Abstract
Gliomas are the most common type of primary brain tumors in adults and a significant cause of cancer-related mortality. Defining glioma subtypes based on objective genetic and molecular signatures may allow for a more rational, patient-specific approach to therapy in the future. Classifications based on gene expression data have been attempted in the past with varying success and with only some concordance between studies, possibly due to inherent bias that can be introduced through the use of analytic methodologies that make a priori selection of genes before classification. To overcome this potential source of bias, we have applied two unsupervised machine learning methods to genome-wide gene expression profiles of 159 gliomas, thereby establishing a robust glioma classification model relying only on the molecular data. The model predicts for two major groups of gliomas (oligodendroglioma-rich and glioblastoma-rich groups) separable into six hierarchically nested subtypes. We then identified six sets of classifiers that can be used to assign any given glioma to the corresponding subtype and validated these classifiers using both internal (189 additional independent samples) and two external data sets (341 patients). Application of the classification system to the external glioma data sets allowed us to identify previously unrecognized prognostic groups within previously published data and within The Cancer Genome Atlas glioblastoma samples and the different biological pathways associated with the different glioma subtypes offering a potential clue to the pathogenesis and possibly therapeutic targets for tumors within each subtype.
Introduction
Primary brain tumors are an important cause of cancer mortality in adults and children in the United States (1). The molecular and genetic heterogeneity of gliomas undoubtedly contributes to the varied and often suboptimal response to treatment that is usually based on standard pathologic diagnoses (2, 3). Glioma diagnosis has been historically based on examining the cellular morphology of the tumor to assess its presumed cell of origin (astrocytic, oligodendroglial, ependymal) and surrogate markers of tumor aggressiveness (necrosis, nuclear pleomorphism, mitoses) to determine the tumor grade (4). Glioblastomas are the most common and aggressive gliomas and are thought to arise de novo (primary glioblastoma) or through the malignant transformation of lower-grade astrocytic and oligodendroglial tumors (secondary glioblastoma; ref. 5). Although some genetic aberrations and clinical characteristics (i.e., age) have been associated with each type of glioblastoma, currently, there are few distinguishable differences in the histopathology or prognosis associated with primary and secondary glioblastomas (6-9).
Histopathologic diagnoses are by nature subjective (10), and the diagnosis of glioma subtypes has historically been associated with significant intraobserver variability. Even when the diagnosis of a distinct subtype of glioma can be agreed upon (i.e., gliosarcoma, small cell glioblastoma), the molecular, genetic, or clinical relevance of such designations remains obscure. Although standard tumor morphologic observations and low throughput genetic studies have revealed some molecular characteristics relevant to particular histologic subtypes, the study of the molecular features of gliomas has only recently come to the forefront with the advent of high-throughput microarray technology (6, 8).
Gene expression profiles provides a transcriptomic snapshot of a biological phenotype and offers the opportunity for quantitative, reproducible evaluation of individual tumor biology (8, 11). Consequently, data derived from genomic-scale gene expression profiling facilitate the characterization of intertumoral variations and similarities (12, 13). Using this approach, several groups have recently attempted to identify glioma subtypes associated with particular molecular features. Although important steps forward, the findings of these studies have been limited by several methodologic constraints, including (a) incomplete coverage of whole-genome expression due to the usage of limited or outdated legacy microarray platforms (2, 3, 14, 15); (b) limited number of samples studied and/or incomplete coverage of the spectrum of glioma subtypes and grades (11, 13, 16, 17); and (c) group stratification using a priori knowledge (traditional histopathologic classification) or use of subjective thresholds when using objective clinical features as the class defining factor (i.e., survival; refs. 2, 3, 11, 13-17).
We believe that further refinement of a rational biology-based classification scheme would optimally be constructed by using the molecular data, obtained from a large number of samples with different histopathologic subtypes to define the classes according to underlying cellular processes without any preconceived investigator biases. To this end, we have now identified a novel group of glioma subtypes based on the most current, full-coverage gene expression profiles available without any a priori class prediction or bias. We have additionally determined classifier sets from the consensus subtypes and extensively validated them in three large independent test data sets (13) to assess the potential for clinical application of these classifiers in a biologically meaningful glioma classification system.
Materials and Methods
Patient selection, tissue acquisition, and sample description
All tumor specimens used in this study were obtained from patients undergoing surgical treatment in several different institutions and hospitals following written consent in accordance with the appropriate clinical protocols (NABTC 01-07).
In total, 471 specimens were profiled via the HG-U133 Plus 2.0 array. The samples were provided as snap frozen sections of areas immediately adjacent to the region used for the histopathologic diagnosis. Each tumor was assigned a WHO glioma histopathologic subtype by the neuropathologist at the tissue contributing institution, and then the pathology slides were rereviewed by two NIH neuropathologists independently (C.O., M.Q.) who were blinded to the original designated diagnosis. The train set used for class discovery and classifier definition consisted of 159 samples. The results obtained were validated in a nonoverlapping, independent internal test set containing 189 samples, including all WHO histologic types and grades, and further validated in two independent external data sets containing 76 published high-grade gliomas and 265 glioblastoma samples taken from The Cancer Genome Atlas (TCGA) data base, respectively (see details in Supplementary Materials and Methods).
