Abstract
Purpose
The DBA/2J (D2) mouse carries mutations in two of its genes, Tyrp1 and Gpnmb. These alterations result in the development of an immune response in the iris, leading to iris atrophy and pigment dispersion. The development of elevated intraocular pressure (IOP) in this model of glaucoma is considered to be a significant factor leading to the death of retinal ganglion cells (RGCs). Changes in gene expression in the retina have already been correlated with the appearance of elevated IOP in the D2 mouse. The purpose of the present study was to determine if any changes in gene expression occur prior to the development of IOP.
Methods
The IOP was measured monthly using a rebound tonometer in D2 and age-matched C57/BL6 (B6) mice (normal controls). D2 animals with normal IOP at 2 and 4 M were used. In addition, mice at the age of 6–7 M were included to look for any trends in gene expression that might develop during the progression of the disease. Separate RNA samples were prepared from each of three individual retinas for each age, and gene expression profiles were determined with the aid of mouse oligonucleotide arrays (Agilent). A subset of genes was examined with the aid of real-time PCR. Immunocytochemistry was used to visualize changes in the retina for some of the gene-products.
Results
Four hundred and thirteen oligonucleotide probes were differentially expressed in the retinas of 4 M versus 2 M old D2 mice. The most significantly up-regulated genes (181) were associated with immune responses including interferon signaling, the complement system and the antigen presentation pathway, whereas the down-regulated genes (232) were linked to pathways related to cell death and known neurological diseases/disorders. These particular changes were not revealed in the age-matched B6 mice. By 6 M, when IOP started to increase in many of the D2 mice, more robust changes of these same genes were observed. Changes in the levels of selected genes, representative of different functions/pathways, were validated with RT-PCR, and changes in glial responses were visualized in the retina with immunocytochemistry.
Conclusions
The results showed that the expression of genes related to the immune response and acute stress were altered independently of the development of elevated IOP, and indicated early involvement of the immune system in the onset of the disease. The later development of elevated IOP, observed in this animal model, was coincident with continued changes in expression of genes observed at earlier time points. Further studies are warranted to identify the roles of specific genes identified here with respect to the death of the RGCs.
Keywords: iris, iris atrophy, pigment dispersion, IOP, glaucoma, retinal ganglion cells
Retinal degeneration characterized by retinal ganglion cell death, is a hallmark of many diseases/disorders, including retinal ischemia and glaucoma. Visual impairment due to glaucoma affects 60 million people worldwide and is a major cause of irreversible blindness.1 It is thought that the elevation of IOP, evident in glaucoma, causes harm to the RGCs and/or their axons in the optic nerve. The principal therapy used in the treatment of glaucoma is a reduction in IOP, however, it is not always successful in halting progression of the disease, and it is now accepted that multiple additional factors likely contribute to pathogenesis of this disease. The elucidation of these other prospective factors would provide us with additional targets for the development of novel treatments.
Animal models are critical for studying molecular mechanisms underlying multi-factorial diseases. One model of glaucoma, the DBA/2J (D2) mouse,2 carries mutations in two of its genes, Tyrp1 and Gpnmb. The presence of these altered genes triggers an immune response in the iris, leading to its atrophy, pigment dispersion and glaucomatous changes with age, including elevation of intraocular pressure (IOP), retinal ganglion cell (RGC) death and optic nerve damage.3–5 D2 mice are therefore useful as an animal model for the study of the interaction of the immune response and increased IOP. High IOP is usually first detected in the eyes of 6–7 M D2 animals.6 There is a tendency for IOP to increase earlier in females than in males, and IOPs are significantly higher in females than in males in 6–7 M mice.6 The period of elevated IOP extends from 6 M to 16 M, with 8–9 M representing an important transition to high IOP for many mice. D2 mice begin to lose RGCs following elevation of IOP. Typically, by 10–12 M, significant RGC loss can be observed in the majority of D2 mice. The prevalence and severity of these retinal lesions increase with age.
Although increased IOP resulting from pigment dispersion and blocking of aqueous humor drainage is thought to be a significant risk factor leading to RGC death in D2 mice,2,6–8 several lines of evidence suggest other factors must contribute to the neurodegeneration. 9,10 For example, high-dose irradiation with X-rays, followed by syngeneic bone marrow transfer almost completely rescues RGC loss, without changing the course of IOP elevation.9 Other studies also have indicated that hypertension is not the only causative factor in the glaucomatous changes that are seen in the retinas of D2 mice.10 These studies raise important questions concerning the possible existence of other contributing factors which may be intrinsic/ extrinsic to the retina.
Previous gene-array studies have focused on the changes in gene expression resulting from elevation of IOP in D2 mice.11 However, as the level of IOP varies among individual D2 mice, we set out to see if there were any changes in retinal gene expression that are IOP-independent. Through a comparison of the gene expression profiles of the retina of D2 mice at early postnatal ages, we have made some novel observations. The expression of genes related to immune and acute stress responses was found to be changed at a very early stage of the disease and independent of the elevation in IOP. The results indicated a possible involvement of the immune system prior to the onset of the normal signs of glaucomatous disease. The study indicated that the increased IOP might act later as an additional aggravating factor to enhance the changes in these and other genes and exacerbate the progression of the disease. In addition, we found that other genes known to be related to neurological damage and cell signaling/death pathways were down-regulated during the earliest stages of the retinal damage in the D2 mice.
Materials and Methods
The DBA/2J (D2) and C57BL/6J (B6) animals were purchased from Jackson Laboratories (Bar Harbor, ME). The animals were handled in accordance with policies and procedures recommended by the Institutional Animal Care and Use Committee at the University of Louisville, and all procedures adhered to the ARVO Statement for the Use and Care of Animals in Ophthalmic and Vision Research. The animals were housed in transparent plastic rodent boxes under a 12-hour-light/12-hour-dark cycle. Food and water were available ad libitum. All animals used in this study were females.
IOP measurement
IOP was measured monthly in D2 (n = 18) and B6 mice (n =9) using the rebound tonometer for rodents (Tonolab Colonial Medical Supply, Franconia, NH) according to the manufacturer’s recommended procedures. The rebound tonometer was easy to use and accurately measured IOP in rats and mice as previously reported.12 All IOP measurements were performed in conscious mice between noon and 3 pm. Animals at the age of 2-months-old (2 M) served as the baseline IOP control. Comparisons between animals at the ages of 2 and 4-months (4 M) old, with normal IOP, were used for detecting age-related changes in gene expression that were not associated with the elevation of IOP. D2 mice at the age of 6~7 months (6~7 M) old were also included to see if the changes evident in gene expression at 4 M continued to change during the development of increased IOP. B6 animals at 2,4 and 8 months of age (2 M, 4 M and 8 M) were used for comparison.
Microarray analysis
RNA Isolation
Whole retinas were isolated from 2, 4, 6–7 M animals with the aid of dissection microscope and rinsed with ice-cold 0.1M phosphate-buffered saline (PBS). Retinal samples from individual animals were homogenized in buffer QIAzol lysis reagent (Qiagen, Valencia, CA) using a motorized rotor-stator homogenizer. The total RNA was isolated using RNeasy Lipid Tissue Kits (Qiagen, Valencia, CA91355) according to the manufacturer’s instructions. The quality of RNAs was measured with the aid of an Agilent 2100 bioanalyzer (Agilent, Wilmington DE 19808). For this study, sharp ribosomal RNA bands were evident with RNA integrity numbers greater than 7.0.
Preparation of labeled cRNA target
300ng total RNA was used to generate fluorescent cRNA with the aid of the Low RNA Input Linear Amplification kit with one-color (Agilent, Wilmington DE 19808). Briefly, this kit uses the T7 promoter primer to synthesize cDNA and T7 RNA polymerase to synthesize cRNA, which simultaneously amplifies target material and incorporates cyanine 3-labeled CTP. The labeled cRNA was purified using a RNeasy Mini Elute kit (Qiagen, Valencia, CA91355). The yields and incorporation efficiencies were measured on a spectrophotometer (NanoDrop technologies). The yields for this project were greater than 1.5 μg, and the specific activities were greater than 9.0 pmol Cy3 per μg cRNA.
Hybridization and scanning
1.65 μg of each labeled cRNA sample was fragmented at 60 °C for 30 min (Agilent Gene Expression Hybridization kit) and then hybridized to Agilent oligonucleotide arrays (4 × 44 K mouse microarray; 60-mer oligonucleotides; G4122F) at 65 °C for 17 hours. After hybridization, the microarray slides were washed with Agilent gene expression wash buffers. The slides were scanned using an Agilent microarray scanner (G2565BA; Agilent Technologies, Wilmington, DE 19808 with settings for one-color, green channel and 5 um resolution. The one-color microarray images (tif) were extracted with the latest protocol and grid design file with the aid of Feature Extraction software (v 9.5.1, Agilent).
