Abstract
Glucuronidation by UDP-glucuronyltransferase 2B enzymes (UGT2Bs) is a major pathway for the elimination of endobiotics and xenobiotics, including therapeutic drugs. Morphine, a probe drug for UGT2B7, is metabolized to morphine-3-β-glucuronide (M3G) and morphine-6-β-glucuronide (M6G) in humans. Morphine has been used in a series of experiments in the baboon to characterize developmental changes in fetal glucuronidation. This study identifies the baboon UGT2B family of enzymes, compares them with that of the human and the monkey (Macaca fascicularis), and measures the activity of the individual baboon UGT2Bs toward morphine. UGT2B cDNAs were cloned from the liver of adult and newborn baboons and expressed in human embryonic kidney 293 cells. The UGT activity toward morphine was assessed by the rate of formation of M3G and M6G by high-performance liquid chromatography. Eight baboon UGT2Bs were cloned and identified: UGT2B41 and UGT2B42, which are 90% homologous to human UGT2B4; UGT2B43, which is 93% homologous to human UGT2B15; and UGT2B39, UGT2B40, UGT2B44, UGT2B45, and UGT2B46, which are 89 to 91% homologous to human UGT2B7. Homology between baboon and monkey UGT2B ranged from 92.6 to 99.1%, with the primary protein structure of UGT2B43 being 99.1% identical to monkey UGT2B20, including a unique R96I substitution. Gene conversion interfered with the phylogenetic signal in the baboon UGT2B7-like and the monkey UGT2B4-like groups and led to concerted evolution of these enzymes. All of the baboon UGT2Bs metabolized morphine to both M3G and M6G. This study lays the foundation for investigating the regulation of UGT2B enzymes during fetal and neonatal development in the baboon.
The major clearance pathway for morphine in humans is glucuronidation. UDP-glucuronosyltransferases (UGTs), the superfamily of genes encoding enzymes that catalyze glucuronidation, undergo considerable regulation during development in humans as well as in other species (Dutton, 1978; Dutton and Leakey, 1981; de Wildt et al., 1999; Strassburg et al., 2002; Caspersen et al., 2007). Morphine has been used in a series of experiments in the baboon to investigate the impact of the developmental changes in glucuronidation on fetal drug and metabolite disposition (Garland et al., 2005, 2006). To support the use of this model in extrapolating results from the baboon to humans, it is important to delineate the differences in these drug-metabolizing enzymes between the species.
UGTs have been classified into two major families, UGT1 and UGT2, based on the similarity of their primary structures. Currently, 19 functional human UGT1 and UGT2 isoforms have been identified and mainly categorized into the two subfamilies, UGT1A and UGT2B (Mackenzie et al., 2005). UGTs from both of these subfamilies of enzymes have been identified as major pathways for the elimination of drugs from all the therapeutic classes in humans (Green et al., 1998; Barbier et al., 2000a; Court et al., 2001, 2003; Soars et al., 2003).
Morphine is considered a probe drug for UGT2B7. In humans, morphine is metabolized to morphine-3-β-glucuronide (M3G) and morphine-6-β-glucuronide (M6G) (Coffman et al., 1997; Court et al., 2003). Although human UGT2B4, UGT2B15, and UGT2B17 have also been shown to glucuronidate morphine to M3G, this reaction is to a much lesser extent than human UGT2B7. Human UGT2B4 was also found to glucuronidate morphine to M6G but again to a lesser extent than UGT2B7 (Court et al., 2003; Ohno et al., 2008).
The recent molecular cloning of the UGT1A family of genes in the baboon revealed considerable structural and functional conservation between the baboon and human UGT1As (Caspersen et al., 2007). In addition, earlier studies have shown that both the baboon and the cynomolgus monkey (Macaca fascicularis) show similar patterns of glucuronidation of drugs and steroids (Garland et al., 1996, 1998a,b; Barbier and Bélanger, 2003). Together, these data provide evidence that the baboon is a clinically relevant model for studying perinatal drug metabolism. However, more recent studies in the baboon show surprisingly high metabolic clearance of morphine by the fetus and seemed at odds with data from human infants, warranting a closer examination of species differences (Chay et al., 1992; Hartley et al., 1993; Scott et al., 1999; Garland et al., 2005, 2006).
The identification of the baboon UGT2B enzymes involved in morphine glucuronidation is an important step toward the understanding of the species differences in drug metabolism. Therefore, the baboon UGT2B isoforms have been cloned and the primary protein structures compared with that of human and monkey (M. fascicularis) UGT2Bs, and the phylogenetic relationship between these species examined. Finally, the activity of the individual baboon UGT2Bs toward morphine has been assessed.
Materials and Methods
Liver Tissue Collection.
Tissues from adult and newborn baboons (Papio anubis) were made available for this study from a breeding colony maintained by the investigators and the Institute of Comparative Medicine at Columbia University (New York, NY). All the collections were done in accordance with animal care protocols approved by the Institutional Animal Care and Use Committee of the Columbia University and in accordance with guidelines of the U.S. Department of Agriculture and the National Institutes of Health. Liver tissues were quickly cut into smaller fragments, immediately immersed in liquid nitrogen, and stored at −80°C until use.
RNA Extraction.
Total RNA was extracted from 50 to 100 mg of liver tissue using the TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions, and the resulting pellet was resuspended in water treated with diethyl pyrocarbonate (Invitrogen). The RNA was quantified by absorbance at 260 nm, and sample integrity was assessed by the appearance of 28S and 18S rRNA on agarose gels.
cDNA Synthesis and Polymerase Chain Reaction.
cDNA was prepared from 3 μg of total RNA in 10 μl of total reaction volume using the Superscript III first-strand synthesis kit (Invitrogen) and oligo(dT) primers. The cDNA (1 μl) was amplified by polymerase chain reaction (PCR) on a PTC-100 (MJ Research, Watertown, MA) using PfuUltra polymerase (Stratagene, La Jolla, CA) in a total reaction volume of 50 μl containing 0.5 μM primers (Invitrogen), 250 μM deoxynucleoside-5′-triphosphates (GE Healthcare Life Sciences, Piscataway, NJ), and PfuUltra reaction buffer. A modified hot-start and step-down PCR protocol was used to minimize nonspecific reactions. The thermal cycling protocol was as follows: preincubation at 95°C for 10 min, hot-start at 80°C, 35 cycles of denaturing at 94°C for 1 min, annealing stepwise down from 60 to 54°C for 1 min, elongation at 72°C for 1 min, followed by a final elongation step at 72°C for 10 min.
PCR primers were designed to amplify UGT2B genes in the baboon based on published mRNA sequences of human and M. fascicularis UGT2B genes. The following GenBank accession numbers were used: human UGT2B4 (BC026264), human UGT2B7 (BC030974), human UGT2B10 (NM001075), human UGT2B11 (NM001073), human UGT2B28 (NM053039), human UGT2B15 (AF180322), M. fascicularis UGT2B19 (AF112112), M. fascicularis UGT2B30 (AF401657), M. fascicularis UGT2B9 (U91582), M. fascicularis UGT2B18 (AF016310), and M. fascicularis UGT2B23 (AF112113).
Primers were designed from the untranslated regions of a consensus sequence using Primer3 software (http://frodo.wi.mit.edu). All the forward primers contained the CACC sequence at the 5′-end to allow for directional cloning. The primers used were as follows: UGT2B4/2B7 (forward), CACCTTGCATTGCAMCAGGATG; UGT2B15 (forward), CACCYGMRTAAGACCAGGATG; UGT2B4 (reverse), YSCAGCTTCCARCCTCA; UGT2B7 (reverse), GTTTTCCAGCTTCAAATCTC; UGT2B15 (reverse), ATTCCACTTCAGGCTTTTGA, where M = C + A, Y = C + T, S = C + G, and R = A + G.
cDNA Cloning.
