Abstract
Cytochrome P450 (P450)1A1 plays a critical role in the metabolic activation and detoxification of polycyclic aromatic hydrocarbons (PAHs), many of which are potent human carcinogens. In this investigation, we tested the hypothesis that MC elicits persistent induction of CYP1A1 expression in human hepatoma cells (HepG2) and that this phenomenon is mediated by sustained transcriptional activation of the CYP1A1 promoter. Treatment of HepG2 cells with MC resulted in marked induction (8–20-fold) of ethoxyresorufin O-de-ethylase activities, CYP1A1 apoprotein contents, and mRNA levels, which persisted for up to 96 h. MC also caused sustained transcriptional activation of the human CYP1A1 promoter for up to 96 h, as inferred from transient transfection experiments. Experiments with deletion constructs indicated that Ah response elements located at −886, −974, and −1047, but not −491, nucleotides from the start site, contributed to the sustained transcriptional activation of the CYP1A1 promoter. Electrophoretic mobility-shift and chromatin immunoprecipitation assays suggested that prolonged CYP1A1 induction was mediated by Ah receptor (AHR)-independent mechanisms. Experiments with [3H]MC and liquid chromatography-tandem mass spectrometry demonstrated rapid elimination of MC and its metabolites from the cells by 12 to 24 h, suggesting that these compounds did not elicit sustained CYP1A1 induction via the classical AHR-mediated pathway. In conclusion, the results of this study support the hypothesis that MC causes persistent induction of CYP1A1 in human hepatoma cells by mechanisms entailing sustained transcriptional activation of the CYP1A1 promoter via AHR-independent mechanisms. These observations have important implications for human carcinogenesis mediated by PAHs.
Polycyclic aromatic hydrocarbons (PAHs) are found in coke ovens, cigarette smoke, diesel exhausts, and charcoal-broiled meats, and numerous PAHs are suspected carcinogens (Hemminki, 1990). 3-Methylcholanthrene (MC) is one of the most potent PAH carcinogens known to date (Harvey, 1982). Metabolism of MC by cytochrome P450 (P450)1 enzymes leads to the formation of highly reactive intermediates that can bind covalently to DNA, a critical step in the initiation of carcinogenesis (Guengerich, 1988). CYP1A enzymes also play an important role in the detoxification of PAHs (Nebert et al., 2004).
The CYP1A subfamily comprises two proteins, CYP1A1 and CYP1A2, that play important roles in carcinogen bioactivation (Guengerich, 1988). The hepatic P450 1A1/1A2 enzymes are inducible by several chemicals, including the hepatocarcinogen 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) (Poland and Glover, 1974; Postlind et al., 1993), MC (Poland and Glover, 1974; Moorthy, 2000, 2008; Kondraganti et al., 2002), benzo[a]pyrene (Conney 1986), and cigarette smoke (Kawajiri et al., 1991).
The molecular mechanisms by which MC or TCDD transcriptionally activates the CYP1A1 gene involve the binding of the ligand to the Ah receptor (AHR), a cytosolic protein. The ligand-bound AHR complex translocates into the nucleus, where it dimerizes with the Ah receptor nuclear translocator (ARNT), and this heterodimer binds to xenobiotic-responsive elements or aryl hydrocarbon responsive elements (AHREs), which are located in enhancer regions of CYP1A1, resulting in enhanced expression of the CYP1A1 gene (Nebert et al., 2004; Fujii-Kuriyama and Mimura, 2005; Nukaya et al., 2009). The core AHRE sequences (5′-TNGCGTG-3′) are present at multiple locations along the promoter/enhancer (Nukaya et al., 2009) regions of the CYP1A1 gene.
The mechanistic events occurring during the induction process have been investigated extensively, but relatively little is known regarding the decline of drug-metabolizing enzymes to preinduction levels after termination of xenobiotic exposure. In rats, a single dose of MC (20–25 mg/kg) induces arylhydrocarbon hydroxylase activity (Poland and Glover, 1974) and CYP1A1 mRNA expression (Bresnick et al., 1981) after 24 h, followed by return of enzyme activity and mRNA levels within 5 to 7 days. Conversely, Boobis et al., (1977) showed that a single dose of MC (80 mg/kg) increases total P450 content and aryl hydrocarbon hydroxylase activities in mice for up to 9 days. Masaki et al. (1984) reported increased total hepatic P450 contents in rats up to 8 days after multiple administrations of MC, and DePierre et al. (1982) showed induction of total hepatic P450 contents that persisted for 2 to 3 weeks.
We reported previously that MC elicits a long-term induction of hepatic CYP1A enzymes in rats for up to 45 days (Moorthy et al., 1993). Furthermore, we demonstrated that the sustained induction of CYP1A enzymes is mediated by mechanisms other than the persistence of the parent compound (Moorthy, 2000). We also showed that MC elicits prolonged induction of CYP1A enzymes in mice (Kondraganti et al., 2002); and by using Cyp1a2-null mice, it was observed that CYP1A2 plays an important role in this phenomenon (Kondraganti et al., 2002; Moorthy, 2002, 2008). In addition, we showed that in rats MC causes sustained induction of phase II enzymes, which are also encoded by the Ah gene battery, presumably by persistent activation of the AHR (Kondraganti et al., 2008). Recently, we reported that MC elicits sustained CYP1A1 and 1A2 induction in vivo via transcriptional activation of the corresponding promoters (Jiang et al., 2009).
CYP1A1 induction has been extensively studied in vitro systems. In earlier studies in fetal cells from hamster, the PAH benz[a]anthracene was shown to elicit induction of arylhydrocarbon hydroxylase linearly for 12 to 16 h, which reached peak level during the next 24 h (Nebert and Gelboin, 1970). MC induces CYP1A1 in rodent (Xu et al., 1993; Riddick et al., 1994) and human (Hines et al., 1988; Postlind et al., 1993; Lekas et al., 2000) hepatoma cell lines for short periods of time (4–36 h), but little is known regarding the persistent induction of CYP1A1 by MC in human cells. Therefore, in this study, we tested the hypothesis that MC elicits persistent induction of CYP1A1 in human hepatoma cells (HepG2) and that this phenomenon is mediated by sustained transcriptional activation of the CYP1A1 promoter.
