Summary
Acid-sensing ion channels are proton-activated, sodium-selective channels composed of three subunits and members of the superfamily of epithelial sodium channels, mechanosensitive and FMRF peptide-gated ion channels. These ubiquitous eukaryotic ion channels play essential roles in biological activities as diverse as sodium homeostasis, taste, and pain. Despite their crucial roles in biology and their unusual trimeric subunit stoichiometry, there is little knowledge of the structural and chemical principles underlying their ion channel architecture and ion binding sites. Here we describe the structure of a functional acid sensing ion channel in a desensitized state at 3 Å resolution, the location and composition of the ~8 Å ‘thick’ desensitization gate, and the trigonal antiprism coordination of cesium ions bound in the extracellular vestibule. Comparison of the acid sensing ion channel structure with the ATP-gated P2×4 receptor reveals similarity in pore architecture and aqueous vestibules, suggesting unanticipated yet common structural and mechanistic principles.
Acid sensing ion channels (ASICs) are voltage-independent, sodium-selective, ligand-gated ion channels expressed in diverse organisms1–3, and are members of the epithelial sodium channel/degenerin (ENaC/Deg) family4. These proton-gated channels form functional homomeric and heteromeric channels with a spectrum of pH sensitivities and desensitization profiles5. The crystal structure of chicken ASIC1 (ΔcASIC1)6, together with subsequent microscopy7 and biochemical8 studies, proved that ASICs are trimers and by extension, ENaCs and Deg channels as well, thus ending the stoichiometry controversy9,10. The structure of ΔcASIC1 defined the architecture of the extracellular domain, the location of a chloride ion and residues implicated in proton binding. Because the ΔcASIC1 construct does not exhibit proton dependent gating6, however, it left unresolved the structure of the ion channel, the location and chemical composition of the ion channel gate, and the stereochemistry and location of ion binding sites.
ASICs and ENaCs are sodium-selective channels, favoring Na+ over K+ by 10-fold11 and 100-fold12,13, respectively. The trimeric subunit arrangement of ASICs and ENaCs, together with their selectivity for sodium, raises questions related to the architecture of the ion channel, the shape and chemical composition of the pore, the location of ion channel gates, and the position(s) of residue(s) implicated in ion permeation and block. In ENaCs, the steric volume of S589 (α subunit) in TM2 has a profound effect on selectivity, with permeation of larger ions correlated with residues of larger steric volume, suggesting that residues at this position modulate the diameter of the selectivity filter10,14,15. To illuminate fundamental principles of ion channel architecture and gating in ASICs and related ion channels, and to define the location of residues such as S589, we have studied the atomic structure of a functional ASIC. Here we describe the crystal structure of a minimal functional chicken ASIC1 construct (cASIC1mfc), show the location and composition of the ion channel’s desensitization gate, define the location and stereochemistry of cesium binding sites in the extracellular vestibule and assign the position of residues implicated in ion selectivity.
The ASIC/ENaC/Deg superfamily is not the only group of trimeric ion channels. P2X receptors are also trimeric ligand-gated ion channels activated by ATP16–20. Although ASICs and P2X receptors share a common transmembrane topology, with intracellular termini and two predicted transmembrane segments, together with large, cysteine-rich extracellular domains, there are no significant amino acid sequence relationships between ASICs and P2X receptors. In a companion paper, we describe the crystal structure of the zebrafish P2×4 receptor21. Comparison of the functional chicken ASIC structure and the P2×4 structure provides unanticipated insights into the structural similarities of ASIC and P2X receptor ion channel domains, the locations and chemical compositions of the ion channel gates of the closed and desensitized states, and the presence of negatively charged aqueous vestibules in the extracellular domains.
Crystallization of a functional ASIC
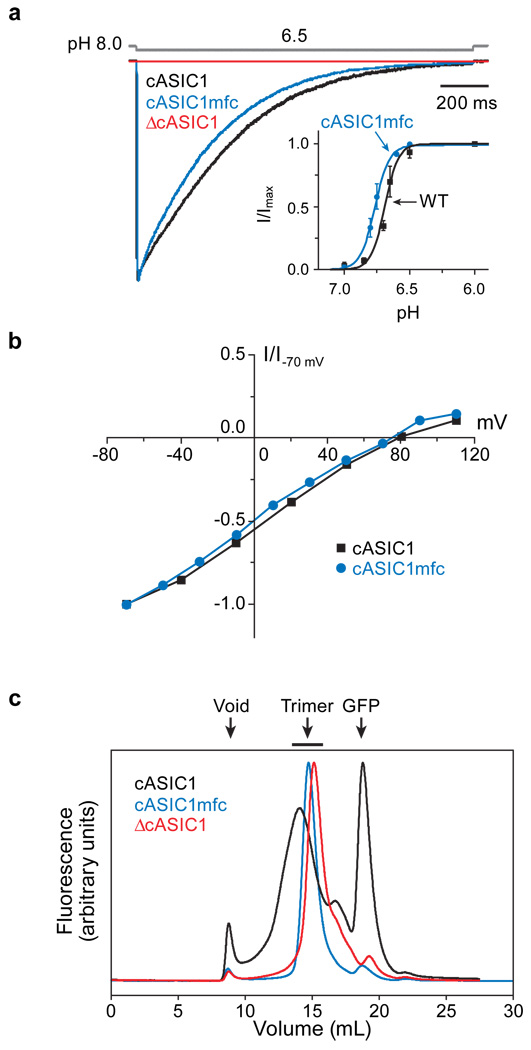
We defined a functional receptor amenable to crystallization by examining cASIC1a deletion mutants by patch-clamp electrophysiology and fluorescence detection, size-exclusion chromatography (FSEC)22. A construct that included the amino terminus, a region of ASICs and ENaCs crucial to activation gating23,24, yet which stopped at residue 466, shortly after the second TM segment, preserved the pH dependent gating and sodium selectivity of the full length channel and yielded a sharp and symmetrical peak by FSEC (Fig. 1a, b, c). The cASIC1mfc produced crystals at pH 6.5 that diffract to 3 Å resolution (Supplementary Table 1 and Supplementary Fig. 1).