The PCR products were extracted from agarose gels and purified using the QIAEX II gel extraction kit (QIAGEN, Valencia, CA). Purified PCR products were cloned into the pcDNA6.2/V5/GW/D-TOPO vector and transformed into TOP10/Escherichia coli-competent cells using the pcDNA Gateway Directional TOPO Expression Kit (Invitrogen). Plasmids were purified using the Plasmid Miniprep Spin Kit (QIAGEN). Inserts were sequenced in the 5′ and 3′ directions at the Columbia University DNA Analysis and Sequencing Facility. cDNA sequences were first screened for quality with the Chromas Lite sequence viewer (http://www.technelysium.com.au) and then assembled with CodonCode Aligner software (CodonCode Corporation, Dedham, MA). The UGT Nomenclature Committee approved the baboon UGT2B names, and the sequences were submitted to GenBank.
Stable Expression of UGT2B cDNA.
Human embryonic kidney (HEK) 293 cells (American Type Culture Collection, Manassas, VA) were grown to 50% confluence in 24-well plates containing Dulbecco's modified Eagle's medium with 10% fetal bovine serum (Invitrogen). The cells were transfected using FuGENE (Roche Diagnostics, Indianapolis, IN) and 1 μg of empty (control) or UGT2B cDNA containing plasmid. Transfected cells were allowed to grow for 2 days, after which the cells were passed to 10-cm diameter plates and continued to be grown in Dulbecco's modified Eagle's medium with 10% fetal bovine serum, 1% penicillin/streptomycin (Invitrogen), and 5 μg/ml blasticidin (Invitrogen) for selective growth of transfected cells. Cells were passed until several plates of viable cells were at least 80% confluent, at which point the cells were harvested.
Heavy Membrane Fraction Isolation.
The cells were harvested into ice-cold lysing buffer consisting of 150 mM KCl, pH 7.2, 10 mM HEPES, 0.5 mM dithiothreitol (Sigma-Aldrich, St. Louis, MO), and 1 mini-tablet of complete protease inhibitor mixture (Roche Diagnostics) per 25 ml of lysing buffer. The harvested cells were lysed with an Ultra-Turrax T8 homogenizer (IKA Works, Inc., Wilmington, NC) in four 1-s bursts on ice. Lysed cells were centrifuged for 7 min at 800g and 4°C. The supernatant fraction was saved, and the pellet resuspended in 5 ml of fresh cold lysing buffer. The lysis procedure and centrifugation were repeated as above. The combined supernatant fractions were then centrifuged for 30 min at 17,000g and 4°C, and the supernatant was removed. The pellet was resuspended in 300 to 500 μl of 150 mM KCl, pH 7.2, containing 10 mM HEPES. Protein concentrations were measured using the Bio-Rad (Bio-Rad Life Science, Hercules, CA) Protein Assay microtiter plate protocol and bovine serum albumin (Sigma-Aldrich) as a standard.
Morphine Glucuronidation Assay.
A glucuronidation assay using morphine as substrate and measuring the metabolites M3G and M6G was adapted from that described by Fisher et al. (2000). Glass incubation tubes, 12 × 17 mm (Fisher Scientific, Pittsburg, PA), were prepared with 10 μg of the pore-forming polypeptide alamethicin (Sigma-Aldrich) just before use by drying down 4 μl of alamethicin (5 mg/2 ml) in methanol under nitrogen. One hundred micrograms of recombinant UGT2B protein in 100 μl of 0.1 M phosphate buffer, pH 7.4, was added to each tube and placed on ice for 15 min. Fifty microliters of 4 mM MgCl2, 20 mM morphine (Spectrum, New Brunswick, NJ), and 0.1 M phosphate buffer, pH 7.4, were then added, and the tubes were preincubated in a water bath at 37°C for 3 min.
The reaction was initiated by the addition of 50 μl of 20 mM UDP-glucuronic acid (Sigma-Aldrich) in 0.1 M phosphate buffer, pH 7.4, and stopped after exactly 30 min for each sample by the addition of 150 μl of ice-cold 100% acetonitrile, high-performance liquid chromatography (HPLC) grade (Thermo Fisher Scientific, Waltham, MA). The tubes were placed on ice for 30 min. After addition of 10 μl of the internal standard nalorphine (Sigma-Aldrich) at 1 mg/ml, the samples were mixed and transferred to 1.5-ml Eppendorf centrifuge tubes and spun at 10,000g for 5 min at 4°C. The supernatants were removed and diluted 1:10 with 24% acetonitrile, HPLC grade, before being assayed by HPLC. All the incubations were performed in duplicate. Preliminary studies indicated that the formation of both M3G and M6G were linear up to 140 μg of protein and up to 50 min of incubation.
HPLC Assay for M3G and M6G.
The samples were analyzed for M3G and M6G by direct injection into a Waters (Milford, MA) Alliance 2695 HPLC system with fluorometric and coulochemical detection as described previously (Garland et al., 2006). In brief, M3G was measured by fluorescence detection on a Linear Model 305 fluorescence detector (Grace, Deerfield, IL) with an excitation wavelength of 210 nm and an emission wavelength of 340 nm. M6G was measured by coulochemical detection on a Coulochem II Electrochemical Detector with a 5011 analytical cell (ESA Inc., Chelmsford, MA) with potentials of +225 mV and +350 mV for E1 and E2, respectively. The internal standard, nalorphine, was measured by both detectors. Separation was performed on a Spherisorb C18 (ODS2, 3 μm, 4.6 × 100 mm i.d.) column (Waters) at ambient temperature. The injection volume was 50 μl. The isocratic elution was performed with a mobile phase consisting of 10 mM sodium phosphate monobasic (Thermo Fisher Scientific), 1.5 mM SDS (Invitrogen), pH 2.1, containing 24% acetonitrile, HPLC grade, at the rate of 1.5 ml/min.
Eight-point external standard curves ranging from 100 to 15,000 ng/100 μl were prepared from M3G and M6G obtained from the National Institute on Drug Abuse, Division of Neuroscience and Behavioral Research (Bethesda, MD). The Waters system used Empower Chromatography Software (Waters) for data collection and analysis. Standard curves were generated using the metabolite/internal standard area ratios. The formation of M3G and M6G was then quantified by comparison of the area ratio (metabolite/internal standard) of the incubated samples with that of the standard curves. Standard curve correlation coefficients (r) were >0.999 for both M3G and M6G. The UGT activity was calculated by dividing the metabolite concentration by the protein concentration and the incubation time and expressed as nanomoles per minute per milligram of protein.
Sequence Analysis.