RNA extraction and array hybridization
Approximately 50 to 80 mg of tissue from each tumor was used to extract total RNA using the Trizol reagent (Invitrogen) following the manufacturer’s instructions. The quality of RNA obtained was verified with the Bioanalyzer System (ref. 18; Agilent Technologies) using the RNA Pico Chips. Six micrograms of RNA were processed for hybridization on the Genechip Human Genome U133 Plus 2.0 Expression arrays (ref. 19; Affymetrix, Inc.). After hybridization, the chips were processed using Fluidics Station 450, High-Resolution Microarray Scanner 3000, and GCOS Workstation Version 1.3. Detailed procedures on the array hybridization and file processing can be found in the supplementary data.
Statistics for classification and prediction
Using specifically designed filters, we generated six independent probeset subsets for glioma stratification containing different numbers of gene probesets according to the variables described in the supplementary data. Two unsupervised machine learning algorithms, k-mean clustering and nonnegative matrix factorization (NMF), were then applied to each of the probeset subsets separately to identify the underlying classes following the workflow illustrated in Supplementary Fig. S1. The k-mean partition was done using Partek version 6.3 and NMF, using Matlab functions implemented by Brunet and colleagues (20). For the purpose of classifier identification and class prediction, six prediction data sets with 15,553 unique genes were initially created by retaining the probeset with the maximum signal intensity. Prediction analysis of microarray (PAM), a supervised machine learning method (21), was then applied to each prediction set to identify classifiers for the glioma subtypes (Supplementary Fig. S1). A detailed explanation of the methodology can be found in Supplementary Materials and Methods.
Kaplan-Meier survival analysis was used to estimate the survival distributions, and the log-rank test was used to assess the statistical significance between stratified survival groups using Prism 4.0.
Gene set enrichment analysis for functional annotation
Gene set enrichment analysis (GSEA) was used to identify up-regulated expression signatures associated with main types and subtypes (22). All 15 pairwise comparisons of the six subtypes were performed in our GSEA analysis by mapping all 1,687 c2 curated gene sets in MsigDB v 2.0 to the ranked gene expression profiles. The enrichment scores were calculated by walking down the ordered list, and the statistical significance of nominal P values of the enrichment scores was estimated using Kolmogorov-Smirnov statistics by constructing a cumulative null distribution with 1,000 permutations (22).
Results
Gliomas are classified into two main types and six hierarchically nested subtypes
Two unsupervised machine learning algorithms, k-mean clustering and NMF, were applied to six independent probeset subsets of the 159 glioma specimens profiled via HG-U133 Plus 2.0 array (Supplementary Fig. S1; Supplementary Tables S1 and S2). After the model selection, samples were separated into two main consensus classes (Fig. 1A): one with 69 samples was designated as the O main type (containing the majority of oligodendroglial tumors) and the other, containing 67 samples, as the G main type (enriched with grade IV glioblastoma tumors) in line with their dominant sample composition (Table 1). These two main types were further stratified by applying the algorithms to each class separately. Hence, the O main type separated into two subtypes, designated as OA and OB (Fig. 1B), whereas the G main type separated into two, three, and four subtypes, respectively (Supplementary Fig. S2A). We found that the tumor samples in the four subtypes of the G main type were hierarchically nested under the two upper level subtypes. Thus, the upper level subtypes were designated as GA and GB and the lower level subtypes nested within them were designated as GA1/GA2 and GB1/GB2 (Supplementary Fig. S2B), respectively. Samples with low percentage agreement scores (<25%) were removed for clarity (Supplementary Fig. S3); however, when assessed using the resulting classifiers, the removed samples were assigned to the types and the subtypes that the class discovery algorithms derived (data not shown).
Figure 1.
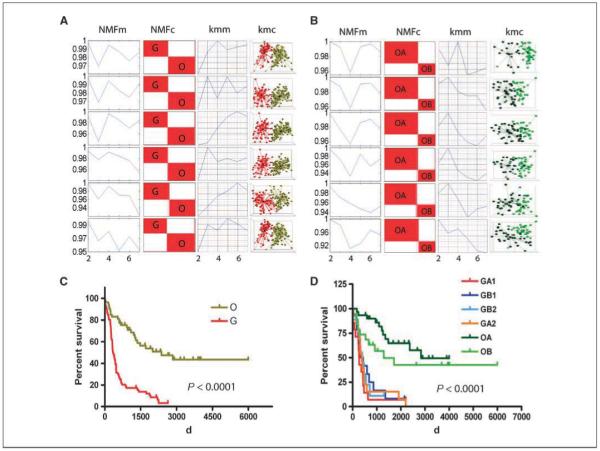
Glioma classification based on two unsupervised machine learning methods: k-mean clustering and nonnegative matrix factorization (NMF) in train set and Kaplan-Meier survival analysis of subtypes. Model selections, NMF consensus matrices, and k-mean clusters (k = 2) of two glioma main types in six probeset subsets (A) and of OA and OB subclasses in six probeset subsets (B). NMFm, NMF model selections based on cophenetic correlation (in a high consensus matrix, the coefficient is close to 1); NMFc, NMF consensus matrices; Kmm, k-mean model selections based on David-Bouldin Index (the smaller the index, the tighter the cluster); Kmc, k-mean clusters. Kaplan-Meier survival analysis for O and G main types (C) and for six subtypes (D). The color scheme representing the six subtypes of glioma throughout the figures is as follows: red, O main type; olive, G main type; dark green, OA subtype; green, OB subtype; dark red, GA1 subtype; orange, GA2 subtype; blue, GB1 subtype; turquoise, GB2 subtype.
