summary
The ability to ectopically control gene expression is a fundamental tool for the study of bacterial physiology and pathogenesis. While many efficient inducible expression systems are available for Gram-negative bacteria, few are useful in phylogenetically distant organisms, such as mycobacteria. We have adapted a highly-inducible regulon of Rhodococcus rhodochrous to artificially regulate gene expression in both rapidly-growing environmental mycobacteria and slow-growing pathogens, such as Mycobacterium tuberculosis. We demonstrate that this artificial regulatory circuit behaves as a bistable switch, which can be manipulated regardless of growth phase in vitro, and during intracellular growth in macrophages. High-level overexpression is also possible, facilitating biochemical and structural studies of mycobacterial proteins produced in their native host.
Keywords: Inducible expression, Gene expression, Nitrile, Overexpression
1. Introduction
Virtually all inducible gene expression systems are based on natural bacterial regulons. However, redesigning these endogenous regulatory circuits is often necessary to produce highly-regulated systems that make the most useful experimental tools. Negatively-regulated promoters often suffer from high basal levels of transcription and are made more inducible by overexpressing the regulatory protein.1,2 Alternatively, optimal inducibility from positive regulators can be achieved by designing the circuit to include positive feedback in which the regulatory protein is induced simultaneously with the target gene.3
The specific structure of a regulatory circuit can have unexpected effects on gene expression at the single cell level. Almost all regulons appear to produce a graded response to different inducer concentrations when a population of bacteria is observed as a whole. However, when expression is assayed in single cells, different circuits produce fundamentally distinct responses. Genes under the control of a highly-expressed repressor generally produce titratable expression in each cell.4 In contrast, circuits that contain a positive feedback loop or those controlled by a regulatory protein present at an extremely low level, can produce bistable behavior in which only two states are possible for a cell, fully induced or fully repressed.5,6 Thus, both the nature of the regulatory element and the overall structure of the circuit can influence the behavior of an artificial regulon.
Despite the importance of regulatable gene expression systems, few have been designed for many of the most important pathogenic species. Mycobacteria are members of a diverse phylum of high G + C bacteria known as Actinobacteria. These bacteria include relatively rapidly-growing environmental organisms, such as M. smegmatis, and slow-growing pathogens, such as M. tuberculosis, the causative agent of tuberculosis. This important human pathogen has been the subject of over a century of research, but only recently have genetic tools been designed to study this organism.
Currently two general types of expression systems are used in mycobacteria. The first to be described is based on an endogenous acetamide-inducible promoter,7 and has been used mostly in fast-growing environmental species. More recently, a number of systems based on the tetracycline repressor, TetR have been developed,1,8,9 which are equally effective in both environmental and pathogenic mycobacteria. While the latter systems have quickly become a mainstay of the field, other regulons with different properties are still needed.
Because most of the commonly used regulatory systems of Gram-negative bacteria are not functional in mycobacteria, we sought to adapt a regulon of a more closely related species. Rhodococcus rhodochrous is a saprophytic organism that is used extensively in the industrial production of acrylamide and nicotinamide and is of interest as a bioremediation agent. Upon encountering nitrile-containing pollutants, this organism produces large amounts of a nitrilase enzyme, encoded by the nitA gene, which detoxifies a broad range of these compounds.10 Under inducing conditions, this single enzyme can account for 35% of total cellular protein.11
The regulatory circuit controlling the nitA gene has been dissected, and appears to be controlled by a single protein, NitR.12 This protein is sufficient to mediate induction from the nitA promoter in several Streptomyces species.13 NitR is homologous to the well-characterized AraC protein of Escherichia coli, which acts as both a positive and negative regulator of the arabinose catabolic regulon and is the basis of the pBAD series E. coli expression plasmids. 14 For AraC, the shift from repression to activation is mediated by allosteric interactions with hexose sugars.15 It has been hypothesized that NitR is similarly regulated by inducer binding,12 but no direct experimental data is available. The high inducibility and simplicity of this system prompted us to adapt it for use in mycobacteria.