Materials and Methods
Chemicals.
MC, calcium chloride, Tris, sucrose, NADP+, ethoxyresorufin, glucose 6-phosphate, and glucose 6-phosphate dehydrogenase were purchased from Sigma-Aldrich (St. Louis, MO). Polyvinylidene difluoride membranes and buffer components for electrophoresis and Western blotting were obtained from Bio-Rad Laboratories (Hercules, CA). The primary monoclonal antibody to CYP1A1 was a gift from Dr. P. E. Thomas (Rutgers University, Piscataway, NJ). Goat anti-mouse IgG conjugated with horseradish peroxidase was obtained from Bio-Rad Laboratories. Anti-AHR antibodies were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). [γ-32P]ATP was obtained from PerkinElmer Life and Analytical Sciences (Boston, MA). CYP1A1-pSV2-CAT promoter construct was kindly provided by Dr. A. K. Jaiswal (University of Maryland, Baltimore, MD). pGL3 enhanced vector constructs and luciferase kits were purchased from Promega (Madison, WI). Plasmids containing various deletions of the CYP1A1 promoter were provided by Dr. X. Coumoul (University of Paris, Paris, France). Randomly ring-labeled [3H]MC (radiochemical purity, >99.9%) was from Moravek Biochemicals (Brea, CA). MC metabolites were purchased from Chemsyn Laboratories (Lawrence, KS).
Cell Culture and Treatments.
HepG2 cells were procured from American Type Culture Collection (Manassas, VA). The cells were maintained in α-minimal Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, 100 mM sodium pyruvate, and 200 mM l-glutamine (Invitrogen, Carlsbad, CA), at 37°C and 5% CO2. The cells were plated overnight at a density of 106 to 107 cells/dish in 100-mm culture dishes. Cells that were at ∼50% confluence were treated in triplicate with MC (2.5 μM) dissolved in DMSO or with DMSO alone as controls and harvested at 6, 12, 24, 48, 72, or 96 h after MC or DMSO exposure. Media were not changed during the course of the treatment. All cell treatment experiments were conducted in the dark to prevent light-induced activation of the AHR. Treated cell lysates were immediately stored at −80°C until analysis. Some MC- or DMSO-treated cells were used for isolation of nuclei. For experiments entailing [3H]MC, HepG2 cells were exposed to [3H]MC (2.5 μM; specific activity, 1.9 mCi/mmol), and levels of parent MC and its metabolites in cell extracts and media were determined by thin-layer chromatography, followed by radiometric analyses of the TLC zone corresponding to the parent MC, as reported previously (Moorthy, 2000). For studies involving MC metabolites, cells were treated with 1 μM concentrations of one of the following metabolites: 1-hydroxy-3-MC; 2-hydroxy-3-MC (2-OH-MC); 11-hydroxy-3-MC; 11,12-dihydro-11,12-dihydroxy-3-MC; 1-one-3-MC; 11,12 epoxy-11,12 dihydro-3-MC; 11,12-dione-3-MC; and 11,12-dialdehyde-3-MC. The cells were harvested after 24 h, followed by determination of ethoxyresorufin O-de-ethylase (EROD) activities as described above. Each experiment was run in triplicate. Cell count and viability were determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) (MTT) reduction assays, as outlined in the MTT Reduction Assay protocol (American Type Culture Collection), as reported previously (Bhakta et al., 2008).
LC-MS/MS Analyses of Parent MC in Cells and Media.
Levels of MC in cells and media were also analyzed by LC-MS/MS using an Acuity TQD mass spectrometer (Waters, Milford, MA) equipped with a diode array UV detector. Treatment of cells with MC was as described under Cell Culture and Treatments. Each experiment was run in triplicate. Quantification standards for MC were prepared with nominal concentrations of MC in methanol and 0.2% formic acid in water, and UV and MS/MS peak areas were established. Controls for the assays were blank samples without MC along with methanol blanks to monitor MC column carryover. Quantification was performed by comparison of sample MC peak areas to quantification curve constructed from nominal MC concentrations and their respective peak areas using QuanLynx software (Waters). The identity of the MC UV peak was confirmed on the basis of retention time and precursor-product ion transition measured by mass spectrometry [m/z 269.2 > 253.9, corresponding to M + 1 and M + 1–15]; retention time at UV was 2.37 min.
Cell culture media and cell pellets were extracted using methyl tertiary butyl ether (MTBE). In brief, the cells were washed with PBS, trypsinized, and centrifuged at 3000 rpm for 15 min. The cell pellets were suspended in 0.5 ml of PBS and transferred to 12-ml glass screw-capped tubes. An additional 0.5 ml of PBS was added, and the cells were sonicated and extracted with 0.5 ml of MTBE. The tubes were centrifuged at 4000 rpm, and the upper organic layer was removed. The aqueous layer was re-extracted with MTBE, and the combined MTBE extracts were evaporated to dryness and reconstituted with 100 μl of methanol and 100 μl of 0.2% formic acid. Samples were than analyzed by LC-MS/MS. MC from the media was analyzed similarly to that described for cell pellets, except that the samples were not sonicated.
Isolation of Nuclei.
Nuclei and nuclear protein extracts from whole cells were prepared using commercially available kits (Pierce Chemical, Rockford, IL), following the manufacturer's instructions. The nuclear protein extracts were stored at −80°C until use.
Enzyme Assays.
EROD (CYP1A1) activities in whole cell lysates were assayed essentially as described previously (Moorthy, 2000; Kondraganti et al., 2002; Bhakta et al., 2008). Each assay was conducted in duplicate using cell extracts from at least three independent experiments.
