Abstract
Initiation of actin polymerization in cells requires nucleation factors. Here we describe an actin-binding protein, leiomodin, which acted as a strong filament nucleator in muscle cells. Leiomodin shared two actin-binding sites with the filament pointed-end capping protein tropomodulin; a flexible N-terminal region and a leucine-rich repeat domain. Leiomodin also contained a C-terminal extension of 150 residues. The smallest fragment with strong nucleation activity included the leucine-rich repeat and C-terminal extension. The N-terminal region enhanced the nucleation activity 3-fold and recruited tropomyosin, which weakly stimulated nucleation and mediated localization of leiomodin to the middle of muscle sarcomeres. Knocking down leiomodin severely compromised sarcomere assembly in cultured muscle cells, suggesting a role for leiomodin in the nucleation of tropomyosin-decorated filaments in muscles.
Actin binding proteins suppress the spontaneous nucleation of actin monomers into filaments, so cells use nucleation factors to initiate actin polymerization. In non-muscle cells, the best-characterized filament nucleators are Arp2/3 complex and formins (1). Less is known about the initiation of actin filaments in striated and smooth muscle cells, where specialized proteins may be used to assemble and remodel the tropomyosin-decorated filaments.
We identified leiomodin (Lmod) as a potential filament nucleator in muscle cells because sequence analysis suggested that it contained at least three actin-binding sites and could possibly recruit three actin monomers to form a polymerization nucleus. Thus, the first ~340 residues of Lmod are ~40% identical to tropomodulin (Tmod) (fig. S1), a protein that caps actin filament pointed ends (2, 3). The N-terminal portion of Tmod is unstructured, except for three helical segments involved in binding tropomyosin (residues 24-35 and 126-135) and actin (residues 65-75) (4). This region of Tmod, caps the pointed end of actin filaments in a tropomyosin-dependent manner (5). Tmod has a second, tropomyosin-independent, actin-binding and capping site within the C-terminal region (residues 160-359) (5), consisting almost entirely of a Leu-rich repeat (LRR) domain (6). Lmod shares this domain organization but has only one of the two tropomyosin-binding sites (Fig. 1A and fig. S1). More importantly, Lmod extends ~150-aa beyond the C-terminus of Tmod. This extension contains a short poly-proline region, two segments with predicted helical structure, and an actin-binding WASP-Homology 2 (WH2) domain.
Fig. 1.
Domain organization and localization of Lmod in cultured rat cardiomyocytes. (A) Modular organization of Lmod and constructs used in this study (TM-h1, TM-h2 and A-h, tropomyosin and actin binding helices; polyP, poly-proline; h1 and h2, predicted helices; B, basic segment). (B) Fluorescent antibody staining shows colocalization of endogenous Lmod (green) and myomesin (red) in the middle of sarcomeres. (C) EGFP-Lmodwt and EGFP-Lmod1-342 constructs (green) also concentrate in the middle of sarcomeres, separated from Z-discs visualized with antibodies to α-actinin, whereas EGFP-Lmod162-495 concentrates in the nucleus (bar, 10μm).
To localize Lmod in cultured neonatal rat cardiomyocytes we used fluorescent antibody staining and expression of EGFP-Lmod fusion proteins. Our rabbit polyclonal antibody against Lmodwt reacted specifically with Lmod in extracts of skeletal muscle and did not cross-react with Tmod (fig. S2). In cardiomyocytes, this antibody concentrated close to the M-line protein myomesin, near the center of the sarcomere (Fig. 1B). EGFP-Lmodwt and EGFP-Lmod1-342, comprising the Tmod-like portion of Lmod, also concentrated in the middle of sarcomeres, well separated from α-actinin in Z-discs (Fig. 1C). Thus, Lmod is located near the pointed-ends of the actin filaments, which were not resolved from M-lines. On the other hand, EGFP-Lmod162-495, a fragment with strong nucleation activity (see below) that included the LRR and C-terminal extension, concentrated in the nucleus. Thus, the N-terminal domain is required for Lmod localization in the center of sarcomeres.
Strong over-expression of all three EGFP-Lmod constructs disrupted sarcomeric organization and induced the formation of aberrant actin structures (fig. S3). Lmod162-495 had the most dramatic phenotype. Even moderate expression of Lmod162-495 induced the formation of abnormal actin bundles in the nucleus, similar to those observed in intranuclear rod myopathy (7).