2. Results and discussion
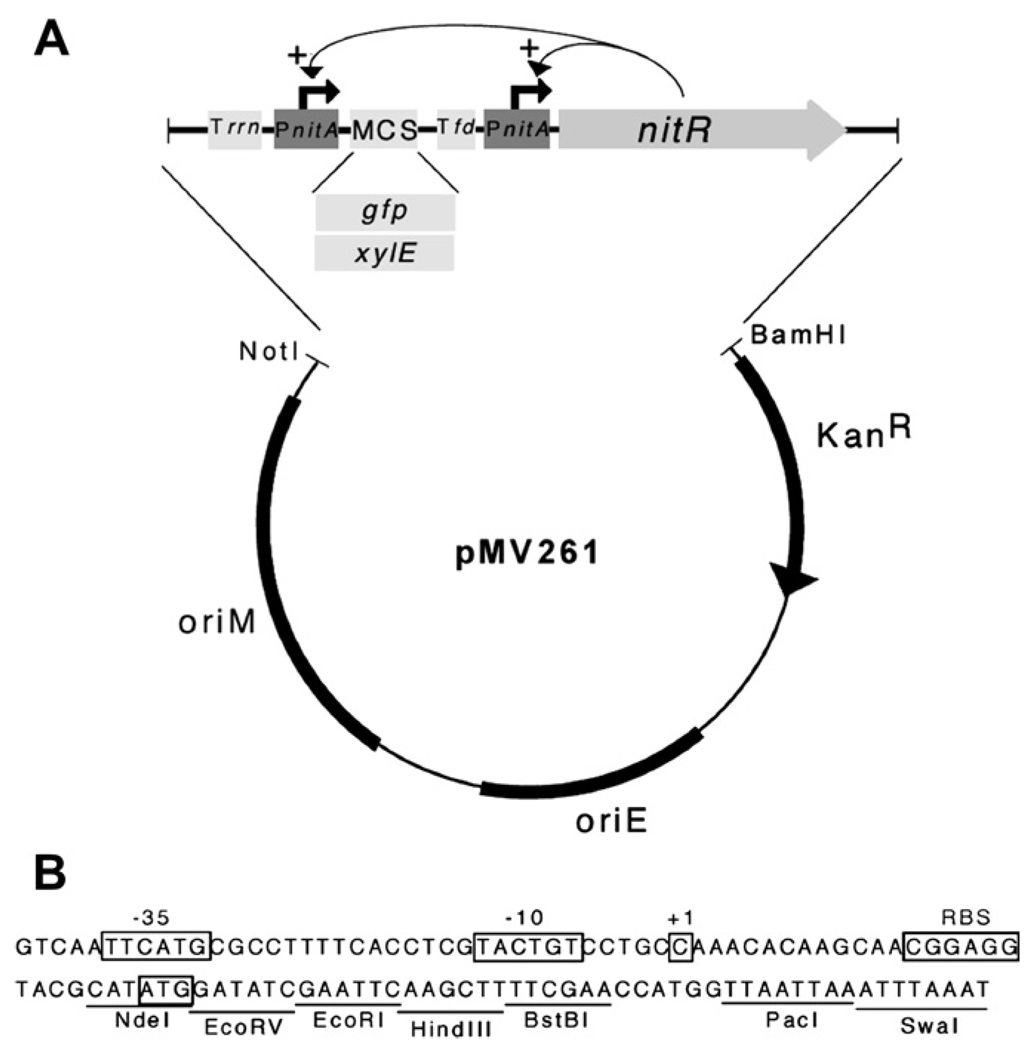
The pNIT series of E. coli-mycobacteria shuttle plasmids contain the artificial regulon depicted in Figure 1. The expression cassette consists of the nitR gene, encoding the regulatory protein, under the control of the inducible nitA promoter. A separate cistron contains a second nitA promoter followed by a multiple cloning site (MCS). The native ribosome binding site of the nitA gene is preserved in the MCS allowing any gene to be cloned in frame using the NdeI site that overlaps the nitA start codon. This arrangement of regulatory elements was designed to maximize the inducibility of the target gene by creating a positive-feedback loop in which the expression of the regulatory protein is simultaneously induced. pNIT-2 contains the same arrangement of regulatory elements and differs largely in the cloning sites available in the MCS. The sequence of pNIT-1 has been deposited in GenBank (accession # FJ173069).
Figure 1.
(A) Design of pNIT-1. The expression cassette contains a multiple cloning site (MCS) into which a target gene is inserted (here shown as the reporters, gfp or xylE) and the gene encoding the regulatory protein (nitR) under the control of the NitR-targeted nitA promoter. This design incorporates a positive feedback loop to increase inducibility. The MCS is transcriptionally isolated by two strong terminator sequences, one from the E. coli rrnAB operon (“Trrn”) and one from the bacteriophage fd (“Tfd”). The vector backbone (pMV261)20 contains elements necessary for replication in both E. coli and mycobacteria. These elements are arranged similarly in pNIT-2. (B) The sequence of the PnitA promoter and MCS is depicted. The probable −35, −10, and +1 (translational start) sites of the promoter, and the ribosome binding site (RBS) and translational start site of the nitA gene are annotated as in Ref. 12. Unique restriction sites in pNIT-1 are indicated.