Electrophoresis and Western Blotting.
Whole cell protein (10 μg) were subjected to SDS-polyacrylamide gel electrophoresis in 10% acrylamide gel followed by Western blotting, as reported in previous articles from our laboratory (Moorthy, 2000; Kondraganti et al., 2002; Bhakta et al., 2008). Detection of CYP1A1 proteins on the Western blots was accomplished by the use of a monoclonal antibody to CYP1A1 that cross-reacts with CYP1A2 as the primary antibody and goat anti-mouse IgG conjugated with horseradish peroxidase as the secondary antibody. The blots were developed with the horseradish peroxidase color reagent 4-chloro-1-naphthlol or by chemiluminescence (Moorthy, 2000; Kondraganti et al., 2002; Bhakta et al., 2008). β-Actin was used as a loading control. The bands corresponding to CYP1A1 were quantified by densitometry and normalized against β-actin controls. The Western blots were conducted in cell extracts from at least three independent experiments.
Real-Time Reverse Transcriptase-Polymerase Chain Reaction.
Total RNA was isolated from cultured HepG2 cells using the TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Total RNA (500 ng) from MC-treated HepG2 cells and DMSO-treated controls was subjected to one-step real-time quantitative TaqMan RT-PCR. The Prism 7700 Sequence Detection System (Applied Biosystems, Foster City, CA) was used for the RT-PCRs. Gene-specific primers in the presence of TaqMan reverse transcription reagents and RT reaction mix (Applied Biosystems) were used to reverse transcribe RNA, and TaqMan Universal PCR Master Mix and Assays-on-Demand Gene Expression probes (Applied Biosystems) were used for PCR amplification. After an RT hold for 30 min at 48°C, the samples were denatured at 95°C for 10 min. The thermal cycling step was for 40 cycles at 95°C for 15 s, and 40 cycles at 60°C for 1 min. Serial dilutions were used to optimize and validate RT-PCR conditions for CYP1A1 and 18S genes. Each data point was repeated three times. Quantitative values were obtained from the threshold PCR cycle number at which the increase in signal was associated with an exponential growth of PCR product after it starts to be detected. The relative mRNA levels for CYP1A1 were normalized to their 18S content. The relative expression levels of the target gene were calculated as reported previously (Jiang et al., 2004; Bhakta et al., 2008).
Construction of Reporter Gene.
The pGL3-1A1 promoter construct was constructed by inserting a 1.6-kilobase human CYP1A1 promoter into the HindIII site of the pGL3 enhancer vector (Promega), followed by ligation and transformation into Escherichia coli, as reported previously (Moorthy et al., 2007).
Construction of Deletion Plasmids.
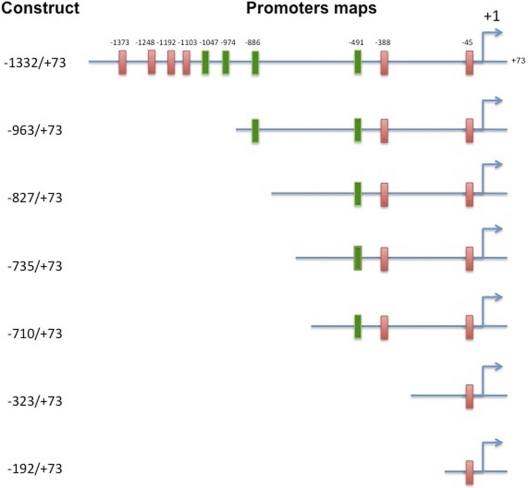
We used exonuclease III, which is only active on a blunt end, 5′-overhang or nick (not active on 3′-overhangs) to perform nested sets of unidirectional random deletions of the original human CYP1A1 plasmid (−1556/+73) (Coumoul et al., 2001). Target DNA was double-restricted to produce one 5′-overhang end (with NdeI) and one 3′-overhang (with KpnI). After purification and removal of each restriction enzyme, exonuclease III (250 units) was added to 5 μg of linearized plasmid (50 μl) at 25°C (digestion rate, 100 bp · min−1). Aliquots of the reaction (3 μl) were transferred to tubes containing fresh nuclease S1 mixture (1 U, 27 μl) (MBI Fermentas, Hanover, MD) on ice to randomly stop the reaction. The samples were transferred to fresh tubes at room temperature, and enzymatic reactions were continued for an additional 30 min to carry out nuclease S1 reaction of single-stranded DNA. After purification, plasmids were recircularized with T4 DNA ligase, produced in large-scale quantities, and sequenced. This resulted in random deletions of plasmids that contained −1332, −963, −827, −735, −710, −323, and −193 bp of the CYP1A1 promoter region.
We also constructed the plasmid pGL3-521 that contains only one functional AHRE (−491) by subjecting the construct pGL3-1332 plasmid to HindIII digestion, purification, and ligation. All the constructs were inserted into the HindIII site of the pGL3 enhancer vector, followed by ligation and transformation as reported previously (Moorthy et al., 2007).
Transient Transfections and Reporter Gene Assays.
Transfection of pGL3-1A1 parent plasmid (2.5 μg) or those carrying deletions into HepG2 cells was performed using Lipofectamine (Invitrogen) when cell density reached 50% confluence. To normalize for transfection efficiency, cells were cotransfected with plasmids (pRL-TK; 1.5 μg) expressing Renilla reniformis luciferase. Dual-luciferase assay (firefly and R. reniformis) was performed using a Promega kit (Promega). The R. reniformis luciferase activity was used to normalize the transfection efficiency in all culture dishes, as reported previously (Moorthy et al., 2007; Bhakta et al., 2008). At least three independent transfections were conducted for each time point.
Electrophoretic Mobility-Shift Assays.