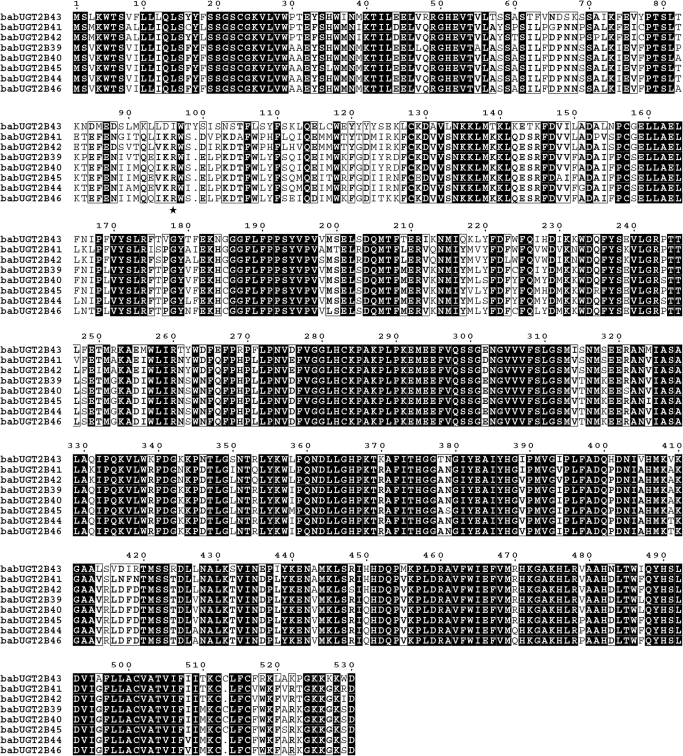
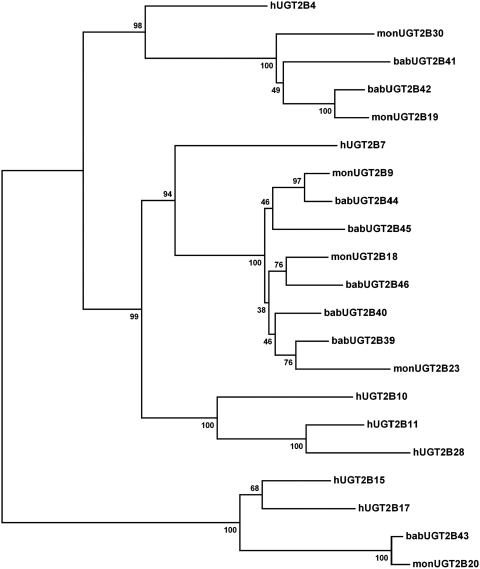
The cloned baboon UGT2B cDNAs were compared with those of both human and monkey (M. fascicularis) UGT2B cDNAs, which were obtained from the GenBank, by multiple alignments of the coding regions using the software suite MEGA4.0 (Tamura et al., 2007). The protein sequences were translated and aligned. The distance matrixes were calculated using the pairwise distance calculation. A distance-based neighbor-joining phylogenetic tree was generated. The reliability of the phylogeny was tested by bootstrap analysis with 1000 replicates. The bootstrap test produced a final reconstructed neighbor-joining consensus tree. Multiple alignments of the baboon UGT2B protein sequences for Fig. 2 were performed using MultAlin software (http://bioinfo.genopole-toulouse.prd.fr/multalin/multalin.html).
Fig. 2.
Multiple alignments of deduced baboon UGT2B protein amino acid sequences were aligned using the Multalin web page. Identical amino acids are shown in black background, and >80% conservation is illustrated by boxes. Residue 96 is indicated by a star.
The baboon, human, and monkey UGT2B cDNA sequences were analyzed for gene conversion events using GENECONV version 1.81 (http://www.math.wustl.edu/∼sawyer/geneconv/) (Sawyer, 1989). GENECONV analyzes multiple alignments and uses statistical methods for inferring conversion events based on the distribution of substitutions. Fragments that potentially originated from gene conversion were identified, and statistical significance (p < 0.05) of the fragments was given based on 10,000 permutations. GENCONV determined both global and pairwise fragments. The global p values were corrected for multiple comparisons and are a stronger predictor of gene conversion than the pairwise.
Results
cDNA Cloning.
Eight novel UGT2Bs clones were isolated from the livers of adult and newborn baboons. The GenBank accession numbers and the official names are listed in Table 1. The cDNAs encoding the baboon UGT2Bs varied in length from 1626 to 1634 base pairs with open reading frames of 1584 to 1590 base pairs, which translates into proteins of 528 to 530 amino acids. A comparison of the baboon UGT2B cDNAs with human UGT2B homologs showed amino acid identities ranging from 89 to 93%.
TABLE 1.
Baboon (Papio anubis) UGT2B cDNAs
The baboon UGT2B gene names are sequentially numbered based on their chronological order of discovery and were approved by the UGT Nomenclature Committee. Human homologs list the number of amino acid identities and percentage of amino acid identity.
| Baboon UGT | GenBank Accession Number | Number of Amino Acids in Coding Region | Human UGT Homologs |
|---|---|---|---|
| UGT2B41 | EU856383 | 528 | UGT2B4 (480/528) 90% |
| UGT2B42 | EU856382 | 528 | UGT2B4 (480/528) 90% |
| UGT2B39 | EU856384 | 529 | UGT2B7 (479/529) 91% |
| UGT2B40 | EU856385 | 529 | UGT2B7 (478/529) 90% |
| UGT2B44 | EU856388 | 528 | UGT2B7 (470/528) 89% |
| UGT2B45 | EU856387 | 529 | UGT2B7 (475/529) 90% |
| UGT2B46 | EU856386 | 528 | UGT2B7 (474/528) 90% |
| UGT2B43 | EU856381 | 530 | UGT2B15 (492/530) 93% |
Phylogenetic Analysis.
The amino acid sequences that were deduced from nucleotides of the corresponding baboon cDNAs were aligned with those of the cynomolgus monkey and human UGT2Bs, and distance matrixes were calculated (Table 2). The baboon UGT2B enzymes share a greater than 74% sequence similarity, which is typical for the UGT2 family of enzymes (Mackenzie et al., 2005). The baboon UGT2Bs fall into three groups: UGT2B43 is the most highly conserved baboon UGT2B with a 92.1 and 90% identity to human UGT2B15 and UGT2B17, respectively; UGT2B41 and UGT2B42 are 88.3 and 88.5% identical to human UGT2B4; and UGT2B39, UGTB240, UGT2B44, UGT2B45, and UGT2B46 are from 88.8 to 90.2% identical to human UGT2B7 (Table 2). These homologies ranging from 88 to 92% are similar to those found in a comparison between monkey and human UGT2B genes (Bélanger et al., 1997, 1999; Beaulieu et al., 1998; Barbier et al., 1999a,b; Girard et al., 2002). No baboon UGT2Bs similar to human UGT2B10, UGT2B11, or UGT2B28 are obvious.
TABLE 2.
Similarities between deduced amino acid sequences of the protein coding regions of UGT2Bs
| bab2B41 | bab2B42 | bab2B39 | bab2B40 | bab2B44 | bab2B45 | bab2B46 | bab2B43 | |
|---|---|---|---|---|---|---|---|---|
| Baboon vs. baboon | ||||||||
| bab2B41 | 100 | |||||||
| bab2B42 | 93.8 | 100 | ||||||
| bab2B39 | 81.8 | 83.1 | 100 | |||||
| bab2B40 | 82.0 | 83.0 | 97.5 | 100 | ||||
| bab2B44 | 81.3 | 83.0 | 95.5 | 96.4 | 100 | |||
| bab2B45 | 80.9 | 82.4 | 94.7 | 95.5 | 95.6 | 100 | ||
| bab2B46 | 81.6 | 82.6 | 95.5 | 96.2 | 96.2 | 93.9 | 100 | |
| bab2B43 | 74.1 | 75.4 | 76.4 | 76.2 | 75.6 | 76.0 | 76.5 | 100 |
| Human vs. baboon | ||||||||
| h2B4 | 88.3 | 88.5 | 86.2 | 86.2 | 86.0 | 85.6 | 85.6 | 76.9 |
| h2B7 | 81.4 | 82.0 | 89.8 | 90.2 | 88.8 | 89.2 | 89.4 | 75.4 |
| h2B10 | 80.3 | 81.0 | 86.7 | 87.1 | 86.7 | 87.5 | 86.0 | 75.6 |
| h2B11 | 81.1 | 81.3 | 85.6 | 86.6 | 86.6 | 86.2 | 86.0 | 74.7 |
| h2B28 | 78.8 | 79.4 | 83.9 | 84.9 | 85.2 | 84.9 | 84.7 | 73.2 |
| h2B15 | 76.5 | 76.5 | 79.0 | 78.8 | 78.0 | 77.7 | 77.7 | 92.1 |
| h2B17 | 76.1 | 76.1 | 77.7 | 78.1 | 77.5 | 77.1 | 77.3 | 90.0 |
| Monkey vs. baboon | ||||||||
| mon2B19 | 93.8 | 97.9 | 82.6 | 82.4 | 82.4 | 81.8 | 82.0 | 75.0 |
| mon2B30 | 92.8 | 93.8 | 82.2 | 82.0 | 82.0 | 81.4 | 81.4 | 75.2 |
| mon2B9 | 81.6 | 83.0 | 95.7 | 96.2 | 98.3 | 95.8 | 95.6 | 76.0 |
| mon2B18 | 82.2 | 83.0 | 96.2 | 96.4 | 95.8 | 95.1 | 96.8 | 75.8 |
| mon2B23 | 79.9 | 81.1 | 95.8 | 94.0 | 93.9 | 92.6 | 93.2 | 74.7 |
| mon2B20 | 74.1 | 75.0 | 76.0 | 75.8 | 75.2 | 75.6 | 75.2 | 99.1 |
bab, baboon/Papio anubis; h, human; mon, cynomolgus monkey/Macaca fascicularis.