Array data analysis
Three arrays for each age were used in the analyses. The data were normalized and analyzed with the aid of GeneSpring (GX 7.3, Agilent Technologies, Santa Clara, CA 95051. The data were transformed to bring any negative value to 0.01. Normalization was performed using a per-chip 50th percentile method that normalizes each chip on its median, allowing comparison among chips. Then a per-gene on median normalization was performed, which normalized the expression of every gene on its median among samples. GeneSpring generates an average value of the three replicates of each gene.
The differentially expressed genes of significance were deduced with the aid of Volcano Plots (p-value versus fold change). Using the data derived from the Volcano Plots, pairwise comparisons of the experimental and control groups were made. Differentially expressed genes with p-values ≤0.05 and with fold changes ≥2 were determined.
Ingenuity pathway analysis
Pathway Analysis software (IPA 6.0; Ingenuity Systems, Mountain View, USA) was used to identify functional groups and canonical pathways for those genes that were differentially expressed at different time points, ie., 2 M vs 4 M, 4 M vs 6–7 M or 2 M vs 6–7 M.
Real time RT-PCR
To validate and confirm the expression levels of selected genes, real time PCR was performed. Following DNase treatment, cDNAs were synthesized from 50 ng RNAs using Taqman reverse transcription reagents (Applied Biosystems, Foster, CA), followed by real time PCR. The primers were designed with primer-express software (Applied Biosystems, Foster, CA) (Table 1). The PCR was performed with 2 μl cDNAs, with a SYBR Green PCR Core reagent kit, and with the aid of the ABI Prism 7000 Sequence Detection System. The SYBR green data were analyzed by the ABI Prism 7000 Sequence Detection System software. The relative expression levels of the target genes were analyzed using the 2−ΔΔCt method13 and by normalizing with GAPDH gene expression (which was not changed in retinas of D2 mice at different ages) and presented as the fold change compared to the controls which were taken as 1. Experiments were performed in triplicate for each gene and then repeated at least three times using independent biological samples.
Table 1.
Primers for real time PCR.
| Gene name | Forward primer | Reverse primer |
|---|---|---|
| BDNF | 5′-TGGAAGCCTGAATGAATGGAC-3′ | 5′-CCCAGGAGAGTAACCACTAACACAT-3′ |
| C4b | 5′-GTTCGTTATCGGGTCTTTGCA-3′ | 5′-TTCTTGAGTACACGGAGGCCA-3′ |
| Edn2 | 5′-TGACTGACATCATGGCTGGC-3′ | 5′-CAGGTTCTGTGCTCCCAAAAGT-3′ |
| GFAP | 5′-AAGAGACAGAGGAGTGGTATCGGT-3′ | 5′-GTCGTTAGCTTCGTGTTTGGC-3′ |
| Gjb6 | 5′-GGACTGCTTCATTTCGAGGC-3′ | 5′-AACACAACTCGGCCACATTG-3′ |
| Lcn2 | 5′-TGCCACTCCATCTTTCCTGTT-3′ | 5′-GGGAGTGCTGGCCAAATAAG-3′ |
| MR1 | 5′-GGATCCTCAGAGCAATGATGTTT-3′ | 5′-TCACTCGAAGGATGTCCCCT-3′ |
| Tnfrsf25 | 5′-AGCTTACTCGGGCAAATGCTAG-3′ | 5′-AACACGTGCAGTTGACCCTG-3′ |
Immunocytochemistry
The expression patterns of selected complement factors and acute response gene-products were assessed in D2 retinas with the aid of immunofluorescence labeling of the relevant proteins. In addition, the activation of microglial cells in D2 retinas was also investigated. Whole eyes from D2 and B6 mice were fixed with 4% paraformaldehyde overnight, followed by cryoprotection in 30% sucrose at 4 °C. Frozen sections were permeabilized using 0.2% Triton-X-100 (Sigma) in PBS. After blocking of non-specific binding sites with donkey serum, tissue sections were incubated with primary antibodies overnight at 4 °C. Antibodies used were: polyclonal antiserum to human C1q (Quidel, San Diego, CA), polyclonal anti-C3 (Calbiochem), polyclonal anti-Factor B (H-95), polyclonal anti-Lcn2 (M-145), rat monoclonal anti-C4 (16D2) (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), and polyclonal anti-Iba1(Wako, Richmond, VA). The primary antibodies were visualized with Cy3- or Alexa 488-conjugated secondary antibody (Molecular Probes, Eugene, OR). The slides were mounted with anti-fade mounting medium (Vector Laboratories, Burlingame, CA) and viewed with the aid of a confocal microscope.
Statistical analysis
All quantitative data from IOP measurements and real time PCR were expressed as means ± S.E.M. A minimum of three independent experiments for each condition were performed. ANOVA was used for multiple comparisons followed by Newman-Keuls paired comparison. A p-value < 0.05 was used as the cut-off value for significance tests.
Results
Measurement of IOP in experimental animals
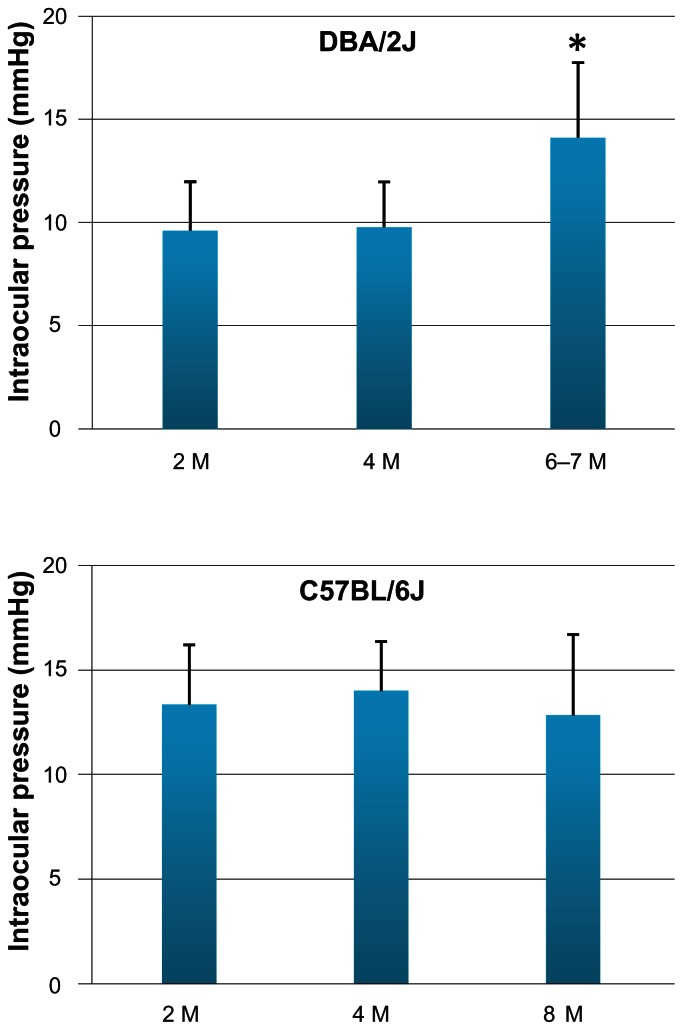
To detect changes in the early gene expression profile that are IOP-independent, we compared 4 M with 2 M D2 mice. Age-matched B6 mice served as controls. Measurements of the IOPs were taken for each eye of all animals. Although there were some individual D2 animals which had an onset of relatively high IOP at a young age, the period when D2 mice consistently start to develop elevated IOP is usually around 6–7 M. (Fig. 1). The 8–9 M period represents an important transition time for the expression of relatively high IOP for the majority of the mice.6 In this study, the average IOP of the D2 and B6 mice at the age of 4 M was 10 mmHg ( range 8–14 mmHg) and 16 mmHg (range 11–17 mmHg), respectively. These were the same levels as that seen in mice at 2 M for both species (Fig. 1). The animals with “normal” IOP at 2 and 4 M were used for microarray analysis and for comparative analyses of retinal gene expression levels which would be independent of changes in IOP levels. In addition, D2 mice at ages of 6–7 M and B6 mice at the age of 8 M were included to see if there were any changes in gene expression that would be consistent with a trend, or that may be associated with changes in IOP. Consistent with prior studies,2,6 many D2 mice started to develop higher IOP (average IOP was14 mmHg, range 8–19 mmHg) by 6–7 M, whereas the IOP of B6 mice at 8 M did not change relative to that observed at younger ages (Fig. 1).