Table 1.
Summary of major clinical features in six glioma subtypes in the train set
| Characteristics | O main type | G main type | P | ||
|---|---|---|---|---|---|
| Histologic type | Oligodendroglioma and astrocytoma-rich (96%) | Glioblastoma-rich (68%) | <0.0001*** | ||
| Histologic grade | WHO grades II and III-rich (96%) | WHO grade IV-rich (68%) | <0.0001*** | ||
| Clinical prognosis | Longer survival (~2,365 d) | Shorter survival (~348 d) | <0.0001*** | ||
| Patient age | Younger (~ 43 y) | Older (~57 y) | <0.0001*** | ||
| OA Subtype | OB Subtype | ||||
|
| |||||
| Histologic type | Astrocytoma-rich (79%) | Astrocytoma-rich (75%) | <0.0001*** | ||
| Histologic grade | WHO grade II-rich (71%) | WHO grade II-rich (65%) | <0.015** | ||
| Clinical prognosis | Longer survival (1,479 d) | Short survival (924 d) | <0.026* | ||
| Patient age | Young (~41.5 y) | Young (~43 y) | ns | ||
| GA1 subtype | GA2 subtype | GB1 subtype | GB2 subtype | ||
|
| |||||
| Histologic type | Glioblastoma-rich (73%) | Glioblastoma-rich (64%) | Glioblastoma-rich (53%) | Glioblastoma-rich (70%) | ns |
| Histologic grade | Grade IV-rich | Grade IV-rich | Grade IV-rich | Grade IV-rich | ns |
| Clinical prognosis | 269.5 d | 389 d | 381 d | 395 d | ns |
| Patient ages | 56.8 y | 59.1 y | 59.3 y | 61.8 y | ns |
Classes defined exhibit association with histologic types, grades, and patient ages
Survival analysis of the two main types shows that the patients in the O main type (median survival, 1,257 d) survive significantly longer than those in the G main type (median survival, 348 d, P < 0.0001; Fig. 1C). Similarly, patients in the OA subtype have a significantly longer survival than those in the OB subtype, suggesting that our unsupervised classification scheme indeed stratify tumors with similar clinical properties (Fig. 1D). Furthermore, there is a strong intragroup association between the histopathology and grade of the tumors within each of the major types and subtypes. The O main type is significantly enriched for a mixture of WHO grade II or grade III tumors (59 of 67), whereas the G main type is predominantly composed of grade IV (GBM) tumors (55 of 69; Table 1 and Supplementary Table S3). In agreement with the known association between patient age and histologic grade, patients with tumors that fall in the O main type are significantly younger than those in the G main type (median age, 43 versus 57 years, respectively; P < 0.0001; Table 1).
Identification of classifiers that assign tumors into the diagnostic subclasses
To apply this objective classification scheme to clinical applications, we proceeded to identify classifiers that can reliably assign an unknown tumor to the defined subtypes. A supervised machine learning method, PAM, was applied to all the prediction data sets resulting in six sets of independent classifiers consisting of 33 to 352 unique genes. These classifiers produced >92% prediction accuracy when assigning samples to the matching subtypes in a 10-fold cross-validation (Supplementary Table S4). All sets of the classifiers were differentially expressed between the pairwise subtypes across the train set and the validation data sets. One set of these classifier sets for predicting the O and the G main types is shown as an example in Table 2; complete classifier sets can be found in Supplementary Table S5.
Table 2.
Gene classifiers for differentiation of the O and G main types
| Probeset ID | Gene symbol | Fold change (O/G) |
|---|---|---|
| 202718_at | IGFBP2 | −5.08959 |
| 218802_at | FLJ20647 | −2.91583 |
| 225799_at | MGC4677 | −2.8173 |
| 208659_at | CLIC1 | −2.61081 |
| 208816_x_at | ANXA2P2 | −2.71187 |
| 202627_s_at | SERPINE1 | −3.19988 |
| 200916_at | TAGLN2 | −2.75073 |
| 211964_at | COL4A2 | −3.15335 |
| 1569003_at | TMEM49 | −2.55704 |
| 211980_at | COL4A1 | −2.54365 |
| 224917_at | MIRN21 | −2.80828 |
| 206157_at | PTX3 | −3.61317 |
| 202237_at | NNMT | −5.25017 |
| 218368_s_at | TNFRSF12A | −3.25914 |
| 201590_x_at | ANXA2 | −2.13874 |
| 203729_at | EMP3 | −4.89802 |
| 207447_s_at | GNTIVH | 3.67803 |
| 200771_at | LAMC1 | −2.13758 |
| 210512_s_at | VEGF | −2.38347 |
| 209360_s_at | RUNX1 | −2.62608 |
| 201012_at | ANXA1 | −3.02947 |
| 217739_s_at | PBEF1 | −2.00338 |
| 209395_at | CHI3L1 | −6.47009 |
| 213418_at | HSPA6 | −2.51082 |
| 205479_s_at | PLAU | −3.02588 |
| 231935_at | ARPP-21 | 2.65251 |