An overall higher level of identity was found, as expected, between the baboon and the cynomolgus monkey UGT2Bs. The average sequence identity was 95.4% (range, 92.6–99.1%) with the highest level of identity (99.1%) found between baboon UGT2B43 and monkey UGT2B20. The same groupings also exist in the baboon and the monkey UGT2Bs. A comparison between the baboon and the two UGT2B isoforms isolated to date from another closely related Macaca species, the rhesus monkey (Macaca mulatta), showed a similar high identity (Dean et al., 2004). These isoforms, UGT2B9*2 and UGT2B33, show an identity ranging from 93 to 94.3% and 95.3 to 99% to the baboon UGT2B7-like isoforms, respectively. On the other hand, mouse and rat Ugt2b isoforms are more distantly related with identities to the baboon UGT2Bs ranging from 61.5 to 72%. (Sequences for rhesus monkey, mouse, and rat proteins were all obtained from the GenBank.)
A phylogenetic tree was reconstructed to further illustrate the relationship between the baboon, cynomolgus monkey, and human UGT2B enzymes (Fig. 1). Overall the baboon and the monkey UGT2Bs interleaf on the phylogenetic tree. Multiple duplications have occurred in both the baboon and the monkey within the UGT2B4-like and the UGT2B7-like groups. These duplications are in contrast to the single branches formed by the human UGT2B4 and UGT2B7. On the other hand, the UGT2B15-like group has a single duplication in the human that is not replicated in the baboon or the monkey. The human UGT2B10, UGT2B11, and UGT2B28 remain a monophyletic clade of paralogs, even though the cloning primers were designed to probe for all the possible baboon UGT2B enzymes.
Fig. 1.
Neighbor-joining (NJ) reconciled phylogenetic tree with baboon, human, and monkey (M. fascicularis) UGT2B enzymes. The sequences for human and monkey UGT2B genes were obtained from the GenBank. The protein coding regions were aligned using MEGA4.0, and an NJ distance-based phylogenetic tree using p distances was generated. Robustness of bifurcations is estimated by bootstrap analysis with 1000 replicates, and the consensus tree is shown. Numbers at bifurcations indicate bootstrap values in percent. Annotation: bab (baboon, P. anubis), h (human), and mon (cynomolgus monkey, M. fascicularis).
The high level of sequence similarity of the UGT2Bs, which is generally true of the UGT2 gene family, complicates identifying true orthologs between species (Table 2). The UGT2 genes are, as a result of this process, sequentially numbered based on the chronological order of discovery (Mackenzie et al., 2005). The within-species duplication/deletion events make it not possible to suggest orthologous relationships between human and Old World monkey UGT2Bs (Fig. 1). Although it is possible to suggest an orthologous relationship within the Old World monkeys between baboon UGT2B43 and monkey UGT2B20 in the UGT2B15-like group, the phylogenetic relationship for the other groups of UGT2Bs is more complex.
The multiple duplications and the high level of similarity within the baboon UGT2Bs and that reported for the monkey UGT2Bs suggest a recombination within the Old World monkey UGT2B gene families (Barbier et al., 1999b; Girard et al., 2002). Gene conversion is a recombination process between highly homologous sequences involving the nonreciprocal transfer of genetic material from one gene to another. The sequences of the two genes have become more similar as a result, and that can lead to concerted evolution (Drouin et al., 1999; Chen et al., 2007). Therefore, the human, baboon, and monkey UGT2Bs were analyzed for gene conversion events using GENECONV software (Table 3). Global p values for potential converted segments reached statistical significance for baboons in the UGT2B7-like group and for monkeys in the UGT2B4-like group. No statistical significance was found for the human UGT2Bs. This finding indicates that gene conversion is a likely mechanism operating within the baboon and the monkey but not in human UGT2Bs. The presence of gene conversion within the Old World monkey UGT2B4-like and UGT2B7-like genes means that orthologous relationships between these genes cannot formally be inferred because different parts of their sequences have different evolutionary histories (Posada et al., 2002).
TABLE 3.
Gene conversion events indicated by GENECONV
Alignments of UGT2B4-like and UGT2B7-like sequences were analyzed with GENECONV. Human, baboon (Papio anubis), and cynomolgus monkey (Macaca fascicularis) sequences were divided into separate groups to limit analysis to within-species gene conversion and reduce the multiple comparison penalty. The p values are based on 10,000 permutations, and only significant (p < 0.05) conversions are included. The different g-scales used to allow for mismatches are given in parentheses after the p value. Only converted sequences >95 nucleotides are listed, and the position is relative to the start codon.
| Sequence Name | Global p Value | Sequence | Pairwise p Values | Sequence |
|---|---|---|---|---|
| babUGT2B39; UGT2B46 | <0.0001 (0) | 676–1193 | <0.0001 (0) | 676–1193 |
| babUGT2B39; UGT2B40 | 0.0117 (0) | 958–1587 | 0.0001 (0) | 958–1587 |
| babUGT2B39; UGT2B40 | 0.0121 (0) | 291–650 | ||
| babUGT2B39; UGT2B45 | 0.0343 (0) | 1131–1305 | 0.0033 (0) | 1131–1305 |
| babUGT2B39; UGT2B45 | 0.0207 (0) | 699–963 | ||
| babUGT2B40; UGT2B44 | 0.0067 (0) | 550–956 | 0.0006 (0) | 550–956 |
| babUGT2B40; UGT2B45 | 0.0321 (0) | 1131–1305 | ||
| babUGT2B44; UGT2B46 | 0.0117 (0) | 1147–1587 | 0.0009 (0) | 1147–1587 |
| babUGT2B41; UGT2B42 | 0.0355 (1) | 596–986 | ||
| monUGT2B19; UGT2B30 | 0.0154 (0) | 334–452 | 0.0095 (0) | 334–452 |
bab, baboon/Papio anubis; mon, cynomolgus monkey/Macaca fascicularis.
Protein Sequence Analysis.