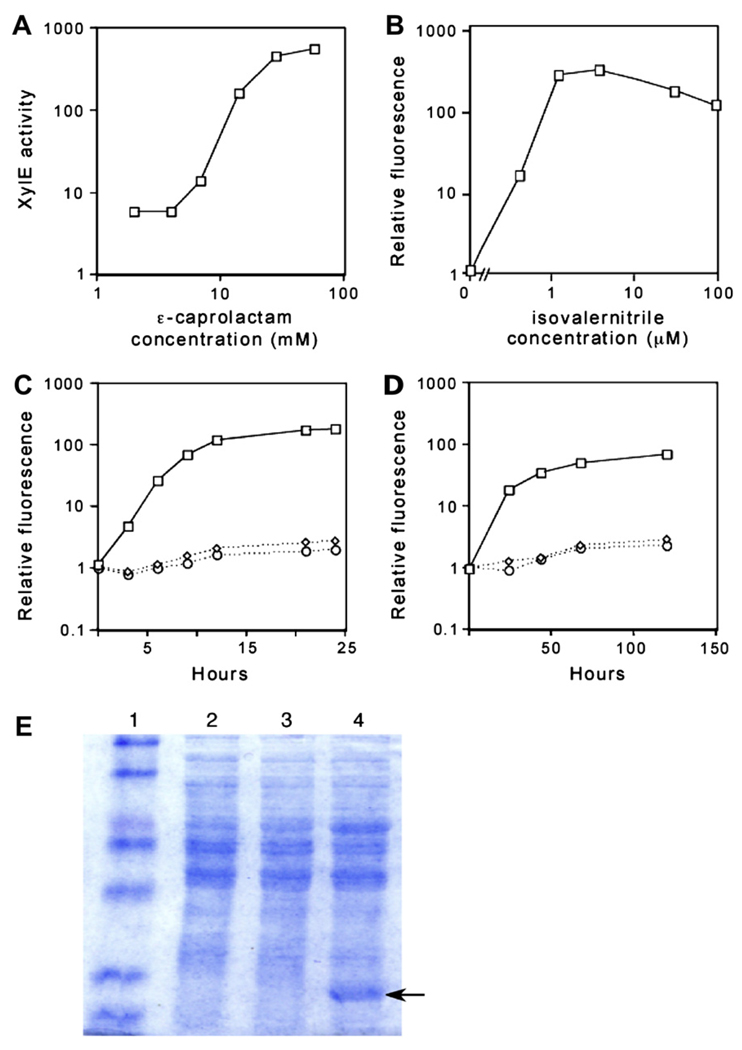
To determine if pNIT plasmids could direct gene expression in mycobacteria, green fluorescent protein (gfp) and catechol-2,3-dioxygenase (xylE) genes were used as reporters. Initially, we used the nitrile analog, ε-caprolactam as an inducer, because this inexpensive and non-toxic compound had been shown to induce nitA expression in both Rhodococcus and Streptomyces spp. expressing NitR.11,13 In M. smegmatis transformed with pNIT-2::xylE, ε-caprolactam addition caused a dose-dependent increase in reporter gene activity (Figure 2A). Optimal induction was achieved with 28 mM ε-caprolactam, which caused a >100-fold increase in reporter gene expression.
Figure 2.
pNIT plasmids direct expression of exogenous reporter genes in mycobacteria. (A, B) Expression from pNIT plasmids depends on the dose of inducer. M. smegmatis transformed with pNIT-2::xylE (A) or pNIT-1::gfp (B) were treated with the indicated concentration of inducer and reporter gene expression was assayed 16 h later. Y-axes are plotted on a log scale. “XylE activity” indicates µmol catechol 2-hydroxymuconic semialdehde/minute/mg total protein. Fluorescence values are reported relative to an uninduced culture. (C, D) Kinetics of pNIT expression in M. smegmatis (C) or M. tuberculosis (D). Strain carrying pNIT-1::gfp treated with 1 µM isovaleronitrile in DMSO (squares). Strain carrying pNIT-1::gfp treated with DMSO (diamonds). Strain carrying pMV261 treated with 1 µM isovaleronitrile (circles). At the indicated times, GFP production was assayed by transferring 0.2 ml of culture to a plate fluorometer. (E) 20 µg of protein from bacterial lysates was separated by 12% SDS PAGE. Lane 1: molecular weight marker, Lane 2: M. smegmatis lysate, Lane 3: M. smegmatis carrying pNIT1::GFP, Lane 4: M. smegmatis carrying pNIT1::GFP induced for 16 h with 1 µM IVN. Arrow indicates a band with the expected migration rate of GFP.