| 223276_at | NID67 | −2.20265 |
| 221577_x_at | GDF15 | −2.97996 |
| 201666_at | TIMP1 | −6.28863 |
| 221729_at | COL5A2 | −2.4534 |
| 202912_at | ADM | −3.24857 |
| 208636_at | ACTN1 | −2.11803 |
| 226722_at | FAM20C | −1.89925 |
| 215223_s_at | SOD2 | −2.17665 |
| 203146_s_at | GABBR1 | 2.43357 |
| 210095_s_at | IGFBP3 | −2.52806 |
| 208394_x_at | ESM1 | −3.3496 |
| 200600_at | MSN | −2.09518 |
| 213308_at | SHANK2 | 2.07505 |
| 227295_at | IKIP | −1.84117 |
| 221898_at | PDPN | −3.90831 |
| 205572_at | ANGPT2 | −3.40265 |
| 212533_at | WEE1 | −2.39671 |
| 203186_s_at | S100A4 | −2.50762 |
| 200650_s_at | LDHA | −1.70141 |
| 229724_at | GABRB3 | 2.33161 |
| 201505_at | LAMB1 | −2.08128 |
| 204465_s_at | INA | 3.98978 |
| 227425_at | REPS2 | 2.13658 |
| 202990_at | PYGL | −1.55722 |
| 212169_at | FKBP9 | −1.96699 |
| 202878_s_at | C1QR1 | −2.13239 |
| 232059_at | DSCAML1 | 2.59214 |
| 214762_at | ATP6V1G2 | 2.20731 |
Validation of the classification system and the classifiers
To ensure that the classification scheme derived from our unsupervised machine learning system are truly representative of the expression signatures in the different tumor classes, we validated our scheme using an independent, nonoverlapping data set. In this analysis, a test set containing 189 specimens (Supplementary Tables S1 and S2) was stratified into two main types using the same methodology as in the train set (Supplementary Fig. S1), leading to one main type containing 78 samples and the other 87 samples (Fig. 2A, middle). We were able to further separate the smaller main type into two and three consensus subtypes (Fig. 2B, middle) and the larger one with 87 samples into two, three, and four consensus subtypes (Fig. 2C, middle). In agreement with the outcome in the train set, a hierarchically nested relationship was detected in the test set (Supplementary Fig. S3E). The reproducibility of the hierarchically nested subtypes in our test set attests to the reliability of our classification scheme derived from the train set.
Figure 2.
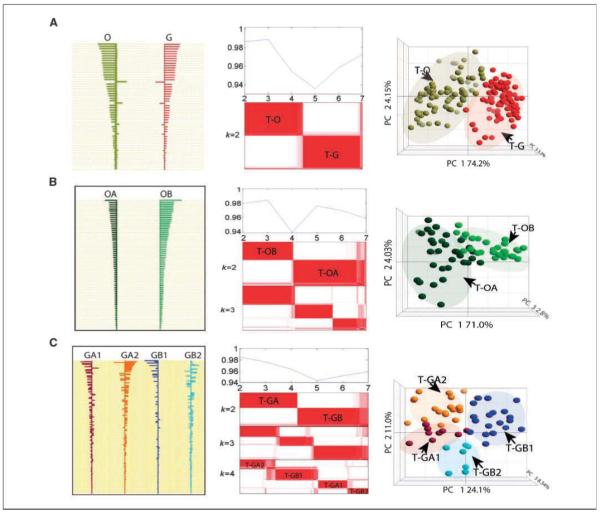
Classifier identification using PAM and their validation in a test set. A, shrunken differences of 54 classifiers for differentiation of O and G types (left); NMF model selections and consensus matrixes (k = 2) of two main types in test set (middle); validation of the 54 classifiers in the test set using PCA (right). B, shrunken differences of the 69 classifiers for differentiation of OA and OB subtypes (left); NMF model selections and consensus matrices (k = 2, k = 3) of OA and OB subtypes in the test set (middle); validation of the 69 classifiers in the test set using PCA (right). C, shrunken differences of the 352 classifiers for differentiation of four G subtypes (GA1, GA2, GB1, and GB2; left); NMF model selections and consensus matrices (k = 2, k = 3, k = 4) of subtypes in GBM-rich type in test set (middle); validation of the 352 classifiers in the test set using PCA (right).
To validate the classifier sets identified from the train set (Fig. 2A–C, left), we applied them to the recently stratified classes in test set using PCA analysis and these classifiers from the train set robustly project the intrinsic data variations of the matching subtypes in the test set (Fig. 2A–C, right). The centroids (fold changes of a given set of classifiers between pairwise subtypes) can be viewed as a prototypical expression pattern of the matching subtypes; the tumor subtypes were then identified according to the resemblance of the classifier expression to the centroids (13). Moreover, the classifier validation using a hierarchical clustering analysis on the stratified classes from the test set indicates clear separations along the subtypes (data not shown).
To ensure that these results are a representation of underlying biological processes and not just a statistical artifact of large data sets, we performed a mathematical validation by classifying six random data sets that were carefully designed with the same dimensions and equivalent scale of variations as our classification probeset subsets using NMF. Our results indicate that none of the six random data sets converged to consensus matrices as did the train set or the test set in our classification (data not shown) and their cophenetic correlation coefficients were much lower (0.45–0.73) than our real probeset subsets (0.90–1).