Figure 1.
IOP measurements in the experimental animals. IOP was measured monthly in D2 (n =6 for each time point) and B6 mice (n =3 for each time point) using the rebound tonometer. The animals with “normal” IOP at the age of 2 and 4 M were used for comparative analyses of retinal gene expression levels which would be independent of changes in IOP levels. D2 mice at ages of 6–7 M and B6 mice at the age of 8 M were included to see if there were any changes in gene expression that may be associated with changes in IOP. Many D2 mice had started to develop higher IOP by 6–7 M, whereas the IOP of B6 mice at 8 M had not changed relative to that observed at younger ages.
*P < 0.05, ANOVA.
Early gene expression profile
The early gene expression that was IOP-independent in D2 retina was identified by comparing the gene expression profile in 4 M mice relative to that in the 2 M mice. Of 44,000 probes on the Agilent oligonucleotide arrays, 413 probes were differentially expressed in the D2 retinas when a twofold cutoff and a p-value < 0.05 were used for analysis. Among these differentially expressed probes, 181 had increased and 232 had decreased expression values at 4 M relative to 2 M. These changes were not revealed in age-matched B6 mice, although changes in genes related to some developmental and metabolism processes in B6 mice were observed.
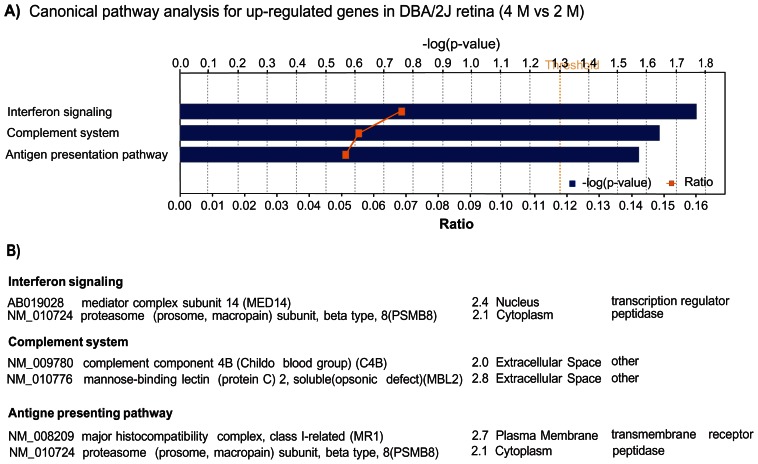
The probes with altered expression values were annotated with the aid of Ingenuity software. Of the 181 up-regulated probes, Pathway Assist software (Ingenuity) analysis was able to recognize 171 IDs, and 81 genes were eligible for function/ pathway annotation. These 81 genes were mainly associated with the following functional groups: immune response/inflammatory disease, neurological disease, cell signaling, gene expression and cell death (Table 2). The three most significant, canonical pathways associated with the up-regulated genes were related to immune responses, including interferon signaling, complement system regulation and antigen presentation (Fig. 2). These results indicated an involvement of immune and inflammatory responses in the earliest stages of retinal damage in the D2 mice.
Table 2.
Up-regulated genes in DBA/2J mouse retina (2 M vs 4 M).
| Accession number | Gene name | Fold change | Location | Type |
|---|---|---|---|---|
| Immune response/Inflammatory disease | ||||
| NM_009780 | Complement component 4B (C4B) | 2.0 | Extracellular space | Other |
| NM_007902 | Endothelin 2 (EDN2) | 3.2 | Extracellular space | Growth factor |
| NM_024406 | Fatty acid binding protein 4, adipocyte (FABP4) | 3.5 | Cytoplasm | Transporter |
| AK011851 | Guanine nucleotide binding protein (G protein), alpha 13 (GNA13) | 2.0 | Plasma membrane | Enzyme |
| NM_008491 | Lipocalin 2 (LCN2) | 4.3 | Extracellular space | Transporter |
| NM_013825 | Lymphocyte antigen 75 (LY75) | 2.0 | Plasma membrane | Other |
| NM_017372 | Lysozyme (renal amyloidosis) (LYZ) | 2.4 | Extracellular space | Enzyme |
| NM_010776 | Mannose-binding lectin (protein C) 2, soluble(opsonic defect)(MBL2) |
2.8 | Extracellular space | Other |
| AK020928 | Melan-A (MLANA) | 2.5 | Plasma membrane | Other |
| NM_023258 | PYD and CARD domain containing (PYCARD) | 2.1 | Cytoplasm | Transcription regulator |
| NM_027852 | Retinoic acid receptor responder (tazarotene induced) 2 (RARRES2) | 2.4 | Plasma Membrane | Transmembrane receptor |
| NM_007734 | Collagen, type IV, alpha 3 (Goodpasture antigen) (COL4A3) | 2.2 | Extracellular Space | Enzyme |
| Neurological disease | ||||
| NM_144784 | Acetyl-Coenzyme A acetyltransferase 1 (ACAT1) | 2.1 | Cytoplasm | Enzyme |
| NM_007670 | Cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) (CDKN2B) | 2.0 | Nucleus | Transcription regulator |
| NM_172119 | Deiodinase, iodothyronine, type III (DIO3) | 4.2 | Plasma membrane | Enzyme |
| K01347 | Glial fibrillary acidic protein (GFAP) | 2.4 | Cytoplasm | Other |
| NM_008128 | Gap junction protein, beta 6, 30 kDa (GJB6) | 5.2 | Plasma membrane | Transporter |
| NM_172778 | Monoamine oxidase B (MAOB) | 2.2 | Cytoplasm | Enzyme |
| NM_198190 | Neurotrophin 5 (NTF5) | 2.2 | Extracellular space | Growth factor |
| NM_009215 | Somatostatin (SST) | 2.1 | Extracellular space | Other |
| Cell signaling/Gene expression | ||||
| AK077820 | Aspartyl-tRNA synthetase (DARS) | 3.0 | Cytoplasm | Enzyme |
| AJ306425 | Hemochromatosis (HFE) | 2.1 | Plasma membrane | Transmembrane receptor |
| NM_010125 | E74-like factor 5 (ets domain transcription factor) (ELF5) | 3.3 | Nucleus | Transcription regulator |
| BC046759 | High-mobility group box 2 (HMGB2) | 2.1 | Nucleus | Transcription regulator |
| AB019029 | Mediator complex subunit 14 (MED14) | 2.4 | Nucleus | Transcription regulator |
| Cell death | ||||
| NM_008183 | Glutathione S-transferase M1 (GSTM1) | 2.3 | Cytoplasm | Enzyme |
| X80339 | SIX homeobox 1 (SIX1) | 2.4 | Nucleus | Transcription regulator |
Figure 2.
A) Significantly affected canonical pathways containing up-regulated genes in the retina of D2 mice at the age of 4 M. Bars represent-log (p-value) for over-representation of affected genes in the selected pathway. The yellow line represents the ratio of affected genes to the total number of genes in a pathway. Threshold (grey line) denotes the p = 0.05 level. B) List of genes in each of the pathways.
Among the 232 probes with decreased expression in 4 M D2 retinas, IPA analysis mapped 141 IDs, and found 65 genes that were eligible for function/pathway annotation. These genes were related to neurological diseases (Atxn2, Bcl2, BDNF, Drd4, MAPK10, etc), cell death/cell compromise (FAP, MYLK, SMOX, TNFRSF25), cell signaling/gene expression (CaMK4, Etv4, SLA) and ophthalmic diseases (Gpnmb) (Table 3). The 3 most significant pathways for these down-regulated genes were labeled by IPA software as: TR/RXR activation (CaMK4, Thrb, Dio2); hepatic fibrosis/hepatic stellate cell activation (Bcl2, Lama1, Myh2); and death receptor signaling (Bcl2, Tnfrsf25) (Fig. 3). Presumably, the etiology for some of the pathological processes in liver and retina share similarly programmed signaling pathways and gene regulatory networks.
Table 3.