Figure 1. Identification of a minimally functional cASIC1 construct.
a, Typical current generated by cASIC1 constructs in outside-out patch recordings (holding potential −60 mV). Channels were activated by stepping from pH 8.0 to 6.5 for the wild-type (black), ΔcASIC1 (red), cASIC1mfc (blue) constructs (open tip junction potential is gray). Typical pH concentration-response relationships are shown in the inset. The wild type channel had pH50 of 6.68 ± 0.01 and n value of 8.35 ± 1.91. The cASIC1mfc had a pH50 and n values of 6.77 ± 0.01 and 7.86 ± 1.22, respectively. Points on the curve are the mean ± s.e.m. of at least four cells. b, Normalized current-voltage relationship. Current observed at low pH and various holding potentials was normalized to the current observed at −70 mV. c, Fluorescence detection size-exclusion chromatography of cASIC1 constructs.
The cASIC1mfc crystal structure was solved by molecular replacement using the extracellular domain of a ΔcASIC1 protomer6 as a search probe. Initial cASIC1mfc electron density maps indicated that the conformation of the transmembrane domains differed from the ΔcASIC1 structure and thus we manually built this portion of the structure (Supplementary Fig. 2 and Supplementary Fig. 3). The model was subjected to cycles of refinement and rebuilding, ultimately yielding a structure with Rwork and Rfree values of 22.5% and 27.4%, respectively, and good stereochemistry (Supplementary Table 1 and Supplementary Table 2). The present structure includes residues 46 through 451; amino (2–45) and carboxyl terminal (452–466) residues were not seen in electron density maps. Unlike in the ΔcASIC crystal form, there are no visible lattice contacts between the transmembrane domains and neighbouring protein molecules. (Supplementary Fig. 1).
Ion channel architecture
In this desensitized state, the cASIC1mfc ion channel has the shape of an hourglass with wide vestibules near the extracellular and cytoplasmic surfaces of the membrane bilayer and a narrow constriction at the middle (Fig. 2a,b,c). The channel domain is defined by six continuous transmembrane helices in which two helical segments (TM1, TM2) are contributed by each of three subunits related by the 3-fold axis of crystallographic symmetry. The TM2 helices are positioned closest to the 3-fold axis, are tilted by ~50° from the membrane normal, and cross each other about halfway across the putative membrane bilayer (Supplementary Fig. 2b). The TM1 helical segments form extensive interactions with the TM2 helix of the same subunit, but also make contacts with TM1 and TM2 helices on adjacent subunits. The TM1 helices therefore make the majority of contacts with the lipid bilayer whereas the TM2 helices line the putative ion channel pore.
Figure 2. Structure of cASIC1mfc.
a, View of the functional, cASIC1mfc trimer. Chloride ions are green spheres. The ‘thumb’, ‘finger’, and ‘wrist’ regions are labelled. b, A vertical slice through a solvent accessible surface representation of the transmembrane domain, the extracellular vestibule and fenestrations. One of the three equivalent fenestrations is indicated by an arrow. Asp433 defines the ‘bottom’ of the extracellular vestibule. c, Key interactions between symmetry related D433 carboxyl and Y425 hydroxyl groups.
Access to the pore
Ions and small molecules access the cASIC1mfc pore through three oval-shaped fenestrations that lead directly to the extracellular vestibule (Supplementary Fig. 4). These portals are located at the 'wrist'6, the juncture between the transmembrane domains and the extracellular domains, near the boundary of the membrane bilayer and the extracellular solution (Fig. 2a). Framing the ~2 Å × ~6 Å fenestrations are the carboxyl terminus of TM1 and the beginning of β1, together with the end of β12 and the amino terminus of TM2. We suggest that ions might also reach the pore by a path along the 3-fold axis of symmetry. Although this pathway is occluded in the present low pH structure, modest conformational changes that might accompany opening of the ion channel may expand the constrictions along the 3-fold axis in the extracellular domain (Fig. 3 and Supplementary Fig. 5).