The multiple alignments of the deduced baboon UGT2B protein sequences reveal primary protein structures characteristic of UGT2B enzymes (Fig. 2). The carboxyl termini of the baboon UGT2B proteins are, as for all the UGT proteins, the most highly conserved region and therefore more homologous than the amino termini. The protein sequences contain hydrophobic signal peptides from amino acids 1 to 23, as well as hydrophobic trans-membrane regions in amino acids ranging from 493 to 511. The consensus sequence for UGT enzymes is also present in amino acids ranging from 371 to 400 (Mackenzie et al., 1997).
Binding studies and NMR spectroscopy, as well as molecular modeling, have shown that the morphine binding site in human UGT2B7 is located within amino acids 96 and 101 (Coffman et al., 2003). Furthermore, the substitution of the negatively charged aspartic acid in position 99 with the neutral alanine led to a marked decrease in morphine binding. The baboon UGT2B7-like proteins all show a substitution in position 99, where aspartic acid has been substituted with the only other negatively charged amino acid, glutamic acid. A substitution in position 98 of the neutral serine with the neutral isoleucine in UGT2B39 and UGT2B44 is the only other difference between the baboon and the human UGT2B7s in this region.
In addition, the alignments of the deduced UGT2B protein sequences show a single amino acid substitution at position 96 in UGT2B43, where a highly conserved arginine has been substituted with isoleucine. The alignment of baboon, monkey, and human amino acid sequences used in the phylogenetic analysis above (sequences not shown) indicate that the arginine in position 96 is conserved in all the other UGT2B proteins except for monkey UGT2B20, which also contains an isoleucine in position 96. Furthermore, the primary protein structures of baboon UGT2B43 and monkey UGT2B20 are identical, except for a difference in only three amino acids (Y16C, G50R, and Q217H), none of which are located in the proposed substrate-binding region between positions 60 to 120 (Tephly and Burchell, 1990).
Post-translational asparagine-linked glycosylation (N-glycosylation) is known to regulate protein function. It has also been found to be essential for the activity of several UGT2B enzymes (Barbier et al., 2000b). The mammalian UGT2B proteins that have been characterized to date have between one and four potential N-glycosylation sites with the sequences of NX(S/T). However, not all the UGT2Bs are glycosylated, nor are all the potential sites actually used. To date, only the human UGT2B7, UGT2B15, and UGT2B17 and the monkey UGT2B20 have been shown to be glycosylated. It has also been shown that UGT2B15 and UGT2B20 are glycosylated at position 65 only (Barbier et al., 2000b). Potential N-glycosylation sites in the primary protein structures of human, baboon, and monkey UGT2Bs were compared (Table 4). Several potential N-glycosylation sites are found in the baboon UGT2B proteins. The location and number of these sites are identical to those found in monkey UGT2B proteins. This finding includes the unique fourth site at position 103 that is only found in monkey UGT2B20 and baboon UGT2B43.
TABLE 4.
Potential N-glycosylation sites in baboon, human, and monkey UGT2B enzymes
Comparison of potential N-glycosylation sites in the primary structures of bab (baboon, Papio anubis), h (human), and mon (cynomolgus monkey, Macaca fascicularis) UGT2B enzymes. The sequences for human and monkey UGT2B genes were obtained from the GenBank. Multiple alignments of deduced UGT2B protein amino acid sequences were aligned using the MEGA4.0 software suite. Amino acid sequences were compared, and the potential N-glycosylation sites were identified and tabulated.
| Mammalian UGT | Number of Potential N-Glycosylation Sites | Amino Acid Position and Sequence of Potential N-Glycosylation Sites |
||||
|---|---|---|---|---|---|---|
| h2B4 | 1 | 315-NTS | ||||
| bab2B41 | 2 | 68-NPS | 315-NMS | |||
| bab2B42 | 2 | 68-NPS | 315-NMS | |||
| mon2B19 | 2 | 68-NPS | 315-NMS | |||
| mon2B30 | 2 | 68-NPS | 315-NMS | |||
| h2B7 | 3 | 67-NNS | 68-NSS | 315-NMT | ||
| bab2B39 | 2 | 67-NNS | 68-NSS | |||
| bab2B40 | 2 | 67-NNS | 68-NSS | |||
| bab2B44 | 2 | 67-NNS | 68-NSS | |||
| bab2B45 | 2 | 67-NNS | 68-NSS | |||
| bab2B46 | 2 | 67-NNS | 68-NSS | |||
| mon2B39 | 2 | 67-NNS | 68-NSS | |||
| mon2B18 | 2 | 67-NNS | 68-NSS | |||
| mon2B23 | 2 | 67-NNS | 68-NSS | |||
| h2B15 | 3 | 65-NAS | 316-NMS | 483-NLT | ||
| h2B17 | 3 | 65-NAS | 316-NMS | 483-NLT | ||
| bab2B43 | 4 | 65-NDS | 103-NST | 316-NMS | 483-NLT | |
| mon2B20 | 4 | 65-NDS | 103-NST | 316-NMS | 483-NLT | |
UGT2B Activity toward Morphine.
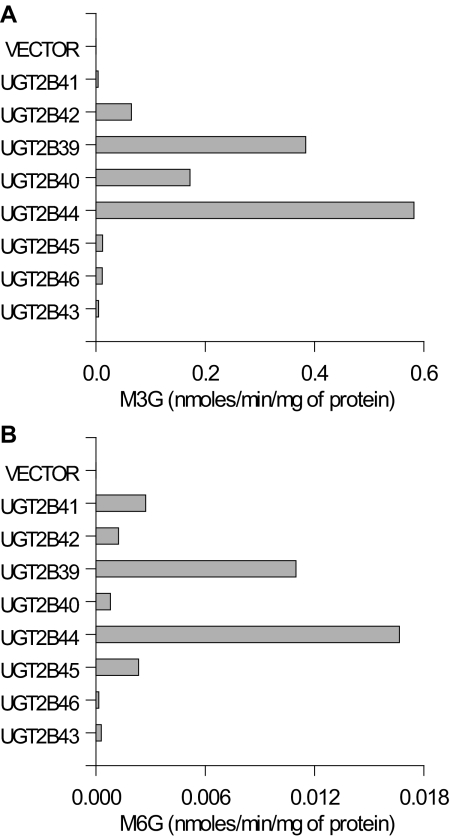
The heavy membrane fractions of the expressed baboon UGT2B cDNAs were tested for activity toward morphine. All the cloned baboon UGT2Bs have the ability to glucuronidate morphine to both M3G and M6G (Fig. 3). Heavy membrane fractions from cells transfected with empty plasmid did not show any activity. This result indicates that the background activity of UGT2Bs in HEK293 cells is either absent or undetectable. Although the assay is only semiquantitative in view of the variable transfection efficiency among clones, the activity was by far highest in the UGT2B7-like group of the baboon UGT2Bs. Of special note is the ability of the UGT2B15-like UGT2B43 to metabolize morphine, not only to M3G but also to M6G.
Fig. 3.
Morphine glucuronidation in heavy membranes from HEK293 cells expressing individual baboon UGT2B isoforms or HEK293 vector control. Morphine-3-UGT and morphine-6-UGT activity was measured at a morphine concentration of 5 mM. Each bar represents the mean of duplicate measurements.
Discussion
Eight novel functioning UGT2B isoforms were expressed in the liver of the baboon that all exhibit some capacity to metabolize morphine to both M3G and M6G. Although there are considerable duplications and deletions among the primates, a close relationship exists among the human, the baboon, and the monkey (M. fascicularis) UGT2B enzyme families.