EMSAs were performed using nuclear extracts as described previously (Moorthy, 2000; Bhakta et al., 2008). In brief, the nuclear proteins (10 μg) were preincubated with poly(dI-dC) (2 μg) on ice for 10 min, followed by incubation at room temperature for 30 min with [γ-32P]ATP-labeled double-stranded oligonucleotides that contain the AHREs (Pendurthi et al., 1993) in the presence of EMSA buffer (25 mM HEPES, 0.5 mM EDTA, 0.5 mM dithiothreitol, 10% glycerol, 50 mM KCl, and 5 mM MgCl2, pH 7.5). In some experiments, the nuclear extracts were incubated with a 50-fold excess of cold AHRE-specific oligonucleotide, before the addition of γ-32P-labeled probe. The labeled samples were separated by nondenaturing polyacrylamide gel electrophoresis (6%) at 200 V for 3 h. The gel was dried, and radioactivity was located by autoradiography using Kodak X-ray film. The EMSAs were performed using nuclear extracts from cells treated in triplicate with DMSO or MC in three independent experiments for each time point.
Chromatin Immunoprecipitation Assays.
ChIP assays were performed to assess specific binding of MC-AHR-ARNT complex to the AHREs on the CYP1A1 promoter. These assays were performed essentially as described by Hestermann and Brown (2003), using the ChIP-IT Express Enzymatic Magnetic Chromatin Immunoprecipitation kits (Active Motif Inc., Carlsbad, CA). HepG2 cells were treated with MC (2.5 μM) or DMSO, and at different time points (6–72 h), the cells were cross-linked with 1% formaldehyde at room temperature for 10 min. The cells were washed twice with ice-cold phosphate-buffered saline and collected in 1 ml of ice-cold phosphate-buffered saline. The cells were pelleted at 700g at 4°C and homogenized with a Dounce homogenizer, and digested with enzymatic shearing cocktail (200 U/ml; Active Motif Inc.) for 10 min at 37°C. Immunoprecipitations were performed with anti-AHR antibodies overnight at 4°C. The chromatin was reverse cross-linked with reverse cross-linking buffer at 95°C for 15 min and then treated with proteinase K at 37°C for 1 h. The reaction was stopped by using proteinase K stop solution (Active Motif Inc., Carlsbad, CA). DNA was recovered by ethanol precipitation and subjected to PCR. The primers were designed to amplify the CYP1A1 enhancer region from 784 to 1156 bp upstream of the CYP1A1 transcription site. The primers sequences were forward primer, 5′-CACCCGCCACCCTTCGACAGTTCT-3′ and reverse primer, 5′-CTCCCGGGGTGGCTAGTAGCTTTGA-3′. The following PCR cycles were used: 95°C for 5 min; 40 cycles of 95°C for 30 s, 58°C for 30 s, 72°C for 45 s, and then 72°C for 5 min. The PCR products were analyzed by agarose gel electrophoresis in the presence of ethidium bromide and visualized under UV light.
Statistical Analyses.
All statistical analyses were conducted by using Student's t test or by two-way ANOVA, followed by modified t tests. P values less than 0.05 are considered significant.
Results
CYP1A1 Enzyme Activity.
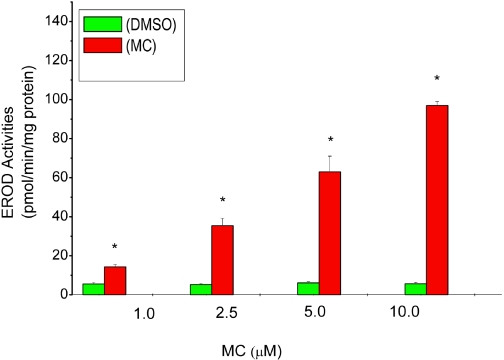
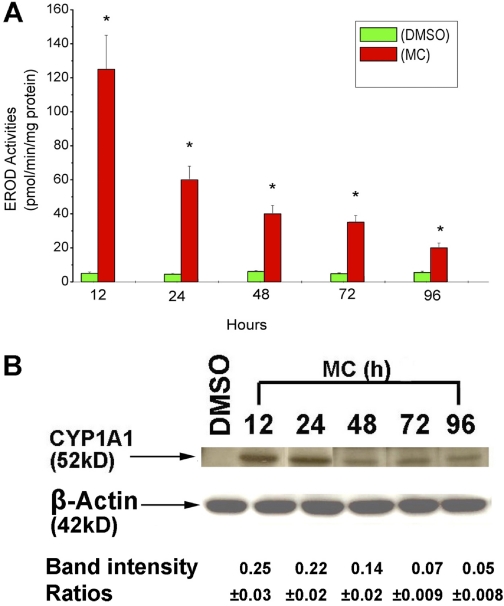
HepG2 cells were treated with DMSO or MC at various concentrations for 24 h, and EROD activity was assayed as an indicator of CYP1A1 activity (Moorthy, 2000). MC elicited a concentration-dependent induction of EROD activities after 24 h of treatment. Maximal induction was noticed after treatment of cells with 10 μM (Fig. 1). To test the hypothesis that MC causes persistent induction of CYP1A1 in HepG2 cells, we used 2.5 μM, which was in the linear dose range for subsequent experiments. At the 12-h time point after MC treatment, EROD activity in HepG2-treated cell lysates was approximately 25-fold higher compared with DMSO-treated controls (Fig. 2A). There was a decline in EROD activities at 24-, 48-, 72-, and 96-h time points, but the activities were still higher compared with corresponding DMSO treated controls. At the 72- and 96-h time points, the EROD activities were persistently higher, being 7- and 3-fold greater, respectively, compared with DMSO-treated controls (Fig. 2A). We did not conduct studies beyond the 96 h due to potential toxicity.
Fig. 1.
Dose-dependent effect of MC on EROD (CYP1A1) activities in HepG2 cells. HepG2 cells were treated with MC at concentrations ranging from 1 to 10 μM or DMSO, as described under Materials and Methods, and EROD activities were determined in the cells 24 h after treatment. Values represent mean ± S.E. (n = 3). *, statistically significant differences between MC- and DMSO-treated cells at P < 0.05.