Figure 3. Vestibules and possible ion permeation pathways.
a, An electrostatic potential surface and cartoon representation of cASIC1mfc sliced along the molecular 3-fold axis of symmetry. The surface is coloured based on electrostatic potential, contoured from −50 kT (red) to +30 kT (blue). White is 0 kT. b, Illustration of the radius of possible pathways along the 3-fold axis, generated using HOLE (red < 1.4 Å < green < 2.3 Å < purple).
Once through the fenestrations, ions move into the broad extracellular vestibule, which is located above residues G432 and D433 of the TM2 domain and is ~8 Å wide and 12 Å high (Fig. 2b,c). If we use F70 of TM1 to define the boundary between the lipid head groups and aqueous solution, the 'lower' half of the extracellular vestibule resides within the membrane plane. Bounding the vestibule on the 'top' are the methyl groups of V75 and on the 'bottom' are three symmetry-related carboxyl moieties of D433, positioned to occlude the ion channel pore (Fig. 2c). On the cytoplasmic side of the closed pore there is another vestibule, shaped like an inverted cone, with a diameter at its base of ~15 Å and a height of ~10 Å, forming the bottom portion of the transmembrane 'hourglass' (Fig. 3).
Vestibules in the extracellular domain have profound negative electrostatic potentials arising from the concentration of 12 acidic residues in each of the central (E80, E374, E412, E417) and extracellular vestibules (D79, E420, E426, D433). We suggest that these vestibules act as a ‘cation reservoir’, concentrating cations near the ion channel pore leading to robust channel conductance25. Attraction of divalent cations within the vestibule may also reduce both the ambient Na+ concentration and resulting Na+ current, as seen in diminished ASIC3 sodium current at high calcium concentration26.
The desensitization gate
There is not a continuous solvent accessible path from one vestibule to the other in the low pH desensitized cASIC1mfc structure and the obvious ion conduction pathway along the molecular 3-fold axis is occluded by a constriction formed by the crossing of the TM2 domains (Fig. 3). This constriction describes the ASIC desensitization gate, an ~8 Å 'thick' occlusion bounded on the extracellular side by the carboxyl groups of D433, on the cytoplasmic side by G436, and in the middle by one turn of the three TM2 α-helices. The conserved G436 residue allows symmetry-related TM2 helices to pack close together, with G436 Cα − Cα distances of ~3.8 Å, thus unambiguously demonstrating that the desensitization gate is a physical block of the transmembrane pore. Beyond G436, toward the cytoplasm, the transmembrane helices diverge, opening up the ion channel to bulk solution.
Residue D433 defines the 'bottom' of the extracellular vestibule, at the boundary of the densensitization gate, and is implicated in low affinity, voltage dependent Ca2+ block27 (Fig. 2b). The close proximity of the three D433 carboxyl groups (~3.4 Å) suggests that their pKa values are perturbed, with either one or more protonated, i.e. neutral, or that there are monovalent or divalent (Ca2+ or Mg2+) cations bound, especially at low proton concentration. Gly432 is 'below' the side chain of D433 and is the site of the degenerin mutation28,29. The substitution of larger amino acid residues at this position in MEC-4 channels results in neuronal death30 due to constitutive channel activation31. In ENaCs and ASICs, similar mutations increase open probability32 and yield constitutively active channels33, respectively. Using simple model building, side chain atoms of non-glycine residues at position 432 sterically clash with symmetry-related TM2 domains, thus providing a mechanism by which mutations at the degenerin site perturb gating.
Ion binding sites
To probe for monovalent cation binding sites, we soaked cASIC1mfc crystals in solutions containing Cs+, a weakly permeant ion (Supplementary Fig. 6) with a substantial anomalous signal (Fig. 4 and Supplementary Fig. 7). The resulting anomalous difference map showed a cylindrical electron density feature located on the crystallographic 3-fold axis stretching from the D433 carboxyl groups to the carbonyl oxygens of G432 (Fig. 4a). The two Cs+ ions manually fit to this elongated peak are separated by ~3.4 Å and thus both sites are probably not simultaneously occupied. The Cs+ ion at position 1, immediately 'above' the base of the extracellular vestibule, is coordinated by three carboxyl oxygen atoms from symmetry-related D433 residues (Fig 4b).
Figure 4. Cs+ binding sites.
a, Electron density peaks (3.5 σ) from anomalous difference Fourier maps calculated using diffraction data measured from crystals soaked in CsCl. b, Key interactions between Cs+ ions at sites 1 and 2 with the main chain and side chain oxygen atoms of Gly432 and Asp433, respectively. c, Stick representation of cASIC1mfc interaction with Cs+ at site 2. d, Trigonal antiprism coordination of the Cs+ ion by the Gly432 carbonyl and Asp433 carboxyl oxygens. Oxygen atoms (red spheres) form the vertices, while solid lines represent the sides of each of the two staggered triangles of the antiprism.