Down-regulated genes in DBA/2J retina (2 M vs 4 M).
| Accession number | Gene name | Fold change | Location | Type |
|---|---|---|---|---|
| Neurological disease | ||||
| AK036209 | Ataxin 2 (ATXN2) | 2.3 | Nucleus | Other |
| AK052418 | Autism susceptibility candidate 2 (AUTS2) | 3.2 | Unknown | Other |
| AK007913 | B-cell CLL/lymphoma 2 (BCL2) | 2.2 | Cytoplasm | Other |
| XM_125706 | Breakpoint cluster region (BCR) | 3.1 | Cytoplasm | Kinase |
| NM_007540 | Brain-derived neurotrophic factor (BDNF) | 2.5 | Extracellular space | Growth factor |
| NM_020263 | Calcium channel, voltage-dependent, alpha 2/delta subunit 2 (CACNA2D2) | 2.3 | Unknown | Ion channel |
| AC115816 | Calcium channel, voltage-dependent, alpha 2/delta subunit 4 (CACNA4D4) | 2.4 | Unknown | Other |
| BC051421 | Dopamine receptor D4 (DRD4) | 2.2 | Plasma membrane | G-protein coupled receptor |
| NM_177273 | Gamma-aminobutyric acid (GABA) A receptor, gamma 3 (GABRG3) |
2.0 | Plasma membrane | Ion channel |
| NM_013559 | Heat shock 105 kDa/110 kDa protein 1 (HSPH1) | 2.1 | Cytoplasm | Other |
| NM_010606 | Potassium inwardly-rectifying channel, subfamily J, member 6 (KCNJ6) | 2.1 | Plasma membrane | Ion channel |
| AK082242 | Mitogen-activated protein kinase 10 (MAPK10) | 2.7 | Cytoplasm | Kinase |
| AK036827 | Nuclear factor of kappa light polypeptide gene enhancer (p105)(NFKB1) | 2.1 | Nucleus | Transcription regulator |
| NM_023333 | Protease, serine, 3 (PRSS3) (includes EG:5646) | 8.3 | Extracellular space | Peptidase |
| NM_011198 | Prostaglandin-endoperoxide synthase 2 (PTGS2) | 2.4 | Cytoplasm | Enzyme |
| AK088003 | SH3 domain containing ring finger 1 (SH3RF1) | 2.9 | Cytoplasm | Other |
| Cell death/Cellular compromise | ||||
| NM_007986 | Fibroblast activation protein, alpha (FAP) | 2.4 | Cytoplasm | Peptidase |
| AK044527 | Myosin light chain kinase (MYLK) | 2.4 | Cytoplasm | Kinase |
| AK087473 | Spermine oxidase (SMOX) | 2.5 | Cytoplasm | Enzyme |
| NM_033042 | Tumor necrosis factor receptor superfamily, member 25 (TNFRSF25) | 2.5 | Plasma membrane | Transmembrane receptor |
| AK051204 | Zinc finger E-box binding homeobox 2 (ZEB2) | 2.0 | Nucleus | Transcription regulator |
| NM_009380 | Thyroid hormone receptor, beta (THRB) | 2.2 | Nucleus | Ligand-dependent nuclear |
| AK046628 | Metastasis suppressor 1 (MTSS1) | 2.0 | Cytoplasm | Other |
| Cell signaling/Gene expression | ||||
| AK043457 | Calcium/calmodulin-dependent protein kinase IV (CAMK4) | 2.0 | Nucleus | Kinase |
| NM_008815 | Ets variant gene 4 (E1A enhancer binding protein, E1AF) (ETV4) | 2.4 | Nucleus | Transcription regulator |
| AK036518 | v-myb myeloblastosis viral oncogene homolog (MYB) | 2.1 | Nucleus | Transcription regulator |
| NM_001029841 | Src-like-adaptor (SLA) | 2.5 | Plasma membrane | Other |
| Ophthalmic disease | ||||
| AK089297 | Glycoprotein (transmembrane) nmb (GPNMB) | 3.6 | Plasma membrane | Enzyme |
Figure 3.
A) Most significant affected canonical pathways containing down-regulated genes in the retina of D2 mice at the age of 4 M. Bars represent -log(p-value) for over-representation of affected genes in the selected pathway. The yellow line represents the ratio of affected genes to the total number of genes in a pathway. Threshold (grey line) denotes the p = 0.05 level. B) List of genes in each of the pathways.
Trend in altered expression of genes
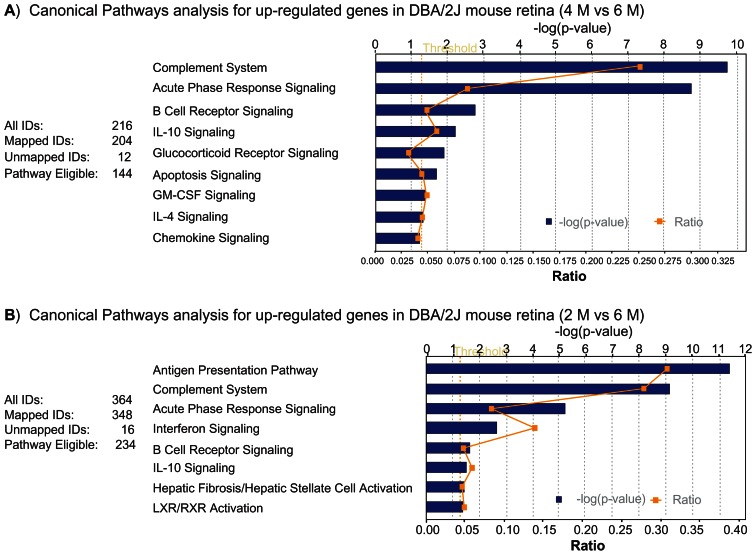
To monitor changes in gene expression values during the progression of the disease, D2 mice at the age of 6–7 M were included. The gene expression profiles were compared with those derived from the 4 M and the 2 M old mice, respectively. With disease progression and the onset of IOP elevation in D2 mice, 263 probes were differentially expressed in 6–7 M vs 4 M, with 216 up-regulated and 47 down-regulated. IPA showed that the top three pathways for up-regulated genes were related to complement regulation, acute phase responses and B cell receptor signaling pathways (Fig. 4A). Down-regulated genes were mainly associated with interferon signaling, complement regulation and coagulation systems.
Figure 4.
Trend in altered gene expression values. A) Canonical pathway for up-regulated genes in D2 retina (4 M vs. 6–7 M); B) Canonical pathway for up-regulated genes in D2 retina (2 M vs. 6–7 M). Bars represent-log (p-value) for over-representation of affected genes in the selected pathway. The yellow line represents the ratio of affected genes to the total number of genes in a pathway. Threshold (grey line) denotes the p = 0.05 level.
When compared with 2 M mice the 6 M mice had 464 differentially expressed probes, including 364 up-regulated and 100 down-regulated. IPA showed that the most significant pathways for the up-regulated genes were related to the antigen presentation pathway, the complement system and the acute phase response signaling pathway (Fig. 4B). The down-regulated genes were mainly involved in dopamine signaling, TR/RXR activation and the endoplasmic reticulum stress pathway.
Many genes already identified as differentially expressed in 4 M vs 2 M retinas showed more robust change in expression level at 6–7 M. Examples of these genes included Lcn2, Edn2 and GFAP.
Some genes that were differentially changed in 4 M vs 2 M, were not changed when 6–7 M vs 4 M, were compared. Examples of these genes included MR1 and C4.
Some genes that were found to be changed in their expression level at an early stage (4 M vs 2 M) decreased/ increased back to their baseline (2 M) level at 6–7 M. Example of these genes included MBL2.
Expression of genes related to the antigen presentation pathway in D2 retina
Antigen presentation is a process in the body’s immune system by which antigen presenting cells (APCs) present autoimmune or invading pathogen antigens to CD4 T cells using MHC class II molecules, and to CD8 T cells using MHC class I molecules on the APC cell surface. IPA showed that two genes (MR1 and PSMB8), related to the antigen presentation pathway, increased at 4 M relative to 2 M. With disease progression at 6–7 M, the expression levels of these two genes remained at their elevated levels. In addition, ten more genes related to the antigen presentation pathway were evidently increased (Table 4), including genes that coded for MHC class I and class II molecules. These data support the idea that there might be an “environment” for triggering adaptive immune responses in the early D2 mouse retina.
Table 4.