Fig. 2.
Time course of CYP1A1 induction by MC in HepG2 cells. A, HepG2 cells were treated with MC (2.5 μM) or DMSO, and EROD activities were determined in the cells at different time points (12–96 h) after treatment. Values represent mean ± S.E. (n = 3). *, statistically significant differences between MC- and DMSO-treated cells at P < 0.05, as determined by two-way ANOVA. B, HepG2 cells were treated with MC or DMSO, and CYP1A1 apoprotein expression was analyzed in whole cell protein (10 μg) at the indicated time points by Western blotting. Quantitative analyses of the apoprotein contents were conducted by densitometry of the CYP1A1 bands, which were normalized against β-actin controls. Data represent mean ± S.E. of at least three independent treatments of the cells with MC.
Effect of MC on CYP1A1 Protein Expression.
As seen in Fig. 2B, MC treatment elicited a marked induction of CYP1A1 protein expression at 12 h, and the induction was maximal at 12 to 24 h. The induction of protein expression declined after 48 h, but augmented expression was maintained for up to 96 h (Fig. 2B). The persistent induction of CYP1A1 protein (Fig. 2B) correlated with enzyme activity (Fig. 2A). Densitometric analyses revealed that MC markedly induced CYP1A1 apoprotein contents at 12 h; and even after 96 h of MC treatment, the extent of induction was approximately 20% of that observed at the 12-h time point (Fig. 2B). In addition, a clear CYP1A1 band was observed at each of the time points studied after MC treatment, whereas no band corresponding to the CYP1A1 protein was detectable in the DMSO-treated controls (Fig. 2B). Quantification of CYP1A1 levels was assessed by densitometry and was normalized against β-actin controls (Fig. 2B).
CYP1A1 mRNA Levels.
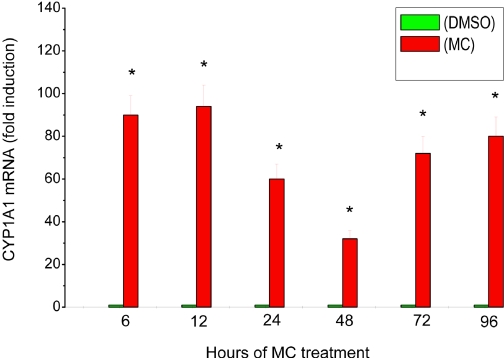
To determine whether the sustained induction of CYP1A1 enzyme activity and apoprotein content was accompanied by corresponding modulation of mRNA expression, we performed real-time RT-PCR analysis on total RNA from HepG2 cells treated with MC. Quantification of mRNA levels revealed that MC induced CYP1A1 mRNAs in the HepG2 cells by 85- and 90-fold, respectively, over the corresponding controls at the 6- and 12-h time points (Fig. 3). It is interesting to note that the induction declined after 48 h, being approximately 30-fold higher than control. At the 72- to 96-h time points, the MC-treated cells showed a much higher induction (70–78-fold) compared with the 48-h time point, suggesting a biphasic induction response (Fig. 3). Our results supported the hypothesis that MC caused sustained induction of CYP1A1 mRNA expression for up to 96 h.
Fig. 3.
Effect of MC on CYP1A1 mRNA in HepG2 cells. HepG2 cells were treated with MC or DMSO, as described under Materials and Methods, and total RNA was isolated in the cells harvested at 6, 12, 24, 48, 72, and 96 h after MC treatment. The RNA was subjected to real-time RT-PCR using CYP1A1-specific primers. Data represent mean ± S.E. of -fold induction versus controls from at least three replicates. *, statistically significant differences between MC- and DMSO-treated cells at P < 0.05.
Effect of MC on CYP1A1 Promoter Activation.
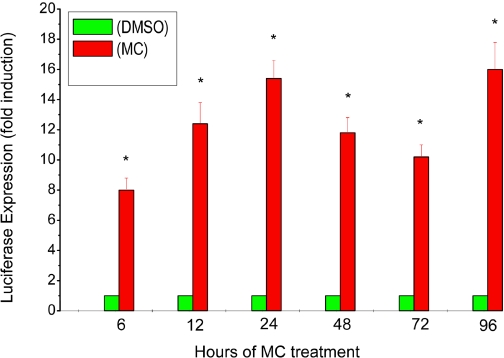
We tested the hypothesis that persistent induction of CYP1A1 gene expression by MC was mediated by sustained transcriptional activation of the CYP1A1 promoter. A plasmid containing the human CYP1A1 promoter (pGL3-1A1) and reporter (luciferase) gene was transiently transfected into HepG2 cells for 16 h, followed by MC or DMSO treatment. MC elicited an 8-fold induction of luciferase activity after 6 h (Fig. 4). The luciferase expression was further enhanced at the 24-h time point (approximately 15-fold), and high levels were maintained through 96 h (Fig. 4).
Fig. 4.
Effect of MC on CYP1A1 promoter activity in HepG2 cells. HepG2 cells were transiently transfected with a plasmid containing the 1.6-kilobase human CYP1A1 promoter (pGL3-1A1) and reporter (luciferase) gene, as described under Materials and Methods, and luciferase expression was determined in the transfected cell lysates at 6 to 96 h after MC or DMSO exposure. Data represent mean ± S.E. of three independent experiments. *, statistically significant differences between MC- and DMSO-treated cells at P < 0.05, as determined by two-way ANOVA.
Role of AHREs in Persistent Activation of the CYP1A1 Promoter.