The Cs+ ion at site 2 is coordinated by three main chain carbonyl oxygen atoms from symmetry-related G432 residues and three oxygens from the D433 side chains (Fig. 4b). The main chain carbonyl groups of G432 can coordinate the Cs+ ion at site 2 because the carbonyl group is turned away from the helix axis, a conformation most accessible to a glycine residue. We suggest that coordination of permeant ions in ASICs, and related ion channels, involves main chain carbonyl oxygen atoms, derived primarily from glycine residues, adopting conformations in which the carbonyl group is tipped off of the helix axis. Thus, main chain carbonyl oxygen atoms at residues G436, G439 and G443 may coordinate permeant ions in the open channel state.
The two Cs+ binding sites in the extracellular domain are within the thumb and finger domains, regions implicated in proton dependent ASIC activation6 (Fig. 4 and Supplementary Fig. 7). One Cs+ site is located near putative proton-binding sites (D238-D350 and E239-D346) in a solvent filled crevice. The other Cs+ binding site is enveloped by backbone carbonyl oxygens beneath the thumb α4-helix. The functional consequence of Cs+ binding to these sites is unknown.
Insights into ion permeation
What can the Cs+ sites tell us about principles of ion binding in a trimeric ion channel? Even though the ionic radius of Cs+ (1.69 Å) is substantially larger than Na+ (0.95 Å)34, analysis of these sites is instructive. The Cs+ ion at site 2 is bound by six ligands arranged in a trigonal antiprism geometry that arises because the three ligands on the 'upper' triangular plane (D433) adopt a staggered arrangement in comparison to the three ligands on the 'lower' triangular plane (G432; Fig. 4c). This coordination geometry is striking because it provides for an appropriate number of ligands for coordination of a Na+ ion (6 partial negative charges)35 while perfectly accommodating the underlying molecular symmetry of the protein. We predict that if permeant ions maintain trigonal antiprism coordination through the open pore, then they would shed most of their hydration shell. Indeed, mutations in the ENaC selectivity filter reduced the Na+ conductance, but became permeable to K+, possibly because of a perturbation of precise channel-ion hydration geometry10,14,36.
The trigonal antiprism coordination of the Cs+ ion at site 2, which is topologically equivalent to octahedral coordination, is reminiscent of potassium coordination by valinomycin37 (Supplementary Fig. 8). We suggest that sodium selective ion channels such as ASICs and ENaCs possess ion binding sites with trigonal antiprism coordination geometry, providing permeant ions with six ligands arranged with an ideal ion to ligand distance (Fig. 4d). Indeed, the trigonal antiprism coordination of the site 2 ion is stereochemically analogous to the 8-fold square antiprism coordination of larger K+ ions in potassium channels38, where in both cases the symmetry and subunit number of the protein matrix is matched to the optimal coordination requirements of the permeant ion35.
By simply translating sites 1 and 2 along the 3-fold axis, toward the cytoplasm, we predict a third ion binding site in the open, conducting state of the channel. This site is defined by the carbonyl oxygens of G432 and G436, all oriented toward the 3-fold axis, and arranged with trigonal antiprism coordination geometry. Additional residues likely contribute, directly and indirectly to the permeation pathway, including S445, which in ENaCs changes ion selectivity upon mutation10,14,15. In cASIC1mfc, however, the side chain of S445 is oriented away from the channel pore and thus it either indirectly affects selectivity or is brought into direct interaction with ions following a significant conformational change39.
ASICs and P2X receptors
The low pH cASIC1mfc crystal structure describes the conformation of a trimeric, proton-gated ion channel in a desensitized state. In a companion paper, the crystal structure of the trimeric ATP-gated P2×4 receptor in an agonist-free, closed state is described21. Both ASICs and P2X receptors have a similar transmembrane topology19,40 and share common functional properties: both are ligand-gated, cation-selective ion channels. However, there is no significant amino acid sequence identity between ASICs and P2X receptors.
Upon solving the structures of cASIC1mfc and the ΔzfP2×4 receptor, we observed that the transmembrane domains of the two proteins were remarkably similar. Superposition of the TM2 helices using residues 427–445 of cASIC1mfc and residues 335–353 of ΔzfP2×4 yielded a root mean square deviation of 2.3 Å, demonstrating that the organization of the TM2 helices are roughly equivalent (Fig. 5a). Thus, the fundamental conformations of the ion channel domains are similar in the resting state of the ΔzfP2×4 receptor and in the desensitized state of cASIC1mfc (Fig. 5a).
Figure 5. ASIC and P2X receptors share a common pore architecture.
a, Stereoview of cASIC1mfc (purple) and ΔzfP2×4 (gold) crystal structure overlay based on superposition of the respective TM2 domains. b, A structure-based sequence alignment of cASIC1 and zfP2×4 TM2 domains is shown. Corresponding residues in cASIC1mfc and zfP2×4 are highlighted by side chain color. A rat ENaCα sequence is shown for comparison. Views of the cASIC1mfc (c) and ΔzfP2×4 (d) ion channel pores from the extracellular side of the membrane. Selected side chain residues of ΔzfP2×4 are in green. The cASIC1mfc Met438 is modelled as an alanine.