Antigen presentation pathway in DBA/2J mice.
| Gene | Fold change (vs 2 M) | |
|---|---|---|
|
|
||
| 4 M | 6 M | |
| Major histocompatibility complex, class I-related (MR1) | 2.7 | 2.5 |
| Beta-2-microglobulin (B2M) | – | 2.7 |
| Major histocompatibility complex, class I, C (HLA-C) | – | 4.1 |
| Major histocompatibility complex, class II, DM beta(HLA-DMB) | – | 4.1 |
| Major histocompatibility complex, class II, DQ beta2(HLA-DQB2) | – | 2.0 |
| Major histocompatibility complex, class I, E(HLA-E) | – | 2.4 |
| Major histocompatibility complex, class I, F(HLA-F) | – | 2.0 |
| Major histocompatibility complex, class I, G(HLA-G) | – | 2.2 |
| Proteasome subunit, beta type 8 (PSMB8) | 2.1 | 3.9 |
| Proteasome subunit, beta type 9 (PSMB9) | – | 3.0 |
| Transporter 1, ATP-binding cassette, subfamilyB (TAP1) | – | 2.0 |
| Transporter 2, ATP-binding cassette, subfamilyB (TAP2) | – | 2.1 |
Expression of complement system genes in the D2 retina
Although D2 mice showed deficiency in the C5 complement component14 which might lead to the compromise of terminal activation, expression of a few genes engaged in the early cascade of complement activation increased in D2 retinas at 4 M as compared with that in 2 M retina, including genes that coded for C2, C4B, complement factor I (CFI), and mannose-binding lectin 2 (MBL2). As the mice aged (6 M), increased expression of more genes in the complement system was observed, including genes that coded for subunits of C1, C2, factor B and factor H. These data indicated an early initiation of the complement system in retinal degeneration in the D2 mice.
Expression of acute phase response genes in D2 retina
Adult mammals typically respond to tissue damage through implementation of an acute phase response, which comprises a series of specific physiological reactions, such as inflammation.
Microarray analysis revealed an increased expression of genes related to an acute phase response at 4 M in the D2 mouse retinas. These genes included Lcn2, Edn2 and Lyz. GFAP was also found to be increased at this early time point. With disease progression, the expression of these early genes was more pronounced at 6–7 M. Moreover, up-regulation of many other acute phase response genes, such as SERPINA3, SERPING1, A2M, SOCS3 and CP, were also observed at this stage (Table 6). Since these genes are thought to be indicators of retinal stress and inflammatory responses, our data implicate an early involvement of inflammation in the retinal degeneration of D2 mice.
Table 6.
Acute phase response genes in DBA/2J mice.
| Gene | Fold change (vs 2 M) | |
|---|---|---|
|
|
||
| 4 M | 6 M | |
| Lipocalin 2 (LCN2) | 4.3 | 22.5 |
| Endothelin 2 (EDN2) | 3.2 | 33.4 |
| Lysozyme (LYZ) | 2.4 | 14.5 |
| Serpin peptidase inhibitor, clade A, member 3(SERPINA3) | – | 8.9 |
| Serpin peptidase inhibitor, clade G, member 1(SERPING1) | 1.5 | 6.4 |
| Alpha-2-macroglobulin (A2M) | – | 3.7 |
| Suppressor of cytokine signaling 3 (SOCS3) | – | 2.5 |
| Cerulopalsmin (CP) | – | 2.0 |
| Glial fibrillary acidic protein (GFAP) | 2.4 | 8.0 |
Validation of early gene expression by real time PCR
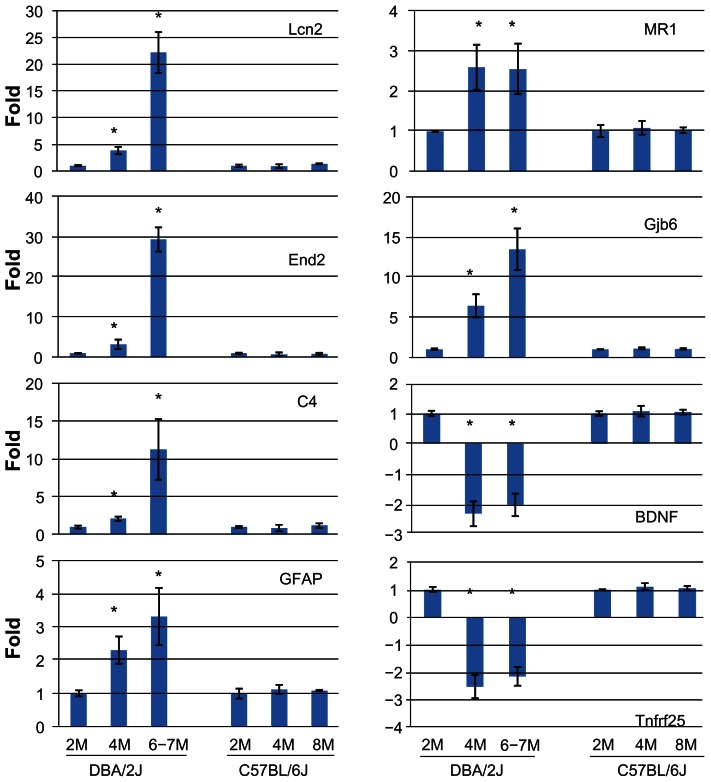
Eight genes, representative of the different implied pathways or functional groups, were selected for validation and confirmation with the aid of real time PCR. The same retinal RNA samples (n =3) as those used for the microarray analysis were used for the PCR. In addition, three individual RNA samples from individual retinas which were independent of those used for microarray were also included for further validation. The expression level of these genes was compared at 2, 4 and 6–7 M in an attempt to identify early changes that were IOP independent. We also looked for possible trends in the expression of these genes as the disease progressed. To control for the possible contribution of age to normal developmentally regulated changes in gene expression, we used retinal samples from age-matched B6 mice for comparison of gene expression values. Real time PCR confirmed the changes identified in the microarray studies (Fig. 5). For all eight selected genes, the level of their upregulation/downregulation detected by real time PCR was very close to that determined from microarray data. No significant changes in the expression of any of these genes were observed in B6 animals at either time point, i.e. 4 or 8 M, suggesting little contribution of age-related changes in expression of these genes.
Figure 5.
Real time PCR confirmation of selected genes identified by microarray analysis. Real time PCR was performed using individual retinal RNA samples from D2 animals at 2, 4, and 6–7 M (n =6 eyes for each time point), or B6 animals at 2, 4 and 8 M (n =3 eyes for each time point). The graphs show the average change relative to gene expression levels at 2 M (whose expression level was taken as 1) for either D2 or B6 mice. *Statistically significant differences (P < 0.05) by ANOVA.
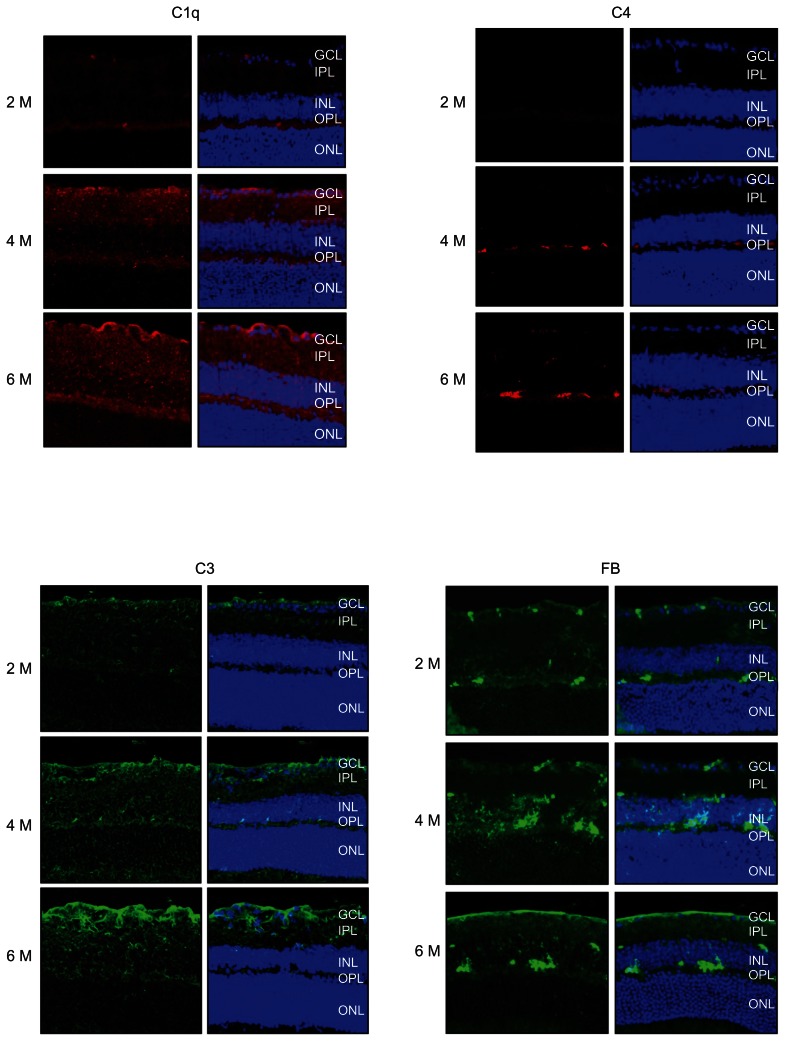
Immunocytochemistry
Immunocytochemical analysis was used to detect the presence and localization of protein products of some of the genes which had altered expression values. These included: C1q, C3, C4, factor B (FB) and Lcn2. As shown in Figure 6, these proteins were all present in the retina of D2 mice. C1q, C3 and FB revealed similar localization patterns, with immunolabled profiles detected from the inner limiting membrane, and the RGC layer through to the outer plexiform layer. Lcn2 and FB shared a similar staining pattern, with a strong presence in the outer plexiform layer and a less uniform expression in the inner plexiform and ganglion cell layers (Fig. 6). In addition, the immunolabeling intensities for all of these proteins were elevated progressively from 2 M to 4 M to 6–7 M.