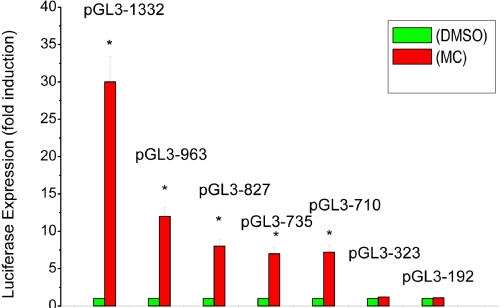
To determine the role of AHREs in the persistent activation of the CYP1A1 promoter, we transfected one of several plasmids containing various deletions (Fig. 5) into HepG2 cells, followed by treatment of the cells with MC, and then we determined luciferase expression at different time points. At first, we studied the effect of different deletion constructs on CYP1A1 induction by MC. As can be seen in Fig. 6, the plasmids containing the whole enhancer region, i.e., pGL3-1332 displayed maximal induction (approximately 30-fold) after 24 h. Transfection of cells with plasmids with broad deletions, i.e., pGL3-963, pGL3-827, and pGL3-735, pGL3-710, followed by MC treatment also showed marked induction (approximately 15-fold), but the extent of induction was approximately 50% of that elicited by transfection the full-length target enhancer (pGL3-1332) (Fig. 6). Transfection of plasmids containing the proximal promoter regions, i.e., pGL3-521, pGL3-323, and pGL3-192 into HepG2 cells, followed by MC treatment did not lead to any significant augmentation of luciferase expression (Fig. 6; data not shown for pGL3-521).
Fig. 5.
Plasmids containing broad deletions of the CYP1A1 enhancer region. Deletion plasmids were conducted by subjecting MC to exonuclease digestion to perform various nested deletions.
Fig. 6.
Effect of MC on luciferase expression using deletion plasmids. HepG2 cells were transiently transfected with a plasmid containing one of several deletions of the human CYP1A1 promoter (pGL3-1A1) and reported (luciferase) gene was determined, as described under Materials and Methods. Data represent mean ± S.E. of three independent experiments. *, statistically significant differences between MC- and DMSO-treated cells at P < 0.05, as determined by two-way ANOVA.
To test whether plasmids with broad deletions would display persistent expression of reporter gene, cells were transfected with pGL3-1332, pGL3-963, pGL3-827, or pGL3-710 into HepG2 cells, followed by MC treatment and determination of luciferase expression at selected time points. As shown in Table 1, MC caused persistent induction of luciferase expression for up to 96 h when cells were transfected with pGL3-1332 or pGL3-963 but not with pGL3-827 or pGL3-710 (Table 1).
TABLE 1.
Effect of MC on luciferase reporter gene expression in deletion plasmids
HepG2 Cells were transfected with plasmids containing the indicated deletions, and after 16 h, the cells were challenged with DMSO or MC. Luciferase reporter gene expression was determined in the cells 24, 72, or 96 h after DMSO or MC treatment. Values mean represent mean ± S.E. of -fold increases in reporter gene expression in the MC-treated cells compared with DMSO controls.
| Deletion Plasmid | Luciferase Expression |
||
|---|---|---|---|
| 24 h | 72 h | 96 h | |
| −1332 | 30.2 ± 3.1* | 18.2 ± 2.2* | 16.0 ± 3.2* |
| −963 | 15.2 ± 2.0* | 11.4 ± 2.0* | 10.2 ± 1.2* |
| −827 | 7.3 ± 0.90* | 2.4 ± 0.3* | 1.4 ± 0.3 |
| −710 | 6.8 ± 0.7* | 2.2 ± 0.3* | 1.2 ± 0.2 |
Statistically significant differences between MC- and DMSO-treated samples at P < 0.05, as determined by one-way ANOVA.
Nuclear MC-AHR Complexes in Vitro.
We conducted EMSA experiments using nuclear extracts of cells treated with MC for different time points to determine whether the MC-mediated sustained transcriptional activation of the CYP1A1 promoter was AHR-dependent. As shown in Fig. 7A, a specific band-shift corresponding to the MC-AHR-ARNT complex was detected as early as 6 h. The binding of MC-AHR-ARNT complex to labeled oligonucleotides containing AHREs was prevalent for up to 24 h (Fig. 7A). The bands shift was not observed in DMSO-treated controls (Fig. 7A, lane 1) or in cells harvested after 48, 72, and 96 h of MC treatment (Fig. 7A, lanes 5–7). The shifted band was competed off in the presence of 50-fold excess cold AHRE-specific oligonucleotide (Fig. 7A, lane 8).
Fig. 7.

A, representative EMSA of nuclear extracts from DMSO- and MC-treated HepG2 cells. Cells were treated in triplicate with MC or DMSO and harvested at 6 to 96 h after MC treatment. Nuclear protein extracts from treated cells were subjected to EMSA, as described under Materials and Methods. The labeled nuclear proteins were separated by polyacrylamide gel electrophoresis, and the gels were dried. The gels were exposed to autoradiography at −80°C for 24 h. Arrow indicates interaction with the AHREs of an MC-specific nuclear protein that appears to be MC-AHR-ARNT complex, which is competed off in the presence of 50-fold excess of cold AHRE-specific oligonucleotide (CP). B, ChIP assays showing complexing of the MC-AHR-ARNT complex to the CYP1A1 enhancer region. Cells were treated with MC or DMSO for the indicated time points, and ChIP assays were conducted using AHR antibodies as described under Materials and Methods.
MC-AHR-ARNT Complex Formation by ChIP Assays.
We tested the hypothesis that MC causes persistent transcriptional activation of CYP1A1 gene by AHR-independent mechanisms. Cells were treated with MC or DMSO, and at different time points (6–72 h), we then studied the binding of MCAHR-ARNT complexes to AHREs on the CYP1A1 enhancer region using ChIP assays using anti-AHR antibodies. As shown in Fig. 7B, MC-AHR-ARNT complexes were detectable at 6, 12, 24, and 48 h but not at 72 h. Bands were not detected when negative control antibodies were used (data not shown).