However, very little reporter gene expression was observed when this compound was added to M. tuberculosis transformed with pNIT2::xylE (not shown). We suspected that this was due to the relatively impenetrable hydrophobic cell wall of the slow-growing pathogenic mycobacterial species. To surmount this barrier, we tested a number of more hydrophobic nitriles. Of the compounds tested, isovaleronitrile (IVN) was the most effective, inducing maximal expression at ~5 µM in both M. smegmatis and M. tuberculosis (Figure 2B and data not shown). Greater than 100-fold induction was routinely achieved in both mycobacterial species (Figures 2C–E). In M. smegmatis, similar kinetics of GFP induction are observed for both compounds (data not shown). At the concentrations used in this study, neither ε-caprolactam nor IVN had a detectable effect on the growth rates of M. smegmatis or M. tuberculosis (not shown).
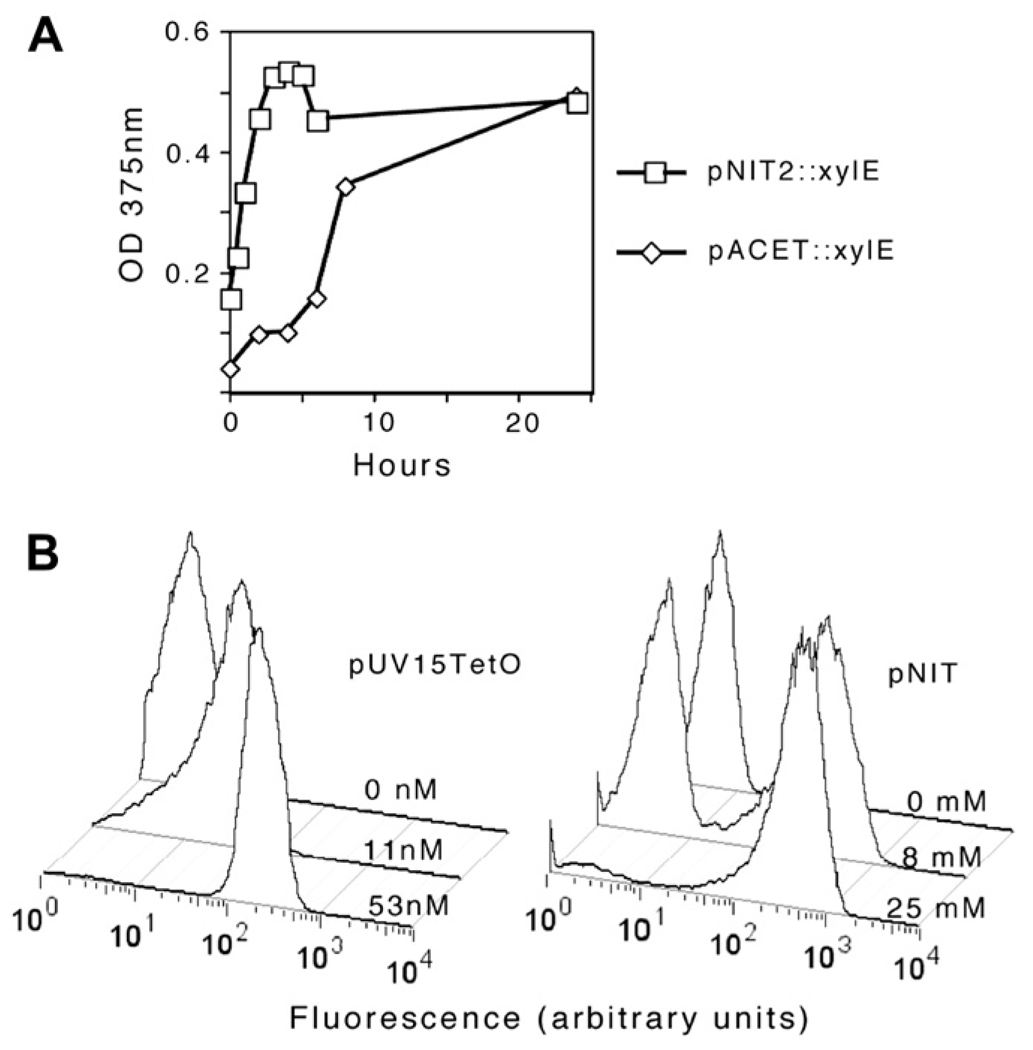
We then compared pNIT with two existing methods for regulating gene expression in mycobacteria. After 24 h of induction, pNIT-2 was found to direct equivalent expression of XylE as the previously described acetamidase-inducible promoter in “pACET” (described in Section 3). However, induction from pNIT-2 was considerably more rapid (Figure 3A). A GFP reporter was used to compare pNIT-1 with a commonly used TetR-based expression plasmid, pUV15TetO.1 Upon induction, both plasmids produced fluorescence with approximately the same kinetics (not shown), but pNIT-1::GFP was found to produce ~3-fold higher fluorescence. For some applications, the most critical parameter is not the maximal induction level, but the absence of basal (uninduced) transcription. While we assume that no expression system can be completely repressed, neither the xylE or gfp reporters used in this study proved sensitive enough to reliably detect this low level of protein production. Thus, while we conclude that the basal transcription from these promoters is reasonably low, these experiments do not allow the direct comparison of these systems.
Figure 3.