The structural superposition of the two receptors allows us to align the amino acid sequences of the TM2 domains, revealing unanticipated relationships (Fig. 5b). For example, we see that there is a common signature sequence associated with the base of the extracellular vestibule, ‘…DIG…’ for cASIC1 and ‘…NIG…’ for P2×4, with the ‘D’27 and ‘N’41 residues both crucial for ion channel function. In the zfP2×4 receptor, however, there is a leucine residue (L340) at the equivalent ASIC degenerin position, yet the receptor is not constitutively active. If the ion channel domains are truly related, one must ask how the zfP2×4 receptor accommodates this bulky amino acid. Leucine is probably tolerated in ΔzfP2×4 due to the presence of a proline (P337), one turn of the helix upstream from L340, which bends the helix, making space for the bulky side chain. In both cases the channels’ respective gates are defined by the close crossing of the TM2 helices, located at cASIC1mfc residue G436 and ΔzfP2×4 amino acid A344, and are composed of a continuous stretch of 5 to 8 amino acid residues (Fig. 5b, c).
The presence of similar cavities and vestibules with deep negative electrostatic potentials in the extracellular domains of the cASIC1mfc (Fig. 3) and ΔzfP2×4 receptors represent additional common features. The ‘upper’ and ‘central’ vestibules are located along the molecular three-fold vertical axis, are similar in size, with the ‘central’ vestibule being especially rich in acidic residues. In ΔzfP2×4 and cASIC1, experimental21 and computational studies42 demonstrate and predict, respectively, the binding of cations in these ‘central’ vestibules. The ‘central’ vestibule is accessible to ions in the resting state ΔzfP2×4 structure, but not in the cASIC1mfc desensitized state structure. Cushman and colleagues have shown that residues in the ‘central’ vestibule of rat ASIC3 are accessible in the resting, but not desensitized state43. We speculate that accessibility to the central vestibule is modulated by conformational changes in the wrist/palm region, near T76, during the transition from the closed to desensitized state.
The cASIC1mfc structure may tell us how permeant cations are coordinated within the P2X receptor channel pore. We speculate that the presence of the glycine residues in the ΔzfP2×4 receptor may also allow main chain carbonyl groups to tip off of the helix axis so as to allow the oxygen atoms to coordinate cations in the pore. Because the cation selectivity of P2X receptors is not as stringent as ASICs, however, there must be features of their respective ion channel pores that are distinct.
Mechanism
Based on the cASIC1mfc and ΔzfP2×4 crystal structures, we propose that proton binding to sites throughout the extracellular domain of acid sensing ion channels results in movements of the thumb and wrist regions that in turn open the activation gate by a rotation of the TM helices. The opening of the pore brings main chain carbonyl oxygens, as well as oxygen atoms from side chain residues such as D433, into an ion conduction pathway along the molecular 3-fold axis of symmetry. Extended sojourns to low pH cause the wrist and adjacent portions of β1 and β12 to draw together, occluding the middle vestibule and triggering closure of the ion channel through the formation of the desensitization gate, as observed in the current cASIC1mfc crystal structure.
METHODS SUMMARY
The cASIC1mfc construct was identified through systematic deletion mutagenesis, patch clamp electrophysiology, and fluorescence detection size-exclusion chromatography studies. The cASIC1mfc protein was purified from baculovirus-infected Sf9 cells using metal ion affinity and size exclusion chromatography. Crystals were obtained in 150–350 mM NaCl, 100 mM HEPES (pH 6.5), 23–28% v/v PEG400, and 10 mM taurine. PHASER44 was employed in molecular replacement, using the extracellular domain of a ΔcASIC1 subunit. Density modification using programs in the CCP4 suite45 improved the initial phases and the structure was refined using REFMAC46 and COOT47. Patch-clamp recording was performed in tsA201 and CHO-K1 cells 24–48 hours post transfection with amino-terminal GFP-tagged cASIC1mfc construct. The endogenous proton-mediated current observed in TsA201 cells was negligible in outside-out patch recording of cASIC1 currents.