External data set validates the scheme and the classifiers
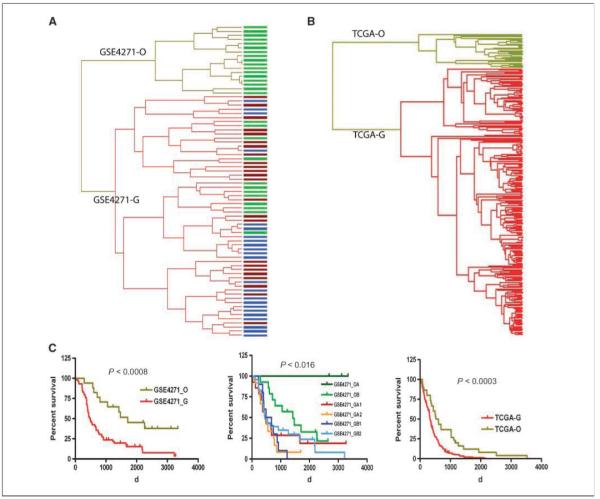
To determine if the classification scheme is independent of the data set used and to determine if the scheme correlates with one of the most comprehensive malignant glioma classification schemes published to date, we used our classifiers to stratify all 76 glioma tumors (GSE4271 data set) from the recent article by Phillips and colleagues (ref. 13; Supplementary Fig. S4). There were several significant differences between our data and the Phillips data. First, the Phillips data was obtained using the HG-U133AB platform, which contains less probesets and genes than the HG-U133 Plus 2.0 array we used. Second, the Phillips data set was comprised only of astrocytic tumors with a high-grade histology. Surprisingly, despite these differences, our classifiers separated the tumors in GSE4271 data set into the six subclasses. Fifty-three of our 54 classifiers for the O-G main types are available on the HG-U133AB array. Hierarchical clustering of all specimens in the GSE4271 data set using these 53 classifiers revealed two dominant tumor clusters, GSE4271-O and GSE4271-G (Fig. 3A). GSE4271-O and GSE4271-G were further assigned according with the centroids of the classifiers (Supplementary Fig. S5). Inspection of the samples in the two main types indicates that the GSE4271-O type is solely composed of tumors that fall into a class that Phillips and colleagues have called “proneural” secondary to the fact that it is characterized by a signature that is enriched for genes involved in neurogenesis (13). By contrast, the GSE4271-G type consists of samples from Phillips’ clusters designated as either “proliferative” or “mesenchymal,” as well as a small number of proneural tumors (Fig. 3A). Further stratification of the GSE4271-O type resolved two distinct subtypes and further stratification of the GSE4271-G type resolved four subtypes using HC (Supplementary Fig. S4A). After the identities of all the subtypes were assigned in terms of the centroids of the classifiers (Supplementary Fig. S5), we found that medium survival of GSE4271-O type (244 weeks) was significantly longer than GSE4271-G type (70 weeks, P < 0.0008; Fig. 3C, left). Possibly more impressively, this unsupervised classification system allowed us to resolve Phillip’s best single prognosis group (proneural) into two distinct subtypes, with the median survival of GSE4271-OA cases (445 weeks) being significantly longer than that in GSE4271-OB (203 weeks, P < 0.016; Fig. 3C, center). No separation in survival was detected among the four subtypes of GSE4271-G (data not shown), consistent with what we found in our own data sets.
Figure 3.
Glioma classification for the external data sets (GSE4271 data set and TCGA date set) using the classifiers. A, hierarchical clustering of GSE4271 data set using 53 classifiers to separate the two main types. Top branch of the dendrogram represents GSE4271-O main type; lower branch represents the GSE4271-G main type. Size of GSE4271-O type is smaller due to the restricted nature of the GSE4271 data set (only high-grade gliomas present). B, hierarchical clustering of TCGA GBM data set using classifiers to separate the two main types. The top branch of the dendrogram represents TCGA-O main type, whereas the lower branch represents the TCGA-G main type. Size of O type is smaller due to the restricted nature of the TCGA GBM data set (only grade IV gliomas present). C, Kaplan-Meier survival analysis of the two main types (left) and the six subtypes (center) derived from GSE4271 data set as well as the two main types of TCGA GBM samples (right). Bar colors in dendrogram represent the three subtypes identified in the original article: green, proneural subtype; dark red, proliferative subtype; blue, mesenchymal subtype.
As a final test of our classification scheme, we classified the expression data of the GBM collection generated by TCGA project (23). Two major differences exist between our train/test data sets and TCGA data set. First, TCGA data set is profiled on HT_HG-U133A (22,268 probesets) rather than on the HG-U133 Plus 2.0 array (54,000 probesets) we used resulting in a smaller gene representation in the TCGA data sets compared with ours (18,400 versus 47,000 transcripts). Second, all of the tumors profiled in TCGA data set are glioblastomas, whereas our original train set and test set represent a wide spectrum of glioma types and grades. Even so, we were able to classify the 265 GBM samples from TCGA data set using our classifiers, stratifying all TCGA GBM samples into two main classes: TCGA-O and TCGA-G. As we found with our other data sets, the TCGA-O and TCGA-G groups could be further separated into two and four subclasses, respectively (Fig. 3B and Supplementary Fig. S4B). The prototypical expression patterns of all the corresponding classes following their hierarchical nested relationship are identical to those subtypes from all the other data sets we used (Supplementary Fig. S5). Significantly, we identified a small group of GBM samples (37 patients) that grouped into the oligo-enriched group. Importantly, Kaplan-Meier analysis of these samples indicated that these patients had significantly prolonged survival compared with the other GBM patients in the TCGA data set (median survival, 561 versus 327 days, P < 0.0003; Fig. 3C, right).
