of the neuropil region containing the axons of known presynaptic neurons (box) increases PA-GFP fluorescence in the dendrites of postsynaptic neurons. Activated GFP molecules diffuse out of the dendrite and into the cell body revealing the location of the somata of postsynaptic neurons. Recordings can be made from these neurons by using the GFP signal to target a patch electrode.
(b) In this approach many different Gal4 lines are used to express a silencing agent such as tetanus toxin. If a behavioral defect is observed this indicates that the cells labeled by the Gal4 line were somehow involved in the behavior. The candidate neuron labeled “?” expresses the silencing agent.
(c) All neurons express the calcium sensor G-CaMP, but only the connected neuron responds to stimulation of the upstream neuron.
Finally, considerable effort has been directed at developing a transsynaptic tracer in Drosophila, but no generally useful tool has yet appeared. The problem stems from the vast evolutionary distance between flies and mammals: plant lectin and viral tracers that work well in mice are not necessarily useful in flies because the relevant endogenous receptor mechanisms are lacking.
Behavioral methods
Historically it has been more common to search for the genes rather than the neurons involved in a behavior. Gene identification via mutant screens, however, can provide a handle for identifying neurons if the population of neurons expressing the gene is not too large or diverse. Cell-specific rescue experiments can be used to identify the specific set of neurons in which the functional gene is required for normal behavior. This method has been used to identify neurons involved in circadian rhythms [29], courtship [30], temperature regulation [22], and learning and memory [31].
A second of type of behavioral screen directly probes neurons rather than genes. In this screen many candidate Gal4 lines are crossed with a UAS-transgene that silences neurons (either by blocking neurotransmitter release, inhibiting spiking, or killing the cells), and progeny are screened for defective behaviors. The goal is to identify neurons that are required for the behavior (Fig. 2b). A silencing screen should be particularly effective at identifying bottlenecks in a neural circuit (e.g., motorneurons). But if neural codes are distributed across large ensembles of central neurons, none of which are strictly required for behavioral performance, this approach might not yield a comprehensive set of important circuit elements. It will be interesting to see what kinds of neurons turn up in these screens. Only a few large-scale screens have been reported so far [21, 32, 33], but already some novel and intriguing circuit elements have emerged from these efforts.
One difficulty is the fact that many Gal4 lines label large and diverse sets of neurons. In this case, the number of neurons expressing the silencing UAS-transgene can be narrowed down using combinatorial methods for transgene expression (for details see ref. [34]). Additionally, a project is underway to produce an enhancer library of Gal4 lines that label relative small subsets of neurons (∼100), but that in total provides comprehensive coverage of all ∼200,000 fly neurons [35].
Physiological methods
A third approach is to use functional imaging. The idea is to express a fluorescent activity reporter (like G-CaMP) under the control of a pan-neuronal Gal4 line, or else a more restrictive Gal4 line arising from a screen. Imaging stimulus-evoked fluorescence in these flies should reveal all the (labeled) neurons that are activated by a particular stimulus (Fig. 2c). However, the available genetically-encoded activity sensors still lack sensitivity [36, 37], meaning that this method will reveal only neurons with high firing rates. This method could also be useful in identifying neurons that are downstream from a genetically-identified set of neurons. For this type of experiment the sensory stimulus could be bypassed with a genetically-encoded stimulation tool such as channelrhodopsin that can be used to activate known upstream neurons while monitoring responses in downstream neurons.
Assigning function to neurons
Once neurons have been genetically identified, we face the challenge of understanding how information is represented by neural activity in this circuit, and how this activity reflects the computations being performed. Until recently, it was considered impossible to monitor neural signals in the Drosophila central nervous system in vivo. As a result, most studies have sought to reveal circuit function by inactivating or stimulating neurons of interest—that is, testing whether neurons are necessary and sufficient for a behavior. Now with the development of functional imaging and single-cell electrophysiology we can also study how Drosophila neurons encode and process the information used to control behavior.
Testing necessity and sufficiency
Demonstrating necessity generally involves silencing a particular neuron or group of neurons. If this disrupts behavioral performance, then the silenced neurons were somehow required. Neurons can be silenced by blocking neurotransmitter release [38], reducing intrinsic excitability [39], or by killing them with a neurotoxin or cell-death gene [40]. Techniques that permit fine temporal control [41, 42] allow the question of necessity to be pinned to a specific point in time. Several recent studies have used this approach to ask if candidate neurons are required for specific visual responses [27], courtship behaviors [43], and circadian rhythms [39, 44], as well as hormonally-driven events in late adult development [45].
Demonstrating sufficiency generally involves artificially triggering activity in a neuron or group of neurons, and asking whether this activity alone is adequate to recapitulate a behavior. If so, then these neurons (and their downstream effector circuits) are adequate to produce this behavior. Neural activity can be boosted by overexpressing sodium channels [46], although this approach lacks temporal control. Temporal specificity can be achieved by placing neural activity under the control of light [47-51] or under the control of an exogenous ligand-gated channel [52, 53]. A different way to test sufficiency is to begin with a mutant fly having a behavioral defect, and then to rescue the defect by rescuing gene expression in a specific population of neurons.