Figure 6.
Immunofluorescence microscopy was used to detect the presence and localization of protein products for some of the genes shown to have altered expression values in the retinas of D2 mice. For each gene-product, 3 ages are shown (2, 4 and 6 M) with the left-hand column of figures, not counter-stained, and the right-hand column counter-stained with propidium iodide, to show cellular nuclei, and for visualization of the retinal cell layers. The gene products examined included: C1q, C3, C4, factor B and Lcn2. C1q, C3 and FB revealed similar patterns of localization, with immunolabeling evident from the inner limiting membrane, the GCL through the OPL. In addition, Lcn2 and C4 shared similar patterns of expression, but these proteins were mainly located in OPL with some scattered presence within the IPL. Bar: 20 μM.
Abbreviations: GCL, ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; OPL, outer plexiform layer; ONL, outer nuclear layer.
Early activation of micro-glial cells
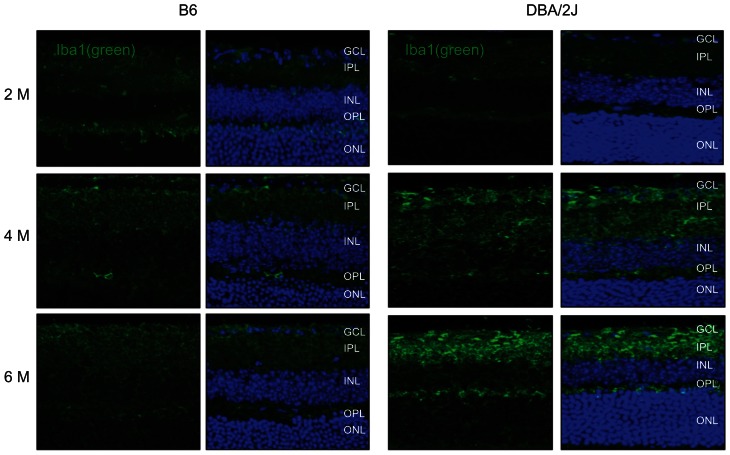
Results from this study pointed to the involvement of immune/inflammatory responses in the onset of retinal degeneration in D2 mice. These results raised the possibility of the involvement of microglial cells. To investigate this possibility, fixed tissue sections of retina from both D2 and B6 mice at the ages of 2 M, 4 M and 6–8 M were collected and immunolabeled for Iba1, a microglia-specific calcium-binding adaptor protein. Iba1 is involved in membrane ruffling through actin-bundling activity15,16 and through cell migration and phagocytosis17 and is up-regulated in activated microglia.
As shown in Figure 7, a more intense level of antibody binding to the Iba1 protein, was detected in the ganglion cell and inner plexiform layers in D2 retinas in 4 M compared to 2 M animals. At 6 M, the staining was even more intense from the GCL to the OPL. Microglia appeared less ramified and more rounded in the retinas of D2 mice at 6 M when compared to 2 M. These data support the notion that microglial cells may become activated in D2 mice. These changes were not evident in the age-matched B6 retinas.
Figure 7.
Activation of micro-glial cells in the D2 retina. Fixed tissue sections of retina from both D2 and B6 mice at the age of 2 M, 4 M and 6 M were collected and immunolabeled for the protein, Iba1 (green). An elevated level of antibody binding to Iba1 was detected in the GCL and IPL in D2 retinas at 4 M when compared to 2 M. At 6 M, the labeling was further elevated and now more extensive from the GCL to the OPL. Compared to 2 M retinas from the D2 mice, Iba1-labeled micro-glial cells were less ramified with round somata at 4 and 6 M of age. These changes were not evident in age-matched B6 retinas. Bar: 20 μM.
Abbreviations: GCL, ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; OPL, outer plexiform layer; ONL, outer nuclear layer.
Discussion
Early onset of retinal disease in D2 mice
This study has shown that there is a developmentally coordinated change in the expression of many genes in the retina of the D2 mice. These genes were altered independently of changes in IOP because they could be detected in 4 M mice with normal IOP when compared with 2 M mice with normal IOP. Further, these changes were not evident in the age matched B6 mice, and were not therefore, associated with normal postnatal development.
Most of the altered genes were shown to be related in gene-families and associated with gene pathways describing immune and stress responses, as well as acute stress responses. With the progression of disease, an increase in the IOP develops in the D2 mice as a result of iris atrophy, pigment dispersion and blockage of the outflow pathways for aqueous humor. The elevation of IOP is thought to be a primary actor in retinal degeneration in this mouse model of glaucoma. However, in view of the results presented here, it seems likely that the elevation in IOP may play a later, and perhaps aggravating role, to the events that are already set in motion at an earlier time point.
In addition to the observed changes in immune/ inflammatory response pathways, there was an altered expression of other genes known to be associated with other neurological diseases and cell signaling/ cell death pathways, and these too, were altered in the retinas of 4 M animals, and prior to the elevation in IOP. The interactions between these various functional groups of genes are of particular interest now. How such interactions may contribute to the full expression of glaucoma would be a worthy topic of future studies.
The early onset of retinal degeneration in D2 mice has been observed in previous studies.8,18 For example, by 3 M, an age at which increased IOP was far from being developed, RGC cell death characterized by the presence of electron-dense karioplasm and cytoplasm in these cells, was observed,18 and RGC-axon degeneration was also found. In addition, Müller cells with ultrastructural signs of enhanced activation, including the presence of fibrillae (presumably made of glial acidic fibrillary protein, GFAP) were found in retinas of 3 M and 6 M mice along with the apoptotic ganglion cells.18 Furthermore, the function of the inner retina of D2 mice also started to decrease at 3 M.8 The mechanism(s) underlying these early retinal degenerative changes is largely unknown, but the results from our current study provide some corroborating molecular evidence of such early events. Thus, the genes related to immune/inflammatory responses, neurological diseases, as well as those involved in cell signaling and death pathways, can be correlated with the onset of the neurodegenerative changes previously observed in the retinas of the D2 mouse model of glaucoma. We know of no clinical or anatomical evidence of an abnormality in the iris at 2, 4 or 6 M prior to the elevation of the IOP. However, we cannot rule out biochemical or molecular changes in the iris which may be associated with altered gene expression in either the anterior or posterior segments at that time.
Early involvement of immune/ inflammatory response genes
Antigen presentation pathway
Class I and II major histocompatibility complex (class I and II MHC) molecules are known to be important for antigen presentation in immune responses. These molecules are implicated in many neurodegenerative diseases, such as Alzheimer’s disease and Parkinson’s disease, etc.19–22 Upregulation of class II MHC molecules was observed in chronically activated glial cells (both microglial and GFAP-positive glial cells) in glaucomatous human eyes.23–25 In addition, class I MHC molecules were also found to be increased in another high IOP animal model.26 Class I MHC molecules are also expressed by neurons that undergo activity-dependent, long-term structural and synaptic modifications. Neurons expressing these molecules are most vulnerable to neurodegeneration in the CNS and Class I MHC molecules are reportedly expressed in RGCs in mouse retina.27 Our data show that Mr1 was up-regulated at early stages of retinal degeneration in D2 mice, and with disease progression, the expression of both class I and II MHC became more significant, suggesting a role for these molecules in both the onset and progression of the disease. However, failure by other studies to detect bone marrow derived cells in the retina raised questions of how and by what mechanisms these molecules play a role in pathogenesis of the retinal disease. This remains largely unknown and needs to be further investigated.