The enzyme activity toward morphine by the baboon UGT2Bs shows that all the baboon cDNA clones encode functional enzymes. In addition, as in humans, all the UGT2B7-like and UGT2B4-like enzymes in the baboon metabolize morphine to M3G and M6G. In contrast, human UGT2B15 and UGT2B17 have only been shown to metabolize morphine to M3G (Court et al., 2003; Ohno et al., 2008). However, the baboon UGT2B43 metabolizes morphine to M3G and M6G, although to a lesser extent than the other baboon UGT2B enzymes. The predominant UGT2B isoforms responsible for morphine glucuronidation in the baboon were, as in the human, the UGT2B7-like enzymes. These results indicate that morphine can also be considered an isoform-selective probe drug in the baboon, even though more UGT2B7-like isoforms are present in this species. The two most active baboon UGT2B7-like isoforms, UGT2B39 and UGT2B44, both have an S98I substitution compared with that of human UGT2B7 and other baboon UGT2B7-like enzymes. However, both serine and isoleucine are neutral amino acids, so this substitution would not be expected to alter the binding capacity of the enzymes.
The total morphine clearance in adult humans, which is almost entirely metabolic, is similar to that found in adult baboons. On the other hand, the metabolic clearance in the baboon fetus is greater than what might have been expected based on enzyme activity comparisons (Garland et al., 2005). The multiple copies of UGT2B7-like isoforms that metabolize morphine could be one possible explanation for the enhanced metabolism of morphine observed in the fetal baboon. In the adult baboon, they would have less of an impact as morphine metabolism is flow-limited.
The phylogenetic analysis indicates that the encoded UGT2B proteins are highly conserved among humans, baboons, and monkeys, although several duplications have taken place in both the human and the Old World monkeys (Table 2; Fig. 1). The major differences are the multiple duplications of the UGT2B7-like genes and a duplication of the UGT2B4-like gene in the Old World monkeys and the single duplication in the human UGT2B15-like genes that is absent in the baboon and the monkey. The UGT2B10, UGT2B11, and UGT2B28 human paralogs are all absent in the Old World monkeys, indicating that these genes most likely have been deleted. Of note, both UGT2B17 and UGT2B28 are frequent deletion polymorphisms among human populations (Ménard et al., 2009).
The multiple duplications make it impossible to infer one-to-one orthologous relationships between the human and any of the baboon or the monkey UGT2Bs. Furthermore, the detection of gene conversion events operating on the primate UGT2B genes interferes with the phylogenetic signal between several of these genes. However, based on bootstrap analysis and the absence of gene conversion, an orthologous relationship is suggested between baboon UGT2B43 and monkey UGT2B20 in the UGT2B15-like group. The UGT2B4-like group shows intermediate bootstrap support, indicating a partial loss of the phylogenetic signal as a result of gene conversions. The human, baboon, and monkey UGT2B within the UGT2B7-like group share a high level of sequence identity, but the phylogenetic signal is obscured by gene conversion events detected in all the baboon genes of this group.
The families of drug-metabolizing enzymes have developed under selective pressure from exogenous substrates throughout the evolution of different species (Nebert and Dieter, 2000; Thomas, 2007). This activity is well illustrated in the cytochrome P450 family, in which enzymes involved in endogenous pathways have retained strong orthologous relationships across species but enzymes involved in xenobiotic metabolism show considerable within-species diversity (Thomas, 2007). In addition, this study concluded that the xenobiotic cytochrome P450 enzymes have evolved primarily through frequent gene duplications and losses, so-called birth-and-death evolution, in a species-specific manner. Therefore, the duplication, diversification, and deletion of the UGT2B enzymes may have occurred in a species-specific manner. The comparison of human, baboon, and monkey UGT2Bs reveals that these enzymes are subject to species-specific duplications and deletions and gene conversion. The gene conversion within the Old World monkeys was extensive enough to break down the orthologous relationship in the UGT2B4-like and the UGT2B7-like groups. This process indicates that duplication and gene conversion in the Old World monkey UGT2B4-like and UGT2B7-like groups most likely happened after speciation. Furthermore, the detection of gene conversion operating within the baboon and monkey UGT2Bs supports that a mixture of concerted and birth-and-death evolution likely contributed to the evolution of these genes in the baboon and the monkey but not in the human (Nei and Rooney, 2005). These observations are consistent with an earlier study of UGT2B genes in several species, indicating that most human, mouse, and rat UGT2Bs were duplicated after speciation and where gene conversion was detected in rats and zebrafish but not in humans (Li and Wu, 2007). However, this study did not include any Old World monkeys in their analysis.
The protein sequence analysis indicated a high level of structural conservation between the human, the baboon, and the monkey UGT2Bs. The sequence identity was especially high between the baboon and the monkey UGT2B proteins. The 99.1% identical protein structure between the baboon UGT2B43 and monkey UGT2B20 is the highest reported with any other UGT2B protein (human or primate) characterized to date (Bélanger et al., 1997, 1999; Beaulieu et al., 1998; Barbier et al., 1999a,b; Girard et al., 2002). As with UGT2B20, baboon UGT2B43 has an isoleucine substitution of a highly conserved arginine at position 96 not found in any other UGT2B (Barbier et al., 2000b). UGT2B20 has been found to be a more labile enzyme than human UGT2B15 because of this isoleucine substitution, which reduces the length of the predicted α-helix between amino acids 84 and 100. The reduced length of the α-helix correlated with decreased enzyme stability (Barbier et al., 2000b).
The identical number and location of N-glycosylation sites known to be essential for the activity of several UGT2B enzymes were found in the baboon and the monkey UGT2Bs. UGT2B20 is the only monkey UGT2B characterized to date that has been shown to be glycosylated (Barbier et al., 2000b). Although the extent of glycosylation or other enzyme functions cannot be predicted by the similarities of protein sequence, the especially high level of identity (99.1%) between baboon UGT2B43 and UGT2B20 makes it reasonable to expect that baboon UGT2B43 would also be glycosylated. Furthermore, the R96I substitution in UGT2B43 and UGT2B20 makes it very likely that both these enzymes are more labile than human UGT2B15. These findings suggest that these two enzymes are true orthologs. This high level of conservation may reflect the specific role of this enzyme in androgen metabolism (Barbier et al., 1999a; Thomas, 2007).
To fully understand the origin of the multiple duplications in the baboon and the monkey UGT2B7-like genes, a comparison of the human, the baboon, and the monkey genomes would be necessary. The genomes for the baboon and M. fascicularis are not yet available for comparison. However, the rhesus monkey (M. mulatta) genome is available in GenBank, although only two UGT2B7 isoforms have been isolated and characterized to date. The remaining UGT2B isoforms on the rhesus genome are predicted from modeling.
An alignment of the monkey (M. fascicularis) UGT2Bs that have been isolated to date with the M. mulatta genome shows a more than 99% identity. However, it did leave one predicted UGT2B7-like gene on the genome without any known expressed isoform (data not shown). The M. fascicularis UGT2Bs have been isolated from cDNA libraries, which leaves the possibility that not all the M. fascicularis UGT2B isoforms have been isolated as of yet. Thus far, it appears there is at least one more UGT2B7-like isoform in the baboon than in Macaca species, consistent with species-specific diversification.
The UGT2B genes are located on the long arm of chromosome 4 below the centromere in humans. In the M. mulatta, the UGT2B genes are located on chromosome 5 above the centromere. The recent constructions of a genetic linkage map of the M. mulatta to the human genome show a pericentric inversion of this region between the Macaca and the human chromosomes (Karere et al., 2008). A pericentric inversion has also been shown in a genetic linkage map of the baboon (Papio hamadryas) chromosome 5 to the human chromosome 4, as well as on chromosome 4 between the human and the chimpanzee (Yunis and Prakash, 1982; Rogers et al., 2000; Chimpanzee Sequencing and Analysis Consortium, 2005; Cox et al., 2006).