Comparison of pNIT with other mycobacterial expression systems. (A) M. smegmatis transformed with pNIT-2::xylE or pACET::xylE were induced (with ε-caprolactam or acetamide, respectively), and XylE activity was assayed at the indicated time points. (B) M. smegmatis transformed with either pNIT-1::gfp or pUV15TetO::gfp were treated with the indicated concentrations of inducers (ε-caprolactam or anhydrotetracycline, respectively) for 16 h and fluorescencewas assayed by FACS. Histograms plot the relative fluorescence (X axis) versus cell number (Y axis).
While the overall induction achieved by pUV15TetO and pNIT-1 appeared roughly similar at the population level, the circuitry controlling these regulons varied significantly. Transcription from the pUV15TetO promoter is controlled by a repressor expressed at constitutively high-levels. Often, regulons designed in this way produce titratable expression in individual cells. In contrast, the pNIT regulatory circuit includes positive feedback component in which NitR induces its own expression. This raised the possibility that the pNIT-encoded circuit may behave like other positive feedback loops and exist only in two distinct states, fully induced or fully repressed. To investigate this question at the single cell level we analyzed GFP production by FACS after induction at a half-maximal inducer concentration. As shown in Figure 3B, pNIT-based expression showed a distinct bistable nature, whereas pUV15TetO expression was titratable in individual cells.
An ideal expression system for studying mycobacterial pathogenesis would be effective during infection, but this introduces several challenges. Due to the intracellular lifestyle of this pathogen, the inducer must permeate eukaryotic membranes. In addition, M. tuberculosis is growing slowly, if at all, during important stages of infection, and it is generally difficult to produce robust induction of gene expression in such slowly-replicating organisms.
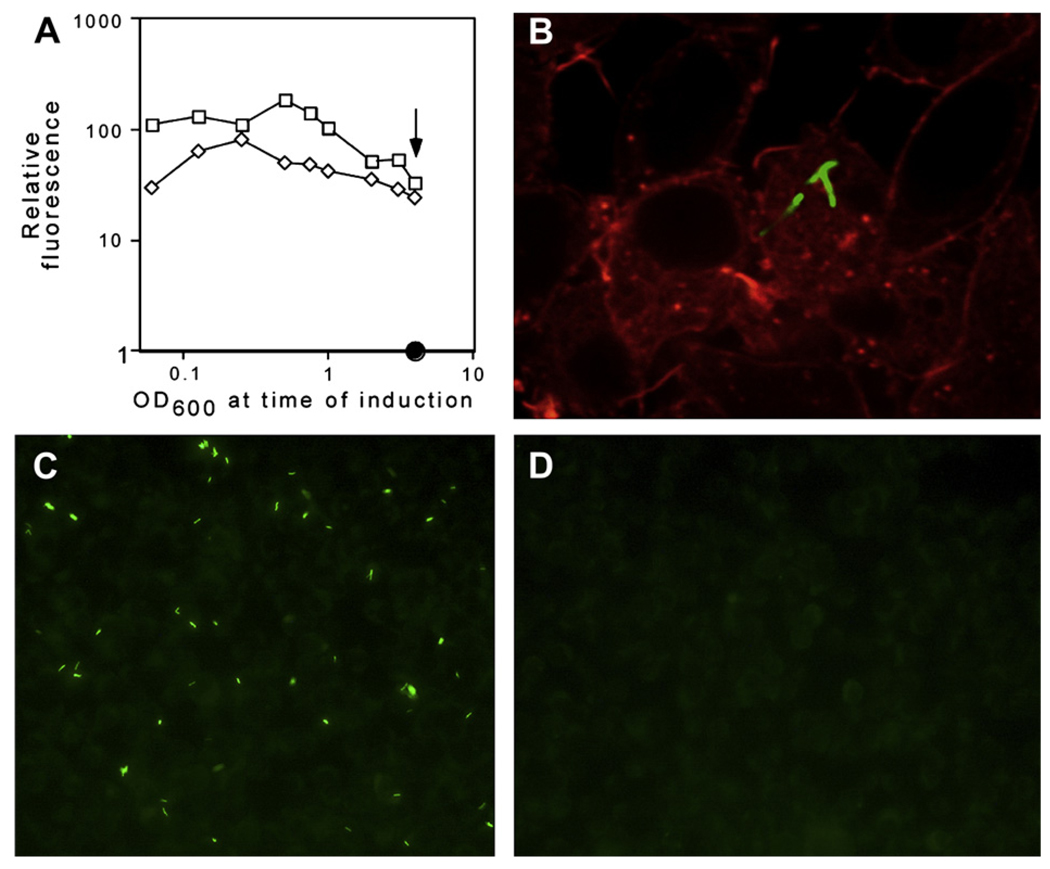
To assess the usefulness of mycobacterial expression systems under these more demanding conditions, we assessed the induction of a GFP reporter by pNIT-1 and pUV15TetO during different growth phases in M. smegmatis (Figure 4). While both systems were less inducible in stationary phase than in exponentially growing cultures, GFP production could be detected in every condition. The higher fluorescence attainable with pNIT1::GFP was evident in all growth phases, indicating that pNIT may be an attractive option for controlling gene expression under a variety of conditions.
Figure 4.