Involvement of complement in retinal degeneration
The complement system is a major component of innate immunity. The functions of complement system include destruction of invading pathogens, antigen opsonization, enhancement of the inflammatory response and facilitation of apoptotic cell removal. Complement has been indicated in neurodegeneration, such as Alzheimer’s disease,28,29 photoreceptor degeneration30 and diabetic retinopathy.31 Elevated expression of multiple complement components has been observed in the retinas of human glaucoma patients and in animal models of glaucoma.26,32,33 Changes in the expression of complement in D2 mice have also been reported in other studies. C1q and C4 have been shown to be up-regulated in D2 mice after the IOP becomes elevated.11 While our study confirms these previously reported findings, we also demonstrated an earlier involvement of complement in D2 mice. The present study showed an increase in genes encoding early complement components and also some of the complement regulatory proteins. This supports the likelihood that activation and regulation of this system occurs at an early stage in the retina of the D2 mice. It should be noted that there is a deficiency in the C5 complement component in the terminal activation pathway of D2 mice.14 Thus, the role of complement in D2 retinal degeneration may be more involved with the enhancement of inflammation and antigen opsonization rather than the formation of the membrane attack complex.
Early stress response genes and retinal degeneration
Previous studies show that stress response genes such as Lcn2, Edn2 and Serpina3 were up-regulated in rat and monkey glaucoma models with elevated IOP.26,32 In D2 mice, up-regulation of Lcn2 and Serpina3 was observed after IOP had increased.11 In addition, these stress genes were also found to be elevated in association with various types of photoreceptor injuries30 and loss.34 The present study demonstrated that these stress response genes increased during an early stage in the D2 mice. Lcn2, Edn2 and Serpina3 are thought to function as general signals in response to inflammatory stresses. Immunocytochemical labeling of fixed tissue sections of retina showed that Lcn2 is mostly located in the inner nuclear layer and also in scattered cells across the inner surface of retina. This localization pattern of Lcn2 in D2 retinas is similar to that observed in various photoreceptor injury models, in which the cells that express Lcn2 are presumed to be Muller cells. Considering that many immune/inflammatory response genes are upregulated in glia in response to retinal stress, it is possible that the glia could themselves be contributing to the precipitation of changes in gene expression in the D2 mice, but this remains any area that requires further investigation.
Activation of micro- and macro-glial cells in DBA/2J retina
The activation of macroglial cells including astrocytes and Müller cells with age and elevation of IOP in the D2 mouse retina has been reported previously.35 A recent study showed that microglial cells also were activated in the D2 retina. Inhibition of this activation with minocycline improved optic nerve integrity even though the intervention did not lower the IOP.36 Our study expanded on these previous studies and demonstrated that both macro-glia (GFAP↑) and micro-glia (IBa1 immunolabeling ↑) were activated at a very early stage (before the IOP increases). The activated glial cells in the retina may contribute to the number and elevation of immune/inflammatory response genes detected in the present study. However, this still leaves open the question of the initial triggering event. The underlying mechanism of this early activation is unclear, but may be related to the immune abnormalities in eyes of D2 mice.
Immune abnormalities in eyes of D2 mice
Eyes of D2 mice exhibit multiple abnormalities in ocular immune privilege, including breakdown of the blood/ocular barrier, an altered immunosuppressive microenvironment and the loss of anterior chamber-associated immune deviation.37 Some of these changes in immune privilege are observed as early as 2–4 M and substantially precede pigment dispersion and IOP elevation. Further, the prevention of iris abnormalities in D2 recipients of B6D2F1 bone marrow indicates a pathogenic role for the immune system. Immune abnormalities may contribute to glaucomatous changes in D2 mice.36,38,39 Recently, the mechanisms underlying the immune abnormalities and its contribution to glaucoma in D2 mice were studied.40 Of the two mutant genes in D2 mice, it is likely that Gpnmb is the candidate gene which influences iris disease and leads to the elevation of IOP. Gpnmb influences the glaucoma phenotype of D2 mice by a bone-marrow derived mechanism that does not require adaptive immunity, suggesting GPNMB deficiency may influence innate immunity.40 GPNMB localizes to pigment producing cells, bone-marrow derived antigen-presenting cells (APCs) of the iris and also dendritic cells. The mutation of Gpnmb in D2 mice creates a premature stop codon, and as a result of nonsense-mediated mRNA decay, the levels of both the Gpnmb transcript and its associated GPNMB protein are significantly decreased in the iris of D2 mice, when compared to age and sex matched wildtype controls.40 It should be noted that GPNMB is also present in the neuronal retina.40 This is consistent with our findings that the Gpnmb transcript was detected in retinal samples of D2 mice. Furthermore, we found that the level of the Gpnmb transcript in the retina decreased at 4M. The results described here imply that a deficiency of GPNMB protein within the retina itself could contribute to the onset of the processes underlying the neurodegeneration. Although the precise mechanism remains to be identified, we have shown that it could involve the stimulation of the inflammatory and immune signaling pathways.
Table 5.
Complement system in DBA/2J mice.
| Gene | Fold change (vs. 2 M) | |
|---|---|---|
|
|
||
| 4 M | 6 M | |
| Complement component 1, q subcomponent, A chain(C1QA) | – | 3.2 |
| Complement component 1, q subcomponent, C chain(C1QC) | – | 3.8 |
| Complement component 1, r subcomponent(C1R) | – | 5.8 |
| Complement component 1, s subcomponent(C1S) | – | 2.7 |
| Complement component 2 | 1.2 | 1.3 |
| Complement component 3(C3) | – | 6.6 |
| Complement component 4B(C4B) | 2.1 | 11.1 |
| Complement factor B(CFB) | – | 10.1 |
| Complement factor H(CFH) | – | 2.3 |
| Complement factor I(CFI) | 1.5 | 4.0 |
| Mannose-binding lectin (protein C) 2 (MBL2) | 2.8 | – |
Acknowledgments
Support from NIH grants R01EY017594 (N.G.F.C.), 7R03EY014578 (J.S.M.) and Research to Prevent Blindness, Inc., NYC, NY and the Kentucky Research Challenge Fund (H.J.K.) are gratefully acknowledged. Thanks also to the University of Louisville Microarray Facility supported by P20RR016481 and the Center for Environmental Genetics and Integrative Biology—Bioinformatics, Biostatistics and Computational Core supported by P30ES014443.
Footnotes
Disclosures
The authors report no conflicts of interest.