The chromosomal rearrangement of human chromosome 4 compared with that of the chimpanzee, such as the pericentric inversion, has been associated with a higher divergence of genes located within the inversion (Marques-Bonet et al., 2007). In addition, this study found that although chromosomal speciation is not common between the human and the chimpanzee lineage, chromosome 4 may be an exception and has potentially been associated with a speciation episode. The pericentric inversion in chromosome 4 of hominoids contains genes involved with biotic stimulus response, such as UGT2Bs, that are likely to be related to adaptation and hence may also be more likely associated with chromosomal speciation. This result is consistent with the findings in a comparative study of the chromosomes from the human, the chimpanzee, the gorilla, and the orangutan, indicating that similar but distinct pericentric inversions occurred on chromosome 4 in each of the speciation events of the orangutan, the gorilla, and the chimpanzee compared with that of the human (Yunis and Prakash, 1982). Thus, it is possible to suggest that the pericentric inversions in the baboon and the monkey chromosome 5, which contains the UGT2B genes, occurred as a result of the speciation event from the common ancestral line. Moreover, the subsequent divergence of the baboon and the monkey from that of the human UGT2B genes would have happened after speciation and under selective pressure from exogenous substances (Nebert and Dieter, 2000).
In summary, this study reports the isolation of eight novel UGT2B isoforms expressed in the baboon liver. These enzymes all showed activity toward morphine and the ability to glucuronidate morphine to M3G and M6G. In addition, the similarities between the primary structures of the human, the baboon, and the monkey UGT2B proteins provided further evidence that there is a close relationship among the UGT2B families of enzymes in these species. Although a one-to-one orthologous relationship cannot be established for human UGT2B7, the UGT2B7-like isoforms in the baboon all have substantial activity toward morphine. This, together with the close relationship shown previously between the human and the baboon UGT1A enzyme families, further substantiates that the baboon is an excellent model for studying clinically relevant aspects of drug metabolism. The data obtained in this study lay the foundation for investigating the regulation of UGT2B enzymes during fetal and neonatal development in the baboon and facilitate the extrapolation of results from the baboon to humans.
This work was supported in part by the National Institutes of Health National Institute on Drug Abuse [Grant R01-DA14215].
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.109.030635.
- UGT
- UDP-glucuronosyltransferase
- M3G
- morphine-3-β-glucuronide
- M6G
- morphine-6-β-glucuronide
- PCR
- polymerase chain reaction
- HEK
- human embryonic kidney
- HPLC
- high-performance liquid chromatography
- N-glycosylation
- asparagine-linked glycosylation.
References
- Barbier O, Bélanger A. (2003) The cynomolgus monkey (Macaca fascicularis) is the best animal model for the study of steroid glucuronidation. J Steroid Biochem Mol Biol 85:235–245 [DOI] [PubMed] [Google Scholar]
- Barbier O, Bélanger A, Hum DW. (1999a) Cloning and characterization of a simian UDP-glucuronosyltransferase enzyme UGT2B20, a novel C19 steroid-conjugating protein. Biochem J 337:567–574 [PMC free article] [PubMed] [Google Scholar]
- Barbier O, Girard C, Breton R, Bélanger A, Hum DW. (2000b) N-Glycosylation and residue 96 are involved in the functional properties of UDP-glucuronosyltransferase enzymes. Biochemistry 39:11540–11552 [DOI] [PubMed] [Google Scholar]
- Barbier O, Lévesque E, Bélanger A, Hum DW. (1999b) UGT2B23, a novel uridine diphosphate-glucuronosyltransferase enzyme expressed in steroid target tissues that conjugates androgen and estrogen metabolites. Endocrinology 140:5538–5548 [DOI] [PubMed] [Google Scholar]
- Barbier O, Turgeon D, Girard C, Green MD, Tephly TR, Hum DW, Bélanger A. (2000a) 3′-azido-3′-deoxythimidine (AZT) is glucuronidated by human UDP-glucuronosyltransferase 2B7 (UGT2B7). Drug Metab Dispos 28:497–502 [PubMed] [Google Scholar]
- Beaulieu M, Lévesque E, Barbier O, Turgeon D, Bélanger G, Hum DW, Bélanger A. (1998) Isolation and characterization of a simian UDP-glucuronosyltransferase UGT2B18 active on 3-hydroxyandrogens. J Mol Biol 275:785–794 [DOI] [PubMed] [Google Scholar]
- Bélanger G, Barbier O, Hum DW, Bélanger A. (1999) Molecular cloning, expression and characterization of a monkey steroid UDP-glucuronosyltransferase, UGT2B19, that conjugates testosterone. Eur J Biochem 260:701–708 [DOI] [PubMed] [Google Scholar]
- Bélanger G, Beaulieu M, Lévesque E, Hum DW, Bélanger A. (1997) Expression and characterization of a novel UDP-glucuronosyltransferase, UGT2B9, from cynomolgus monkey. DNA Cell Biol 16:1195–1205 [DOI] [PubMed] [Google Scholar]
- Caspersen CS, Reznik B, Weldy PL, Abildskov KM, Stark RI, Garland M. (2007) Molecular cloning of the baboon UDP-glucuronosyltransferase 1A gene family: evolution of the primate UGT1 locus and relevance for models of human drug metabolism. Pharmacogenet Genomics 17:11–24 [DOI] [PubMed] [Google Scholar]
- Chay PC, Duffy BJ, Walker JS. (1992) Pharmacokinetic-pharmacodynamic relationships of morphine in neonates. Clin Pharmacol Ther 51:334–342 [DOI] [PubMed] [Google Scholar]
- Chen JM, Cooper DN, Chuzhanova N, Férec C, Patrinos GP. (2007) Gene conversion: mechanisms, evolution and human disease. Nat Rev Genet 8:762–775 [DOI] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium (2005) Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437:69–87 [DOI] [PubMed] [Google Scholar]
- Coffman BL, Kearney WR, Goldsmith S, Knosp BM, Tephly TR, Dean B, Arison B, Chang S, Thomas PE, King C. (2003) Opioids bind to the amino acids 84 to 118 of UDP-glucuronosyltransferase UGT2B7. Mol Pharmacol 63:283–288 [DOI] [PubMed] [Google Scholar]
- Coffman BL, Rios GR, King CD, Tephly TR. (1997) Human UGT2B7 catalyzes morphine glucuronidation. Drug Metab Dispos 25:1–4 [PubMed] [Google Scholar]
- Court MH, Duan SX, von Moltke LL, Greenblatt DJ, Patten CJ, Miners JO, Mackenzie PI. (2001) Interindividual variability in acetaminophen glucuronidation by human liver microsomes: identification of relevant acetaminophen UDP-glucuronosyltransferase isoforms. J Pharmacol Exp Ther 299:998–1006 [PubMed] [Google Scholar]
- Court MH, Krishnaswamy S, Hao Q, Duan SX, Patten CJ, Von Moltke LL, Greenblatt DJ. (2003) Evaluation of 3′-azido-3′-deoxythymidine, morphine, and codeine as probe substrates for UDP-glucuronosyltransferase 2B7 (UGT2B7) in human liver microsomes: specificity and influence of the UGT2B7*2 polymorphism. Drug Metab Dispos 31:1125–1133 [DOI] [PubMed] [Google Scholar]