Monitoring neural activity
Neurons rarely respond to only one stimulus and their responses are rarely binary. Thus, the question is not ‘which neurons underlie a specific behavior?’, but rather, ‘which activity patterns produce behavior?’ Answering this question requires measuring neural activity in vivo. Functional imaging and single-cell recordings are complementary tools for making such measurements.
The advantage of functional imaging is the ability to monitor the activity of many neurons simultaneously. Several genetically-encoded fluorescent activity sensors have been developed in Drosophila [54, 55], and permit recording from genetically-specified neurons. Of these, the most widely used is probably G-CaMP, which has a high signal-to-noise ratio. Functional imaging has been used to examine the representation of odors and taste in the brain [40, 56-59], and also to visualize memory-related representations in the mushroom bodies[60, 61] and plasticity in the antennal lobes[62, 63].
However, G-CaMP (and other genetically-encoded calcium indicators) have some disadvantages including limited sensitivity, small dynamic range, and slow kinetics [36, 37, 64]. Also, the level of stimulus-evoked fluorescence in a neuron will depend on its Gal4 expression level. This limits quantitative comparisons between the responses of different neurons.
By comparison with functional imaging, the advantage of electrophysiology is the ability to monitor activity with high sensitivity and high temporal resolution. Recordings from Drosophila peripheral neurons can be performed extracellularly with fine tungsten or glass electrodes [65-68]. Extracellular recordings have been used to map the odor response profiles of most primary receptor neurons in the olfactory system of the Drosophila adult and larva [69, 70]. In the Drosophila brain extracellular currents are generally too small to be resolved, so recordings from central neurons are performed with a patch electrode at the soma in cell-attached or whole-cell mode. Recordings can be targeted to genetically-specified neurons by marking these cells with a fluorescent protein. In the fly brain, whole-cell recordings have been used to map the receptive fields of both second- and third-order olfactory neurons [14, 71-76]. Whole-cell recordings have also been used to characterize the receptive fields of visual neurons [77], and to probe circadian fluctuations in the intrinsic excitability of identified neurons [78].
A disadvantage of this approach is the inability to record from many neurons simultaneously. Moreover, whole-cell and cell-attached recordings are necessarily limited to neurons with large somata, although this limit is being pushed to ever-smaller cells [75, 79].
An integrative approach
Necessity and sufficiency experiments help define causal relationships between neurons and behavior, but this kind of experiment can be difficult to interpret. Most tasks will probably involve neural activity patterns that are distributed across large populations of neurons. In a distributed neural representation, which elements can meaningfully be considered necessary or sufficient? The answer likely depends critically on the demands of the task, but measurements of neural activity provide useful clues. For example, consider a simple example from a recent study of olfactory behavior in Drosophila larvae [70]. Larvae are attracted to the odor ethyl acetate, and information about this odor is carried by at least two different odor receptors. Depending on the concentration of ethyl acetate, either one of these receptors can be un/necessary or in/sufficient. This apparent inconsistency was resolved by asking how these two receptors respond to this odor. Electrophysiological measurements showed that one receptor has a higher affinity for ethyl acetate compared to the other. At low concentrations, the high-affinity receptor is necessary for ethyl acetate attraction, while the low-affinity receptor is dispensable. The reverse is true for the low-affinity receptor (Fig. 3). This example shows how seemingly contradictory results can be obtained if the behavioral space is not probed in fine detail, and if the selectivity of the neurons encoding the information used for the behavior is not known. The difficulty in interpreting necessity and sufficiency experiments increases for more complex behaviors and more distributed neural representations. In this case, measurements of neural activity provide a context for effectively designing and interpreting these types of experiments.
Figure 3. Monitoring neural activity helps interpret necessity and sufficiency experiments.
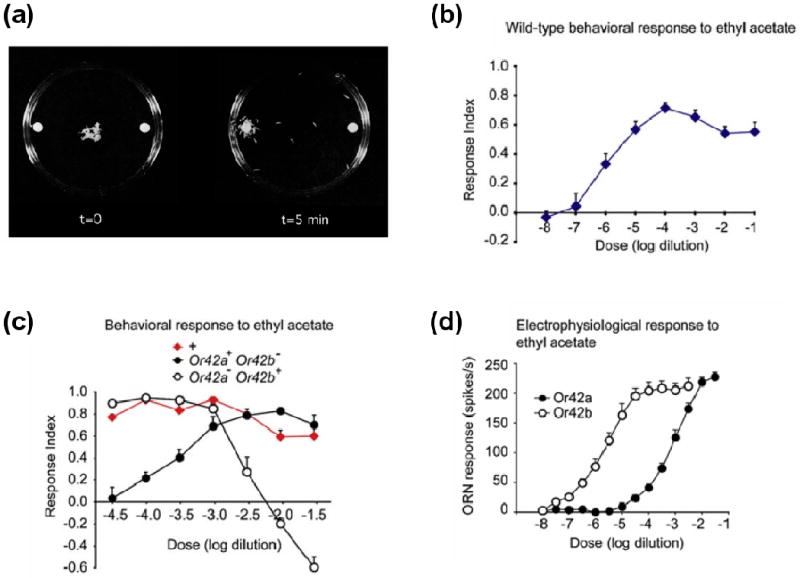
(b) Assay for larval chemotaxis behavior.