FULL METHODS
Expression and purification
The cASIC1mfc construct, consisting of residues M2-R466, was used for crystallographic trials. This minimal functional construct was expressed as an amino terminal fusion with GFP from baculovirus-infected Sf9 cells. Infected cells were incubated at 27° C in serum-free media for 48 hours prior to harvesting. Cells were collected by centrifugation and sonicated in the presence of 150 mM NaCl, 20 mM Tris (pH 8.0), 4 mM phenylmethylsuphonyl fluoride and protease inhibitor cocktail (Roche cOmplete tablets). Crude membranes were solubilized in 15 ml of 150 mM NaCl, 20 mM Tris (pH 8.0) and 40 mM n-dodecyl-β-D-maltoside (DDM) per gram of membrane for 1 hour at 4° C. The resulting solution was clarified by centrifugation at 40,000 rpm at 4° C for 1 hour (Ti45 rotor). Protein was bound to TALON resin and eluted with 150 mM NaCl, 20 mM Tris (pH 8.0), 1 mM DDM, and 250 mM imidazole. The 8x histidine, GFP tag was cleaved with a 1 hour thrombin digestion at room temperature and the receptor was further purified via size exclusion chromatography using a mobile phase composed of 150 mM NaCl, 20 mM Tris (pH 8.0), 1 mM DDM, 1 mM dithiothreitol, and 1 mM EDTA. Peak fractions were pooled and concentrated to 1.8∓2.2 mg ml−1 (calculated molar extinction of 53,570 at 280 nm) prior to crystallization.
Crystallization
Crystal trials were carried out using the hanging-drop vapor diffusion method by mixing a 1:1 ratio of protein solution and reservoir solution consisting of: 150–350 mM NaCl, 100 mM HEPES (pH 6.5), 23–28% PEG400 and 10 mM taurine. Crystals were grown and harvested at 4° C. Cryoprotection was achieved by soaking crystals in reservoir solutions supplemented with DDM and with increasing concentrations of PEG 400 (final concentration 38–40%). The crystals were then flash cooled in liquid nitrogen. For Cs+ soaked crystals, 250 mM CsCl was substituted for NaCl in the cryo-protection solution.
Structure determination
X-ray diffraction data sets were collected at the Advanced Light Source (beam line 5.0.2). Cs+ soaked crystal diffraction data diffraction data were measured using low energy x-rays (9000 eV). Collected data were processed using HKL200048. Molecular replacement using the extracellular domain of the ΔcASIC1 structure was used to obtain the initial phases. The phases were improved by density modification using DM. The resulting electron density maps demonstrated that whereas the ΔcASIC1 extracellular domain was a faithful structure for the cASIC1mfc extracellular domain, the transmembrane domains associated with the cASIC1mfc maps adopted a significantly different conformation that the transmembrane domains of the ΔcASIC1 structure. Therefore, the cASIC1mfc transmembrane domains were manually rebuilt. Iterative model building and refinement was performed using COOT47 and REFMAC546, respectively. The stereochemical quality of the final structure validity was assessed using PROCHECK and Molprobity49.
Electrophysiology
Whole-cell and outside-out patch recording were carried out using tsA201 and CHO-K1 cells 24–48 hours post-transfection using plasmids encoding the fluorescently tagged cASIC1mfc construct. We did not observe endogenously expressed ASIC current in outside-out patches from tsA201 cells. Patch pipettes were pulled and polished to 2–5 MΩ resistance and filled with the following internal solution (in mM): KCl (100), NaCl (5), HEPES (40), EGTA (10), MgCl2 (5) adjusted to pH 7.4 with KOH. External solutions for whole-cell recording consisted of (in mM): NaCl (150), KCl (5), HEPES (5), MES (5), and calcium chloride (1.8). In outside-out patch recording, the external solution contained 168 mM NaCl and 5 mM sucrose to enhance current and improve visibility between the two solutions, respectively. For CsCl substitution, the external solution contained 168 mM CsCl instead of NaCl. The pH values of all external solutions were adjusted with N-methyl-D-glucamine. Rapid solution exchange was achieved by placing outside-out patches containing cASIC1mfc receptors at the interface between high pH (8.0) and test solutions streaming from a theta-tube attached to a piezo-electric stack. Test-solution was applied for 1 sec at a holding potential of −60 mV. The open tip junction potential was obtained after each experiment. Concentration-response profiles were recorded, normalized to maximal current (Imax), and fitted to the Hill equation.