- Cox LA, Mahaney MC, Vandeberg JL, Rogers J. (2006) A second-generation genetic linkage map of the baboon (Papio hamadryas) genome. Genomics 88:274–281 [DOI] [PubMed] [Google Scholar]
- de Wildt SN, Kearns GL, Leeder JS, van den Anker JN. (1999) Glucuronidation in humans. Pharmacogenetic and developmental aspects. Clin Pharmacokinet 36:439–452 [DOI] [PubMed] [Google Scholar]
- Dean B, Arison B, Chang S, Thomas PE, King C. (2004) Identification of UGT2B9*2 and UGT2B33 isolated from female rhesus monkey liver. Arch Biochem Biophys 426:55–62 [DOI] [PubMed] [Google Scholar]
- Drouin G, Prat F, Ell M, Clarke GD. (1999) Detecting and characterizing gene conversions between multigene family members. Mol Biol Evol 16:1369–1390 [DOI] [PubMed] [Google Scholar]
- Dutton GJ. (1978) Developmental aspects of drug conjugation, with special reference to glucuronidation. Annu Rev Pharmacol Toxicol 18:17–35 [DOI] [PubMed] [Google Scholar]
- Dutton GJ, Leakey JE. (1981) The perinatal development of drug-metabolizing enzymes: what factors trigger their onset? Prog Drug Res 25:189–273 [PubMed] [Google Scholar]
- Fisher MB, Campanale K, Ackermann BL, VandenBranden M, Wrighton SA. (2000) In vitro glucuronidation using human liver microsomes and the pore-forming peptide alamethicin. Drug Metab Dispos 28:560–566 [PubMed] [Google Scholar]
- Garland M, Abildskov KM, Kiu TW, Daniel SS, Stark RI. (2005) The contribution of fetal metabolism to the disposition of morphine. Drug Metab Dispos 33:68–76 [DOI] [PubMed] [Google Scholar]
- Garland M, Abildskov KM, Taylor S, Benzeroual K, Caspersen CS, Arroyo SE, Kiu TW, Reznik B, Weldy P, Daniel SS, et al. (2006) Fetal morphine metabolism and clearance are constant during late gestation. Drug Metab Dispos 34:636–646 [DOI] [PubMed] [Google Scholar]
- Garland M, Szeto HH, Daniel SS, Tropper PJ, Myers MM, Stark RI. (1996) Zidovudine kinetics in the pregnant baboon. J Acquir Immune Defic Syndr Hum Retrovirol 11:117–127 [DOI] [PubMed] [Google Scholar]
- Garland M, Szeto HH, Daniel SS, Tropper PJ, Myers MM, Stark RI. (1998a) Placental transfer and fetal metabolism of zidovudine in the baboon. Pediatr Res 44:47–53 [DOI] [PubMed] [Google Scholar]
- Garland M, Szeto HH, Daniel SS, Tropper PJ, Myers MM, Stark RI. (1998b) Implications of the kinetics of zidovudine in the pregnant baboon following oral administration. J Acquir Immune Defic Syndr Hum Retrovirol 19:433–440 [DOI] [PubMed] [Google Scholar]
- Girard C, Barbier O, Turgeon D, Bélanger A. (2002) Isolation and characterization of the monkey UGT2B30 gene that encodes a uridine diphosphate-glucuronosyltransferase enzyme active on mineralocorticoid, glucocorticoid, androgen and oestrogen hormones. Biochem J 365:213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MD, King CD, Mojarrabi B, Mackenzie PI, Tephly TR. (1998) Glucuronidation of amines and other xenobiotics catalyzed by expressed human UDP-glucuronosyltransferase 1A3. Drug Metab Dispos 26:507–512 [PubMed] [Google Scholar]
- Hartley R, Green M, Quinn M, Levene MI. (1993) Pharmacokinetics of morphine infusion in premature neonates. Arch Dis Child 69:55–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karere GM, Froenicke L, Millon L, Womack JE, Lyons LA. (2008) A high-resolution radiation hybrid map of rhesus macaque chromosome 5 identifies rearrangements in the genome assembly. Genomics 92:210–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wu Q. (2007) Adaptive evolution of multiple-variable exons and structural diversity of drug-metabolizing enzymes. BMC Evol Biol 7:69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie PI, Bock KW, Burchell B, Guillemette C, Ikushiro S, Iyanagi T, Miners JO, Owens IS, Nebert DW. (2005) Nomenclature update for the mammalian UDP glycosyltransferase (UGT) gene superfamily. Pharmacogenet Genomics 15:677–685 [DOI] [PubMed] [Google Scholar]
- Mackenzie PI, Owens IS, Burchell B, Bock KW, Bairoch A, Bélanger A, Fournel-Gigleux S, Green M, Hum DW, Iyanagi T, et al. (1997) The UDP glycosyltransferase gene superfamily: recommended nomenclature update based on evolutionary divergence. Pharmacogenetics 7:255–269 [DOI] [PubMed] [Google Scholar]
- Marques-Bonet T, Sànchez-Ruiz J, Armengol L, Khaja R, Bertranpetit J, Lopez-Bigas N, Rocchi M, Gazave E, Navarro A. (2007) On the association between chromosomal rearrangements and genic evolution in humans and chimpanzees. Genome Biol 8:R230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménard V, Eap O, Harvey M, Guillemette C, Lévesque E. (2009) Copy-number variations (CNVs) of the human sex steroid metabolizing genes UGT2B17 and UGT2B28 and their associations with a UGT2B15 functional polymorphism. Hum Mutat 30:1310–1319 [DOI] [PubMed] [Google Scholar]
- Nebert DW, Dieter MZ. (2000) The evolution of drug metabolism. Pharmacology 61:124–135 [DOI] [PubMed] [Google Scholar]
- Nei M, Rooney AP. (2005) Concerted and birth-and-death evolution of multigene families. Annu Rev Genet 39:121–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S, Kawana K, Nakajin S. (2008) Contribution of UDP-glucuronosyltransferase 1A1 and 1A8 to morphine-6-glucuronidation and its kinetic properties. Drug Metab Dispos 36:688–694 [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA, Holmes EC. (2002) Recombination in evolutionary genomics. Annu Rev Genet 36:75–97 [DOI] [PubMed] [Google Scholar]
- Rogers J, Mahaney MC, Witte SM, Nair S, Newman D, Wedel S, Rodriguez LA, Rice KS, Slifer SH, Perelygin A, et al. (2000) A genetic linkage map of the baboon (Papio hamadryas) genome based on human microsatellite polymorphisms. Genomics 67:237–247 [DOI] [PubMed] [Google Scholar]
- Sawyer S. (1989) Statistical tests for detecting gene conversion. Mol Biol Evol 6:526–638 [DOI] [PubMed] [Google Scholar]
- Scott CS, Riggs KW, Ling EW, Fitzgerald CE, Hill ML, Grunau RV, Solimano A, Craig KD. (1999) Morphine pharmacokinetics and pain assessment in premature newborns. J Pediatr 135:423–429 [DOI] [PubMed] [Google Scholar]
- Soars MG, Ring BJ, Wrighton SA. (2003) The effect of incubation conditions on the enzyme kinetics of udp-glucuronosyltransferases. Drug Metab Dispos 31:762–767 [DOI] [PubMed] [Google Scholar]
- Strassburg CP, Strassburg A, Kneip S, Barut A, Tukey RH, Rodeck B, Manns MP. (2002) Developmental aspects of human hepatic drug glucuronidation in young children and adults. Gut 50:259–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- Tephly TR, Burchell B. (1990) UDP-glucuronosyltransferases: a family of detoxifying enzymes. Trends Pharmacol Sci 11:276–279 [DOI] [PubMed] [Google Scholar]
- Thomas JH. (2007) Rapid birth-death evolution specific to xenobiotic cytochrome P450 genes in vertebrates. PLoS Genet 3:e67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunis JJ, Prakash O. (1982) The origin of man: a chromosomal pictorial legacy. Science 215:1525–1530 [DOI] [PubMed] [Google Scholar]