pNIT induction can be achieved in stationary phase and during intracellular growth. (A) M. smegmatis transformed with pNIT-1::gfp or pUV15tetO::gfp were grown to late stationary phase (OD600 >4) and either induced directly (arrow) or diluted to the indicated optical density (X axis, log scale) before 16 h of induction with 1 µM isovaleronitrile. The resulting fluorescence (relative to an uninduced culture) is plotted on a log scale. (B–D) M. bovis BCG transformed with pNIT-1::gfp was used to infect RAW264.7 macrophages at an MOI of 3 for 4 h. 250 nM isovaleronitrile in DMSO (B and C) or DMSO alone (D) was added for an additional 24 h. GFP (green) and phalloidin-labeled actin (red) were visualized by microscopy. The image in panel B represents a single optical section obtained by confocal microscopy, which demonstrates the colocalization of the bacteria with macrophage actin.
We suspected that the small and relatively hydrophobic inducer of NitR, IVN, was likely to diffuse through mammalian membranes. Therefore, we determined if it was possible to induce gene expression from pNIT-1::GFP during intracellular growth in macrophages. Indeed, the addition of 250 nM IVN to infected macrophages caused robust expression of the GFP reporter that could be detected by microscopy (Figures 4B–D). Confocal microscopy was then used to verify the intracellular location of these bacteria. The colocalization of GFP-expressing bacteria with host actin in multiple optical sections (Figure 4) verified that these bacteria were inside of the macrophages. The relatively low concentration of IVN used for gene induction did not appear to be toxic to or alter the morphology of the macrophage cell line used in these experiments. However the long-term effects of this concentration of IVN on mammalian cells or whole animals remains to be rigorously tested.
In summary, we have demonstrated that pNIT can direct nitrile-inducible gene expression in both saprophytic and pathogenic mycobacteria. This complements existing expression systems and may provide a number of advantages under certain circumstances, such as when high-level overexpression or induction in slowly-replicating cells is desirable. In addition, the availability of multiple compatible inducible systems should allow the independent regulation of multiple genes in the same cell.