(c) Wild-type behavioral response to increasing concentrations of the odor ethyl acetate. The response index measures the degree of attractive behavior. Positive values indicate attraction and negative values indicate repulsion.
(d) Comparison of behavioral responses between wild-type flies and two different odor receptor mutants, Or42a and Or42b. Wild-type flies display uniform attractive behavior to concentrations of ethyl acetate across four orders of magnitude. The odor receptor Or42b is necessary for attraction to low concentrations of ethyl acetate, but is unnecessary for high concentrations. The reverse is true for Or42a.
(e) Electrophysiological analysis of olfactory receptor neurons (ORNs) expressing either Or42a or Or42b. Or42b is sensitive to changes in low concentrations of ethyl acetate but saturates at high concentrations. Or42a is only activated by high concentrations of ethyl acetate. Modified with permission from Ref. 70.
Mechanisms of circuit processing
The next step is to uncover the synaptic and cellular mechanisms underlying sensorimotor transformations. This means mapping synaptic connectivity and investigating the functional properties of these connections.
The gold standard for demonstrating a synaptic connection is to directly visualize both pre- and postsynaptic specializations with electron microscopy. Systematic ultrastructural studies of the Drosophila brain are currently underway in several laboratories, but full 3D reconstructions of any brain region are probably not imminent. What is urgently needed is a high-throughput method for establishing whether two neurons are connected. A recent study in C. elegans describes a promising new strategy [80]. A pair of complementary GFP fragments is expressed in different neurons, each tethered to the extracellular domain of a transmembrane carrier protein. In locations where the membranes of the two neurons are closely apposed, fluorescence is produced. When the GFP fragments are localized to synapses, this method is a reliable reporter of whether (and where) two neurons in the worm are synaptically connected.
Another litmus test for synaptic connectivity is electrophysiological: if one can demonstrate that a precisely-timed depolarization of one neuron evokes a short-latency synaptic response in the other neuron, then a direct connection is unequivocal. Paired intracellular recordings are feasible [71] but heroic. It may be faster to perform a single intracellular recording while mapping candidate channelrhodopsin-expressing presynaptic inputs using brief localized pulses of light [81]. Electrophysiological approaches to mapping connectivity have the advantage of revealing some of the functional properties of the synaptic connections between neurons including the relative strength of the synapse and whether it is inhibitory or excitatory. In the antennal lobe, a recent study explored the properties of the connection between olfactory receptor neurons and postsynaptic projection neurons, and found that the unique properties of this synapse help explain many features of projection neuron odor responses [82].
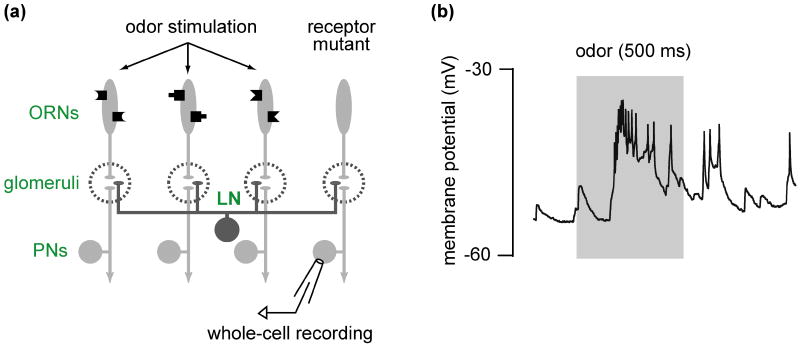
Genetic perturbation experiments can be coupled with physiological measurements to reveal the properties of the various synaptic inputs to a neuron. The logic of these experiments is to selectively silence or activate identified neuronal connections, and then measure the effects of this manipulation on the postsynaptic neurons. For example, in the antennal lobe, silencing the direct receptor neuron input to projection neurons has revealed the functional organization of local excitatory circuits in the antennal lobe (Fig. 4) [73, 74, 83].
Figure 4. Functional dissection of synaptic inputs to a neuron.
(a) Experimental design for silencing one synaptic input in order to reveal others. The odor receptor gene expressed in one particular type of olfactory receptor neuron (ORN) is mutated, and therefore these neurons don't respond to odors. A whole-cell recording is made from a projection neuron directly postsynaptic to the mutant ORNs. An odor is used to stimulate the remaining functional ORN types. Local neurons (LN) are anatomically poised to mediate communication between the glomerular compartments.
(b) Example voltage trace from a PN postsynaptic to the silenced ORNs. Odor stimulation (gray bar) produces a depolarization leading to a train of spikes. Because this PN's direct presynaptic ORNs are silent, this excitation must derive from the activation of surrounding glomeruli, and therefore implicates local excitatory circuits in contributing to PN odor responses. (b) is modified with permission from Ref. 73.