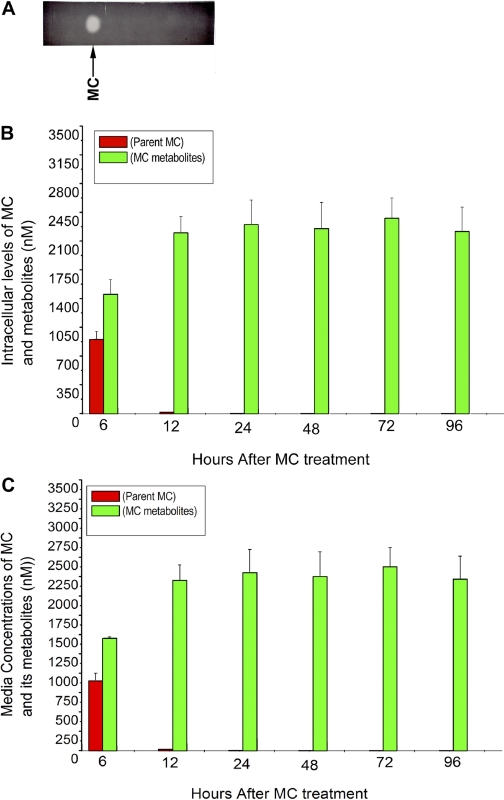
Levels of [3H]MC and Its Metabolites in HepG2 Cells.
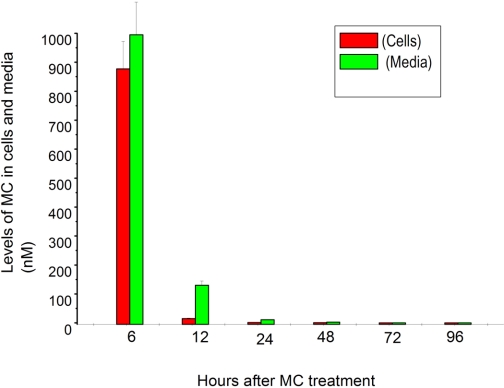
To ascertain whether the sustained induction of CYP1A1 was due to persistence of the parent MC or its metabolites in the cells, HepG2 cells were exposed to [3H]MC (2.5 μM), as described under Materials and Methods, and then at selected time points, levels of parent MC in the cell extracts and media were measured by radiometric TLC (Fig. 8A) and scintillation counting. As shown in Fig. 8B, MC was taken up rapidly by the cells and extensively metabolized. The recovery of radioactivity in the cells and media at any given time was greater than 98.4% (Fig. 8, B and C). Although the cells were exposed to 2.5 μM MC (62.5 nmol), at the 24-h time point, when maximal level of CYP1A1 induction was observed, the intracellular concentration of the parent MC was approximately 5 nM, representing only approximately 0.2% of the MC that was used to treat the cells (Fig. 8B). Most of the parent MC was metabolized by 72 h; and at 96 h, the total concentration of MC was only approximately 0.4 nM, representing 0.016% of the concentration used in cell treatments.
Fig. 8.
Metabolism of [3H]MC and its metabolites by radiometric TLC (A) in HepG2 cells (B) and media (C). A, HepG2 cells were treated with [3H]MC (2.5 μM; 1.9 Ci/mmol), and at different time points (6–96 h), the cells were harvested, extensively washed, and then MC and its metabolites were extracted with chloroform/methanol. The total radioactivity, representing MC and its metabolites, was measured, and the material was then subjected to radiometric TLC. B, the parent MC and metabolite zones were then excised from TLC plates, counted, and intracellular levels of MC and its metabolites were estimated. Values represent mean ± S.E. of at least three independent treatments of the cells. C, HepG2 cells were treated with [3H]MC, as described in legend to B, and at the indicated time points, the media were extracted with chloroform/methanol, and MC and its metabolites were quantified by radiometric TLC, as described under Materials and Methods. Values represent mean ± S.E. of at least three independent treatments of the cells.
We also analyzed the concentrations of MC and its metabolites in the media by extraction of the media with chloroform/methanol and radiometric TLC analyses (Fig. 8C). A significant amount of unchanged MC was present in the media at 6 h, but at subsequent time points, most of the radioactivity represented MC metabolites with very little parent MC (Fig. 8C).
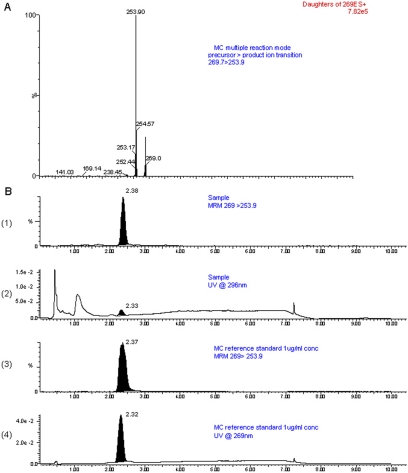
In an independent experiment, intracellular concentrations of MC were determined using LC-MS/MS analyses after exposure of the cells to MC (2.5 μM), followed by determination of parent MC at 6 to 96 h. Sample and reference LC-MS/MS chromatograms are shown in Fig. 9. Six hours after treatment of the cells with MC, we were able to recover approximately 37% of the MC in the cells, suggesting rapid uptake into HepG2 cells after treatment (Fig. 10). By 12 h, the intracellular concentration of MC was approximately 20 nM, representing approximately 0.8% of the MC that the cells received (Fig. 10). After 24 h, the concentration of parent MC in the cells was approximately 8 nM, representing 0.32% of the MC that was used to treat the cells (Fig. 10). This level was in the range that we obtained by radiometric TLC experiments (Fig. 8B), suggesting that our radiometric TLC experiments were indeed reliable and accurate. By 72 h, the levels of MC were below the level of detection (Fig. 10).
Fig. 9.