Potential biological characteristics of the six subtypes according to enriched expression signatures
GSEA detects cellular pathways and/or mechanism that are differentially expressed between two phenotypes in functional units of genes (22). To begin to elucidate some of the basic underlying biological differences between the subtypes of gliomas identified through our analyses, we performed pairwise comparisons between types and subtypes that were located at the same level in the nested hierarchical structure (Supplementary Table S6). A summary of the results obtained is presented in Fig. 4. When comparing the two main types, we found that the O main type exhibited enhanced activities of exogenous hormone stimulated growth and PAR1 signaling activity whereas up-regulated profiles associated with the G main type included a large number of cell cycle/mitotic pathways, hypoxia, tumor necrosis, and nuclear factor-κB (NF-κB) pathway signaling (Fig. 4; Supplementary Table S7), consistent with a more malignant and aggressive tumor type; a supposition supported by the poorer survival of the G main type.
Figure 4.
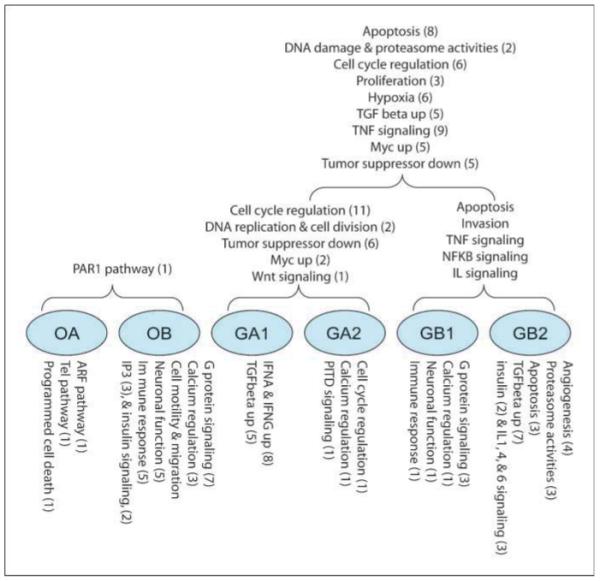
Overview of the biological functions found enriched in six subtypes based on the significantly up-regulated gene sets from GSEA analysis (nominal P < 0.05) as compared pairwise according to their hierarchically nested relationship. The numbers in parenthesis represent the number of gene sets in the categories found to be significant in the GSEA analysis.
In the case of the OA/OB comparison, we found that three gene sets (ARF pathway, TEL pathway, and programmed cell death signaling) were up-regulated signatures in the OA subtype (Fig. 4). These pathways are suggestive of tumors with relatively intact cell cycle regulation, maintenance of chromosome ends, and apoptosis, consistent with a less malignant phenotype, as supported by the longer survival of patients in this subtype (Fig. 4). By contrast, the OB subtype is notable for genes involved in plateletderived growth factor (PDGF) and epidermal growth factor (EGF) signaling, a common feature of more aggressive tumors (Supplementary Table S8).
The expression profiles in GA subtype are dominated by pathways involved in mitosis as indicated by 13 up-regulated gene sets in cell cycle regulation, DNA replication, and proliferation (Supplementary Table S9), making the GA subtype one of high proliferative potential. Interestingly, the proliferative subtype of the GSE4271 data set maps overwhelmingly to the GA subtype, indicating concordance between the two different schemes. Meanwhile, the GB subtype is notable for overexpressed signatures of the NF-κB pathway, tumor necrosis factor pathway, transforming growth factor-β signaling pathway, interleukin (IL-1, IL-10, IL-22, IL-6) signaling pathways, and up-regulated vascular endothelial growth factor signatures (Supplementary Table S9), all consistent with a mesenchymal phenotype, in agreement with the observation that the majority of the tumors designated as mesenchymal in the GSE4271 data set map to the GB subtype. Further comparisons of the functional annotations between the GA1/GA2 and GB1/GB2 subtypes are detailed in Supplementary Tables S10 and S11.
Discussion
The need for an objective, biological-based classification of gliomas is exemplified by the high rate of divergent diagnoses, the inexact prognostic capabilities, and poor therapeutic predictive properties of the current histopathologic classification schemes (10). Here, we report the development of an unbiased, gene expression–based, histology-independent glioma classification system by applying unsupervised machine learning algorithms to the gene expression profiles using the largest collection of gliomas published to-date, covering the main WHO histologic types and grades. Our analyses show that gliomas can be separated into two main types, O and G. The O main type is further divided into OA and OB subtypes, and the G can be divided into four subtypes, showing hierarchically nested relationships designated as GA1, GA2, GB1, and GB2 (Supplementary Fig. S2B).
Our classification system differs from previously reported ones in several important aspects. First, the classes were discovered based on the expression profiles of all glioma histologic types and grades without any a priori exclusion of the samples based on some clinical variable. Second, we did not select the genes to be analyzed from a manually curated list of genes thought to be relevant but rather to include all informative genes to represent the entire transcriptomic profiles of individual glioma patients. Third, we selected the subtype-defining classifiers based solely on their statistical ability to separate the classes rather than our preconceived notion of their functionality. That is, we elected to allow the computational analyses determine the appropriate classification scheme based totally on the biology of a wide spectrum of gliomas, as manifested by their genomic-scale transcriptomic profiles, in the hope that the biology would translate to a clinically relevant classification. By respecting the concordant molecular signature behavior of the tumors in the analyses, we have achieved a truly unsupervised solution. The difference in how this analysis was performed compared with prior studies is not trivial because the classification scheme we have constructed is based purely on observed gene expression profiles, without predefined genetic, pathologic, or clinical assumptions, and thereby represents a glioma classification scheme based solely on unbiased biological data. The power and reliability of the classification system described here rest not only on the purely statistical and objective way in which the analyses were performed but also on the fact that all six glioma subtypes and their corresponding classifiers were derived from a train set, confirmed and validated using an in-house and totally independently generated test set larger than the train set, and further validated using two independent, externally generated data sets.