Local connections may actually be more difficult to map than long-range connections. There are almost no studies of local interneurons in the Drosophila brain. It seems likely that local interneurons are as numerous (and as important) as projection neurons, but almost nothing is known about them. The scale of the problem is suggested by a recent study documenting 22 morphological types of local interneurons in a single layer of the Drosophila optic lobe [26]. Are there recurring connectivity motifs [84] that tend to describe the interactions of local interneurons with projection neurons in different brain regions? Such general principles—if they exist—would simplify the challenge of local connectivity.
Another potential complication is that in an insect neuron, it is not unusual for pre- and postsynaptic machinery to be localized to the same axonal or dendritic compartment. For example, in the antennal lobe, the postsynaptic dendrites of projection neurons are actually sites of neurotransmitter release [71, 85]. Additionally, the axon terminals of olfactory receptor neurons express GABA receptors that can inhibit neurotransmitter release [76]. Similarly, in the protocerebrum, the axons of lobula plate tangential cells are rich in neurotransmitter receptors [86]. To understand the microcircuitry of any particular region in the fly brain, it may be important to understand how pre- and postsynaptic proteins are segregated into different compartments of the same cell. Several laboratories have developed tagged pre- and postsynaptic markers that should make these experiments feasible [86, 87], although the trafficking of overexpressed proteins should always be interpreted with caution.
Mathematical models of circuits and behavior
The final output of even small circuits represents a complex interplay between circuit elements. Mathematical models can help reveal the logic of circuit operation by organizing anatomical and neurophysiological data, and can guide experimental studies by providing testable predications about both neural activity and behavior. So far, Drosophila has inspired few modeling efforts, but studies in other relatively simple animals suggest ways in which these techniques might be useful. For example, models of central pattern generator circuits in the crustacean stomatogastric nervous system have helped define how dynamic motor patterns arise from simple circuits [88, 89]. In C. elegans quantitative models of chemotaxis behavior have suggested algorithms by which this behavior is produced, and have helped direct physiological investigation of the circuits performing these operations [90, 91]. Likewise, analysis of visual motion processing in the blow fly have inspired mathematical models of specific neural computations [92]; the synaptic and cellular correlates of these computations can now be studied in Drosophila [77]. These studies illustrate how mathematical models can be helpful in understanding the relationship between circuits, computation, and behavior.
Limitations of the model organism
The virtues of Drosophila as a model for systems neuroscience are easy to grasp. However, the limitations of this model organism receive less public attention. Some of these limitations may be swept aside by future breakthroughs, but others may be intrinsic to the fly.
One problem is that the small size of the Drosophila brain will make it extremely challenging to perform electrophysiological measurements in the behaving fly. Functional imaging might offer a solution to this problem, but imaging is even more sensitive than electrophysiology to movements of the preparation. Because neural circuits can be highly stereotyped in the fly [see e.g. 25 for a review], we should be able to learn an enormous amount simply by comparing neural activity in one individual fly with the behavior of a different individual. However, it will be difficult to correlate trial-to-trial fluctuations in neural activity with variations in behavioral performance.
We should also consider whether the fly is a good model for how neural activity sculpts neural circuits, especially during development. Certainly flies do learn, and this learning must correspond to experience-dependent changes somewhere in the nervous system. But connectivity in many fly neural circuits may be completely determined by the genome. This idea is supported by several studies in the olfactory system which demonstrate that activity plays no discernable role in instructing connectivity [93-98]. One recent study, however, provides some evidence that local connectivity in the antennal lobe may be activity-dependent [62]. The “hard-wired” nature of the fly brain may limit our ability to generalize our findings to more plastic brains.
To a large extent the motivation for circuit analysis of fly behavior is based on the promise of rapid progress and complete understanding. It is worth keeping in mind that there may be circuits in the fly brain that are more difficult to crack than others. For some circuits a slow rate of advancement may be justified by the insight that will ultimately be gained, perhaps through a comparative analysis of similar systems across animals; for others it may be wiser to take a more opportunistic approach in another model organism.
Finally, we might consider how much circuits in a simple brain can really teach us about circuits in a big brain. Perhaps the sheer volume of synapses in the human cortex produces a kind of magic which cannot occur in a radically simpler brain. If so, the cognitive limitations of the fly may prove as interesting as its capabilities.
In summary, Drosophila displays complex behaviors but accomplishes this with relatively few neurons. The confluence of genetic tools for mapping circuits and perturbing neurons, along with the ability to record neural activity, means that a relatively complete understanding of some circuits is likely feasible in less than 10 years. This would be a real achievement: at the very least, it would suggest how larger brains (and even robotic devices [99]) might implement these computations. Whether many Drosophila circuits can be cracked in this way, and whether fly circuits bear a fundamental resemblance to vertebrate neural circuits—only time will tell.