3. Materials and methods
3.1. Bacterial strains and culture conditions
Mycobacterium tuberculosis, H37Rv and M. smegmatis, mc2155,16 were cultured on Middlebrook 7H10 agar or 7H9 broth supplemented with 0.05% Tween-80 and 10% OADC (for M. tuberculosis), or 0.5% bovine serum albumin and 0.15% sodium chloride (for M. smegmatis). Kanamycin was added at 20 µg/ml. Isovaleronitrile and ε-caprolactam were purchased from Sigma (St. Louis, MO).
3.2. Plasmids
pNIT-1: A DNA fragment encompassing a nitA promoter, MCS, phage fd terminator, and nitR gene was synthesized (GenScript Corp, Piscataway, NJ). This fragment was cloned downstream of the rrn transcriptional terminator of pJEB402.17 This terminator/expression cassette was excised and ligated into pMV261 as shown in Figure 1. Plasmid pNIT-2 was constructed by cloning nitA–nitR cassette from plasmid pSH03112 into the replicating shuttle vector pMV261, replacing its original hsp60 promoter. The nitA–nitR cassette was excised from pSH031 by digesting with BsrGI, blunting and then digesting with SpeI. The regulatory cassette was then cloned into pMV261 digested with Dra1 and Nhe1. The egfp and xylE genes were PCR amplified from pEGFP-N1 (Clontech, Mountain View, CA) or pRH1351,18 and cloned into the MCS of pNIT-1 or pNIT- 2, respectively. Similarly, pACET::xylE was made by inserting the xylE gene downstream the acetamidase-inducible ami promoter7 in pMV261.
3.3. Reporter assays
For GFP quantification, bacteria were diluted to an optical density at 600 nm of 0.2 at the beginning of each experiment unless otherwise noted. Inducer was added at the indicated concentrations and the resulting fluorescence was quantified by two methods. Total fluorescence of the bacterial population was determined for 0.2 ml of culture using a plate fluorometer (Molecular Devices, Sunnyvale, CA). Single cell fluorescence was determined after fixation with 1% paraformaldehyde (PFA) using a FACSCalibur instrument (Becton Dickinson, San Jose, CA). XylE assays were performed using cell free extracts of M. smegmatis cultures harvested at serial time points following induction at OD600 of 0.5. Briefly, the cell pellet was washed with 100 mM potassium phosphate (pH 7.5) and resuspended in 100 mM potassium phosphate-10% acetone. The cells were lysed by short pulse sonication. The cell debris was pelleted by centrifugation and the clarified lysates were assayed for protein concentration using Bio-Rad protein assay reagent, with bovine serum albumin as a standard. XylE assays were performed in assay buffer containing 100 mM potassium phosphate and 0.5 mM catechol.19 Linear change in absorbance at 375 nm was recorded using a plate reader (HTS 7000 plus, Perkin Elmer). The “specific activity” of XylE was expressed as ΔOD375 nm/minute/mg of total protein.
3.4. Macrophage infection and microscopy
Bone marrow-derived macrophages (BMM) were isolated from C57BL6 mice and were plated on a Lab-Tek tissue culture chamber slide at 2 × 105 cells/well. The BMM’s were then infected with GFP-expressing BCG at an MOI of 3 for 4 h. Subsequently, the cells were washed and replenished with media containing 250 nM IVN in DMSO or DMSO alone and incubated for 24 h. Infected cells were fixed in 4% PFA for 1 h at 25 °C and permeabilized with 0.1% Triton X-100. F-actin was stained with Alexa 568-phalloidin (1:500; Molecular Probes, Eugene OR). Images were acquired using either standard epifluorescence or spinning disk confocal microscopy.
Acknowledgements
We thank Sabine Ehrt and Michihiko Kobayashi for their kind gifts of pUV15TetO and pSH19, respectively. This work was supported by grants from the National Institutes of Health to CMS (AI073509) and RNH (AI066978).
Funding. Please see Acknowledgements.
Footnotes
Competing interests. None declared.
Ethical approval. Not required.