We investigated the role of Lmod in sarcomere assembly using RNAi to knockdown Lmod expression in cardiomyocytes. Cells transfected with Lmod-siRNA had drastically lower levels of Lmod protein than control cells (fig. S4). Fluorescent antibody staining for Lmod was strongly reduced in Lmod-siRNA cells (Fig. 2), which also lacked the striated pattern of Lmod observed in control cells transfected with a scrambled oligonucleotide. Lmod knockdown cells lacked organized sarcomeres (Fig. 2 and S5). In Lmod-siRNA cells, the Z-disc protein α-actinin concentrated in small spots, contrasting with the typical striated pattern observed in control cells (fig. S5). Lmod knockdown cells were also less adherent and spread than wild-type cells. Thus, Lmod plays a role in sarcomere assembly and organization.
Fig. 2.
Lmod knockdown disrupts sarcomere assembly in rat cardiomyocytes. Lmod antibody staining is reduced and the striated Lmod pattern is lost in cardiomyocytes transfected with Lmod specific siRNA as compared to control cells (transfected with scrambled-siRNA). The micrographs of Alexa 568-phalloidin show that sarcomeric actin structures are severely disorganized in knockdown cells (bar, 10μm).
We studied the effect of Lmod on actin assembly by monitoring the fluorescence increase of 6% pyrene-labeled actin upon polymerization. Nanomolar concentrations of Lmodwt dramatically stimulated polymerization compared with spontaneous actin assembly (Fig. 3A). Lmod1-342 was significantly less effective, although more active than Tmod, which has weak nucleation activity (5). Lmod162-342, comprising the LRR and an additional 14 residues necessary for pointed-end capping in Tmod (5), and Lmod343-495, corresponding to the C-terminal extension, both had low polymerization-inducing activity. However, the combination of these two constructs in Lmod162-495 retained one third of the activity of Lmodwt (Fig. 3D). Deletions and substitutions in Lmod162-495 suggested that each of the modules of the C-terminal extension contributed to this activity; some seemed to bind actin directly whereas others seemed to help properly position the actin-binding sites for assembly (fig. S6).
Fig. 3.

Lmod nucleates actin filaments. (A) Time course of fluorescence increase upon polymerization of 2μM Mg-ATP-actin (6% pyrene-labeled) alone (black) or with 25nM Lmod constructs (colored according to Fig. 1A) in polymerization buffer (10mM Tris pH7.5, 1mM MgCl2, 50mM KCl, 1mM EGTA, 0.1mM NaN3, 0.02mg/mL BSA, 0.2mM ATP). (B) Number of barbed-ends formed after 4min of polymerization with varying Lmod concentration measured by TIRF. For each construct, the average number of actin seeds was measured from 20 images (150 × 120 μm). Data reported are mean ± SD. (C) TIRF micrographs of fields of actin filaments prepared 4min after initiating of polymerization of 1μM Mg-ATP-actin with a range of Lmodwt concentrations. (D) Time course of barbed end formation in the presence of Lmod constructs calculated from the polymerization rate, concentration of actin monomers remaining, and rate constant for barbed-end elongation (10 μM-1s-1) (fig. S7A). The table gives the nucleation rates.
To determine if Lmod promoted assembly by stimulating nucleation or elongation, we observed Lmod-induced actin polymerization directly by total internal reflection fluorescence (TIRF) microscopy. The number of filaments increased exponentially with Lmodwt concentration (Fig. 3B and 3C and movies S1), whereas Lmodwt had no effect on the elongation rate of either filament end (fig. S7). We obtained similar results with Lmod162-495. In agreement with the bulk polymerization assay (fig. S6), constructs with deletions or substitutions of the modules of the C-terminal extension formed fewer filaments than Lmod162-495 (Fig. 3B). Thus, Lmod stimulates polymerization by nucleating filaments rather than increasing their elongation rates.
From the time course of polymerization (Fig. 3A and fig. S6) and the elongation rates established by TIRF (fig. S7), we determined that 2μM actin formed 3.4nM barbed-ends per minute in the presence of 25nM Lmodwt (Fig. 3D). This high nucleation rate is comparable to that of Arp2/3 complex (8). Lmod162-495 retained one third of this activity (Fig. 3D).
We tested the effect of tropomyosin on polymerization induced by Lmodwt and Lmod162-495, which lacks the tropomyosin-binding site (fig. S1). All concentrations of tropomyosin inhibited polymerization by Lmod162-495 (Fig. 4A). In contrast, polymerization by Lmodwt was stimulated at low tropomyosin concentrations, but inhibited at tropomyosin concentrations higher than 1μM. This biphasic effect of tropomyosin can be explained by two opposite mechanisms: tropomyosin weakly promotes nucleation by Lmodwt, but reduces the formation of ends via spontaneous fragmentation (9). At 1μM concentration, tropomyosin promoted polymerization of 2μM actin with a range of concentrations of Lmodwt and Lmod1-342, but not Tmod, all of which were expected to bind tropomyosin (Fig. 4B). This stimulatory effect of tropomyosin on Lmod-induced nucleation occurred within a physiologically relevant range, because the ratio of tropomyosin to actin in muscle thin filaments is 1:7.