References
- 1.Holmes TC, et al. Circuit-breaking and behavioral analysis by molecular genetic manipulation of neural activity in Drosophila. In: North G, Greenspan RJ, editors. Invertebrate Neurobiology. Cold Spring Harbor Laboratory Press; 2007. pp. 19–52. [Google Scholar]
- 2.Luo L, et al. Genetic dissection of neural circuits. Neuron. 2008;57:634–660. doi: 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choe KM, Clandinin TR. Thinking about visual behavior; learning about photoreceptor function. Curr Top Dev Biol. 2005;69:187–213. doi: 10.1016/S0070-2153(05)69007-2. [DOI] [PubMed] [Google Scholar]
- 4.Heisenberg M. Pattern recognition in insects. Curr Opin Neurobiol. 1995;5:475–481. doi: 10.1016/0959-4388(95)80008-5. [DOI] [PubMed] [Google Scholar]
- 5.Tang S, et al. Visual pattern recognition in Drosophila is invariant for retinal position. Science. 2004;305:1020–1022. doi: 10.1126/science.1099839. [DOI] [PubMed] [Google Scholar]
- 6.Liu G, et al. Distinct memory traces for two visual features in the Drosophila brain. Nature. 2006;439:551–556. doi: 10.1038/nature04381. [DOI] [PubMed] [Google Scholar]
- 7.Neuser K, et al. Analysis of a spatial orientation memory in Drosophila. Nature. 2008;453:1244–1247. doi: 10.1038/nature07003. [DOI] [PubMed] [Google Scholar]
- 8.Louis M, et al. Bilateral olfactory sensory input enhances chemotaxis behavior. Nat Neurosci. 2008;11:187–199. doi: 10.1038/nn2031. [DOI] [PubMed] [Google Scholar]
- 9.Faucher C, et al. Behavioral responses of Drosophila to biogenic levels of carbon dioxide depend on life-stage, sex and olfactory context. J Exp Biol. 2006;209:2739–2748. doi: 10.1242/jeb.02297. [DOI] [PubMed] [Google Scholar]
- 10.Frye MA, Dickinson MH. Motor output reflects the linear superposition of visual and olfactory inputs in Drosophila. J Exp Biol. 2004;207:123–131. doi: 10.1242/jeb.00725. [DOI] [PubMed] [Google Scholar]
- 11.Guo J, Guo A. Crossmodal interactions between olfactory and visual learning in Drosophila. Science. 2005;309:307–310. doi: 10.1126/science.1111280. [DOI] [PubMed] [Google Scholar]
- 12.Chow DM, Frye MA. Context-dependent olfactory enhancement of optomotor flight control in Drosophila. J Exp Biol. 2008;211:2478–2485. doi: 10.1242/jeb.018879. [DOI] [PubMed] [Google Scholar]
- 13.Duistermars BJ, Frye MA. Crossmodal visual input for odor tracking during fly flight. Curr Biol. 2008;18:270–275. doi: 10.1016/j.cub.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 14.Schlief ML, Wilson RI. Olfactory processing and behavior downstream from highly selective receptor neurons. Nat Neurosci. 2007;10:623–630. doi: 10.1038/nn1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ejima A, et al. Sequential learning of pheromonal cues modulates memory consolidation in trainer-specific associative courtship conditioning. Curr Biol. 2005;15:194–206. doi: 10.1016/j.cub.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ejima A, et al. Generalization of courtship learning in Drosophila is mediated by cis-vaccenyl acetate. Curr Biol. 2007;17:599–605. doi: 10.1016/j.cub.2007.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bennet-Clark HC, Ewing AW. The love song of the fruit fly. Sci Am. 1970;223:85–90. passim. [PubMed] [Google Scholar]
- 18.Ritchie MG, et al. Drosophila song as a species-specific mating signal and the behavioural importance of Kyriacou & Hall cycles in D. melanogaster song. Anim Behav. 1999;58:649–657. doi: 10.1006/anbe.1999.1167. [DOI] [PubMed] [Google Scholar]
- 19.Yurkovic A, et al. Learning and memory associated with aggression in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2006;103:17519–17524. doi: 10.1073/pnas.0608211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maimon G, et al. A simple vision-based algorithm for decision making in flying Drosophila. Curr Biol. 2008;18:464–470. doi: 10.1016/j.cub.2008.02.054. [DOI] [PubMed] [Google Scholar]
- 21.Katsov A, Clandinin TR. Motion processing streams in Drosophila are behaviorally specialized. Neuron. 2008 doi: 10.1016/j.neuron.2008.05.022. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamada FN, et al. An internal thermal sensor controlling temperature preference in Drosophila. Nature. 2008;454:217–220. doi: 10.1038/nature07001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phelps CB, Brand AH. Ectopic gene expression in Drosophila using GAL4 system. Methods. 1998;14:367–379. doi: 10.1006/meth.1998.0592. [DOI] [PubMed] [Google Scholar]
- 24.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 25.Jefferis GS, et al. Development of neuronal connectivity in Drosophila antennal lobes and mushroom bodies. Curr Opin Neurobiol. 2002;12:80–86. doi: 10.1016/s0959-4388(02)00293-3. [DOI] [PubMed] [Google Scholar]