References
- 1.Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 2006;90:262–7. doi: 10.1136/bjo.2005.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.John SW, Smith RS, Savinova OV, et al. Essential iris atrophy, pigment dispersion, and glaucoma in DBA/2J mice. Invest Ophthalmol Vis Sci. 1998;39:951–62. [PubMed] [Google Scholar]
- 3.Anderson MG, Smith RS, Hawes NL, et al. Mutations in genes encoding melanosomal proteins cause pigmentary glaucoma in DBA/2J mice. Nat Genet. 2002;30:81–5. doi: 10.1038/ng794. [DOI] [PubMed] [Google Scholar]
- 4.Anderson MG, Libby RT, Mao M, et al. Genetic context determines susceptibility to intraocular pressure elevation in a mouse pigmentary glaucoma. BMC Biol. 2006;4:20. doi: 10.1186/1741-7007-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang B, Smith RS, Hawes NL, et al. Interacting loci cause severe iris atrophy and glaucoma in DBA/2J mice. Nat Genet. 1999;21:405–9. doi: 10.1038/7741. [DOI] [PubMed] [Google Scholar]
- 6.Libby RT, Anderson MG, Pang IH, et al. Inherited glaucoma in DBA/2J mice: pertinent disease features for studying the neurodegeneration. Vis Neurosci. 2005;22:637–48. doi: 10.1017/S0952523805225130. [DOI] [PubMed] [Google Scholar]
- 7.Libby RT, Li Y, Savinova OV, et al. Susceptibility to neurodegeneration in a glaucoma is modified by bax gene dosage. PLoS Genet. 2005;1:e4. doi: 10.1371/journal.pgen.0010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saleh M, Nagaraju M, Porciatti V. Longitudinal evaluation of retinal ganglion cell function and IOP in the DBA/2J mouse model of glaucoma. Invest Ophthalmol Vis Sci. 2007;48:4564–72. doi: 10.1167/iovs.07-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson MG, Libby RT, Gould DB, Smith RS, John SW. High-dose radiation with bone marrow transfer prevents neurodegeneration in an inherited glaucoma. Proc Natl Acad Sci U S A. 2005;102:4566–71. doi: 10.1073/pnas.0407357102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scholz M, Buder T, Seeber S, Adamek E, Becker CM, Lutjen-Drecoll E. Dependency of Intraocular Pressure Elevation and Glaucomatous Changes in DBA/2J and DBA/2J-Rj Mice. Invest Ophthalmol Vis Sci. 2008;49:613–21. doi: 10.1167/iovs.07-0745. [DOI] [PubMed] [Google Scholar]
- 11.Steele MR, Inman DM, Calkins DJ, Horner PJ, Vetter ML. Microarray analysis of retinal gene expression in the DBA/2J model of glaucoma. Invest Ophthalmol Vis Sci. 2006;47:977–85. doi: 10.1167/iovs.05-0865. [DOI] [PubMed] [Google Scholar]
- 12.Wang WH, Millar JC, Pang IH, Wax MB, Clark AF. Noninvasive measurement of rodent intraocular pressure with a rebound tonometer. Invest Ophthalmol Vis Sci. 2005;46:4617–21. doi: 10.1167/iovs.05-0781. [DOI] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-[Delta][Delta]CT Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Nilsson UR, Muller-Eberhard HJ. Deficiency of the fifth componenet in mice with an inherited complement defect. J Exp Med. 1967;125:1–16. doi: 10.1084/jem.125.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito D, Imai Y, Ohsawa K, Nakajima K, Fukuuchi Y, Kohsaka S. Microgliaspecific localisation of a novel calcium binding protein, Iba1. Brain Res Mol Brain Res. 1998;57:1–9. doi: 10.1016/s0169-328x(98)00040-0. [DOI] [PubMed] [Google Scholar]
- 16.Sasaki Y, Ohsawa K, Kanazawa H, Kohsaka S, Imai Y. Iba1 is an actin-cross- linking protein in macrophages/microglia. Biochem Biophys Res Commun. 2001;286:292–7. doi: 10.1006/bbrc.2001.5388. [DOI] [PubMed] [Google Scholar]
- 17.Ohsawa K, Imai Y, Kanazawa H, Sasaki Y, Kohsaka S. Involvement of Iba1 in membrane ruffling and phagocytosis of macrophages/microglia. J Cell Sci. 2000;113(Pt 17):3073–84. doi: 10.1242/jcs.113.17.3073. [DOI] [PubMed] [Google Scholar]
- 18.Schuettauf F, Rejdak R, Walski M, et al. Retinal neurodegeneration in the DBA/2J mouse-a model for ocular hypertension. Acta Neuropathol. 2004;107:352–8. doi: 10.1007/s00401-003-0816-9. [DOI] [PubMed] [Google Scholar]
- 19.Candore G, Balistreri CR, Colonna-Romano G, Lio D, Caruso C. Major histocompatibility complex and sporadic Alzheimer’s disease: a critical reappraisal. Exp Gerontol. 2004;39:645–52. doi: 10.1016/j.exger.2003.10.027. [DOI] [PubMed] [Google Scholar]
- 20.Kadish I, Thibault O, Blalock EM, et al. Hippocampal and cognitive aging across the lifespan: a bioenergetic shift precedes and increased cholesterol trafficking parallels memory impairment. J Neurosci. 2009;29:1805–16. doi: 10.1523/JNEUROSCI.4599-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benner EJ, Banerjee R, Reynolds AD, et al. Nitrated alpha-synuclein immunity accelerates degeneration of nigral dopaminergic neurons. PLoS ONE. 2008;3:e1376. doi: 10.1371/journal.pone.0001376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orr CF, Rowe DB, Mizuno Y, Mori H, Halliday GM. A possible role for humoral immunity in the pathogenesis of Parkinson’s disease. Brain. 2005;128:2665–74. doi: 10.1093/brain/awh625. [DOI] [PubMed] [Google Scholar]
- 23.Hernandez MR. The optic nerve head in glaucoma: role of astrocytes in tissue remodeling. Prog Retin Eye Res. 2000;19:297–321. doi: 10.1016/s1350-9462(99)00017-8. [DOI] [PubMed] [Google Scholar]
- 24.Tezel G, Chauhan BC, LeBlanc RP, Wax MB. Immunohistochemical Assessment of the Glial Mitogen-Activated Protein Kinase Activation in Glaucoma. Invest Ophthalmol Vis Sci. 2003;44:3025–33. doi: 10.1167/iovs.02-1136. [DOI] [PubMed] [Google Scholar]
- 25.Yang J, Yang P, Tezel G, Patil RV, Hernandez MR, Wax MB. Induction of HLA-DR Expression in Human Lamina Cribrosa Astrocytes by Cytokines and Simulated Ischemia. Invest Ophthalmol Vis Sci. 2001;42:365–371. [PubMed] [Google Scholar]
- 26.Ahmed F, Brown KM, Stephan DA, Morrison JC, Johnson EC, Tomarev SI. Microarray analysis of changes in mRNA levels in the rat retina after experimental elevation of intraocular pressure. Invest Ophthalmol Vis Sci. 2004;45:1247–58. doi: 10.1167/iovs.03-1123. [DOI] [PubMed] [Google Scholar]
- 27.Huh GS, Boulanger LM, Du H, Riquelme PA, Brotz TM, Shatz CJ. Functional requirement for class I MHC in CNS development and plasticity. Science. 2000;290:2155–9. doi: 10.1126/science.290.5499.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fonseca MI, Ager RR, Chu SH, et al. Treatment with a C5aR antagonist decreases pathology and enhances behavioral performance in murine models of Alzheimer’s disease. J Immunol. 2009;183:1375–83. doi: 10.4049/jimmunol.0901005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sjoberg AP, Trouw LA, Blom AM. Complement activation and inhibition: a delicate balance. Trends Immunol. 2009;30:83–90. doi: 10.1016/j.it.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 30.Rattner A, Nathans J. The Genomic Response to Retinal Disease and Injury: Evidence for Endothelin Signaling from Photoreceptors to Glia. J Neurosci. 2005;25:4540–9. doi: 10.1523/JNEUROSCI.0492-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerhardinger C, Costa MB, Coulombe MC, Toth I, Hoehn T, Grosu P. Expression of acute-phase response proteins in retinal Muller cells in diabetes. Invest Ophthalmol Vis Sci. 2005;46:349–57. doi: 10.1167/iovs.04-0860. [DOI] [PubMed] [Google Scholar]
- 32.Miyahara T, Kikuchi T, Akimoto M, Kurokawa T, Shibuki H, Yoshimura N. Gene microarray analysis of experimental glaucomatous retina from cynomologous monkey. Invest Ophthalmol Vis Sci. 2003;44:4347–56. doi: 10.1167/iovs.02-1032. [DOI] [PubMed] [Google Scholar]
- 33.Stasi K, Nagel D, Yang X, et al. Complement Component 1Q (C1Q) Upregulation in Retina of Murine, Primate, and Human Glaucomatous Eyes. Invest Ophthalmol Vis Sci. 2006;47:1024–9. doi: 10.1167/iovs.05-0830. [DOI] [PubMed] [Google Scholar]
- 34.Swiderski RE, Nishimura DY, Mullins RF, et al. Gene expression analysis of photoreceptor cell loss in bbs4-knockout mice reveals an early stress gene response and photoreceptor cell damage. Invest Ophthalmol Vis Sci. 2007;48:3329–40. doi: 10.1167/iovs.06-1477. [DOI] [PubMed] [Google Scholar]
- 35.Inman DM, Horner PJ. Reactive nonproliferative gliosis predominates in a chronic mouse model of glaucoma. Glia. 2007;55:942–53. doi: 10.1002/glia.20516. [DOI] [PubMed] [Google Scholar]
- 36.Bosco A, Inman DM, Steele MR, et al. Reduced retina microglial activation and improved optic nerve integrity with minocycline treatment in the DBA/2J mouse model of glaucoma. Invest Ophthalmol Vis Sci. 2008;49:1437–46. doi: 10.1167/iovs.07-1337. [DOI] [PubMed] [Google Scholar]
- 37.Mo JS, Anderson MG, Gregory M, et al. By altering ocular immune privilege, bone marrow-derived cells pathogenically contribute to DBA/2J pigmentary glaucoma. J Exp Med. 2003;197:1335–44. doi: 10.1084/jem.20022041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou X, Li F, Kong L, Tomita H, Li C, Cao W. Involvement of inflammation, degradation, and apoptosis in a mouse model of glaucoma. J Biol Chem. 2005;280:31240–8. doi: 10.1074/jbc.M502641200. [DOI] [PubMed] [Google Scholar]
- 39.Zhou X, Li F, Kong L, Chodosh J, Cao W. Anti-inflammatory effect of pigment epithelium-derived factor in DBA/2J mice. Mol Vis. 2009;15:438–50. [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson MG, Nair KS, Amonoo LA, et al. GpnmbR150X allele must be present in bone marrow derived cells to mediate DBA/2J glaucoma. BMC Genet. 2008;9:30. doi: 10.1186/1471-2156-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]