Supplementary Material
Acknowledgements
We thank the personnel at beamlines 5.0.2 of the Advanced Light Source. We also thank C. Canessa for chicken ASIC1 DNA, L. Vaskalis for assistance with illustrations, and Gouaux lab members for discussion. This work was supported by an NIGMS-NRSA to E.B.G. and the NIH (E.G.). E.G. is an investigator with the Howard Hughes Medical Institute.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Coordinates and structure factors have been deposited with the Protein Data Bank under code 3HGC. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Waldmann R, Champigny G, Bassilana F, Heurteaux C, Lazdunski M. A proton-gated cation channel involved in acid-sensing. Nature. 1997;386:173–177. doi: 10.1038/386173a0. [DOI] [PubMed] [Google Scholar]
- 2.Coric T, Zheng D, Gerstein M, Canessa CM. Proton sensitivity of ASIC1 appeared with the rise of fishes by changes of residues in the region that follows TM1 in the ectodomain of the channel. J Physiol. 2005;568:725–735. doi: 10.1113/jphysiol.2005.087734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coric T, Passamaneck YJ, Zhang P, Di Gregorio A, Canessa CM. Simple chordates exhibit a proton-independent function of acid-sensing ion channels. FASEB J. 2008;22:1914–1923. doi: 10.1096/fj.07-100313. [DOI] [PubMed] [Google Scholar]
- 4.Kellenberger S, Schild L. Epithelial sodium channel/degenerin family of ion channels: a variety of functions for a shared structure. Physiol Rev. 2002;82:735–767. doi: 10.1152/physrev.00007.2002. [DOI] [PubMed] [Google Scholar]
- 5.Hesselager M, Timmermann DB, Ahring PK. pH Dependency and desensitization kinetics of heterologously expressed combinations of acid-sensing ion channel subunits. J Biol Chem. 2004;279:11006–11015. doi: 10.1074/jbc.M313507200. [DOI] [PubMed] [Google Scholar]
- 6.Jasti J, Furukawa H, Gonzales EB, Gouaux E. Structure of acid-sensing ion channel 1 at 1.9 Å resolution and low pH. Nature. 2007;449:316–323. doi: 10.1038/nature06163. [DOI] [PubMed] [Google Scholar]
- 7.Carnally S, et al. Direct visualization of the trimeric structure of the ASIC1a channel, using AFM imaging. Biochem Biophys Res Commun. 2008;372:752–755. doi: 10.1016/j.bbrc.2008.05.100. [DOI] [PubMed] [Google Scholar]
- 8.Zha XM, et al. Oxidant regulated inter-subunit disulfide bond formation between ASIC1a subunits. Proc Natl Acad Sci U S A. 2009;106:3573–3578. doi: 10.1073/pnas.0813402106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coscoy S, Lingueglia E, Lazdunski M, Barbry P. The Phe-Met-Arg-Phe-amide-activated sodium channel is a tetramer. J Biol Chem. 1998;273:8317–8322. doi: 10.1074/jbc.273.14.8317. [DOI] [PubMed] [Google Scholar]
- 10.Kellenberger S, Hoffmann-Pochon N, Gautschi I, Schneeberger E, Schild L. On the molecular basis of ion permeation in the epithelial Na+ channel. J Gen Physiol. 1999;114:13–30. doi: 10.1085/jgp.114.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bassler EL, Ngo-Anh TJ, Geisler HS, Ruppersberg JP, Gründer S. Molecular and functional characterization of acid-sensing ion channel (ASIC) 1b. J Biol Chem. 2001;276:33782–33787. doi: 10.1074/jbc.M104030200. [DOI] [PubMed] [Google Scholar]
- 12.Benos DJ, Mandel LJ, Simon SA. Cationic selectivity and competition at the sodium entry site in frog skin. J Gen Physiol. 1980;76:233–247. doi: 10.1085/jgp.76.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palmer LG. Ion selectivity of the apical membrane Na channel in the toad urinary bladder. J Membr Biol. 1982;67:91–98. doi: 10.1007/BF01868651. [DOI] [PubMed] [Google Scholar]
- 14.Kellenberger S, Gautschi I, Schild L. A single point mutation in the pore region of the epithelial Na+ channel changes ion selectivity by modifying molecular sieving. Proc Natl Acad Sci U S A. 1999;96:4170–4175. doi: 10.1073/pnas.96.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Snyder PM, Olson DR, Bucher DB. A pore segment in DEG/ENaC Na(+) channels. J Biol Chem. 1999;274:28484–28490. doi: 10.1074/jbc.274.40.28484. [DOI] [PubMed] [Google Scholar]
- 16.Brake AJ, Wagenbach MJ, Julius D. New structural motif for ligand-gated ion channels defined by an ionotropic ATP receptor. Nature. 1994;371:519–523. doi: 10.1038/371519a0. [DOI] [PubMed] [Google Scholar]
- 17.Valera S, et al. A new class of ligand-gated ion channel defined by P2x receptor for extracellular ATP. Nature. 1994;371:516–519. doi: 10.1038/371516a0. [DOI] [PubMed] [Google Scholar]
- 18.Nicke A, et al. P2X1 and P2X3 receptors form stable trimers: a novel structural motif of ligand-gated ion channels. EMBO J. 1998;17:3016–3028. doi: 10.1093/emboj/17.11.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aschrafi A, Sadtler S, Niculescu C, Rettinger J, Schmalzing G. Trimeric architecture of homomeric P2X2 and heteromeric P2X1+2 receptor subtypes. J Mol Biol. 2004;342:333–343. doi: 10.1016/j.jmb.2004.06.092. [DOI] [PubMed] [Google Scholar]
- 20.Surprenant A, North RA. Signaling at Purinergic P2X Receptors. Annu Rev Physiol. 2008 doi: 10.1146/annurev.physiol.70.113006.100630. [DOI] [PubMed] [Google Scholar]