- 26.Morante J, Desplan C. The color vision circuit in the medulla of Drosophila. Curr Biol. 2008;18 doi: 10.1016/j.cub.2008.02.075. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rister J, et al. Dissection of the peripheral motion channel in the visual system of Drosophila melanogaster. Neuron. 2007;56:155–170. doi: 10.1016/j.neuron.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 28.Datta SR, et al. The Drosophila pheromone cVA activates a sexually dimorphic neural circuit. Nature. 2008;452:473–477. doi: 10.1038/nature06808. [DOI] [PubMed] [Google Scholar]
- 29.Nitabach MN, Taghert PH. Organization of the Drosophila circadian control circuit. Curr Biol. 2008;18:R84–93. doi: 10.1016/j.cub.2007.11.061. [DOI] [PubMed] [Google Scholar]
- 30.Stockinger P, et al. Neural circuitry that governs Drosophila male courtship behavior. Cell. 2005;121:795–807. doi: 10.1016/j.cell.2005.04.026. [DOI] [PubMed] [Google Scholar]
- 31.Davis RL. Olfactory memory formation in Drosophila: from molecular to systems neuroscience. Annu Rev Neurosci. 2005;28:275–302. doi: 10.1146/annurev.neuro.28.061604.135651. [DOI] [PubMed] [Google Scholar]
- 32.Baker DA, et al. Functional dissection of the neural substrates for gravitaxic maze behavior in Drosophila melanogaster. J Comp Neurol. 2007;501:756–764. doi: 10.1002/cne.21257. [DOI] [PubMed] [Google Scholar]
- 33.Gordon MD, et al. Fly neurobiology: development and function of the brain. Meeting on the Neurobiology of Drosophila. EMBO Rep. 2008;9:239–242. doi: 10.1038/embor.2008.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luan H, White BH. Combinatorial methods for refined neuronal gene targeting. Curr Opin Neurobiol. 2007;17:572–580. doi: 10.1016/j.conb.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 35.Pfeiffer BD, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci U S A. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mao T, et al. Characterization and subcellular targeting of GCaMP-type genetically-encoded calcium indicators. PLoS ONE. 2008;3:e1796. doi: 10.1371/journal.pone.0001796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jayaraman V, Laurent G. Evaluating a genetically encoded optical sensor of neural activity using electrophysiology in intact adult fruit flies. Front Neural Circuits. 2007;1:3. doi: 10.3389/neuro.3304/3003.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sweeney ST, et al. Targeted expression of tetanus toxin light chain in Drosophila specifically eliminates synaptic transmission and causes behavioral defects. Neuron. 1995;14:341–351. doi: 10.1016/0896-6273(95)90290-2. [DOI] [PubMed] [Google Scholar]
- 39.Nitabach MN, et al. Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell. 2002;109:485–495. doi: 10.1016/s0092-8674(02)00737-7. [DOI] [PubMed] [Google Scholar]
- 40.Wang Z, et al. Taste representations in the Drosophila brain. Cell. 2004;117:981–991. doi: 10.1016/j.cell.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 41.Kitamoto T. Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J Neurobiol. 2001;47:81–92. doi: 10.1002/neu.1018. [DOI] [PubMed] [Google Scholar]
- 42.Zhang F, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 43.Sakai T, Kitamoto T. Differential roles of two major brain structures, mushroom bodies and central complex, for Drosophila male courtship behavior. J Neurobiol. 2006;66:821–834. doi: 10.1002/neu.20262. [DOI] [PubMed] [Google Scholar]
- 44.Joiner WJ, et al. Sleep in Drosophila is regulated by adult mushroom bodies. Nature. 2006;441:757–760. doi: 10.1038/nature04811. [DOI] [PubMed] [Google Scholar]
- 45.Luan H, et al. Functional dissection of a neuronal network required for cuticle tanning and wing expansion in Drosophila. J Neurosci. 2006;26:573–584. doi: 10.1523/JNEUROSCI.3916-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nitabach MN, et al. Electrical hyperexcitation of lateral ventral pacemaker neurons desynchronizes downstream circadian oscillators in the fly circadian circuit and induces multiple behavioral periods. J Neurosci. 2006;26:479–489. doi: 10.1523/JNEUROSCI.3915-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schroll C, et al. Light-induced activation of distinct modulatory neurons triggers appetitive or aversive learning in Drosophila larvae. Curr Biol. 2006;16:1741–1747. doi: 10.1016/j.cub.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 48.Zhang W, et al. A toolbox for light control of Drosophila behaviors through Channelrhodopsin 2-mediated photoactivation of targeted neurons. Eur J Neurosci. 2007;26:2405–2416. doi: 10.1111/j.1460-9568.2007.05862.x. [DOI] [PubMed] [Google Scholar]
- 49.Hwang RY, et al. Nociceptive neurons protect Drosophila larvae from parasitoid wasps. Curr Biol. 2007;17:2105–2116. doi: 10.1016/j.cub.2007.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suh GS, et al. Light activation of an innate olfactory avoidance response in Drosophila. Curr Biol. 2007;17:905–908. doi: 10.1016/j.cub.2007.04.046. [DOI] [PubMed] [Google Scholar]
- 51.Clyne JD, Miesenbock G. Sex-specific control and tuning of the pattern generator for courtship song in Drosophila. Cell. 2008;133:354–363. doi: 10.1016/j.cell.2008.01.050. [DOI] [PubMed] [Google Scholar]