Despite the use for the first time of a purely unsurpervised methodology, our results are generally consistent with and extend the findings from previously published smaller glioma classification studies (11, 13, 24). For example, our O main type is very similar to GSE4271 proneural class and our G main type extensively overlaps with the GSE4271 mesenchymal and GSE4271 proliferative classes. More specifically, the GSE4271 proliferative class largely overlaps with our GA subtype, both being represented by up-regulated cell proliferation–associated genes, whereas the mesenchymal class largely overlaps with our GB subtype, both characterized by invasive and mesenchymal tissue–associated genes. Our subtypes, however, further refine this classification system as exemplified by the fact that the OA and OB classification separates the GSE4271 proneural tumors into two different subtypes with significantly different overall survivals. Furthermore, our analyses show that the GSE4271 proliferative and GSE4271 mesenchymal classes can be broken into four different subtypes based on the differences in their classifier signatures.
It should be noted that the lack of survival difference between the most aggressive tumors that fell into the GBM-rich subtypes should not be seen as a weakness in or failure of our classification system. In contrast to diseases, such as breast cancer and lymphoma, that tend to have much greater variability in the natural history of the disease (i.e., survival), the vast majority of glioblastoma patients have a rather uniform and poor survival. Our classification system is consistent with those by Phillips and others and with data from large clinical trials conducted over the last two decades that consistently show a small subtype of GBM patients with better-than-expected survival (as represented by the GBMs in our O main type) but with a majority of GBM patients having uniformly short survival. Thus, although survival can be a correlation of biology, biology is not necessarily a correlation of survival and is the reason that we intentionally chose to perform an unsupervised analysis that was not based on survival, as has been done so often in the past. Certainly, patients with aggressive tumors with vastly different biologies can have roughly equivalent survivals (i.e., GBM, pancreatic cancer, metastatic lung cancer). In the case of tumors within the brain, survival may, in particular, be more a surrogate of tumor growth rate and lack of therapeutic responsiveness than biology given the physiologic constraints of a mass growing within the closed compartment of the cranium and the vital and sensitive nature of the underlying brain tissue. This is likely why nearly all malignant tumors within the brain, whether they be primary tumors (gliomas, medulloblastomas) or secondary tumors (metastatic lung cancer, melanoma), tend to have a uniformly poor and homogenous survival (6–12 months) if the specific tumor is not inherently sensitive to therapy. Nevertheless, the accumulating molecular data clearly shows that, despite survival homogeneity, GBMs are heterogeneous at the molecular and genetic level (11, 16, 25). Our classification scheme defines and categorizes this heterogeneous biology that will be crucial for designing clinical trials of molecularly target agents that are enriched for patients most likely to respond to therapy and ultimately for the practice of patient-specific or “personalized medicine.” Thus, the clinical utility of these GBM-rich subtypes will only be realized with the acquisition and the analyses of much more corollary clinical data.
We entered into this project with the bias that the standard WHO classification system would be very poorly representative of the underlying tumor biology. For the most part, we found this to be true, for although the WHO classification of GBM versus non-GBM pathology was largely upheld in our two major expression groups (96% of all GBMs in GBM-rich group versus 75% of non-GBMs in the oligo-rich group), tumors designated as WHO grade 2 or grade 3 astrocytomas, oligodendrogliomas, or mixed gliomas randomly distributed between the expression subgroups whether that designation was based on the original pathologic diagnosis (home institution) or by our central pathology review (data not shown). The fact that there was nearly a 30% to 40% discrepancy in the designation of grade and glioma subtype between the original and central pathology review of non-GBM tumors (data not shown) reinforces the subjective nature of the currently used classification system and testifies to the potential problem of using the WHO system to group specific patients into particular biological strata. By contrast, our classification system, derived and based purely on computational analyses of gene expression, consistently groups tumors into one of six groups across four very divergent data sets constituting over 700 gliomas.
The classifier gene sets described here showed >92% prediction accuracy in a 10-fold cross-validation (Supplementary Table S4). Additionally, the classifiers were successfully used to predict and to assign the derived subtypes in a large independent test set (Fig. 2) and further to stratify two external data sets into six subtypes (Fig. 3 and Supplementary Fig. S4). The robustness of our gene classifiers suggested a potential for a useful clinical tool for glioma diagnosis once more extensive validation is undertaken. Indeed, our classification system allowed us to identify a subgroup of GBMs in the TCGA data base that fell into the oligo-rich group and had markedly better survival than the remainder of the group, demonstrating the potential power of this biologically based classification system.
Finally, our GSEA analysis points to potential functional properties of the different subtypes identified in this analysis. For example, the survival advantages of OA subtype might in part be explained by an intact p53 regulatory pathway as represented by the activity of the ARF pathway and by a tendency toward genomic stability through the maintenance of chromosome ends, as suggested by activity of the Tel pathway. In contrast, the protooncogene signaling of the PDGF pathway (known to be aberrantly regulated in a significant percentage of astrocytomas) may confer higher proliferative properties to tumors in the OB subtype. Needless to say, the true functional significance of pathway activation within tumors in each of these subtypes remains to be elucidated through biological studies. Nevertheless, this analysis, as well as others like it, begins to build a new framework by which basic and clinical scientists can investigate the biological, functional, and clinical significance of these novel molecular classes with the hope of ultimately deriving a tumor classification system that will have both biological and therapeutic predictive value.