Fig. 4.
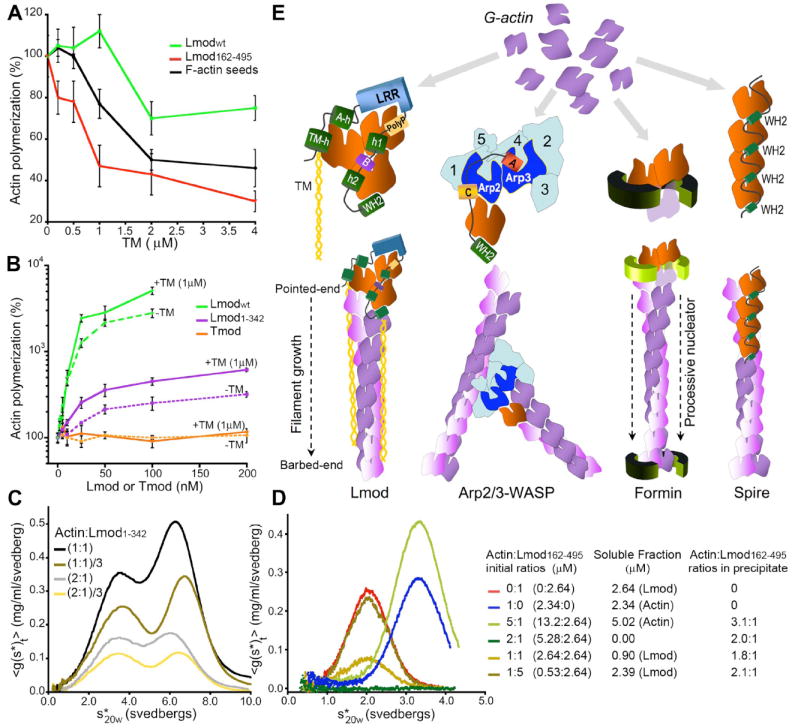
Mechanism of actin nucleation by Lmod. (A) Effect of tropomyosin on the polymerization of 2μM actin induced by 0.5μM F-actin seeds, 25nM Lmodwt or 25nM Lmod162-495. Rates were determined from the slope of the curves at 50% polymerization and expressed as percentage of the rates without tropomyosin. Data reported are mean ± SD of three or more experiments. (B) Polymerization rates of 2μM actin with increasing concentrations of Lmodwt, Lmod1-342 or Tmod in the absence (dashed lines) or the presence (solid lines) of 1μM tropomyosin. Rates are expressed as percentages of the polymerization rate of actin alone. Data reported are mean ± SD of three or more experiments. (C) Sedimentation velocity dilution series of complexes of actin:Lmod1-342 at two different molar ratios. The samples contained 50μM LatB to prevent polymerization. The first peak corresponds to actin-LatB. The main species in the second peak is a 2:1 actin:Lmod1-342 complex. (D) Sedimentation velocity analysis of actin:Lmod162-495 mixed at different ratios (right). In the presence of LatB, actin:Lmod162-495 complexes precipitated. The stoichiometries of the complexes in the precipitate were determined by difference from the amount of actin or Lmod162-495 remaining in solution, using the interference optics of the ultracentrifuge. (E) Comparison of models for actin filament nucleation. We propose that Lmod stabilizes a trimeric actin nucleus and that tropomyosin enhances nucleation and mediates the localization of Lmod to filament pointed-ends. Arp2 and Arp3 of Arp2/3 complex form a trimeric seed with an actin monomer bound to the WH2 of a nucleation-promoting factor. Formin dimers form nuclei and remain associated with the barbed-end of the growing filament. Spire stabilizes a nucleus consisting of four actin subunits along a long-pitch filament strand.
The C-terminal extension of Lmod contains a short poly-Pro sequence, a potential binding site for profilin (fig. S1). However, profilin inhibited to the same extent polymerization induced by Lmod162-495 and Lmod162-495(polyP-GS), a construct that lacks the poly-Pro sequence (fig. S8). Thus, profilin does not seem to play a role in filament nucleation by Lmod.
We used sedimentation velocity to determine the stoichiometry of the Lmod-actin polymerization nucleus (fig. S9). We included latrunculin B (LatB) in the buffer to prevent polymerization. Separately, actin-LatB and Lmod constructs Lmodwt, Lmod162-495 and Lmod1-342 were each monomeric and soluble (fig. S9). Complexes of Lmodwt with actin formed a reaction boundary (multiple species) around 12S, whose stoichiometry could not be determined by fitting. Mixtures of Lmod1-342 with actin formed reaction boundaries around 3.5S (corresponding to actin-LatB) and 6.5S (Fig. 4C). The predominant species in the second peak was a 2:1 complex of actin:Lmod1-342. Mixtures of Lmod162-495 with actin-LatB precipitated. However, by quantitating the proteins in the supernatant using the interference optics of the ultracentrifuge, we determined the composition of the complexes in the pellets (Fig. 4D). When actin was present in 2-fold molar excess over Lmod162-495, all of both proteins precipitated, indicating that Lmod162-495 binds two actins. Actin may bind cooperatively to the two sites, because actin:Lmod162-495 pelleted as a 2:1 complex even when Lmod162-495 was present in excess.