- 52.Lima SQ, Miesenbock G. Remote control of behavior through genetically targeted photostimulation of neurons. Cell. 2005;121:141–152. doi: 10.1016/j.cell.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 53.Marella S, et al. Imaging taste responses in the fly brain reveals a functional map of taste category and behavior. Neuron. 2006;49:285–295. doi: 10.1016/j.neuron.2005.11.037. [DOI] [PubMed] [Google Scholar]
- 54.Griesbeck O. Fluorescent proteins as sensors for cellular functions. Curr Opin Neurobiol. 2004;14:636–641. doi: 10.1016/j.conb.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 55.Miesenbock G, Kevrekidis IG. Optical imaging and control of genetically designated neurons in functioning circuits. Annu Rev Neurosci. 2005;28:533–563. doi: 10.1146/annurev.neuro.28.051804.101610. [DOI] [PubMed] [Google Scholar]
- 56.Wang JW, et al. Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell. 2003;112:271–282. doi: 10.1016/s0092-8674(03)00004-7. [DOI] [PubMed] [Google Scholar]
- 57.Wang Y, et al. Stereotyped odor-evoked activity in the mushroom body of Drosophila revealed by green fluorescent protein-based Ca2+ imaging. J Neurosci. 2004;24:6507–6514. doi: 10.1523/JNEUROSCI.3727-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fischler W, et al. The detection of carbonation by the Drosophila gustatory system. Nature. 2007;448:1054–1057. doi: 10.1038/nature06101. [DOI] [PubMed] [Google Scholar]
- 59.Silbering AF, Galizia CG. Processing of odor mixtures in the Drosophila antennal lobe reveals both global inhibition and glomerulus-specific interactions. J Neurosci. 2007;27:11966–11977. doi: 10.1523/JNEUROSCI.3099-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu D, et al. Drosophila alpha/beta mushroom body neurons form a branch-specific, long-term cellular memory trace after spaced olfactory conditioning. Neuron. 2006;52:845–855. doi: 10.1016/j.neuron.2006.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu D, et al. Drosophila DPM neurons form a delayed and branch-specific memory trace after olfactory classical conditioning. Cell. 2005;123:945–957. doi: 10.1016/j.cell.2005.09.037. [DOI] [PubMed] [Google Scholar]
- 62.Sachse S, et al. Activity-dependent plasticity in an olfactory circuit. Neuron. 2007;56:838–850. doi: 10.1016/j.neuron.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 63.Yu D, et al. Altered representation of the spatial code for odors after olfactory classical conditioning; memory trace formation by synaptic recruitment. Neuron. 2004;42:437–449. doi: 10.1016/s0896-6273(04)00217-x. [DOI] [PubMed] [Google Scholar]
- 64.Pologruto TA, et al. Monitoring neural activity and [Ca2+] with genetically encoded Ca2+ indicators. J Neurosci. 2004;24:9572–9579. doi: 10.1523/JNEUROSCI.2854-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ikeda K, Kaplan WD. Patterned neural activity of a mutant Drosophila melanogaster. Proc Natl Acad Sci U S A. 1970;66:765–772. doi: 10.1073/pnas.66.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fujishiro N, et al. Impulse frequency and action potential amplitude in labellar chemosensory neurones of Drosophila melanogaster. J Insect Physiol. 1984;30:317–325. [Google Scholar]
- 67.Corfas G, Dudai Y. Adaptation and fatigue of a mechanosensory neuron in wild-type Drosophila and in memory mutants. J Neurosci. 1990;10:491–499. doi: 10.1523/JNEUROSCI.10-02-00491.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Clyne P, et al. Odorant response of individual sensilla on the Drosophila antenna. Invert Neurosci. 1997;3:127–135. doi: 10.1007/BF02480367. [DOI] [PubMed] [Google Scholar]
- 69.Hallem EA, Carlson JR. Coding of odors by a receptor repertoire. Cell. 2006;125:143–160. doi: 10.1016/j.cell.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 70.Kreher SA, et al. Translation of sensory input into behavioral output via an olfactory system. Neuron. 2008;59:110–124. doi: 10.1016/j.neuron.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wilson RI, et al. Transformation of olfactory representations in the Drosophila antennal lobe. Science. 2004;303:366–370. doi: 10.1126/science.1090782. [DOI] [PubMed] [Google Scholar]
- 72.Bhandawat V, et al. Sensory processing in the Drosophila antennal lobe increases the reliability and separability of ensemble odor representations. Nat Neurosci. 2007;10:1474–1482. doi: 10.1038/nn1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Olsen SR, et al. Excitatory interactions between olfactory processing channels in the Drosophila antennal lobe. Neuron. 2007;54:89–103. doi: 10.1016/j.neuron.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Root CM, et al. Propagation of olfactory information in Drosophila. Proc Natl Acad Sci U S A. 2007;104:11826–11831. doi: 10.1073/pnas.0704523104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Turner GC, et al. Olfactory representations by Drosophila mushroom body neurons. J Neurophysiol. 2007 doi: 10.1152/jn.01283.2007. [DOI] [PubMed] [Google Scholar]