Supplementary Material
Acknowledgments
Grant support: Intramural Research Program of NIH National Cancer Institute Center for Cancer Research.
Footnotes
J.C. Zenklusen and H.A. Fine contributed equally.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Cancer Statistics Branch N. NIH . Cancer Survival rates. In: Harras A, editor. Cancer: Rates & Risks. US Dept of Health & Human Services, National Institutes of Health; Washington (DC): 1996. pp. 28–34. [Google Scholar]
- 2.Godard S, Getz G, Delorenzi M, et al. Classification of human astrocytic gliomas on the basis of gene expression: a correlated group of genes with angiogenic activity emerges as a strong predictor of subtypes. Cancer Res. 2003;63:6613–25. [PubMed] [Google Scholar]
- 3.Shai R, Shi T, Kremen TJ, et al. Gene expression profiling identifies molecular subtypes of gliomas. Oncogene. 2003;22:4918–23. doi: 10.1038/sj.onc.1206753. [DOI] [PubMed] [Google Scholar]
- 4.Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol (Berl) 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohgaki H, Dessen P, Jourde B, et al. Genetic pathways to glioblastoma: a population-based study. Cancer Res. 2004;64:6892–9. doi: 10.1158/0008-5472.CAN-04-1337. [DOI] [PubMed] [Google Scholar]
- 6.Kitange GJ, Templeton KL, Jenkins RB. Recent advances in the molecular genetics of primary gliomas. Curr Opin Oncol. 2003;15:197–203. doi: 10.1097/00001622-200305000-00003. [DOI] [PubMed] [Google Scholar]
- 7.Rich JN, Bigner DD. Development of novel targeted therapies in the treatment of malignant glioma. Nat Rev. 2004;3:430–46. doi: 10.1038/nrd1380. [DOI] [PubMed] [Google Scholar]
- 8.Rich JN, Hans C, Jones B, et al. Gene expression profiling and genetic markers in glioblastoma survival. Cancer Res. 2005;65:4051–8. doi: 10.1158/0008-5472.CAN-04-3936. [DOI] [PubMed] [Google Scholar]
- 9.Okada Y, Hurwitz EE, Esposito JM, Brower MA, Nutt CL, Louis DN. Selection pressures of TP53 mutation and microenvironmental location influence epidermal growth factor receptor gene amplification in human glioblastomas. Cancer Res. 2003;63:413–6. [PubMed] [Google Scholar]
- 10.Coons SW, Johnson PC, Scheithauer BW, Yates AJ, Pearl DK. Improving diagnostic accuracy and inter-observer concordance in the classification and grading of primary gliomas. Cancer. 1997;79:1381–93. doi: 10.1002/(sici)1097-0142(19970401)79:7<1381::aid-cncr16>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 11.Liang Y, Diehn M, Watson N, et al. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc Natl Acad Sci U S A. 2005;102:5814–9. doi: 10.1073/pnas.0402870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–7. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 13.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–73. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 14.Mischel PS, Shai R, Shi T, et al. Identification of molecular subtypes of glioblastoma by gene expression profiling. Oncogene. 2003;22:2361–73. doi: 10.1038/sj.onc.1206344. [DOI] [PubMed] [Google Scholar]
- 15.Nutt CL, Betensky RA, Brower MA, Batchelor TT, Louis DN, Stemmer-Rachamimov AO. YKL-40 is a differential diagnostic marker for histologic subtypes of high-grade gliomas. Clin Cancer Res. 2005;11:2258–64. doi: 10.1158/1078-0432.CCR-04-1601. [DOI] [PubMed] [Google Scholar]
- 16.Nigro JM, Misra A, Zhang L, et al. Integrated array-comparative genomic hybridization and expression array profiles identify clinically relevant molecular subtypes of glioblastoma. Cancer Res. 2005;65:1678–86. doi: 10.1158/0008-5472.CAN-04-2921. [DOI] [PubMed] [Google Scholar]
- 17.Tso CL, Shintaku P, Chen J, et al. Primary glioblastomas express mesenchymal stem-like properties. Mol Cancer Res. 2006;4:607–19. doi: 10.1158/1541-7786.MCR-06-0005. [DOI] [PubMed] [Google Scholar]
- 18.Miller CL, Diglisic S, Leister F, Webster M, Yolken RH. Evaluating RNA status for RT-PCR in extracts of postmortem human brain tissue. Biotechniques. 2004;36:628–33. doi: 10.2144/04364ST03. [DOI] [PubMed] [Google Scholar]
- 19.Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nat Genet. 1999;21:20–4. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- 20.Brunet JP, Tamayo P, Golub TR, Mesirov JP. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci U S A. 2004;101:4164–9. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99:6567–72. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLendon R, Friedman A, Bigner D, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freije WA, Castro-Vargas FE, Fang Z, et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 2004;64:6503–10. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- 25.Kotliarov Y, Steed ME, Christopher N, et al. High-resolution global genomic survey of 178 gliomas reveals novel regions of copy number alteration and allelic imbalances. Cancer Res. 2006;66:9428–36. doi: 10.1158/0008-5472.CAN-06-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.