- 21.Kawate T, Michel JC, Birdsong WT, Gouaux E. Crystal structure of the ATP-gated P2X4 ion channel in the closed state. Nature. 2009;XXX:XXX. doi: 10.1038/nature08198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14:673–681. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 23.Gründer S, et al. A mutation causing pseudohypoaldosteronism type 1 identifies a conserved glycine that is involved in the gating of the epithelial sodium channel. EMBO J. 1997;16:899–907. doi: 10.1093/emboj/16.5.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfister Y, et al. A gating mutation in the internal pore of ASIC1a. J Biol Chem. 2006;281:11787–11791. doi: 10.1074/jbc.M513692200. [DOI] [PubMed] [Google Scholar]
- 25.Dani JA. Ion-channel entrances influence permeation. Net charge, size, shape, and binding considerations. Biophys J. 1986;49:607–618. doi: 10.1016/S0006-3495(86)83688-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Immke DC, McCleskey EW. Protons open acid-sensing ion channels by catalyzing relief of Ca2+ blockade. Neuron. 2003;37:75–84. doi: 10.1016/s0896-6273(02)01130-3. [DOI] [PubMed] [Google Scholar]
- 27.Paukert M, Babini E, Pusch M, Gründer S. Identification of the Ca2+ blocking site of acid-sensing ion channel (ASIC) 1: implications for channel gating. J Gen Physiol. 2004;124:383–394. doi: 10.1085/jgp.200308973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chalfie M, Wolinsky E. The identification and suppression of inherited neurodegeneration in Caenorhabditis elegans . Nature. 1990;345:410–416. doi: 10.1038/345410a0. [DOI] [PubMed] [Google Scholar]
- 29.Driscoll M, Chalfie M. The mec-4 gene is a member of a family of Caenorhabditis elegans genes that can mutate to induce neuronal degeneration. Nature. 1991;349:588–593. doi: 10.1038/349588a0. [DOI] [PubMed] [Google Scholar]
- 30.Hong K, Driscoll M. A transmembrane domain of the putative channel subunit MEC-4 influences mechanotransduction and neurodegeneration in C. elegans. Nature. 1994;367:470–473. doi: 10.1038/367470a0. [DOI] [PubMed] [Google Scholar]
- 31.Brown AL, Fernandez-Illescas SM, Liao Z, Goodman MB. Gain-of-function mutations in the MEC-4 DEG/ENaC sensory mechanotransduction channel alter gating and drug blockade. J Gen Physiol. 2007;129:161–173. doi: 10.1085/jgp.200609672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snyder PM, Bucher DB, Olson DR. Gating induces a conformational change in the outer vestibule of ENaC. J Gen Physiol. 2000;116:781–790. doi: 10.1085/jgp.116.6.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Champigny G, Voilley N, Waldmann R, Lazdunski M. Mutations causing neurodegeneration in Caenorhabditis elegans drastically alter the pH sensitivity and inactivation of the mammalian H+-gated Na+ channel MDEG1. J Biol Chem. 1998;273:15418–15422. doi: 10.1074/jbc.273.25.15418. [DOI] [PubMed] [Google Scholar]
- 34.Hille B. Ion Channels of Excitable Membranes. 3rd edn. Sinauer Associates; 2001. [Google Scholar]
- 35.Harding MM. Metal-ligand geometry relevant to proteins and in proteins: sodium and potassium. Acta Crystallogr D Biol Crystallogr. 2002;58:872–874. doi: 10.1107/s0907444902003712. [DOI] [PubMed] [Google Scholar]
- 36.Kellenberger S, Auberson M, Gautschi I, Schneeberger E, Schild L. Permeability properties of ENaC selectivity filter mutants. J Gen Physiol. 2001;118:679–692. doi: 10.1085/jgp.118.6.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neupert-Laves K, Dobler M. The crystal structure of a K+ complex of valinomycin. Helv Chim Acta. 1975;58:432–442. doi: 10.1002/hlca.19750580212. [DOI] [PubMed] [Google Scholar]
- 38.Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 Å resolution. Nature. 2001;414:43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 39.Askwith CC, Benson CJ, Welsh MJ, Snyder PM. DEG/ENaC ion channels involved in sensory transduction are modulated by cold temperature. Proc Natl Acad Sci U S A. 2001;98:6459–6463. doi: 10.1073/pnas.111155398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang L, et al. Subunit arrangement in P2X receptors. J Neurosci. 2003;23:8903–8910. doi: 10.1523/JNEUROSCI.23-26-08903.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silberberg SD, Chang TH, Swartz KJ. Secondary structure and gating rearrangements of transmembrane segments in rat P2×4 receptor channels. J Gen Physiol. 2005;125:347–359. doi: 10.1085/jgp.200409221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaikh SA, Tajkhorshid E. Potential cation and H+ binding sites in acid sensing ion channel-1. Biophys J. 2008;95:5153–5164. doi: 10.1529/biophysj.108.141606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cushman KA, Marsh-Haffner J, Adelman JP, McCleskey EW. A conformation change in the extracellular domain that accompanies desensitization of acid-sensing ion channel (ASIC) 3. J Gen Physiol. 2007;129:345–350. doi: 10.1085/jgp.200709757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCoy AJ, et al. Phaser crystallographic software. J Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 46.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 47.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 48.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter CW, Sweet RM, editors. Methods in Enzymology. Vol. 276. Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 49.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.