- 76.Olsen SR, Wilson RI. Lateral presynaptic inhibition mediates gain control in an olfactory circuit. Nature. 2008;452:956–960. doi: 10.1038/nature06864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Joesch M, et al. Response properties of motion-sensitive visual interneurons in the lobula plate of Drosophila melanogaster. Curr Biol. 2008;18:368–374. doi: 10.1016/j.cub.2008.02.022. [DOI] [PubMed] [Google Scholar]
- 78.Sheeba V, et al. Circadian- and light-dependent regulation of resting membrane potential and spontaneous action potential firing of Drosophila circadian pacemaker neurons. J Neurophysiol. 2007 doi: 10.1152/jn.00930.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gu H, O'Dowd DK. Cholinergic synaptic transmission in adult Drosophila Kenyon cells in situ. J Neurosci. 2006;26:265–272. doi: 10.1523/JNEUROSCI.4109-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Feinberg EH, et al. GFP Reconstitution Across Synaptic Partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron. 2008;57:353–363. doi: 10.1016/j.neuron.2007.11.030. [DOI] [PubMed] [Google Scholar]
- 81.Petreanu L, et al. Channelrhodopsin-2-assisted circuit mapping of long-range callosal projections. Nat Neurosci. 2007;10:663–668. doi: 10.1038/nn1891. [DOI] [PubMed] [Google Scholar]
- 82.Kazama H, Wilson RI. Homeostatic matching and nonlinear amplification at genetically-identified central synapses. Neuron. 2008;58:401–413. doi: 10.1016/j.neuron.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shang Y, et al. Excitatory local circuits and their implications for olfactory processing in the fly antennal lobe. Cell. 2007;128:601–612. doi: 10.1016/j.cell.2006.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Milo R, et al. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- 85.Ng M, et al. Transmission of olfactory information between three populations of neurons in the antennal lobe of the fly. Neuron. 2002;36:463–474. doi: 10.1016/s0896-6273(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 86.Raghu SV, et al. Synaptic organization of lobula plate tangential cells in Drosophila: gamma-aminobutyric acid receptors and chemical release sites. J Comp Neurol. 2007;502:598–610. doi: 10.1002/cne.21319. [DOI] [PubMed] [Google Scholar]
- 87.Estes PS, et al. Synaptic localization and restricted diffusion of a Drosophila neuronal synaptobrevin--green fluorescent protein chimera in vivo. J Neurogenet. 2000;13:233–255. doi: 10.3109/01677060009084496. [DOI] [PubMed] [Google Scholar]
- 88.Soto-Trevino C, et al. Activity-dependent modification of inhibitory synapses in models of rhythmic neural networks. Nat Neurosci. 2001;4:297–303. doi: 10.1038/85147. [DOI] [PubMed] [Google Scholar]
- 89.Nusbaum MP, Beenhakker MP. A small-systems approach to motor pattern generation. Nature. 2002;417:343–350. doi: 10.1038/417343a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ferree TC, Lockery SR. Computational rules for chemotaxis in the nematode C. elegans. J Comput Neurosci. 1999;6:263–277. doi: 10.1023/a:1008857906763. [DOI] [PubMed] [Google Scholar]
- 91.Suzuki H, et al. Functional asymmetry in Caenorhabditis elegans taste neurons and its computational role in chemotaxis. Nature. 2008;454:114–117. doi: 10.1038/nature06927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Borst A, Haag J. Neural networks in the cockpit of the fly. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2002;188:419–437. doi: 10.1007/s00359-002-0316-8. [DOI] [PubMed] [Google Scholar]
- 93.Wong AM, et al. Spatial representation of the glomerular map in the Drosophila protocerebrum. Cell. 2002;109:229–241. doi: 10.1016/s0092-8674(02)00707-9. [DOI] [PubMed] [Google Scholar]
- 94.Dobritsa AA, et al. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron. 2003;37:827–841. doi: 10.1016/s0896-6273(03)00094-1. [DOI] [PubMed] [Google Scholar]
- 95.Tanaka NK, et al. Integration of chemosensory pathways in the Drosophila second-order olfactory centers. Curr Biol. 2004;14:449–457. doi: 10.1016/j.cub.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 96.Berdnik D, et al. Wiring stability of the adult Drosophila olfactory circuit after lesion. J Neurosci. 2006;26:3367–3376. doi: 10.1523/JNEUROSCI.4941-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hiesinger PR, et al. Activity-independent prespecification of synaptic partners in the visual map of Drosophila. Curr Biol. 2006;16:1835–1843. doi: 10.1016/j.cub.2006.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ting CY, Lee CH. Visual circuit development in Drosophila. Curr Opin Neurobiol. 2007;17:65–72. doi: 10.1016/j.conb.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 99.Webb B. Robots in invertebrate neuroscience. Nature. 2002;417:359–363. doi: 10.1038/417359a. [DOI] [PubMed] [Google Scholar]