References
- 1.Ehrt S, Guo XV, Hickey CM, Ryou M, Monteleone M, Riley LW, et al. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 2005;33:e21. doi: 10.1093/nar/gni013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann E, Brosius J, Ptashne M. Vectors bearing a hybrid trp-lac promoter useful for regulated expression of cloned genes in Escherichia coli. Gene. 1983;25:167–178. doi: 10.1016/0378-1119(83)90222-6. [DOI] [PubMed] [Google Scholar]
- 3.Schultz LD, Hofmann KJ, Mylin LM, Montgomery DL, Ellis RW, Hopper JE. Regulated overproduction of the GAL4 gene product greatly increases expression from galactose-inducible promoters on multi-copy expression vectors in yeast. Gene. 1987;61:123–133. doi: 10.1016/0378-1119(87)90107-7. [DOI] [PubMed] [Google Scholar]
- 4.Karmakar R, Bose I. Graded and binary responses in stochastic gene expression. Phys Biol. 2004;1:197–204. doi: 10.1088/1478-3967/1/4/001. [DOI] [PubMed] [Google Scholar]
- 5.Ingolia NT, Murray AW. Positive-feedback loops as a flexible biological module. Curr Biol. 2007;17:668–677. doi: 10.1016/j.cub.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ozbudak EM, Thattai M, Lim HN, Shraiman BI, Van Oudenaarden A. Multi-stability in the lactose utilization network of Escherichia coli. Nature. 2004;427:737–740. doi: 10.1038/nature02298. [DOI] [PubMed] [Google Scholar]
- 7.Parish T, Stoker NG. Development and use of a conditional antisense mutagenesis system in mycobacteria. FEMS Microbiol Lett. 1997;154:151–157. doi: 10.1111/j.1574-6968.1997.tb12637.x. [DOI] [PubMed] [Google Scholar]
- 8.Blokpoel MC, Murphy HN, O’Toole R, Wiles S, Runn ES, Stewart GR, et al. Tetracycline-inducible gene regulation in mycobacteria. Nucleic Acids Res. 2005;33:e22. doi: 10.1093/nar/gni023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carroll P, Muttucumaru DG, Parish T. Use of a tetracycline-inducible system for conditional expression in Mycobacterium tuberculosis and Mycobacterium smegmatis. Appl Environ Microbiol. 2005;71:3077–3084. doi: 10.1128/AEM.71.6.3077-3084.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi M, Nagasawa T, Yamada H. Nitrilase of Rhodococcus rhodochrous J1. Purification and characterization. Eur J Biochem. 1989;182:349–356. doi: 10.1111/j.1432-1033.1989.tb14837.x. [DOI] [PubMed] [Google Scholar]
- 11.Nagasawa T, Nakamura T. Yamada N. ε-caprolactam, a new powerful inducer for the formation of Rhodococcus rhodochrous J1 nitrilase. Arch Microbiol. 1990;155:13–17. [Google Scholar]
- 12.Komeda H, Hori Y, Kobayashi M, Shimizu S. Transcriptional regulation of the Rhodococcus rhodochrous J1 nitA gene encoding a nitrilase. Proc Natl Acad Sci U S A. 1996;93:10572–10577. doi: 10.1073/pnas.93.20.10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herai S, Hashimoto Y, Higashibata H, Maseda H, Ikeda H, Omura S, et al. Hyper-inducible expression system for streptomycetes. Proc Natl Acad Sci U S A. 2004;101:14031–14035. doi: 10.1073/pnas.0406058101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee NL, Gielow WO, Wallace RG. Mechanism of araC autoregulation and the domains of two overlapping promoters, Pc and PBAD, in the L-arabinose regulatory region of Escherichia coli. Proc Natl Acad Sci U S A. 1981;78:752–756. doi: 10.1073/pnas.78.2.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WRJ. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Molecular Microbiology. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 17.Guinn KM, Hickey MJ, Mathur SK, Zakel KL, Grotzke JE, Lewinsohn DM, et al. Individual RD1-region genes are required for export of ESAT-6/CFP-10 and for virulence of Mycobacterium tuberculosis. Mol Microbiol. 2004;51:359–370. doi: 10.1046/j.1365-2958.2003.03844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raman S, Hazra R, Dascher CC, Husson RN. Transcription regulation by the Mycobacterium tuberculosis alternative sigma factor SigD and its role in virulence. J Bacteriol. 2004;186:6605–6616. doi: 10.1128/JB.186.19.6605-6616.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zukowski MM, Gaffney DF, Speck D, Kauffmann M, Findeli A, Wisecup A, et al. Chromogenic identification of genetic regulatory signals in Bacillus subtilis based on expression of a cloned Pseudomonas gene. Proc Natl Acad Sci U S A. 1983;80:1101–1105. doi: 10.1073/pnas.80.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stover CK, Bansal GP, Hanson MS, Burlein JE, Palaszynski SR, Young JF, et al. Protective immunity elicited by recombinant bacille Calmette-Guerin (BCG) expressing outer surface protein A (OspA) lipoprotein: a candidate Lyme disease vaccine. J Exp Med. 1993;178:197–209. doi: 10.1084/jem.178.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]