Lmod162-495 and Lmod1-342 share one actin-binding site (LRR domain) and each binds two actin subunits, suggesting that full-length Lmod stabilizes a nucleus of three actin subunits, bound to the flexible N-terminal, LRR and WH2 domains. Information about the affinities of actin for the three binding sites and potential cooperative binding effects will be required to trace the actual nucleation pathway (SOM text). Because Lmod162-495 is a strong nucleator, we propose that assembly of the nucleus begins with actin monomers binding to the LRR and WH2 domains, followed by actin binding to the flexible N-terminus. The N-terminus is also necessary for Lmod localization near the center of sarcomeres, suggesting that Lmod (like Tmod) binds to filament pointed-ends, since actin and tropomyosin are the only known binding partners of this region of Lmod. Due to steric hindrance, the WH2 domain may dissociate as the nucleus grows from the barbed end. After nucleation the fates of the other interactions of Lmod with actin and tropomyosin are unknown, but they may serve to localize Lmod to actin filament pointed ends near the center of sarcomeres. It is unknown if Lmod can coexist with Tmod at these sites.
The nucleation mechanism that we propose for Lmod has features in common with nucleation by Arp2/3 complex together with WH2-containing proteins such as WASP (Fig. 4E). In both cases a WH2 domain delivers an actin monomer to complete a trimeric seed consisting of three actin subunits (Lmod) or one actin subunit and the Arp2 and Arp3 subunits of Arp2/3 complex. However, Arp2/3 complex depends on an extrinsic WH2 domain, while Lmod has an intrinsic WH2 and is active on its own. Arp2/3 complex anchors the pointed-end of the new filament to the side of another filament, whereas Lmod does not form branches. Lmod is also distinct from formins and spire. Formins remain bound to elongating barbed-ends after initiating a filament (1). Spire is thought to stabilize nuclei by bridging actin subunits along a filament strand (10). A unique feature of Lmod is its ability to work in conjunction with tropomyosin, which potentiates its nucleation activity and mediates its localization to the middle of sarcomeres. The strong nucleation activity and severe knockdown phenotype make Lmod a strong candidate to nucleate tropomyosin-decorated actin filaments in muscle sarcomeres.
Supplementary Material
Footnotes
Supporting Online Material
Materials and Methods
Simulations of Actin Polymerization
figs. S1 to S9
References
Movie legends
Reference and Notes
- 1.Pollard TD. Annu Rev Biophys Biomol Struct. 2007;36:451. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- 2.Fischer RS, Fowler VM. Trends Cell Biol. 2003;13:593. doi: 10.1016/j.tcb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 3.Conley CA, Fritz-Six KL, Almenar-Queralt A, Fowler VM. Genomics. 2001;73:127. doi: 10.1006/geno.2000.6501. [DOI] [PubMed] [Google Scholar]
- 4.Kostyukova AS, Hitchcock-Degregori SE, Greenfield NJ. J Mol Biol. 2007;372:608. doi: 10.1016/j.jmb.2007.05.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fowler VM, Greenfield NJ, Moyer J. J Biol Chem. 2003;278:40000. doi: 10.1074/jbc.M306895200. [DOI] [PubMed] [Google Scholar]
- 6.Krieger I, Kostyukova A, Yamashita A, Nitanai Y, Maeda Y. Biophys J. 2002;83:2716. doi: 10.1016/S0006-3495(02)75281-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarkson E, Costa CF, Machesky LM. J Pathol. 2004;204:407. doi: 10.1002/path.1648. [DOI] [PubMed] [Google Scholar]
- 8.Mahaffy RE, Pollard TD. Biophys J. 2006;91:3519. doi: 10.1529/biophysj.106.080937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hitchcock-DeGregori SE, Sampath P, Pollard TD. Biochemistry. 1988;27:9182. doi: 10.1021/bi00426a016. [DOI] [PubMed] [Google Scholar]
- 10.Quinlan ME, Heuser JE, Kerkhoff E, Mullins RD. Nature. 2005;433:382. doi: 10.1038/nature03241. [DOI] [PubMed] [Google Scholar]
- 11.Supported by NIH grants HL086655, GM073791 and GM026338.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