LC-MS/MS analyses of MC. A, multiple reaction monitoring (MRM) transition showing the primary product ion MC at m/z 253.9, which represents M+ + 1 −15 (loss of methyl group) fragment. B, MS/MS (B1, B3) and UV (B2, B4) chromatograms of MC sample (B1 and B2) and reference standards (B3 and B4) showing the presence of MC by both UV and MS/MS detection.
Fig. 10.
Determination of intracellular and media levels of MC by LC-MS/MS analyses. HepG2 cells were treated with cold MC (2.5 μM), and at the indicated time points, the cells and media were used to analyze the levels of parent MC by LC-MS/MS, as described under Materials and Methods. Values represent mean ± S.E. of at least three independent treatments of the cells with MC.
Effect of MC Metabolites on CYP1A1 Induction.
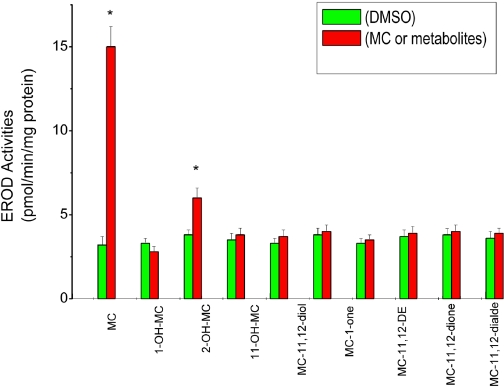
The goal of this experiment was to test whether persistent induction of CYP1A1 by MC was due to a MC metabolite that may be persisting in the cell or media. Cells were treated with one of several metabolites for 24 h. With the exception of 2-OH-MC, which induced CYP1A, albeit modestly (Fig. 11), none of the other major MC metabolites caused significant induction of CYP1A1 in the human hepatoma cells.
Fig. 11.
Effect of MC metabolites on EROD (CYP1A1) activities. HepG2 cells were treated with each of the MC metabolites (1 μM), i.e., 1-hydroxy-3-MC (1-OH-MC), 2-hydroxy-3-MC (2-OH-MC), 11-hydroxy-3-MC (11-OH-MC), 11,12-dihydro-11,12-dihydroxy-3-MC (MC-11,12-diol), 1-one-3-MC (MC-1-one), 11,12 epoxy-11,12 dihydro-3-MC (MC-11,12-DE), 11,12-dione-3-MC (MC-11,12-dione), and 11,12-dialdehyde-3-MC (MC-11,12-dialde). The cells were harvested after 24 h, and EROD activities were measured. Values represent mean ± S.E. (n = 3). *, statistically significant differences between MC metabolites and DMSO-treated cells at P < 0.05, as determined by Student's t test.
Cell Viability.
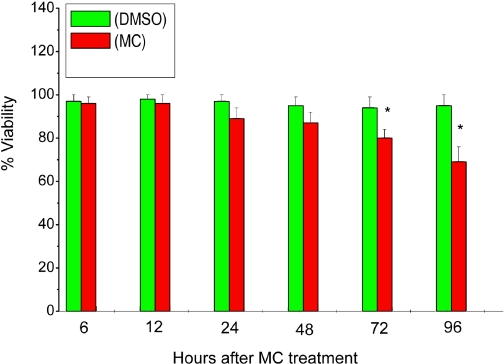
We exposed the cells to MC and measured cell viability by MTT reduction to assess whether MC was toxic to cells. As shown in the Fig. 12, MC did not cause any appreciable toxicity to the cells for up to 48 h. However, at the 72- and 96-h time points, there was a 14 to 22% cell death in the MC-treated cells, compared with DMSO-treated controls.
Fig. 12.
Cell viability by MTT reduction. HepG2 cells were treated with MC as described under Materials and Methods, and at the indicated time points, the viabilities of the cells were determined by MTT reduction. Values represent mean ± S.E. (n = 3). *, statistically significant differences between MC- and DMSO-treated cells at P < 0.05, as determined by Student's t test.
Discussion
In the present study, we investigated the mechanisms of sustained CYP1A1 induction by MC in HepG2 cells. The dose response of EROD (CYP1A1) induction by MC suggested that the kinetics of induction correlated with the cellular concentration of the ligand. Our results showing marked induction of EROD activities and apoproteins for up to 96 h strongly suggested that MC caused persistent CYP1A1 induction in human hepatoma cells for up to 96 h. This finding was in agreement with our previous findings showing long-term induction of CYP1A1 enzymes by MC in rodent models (Moorthy et al., 1993; Moorthy, 2000; Kondraganti et al., 2002; Jiang et al., 2009).
The fact that sustained induction of CYP1A1 activities and apoproteins for up to 96 h post-MC treatment was preceded by persistent induction of the CYP1A1 mRNA expression supported the idea that the prolonged induction of CYP1A1 activity was a result of persistent induction of CYP1A1 gene expression. Lekas et al. (2000) showed previously that human TCDD-inducible CYP1A1 mRNA is rapidly degraded in HepG2 cells, with a half-life of 2.4 h. Thus, our observations showing continued induction of CYP1A1 mRNA expression by MC supported the hypothesis that this phenomenon was mediated by new synthesis of mRNA, which in turn was due to up-regulation of the transcriptional activation of the CYP1A1 promoter, as revealed by our data on the effect of MC on luciferase reporter gene expression.
The fact that plasmids with broad deletions, i.e., pGL3-963 elicited approximately 50% induction of luciferase expression, compared with that observed upon transfection of pGL3-1604 or pGL3-1332 supported the hypothesis that functional AHREs at −974 and −1047 were necessary for maximal induction of promoter-driven reporter expression. The fact that transfection of pGL3-837 or pGL3-710 resulted in approximately 25% of the induction of luciferase expression by MC compared with that obtained with pGL3-1332 suggested that the AHRE located at −491 contributed to approximately 25% of the maximal induction of promoter activity. It is interesting to note that transfection of pGL3-521 did not result in induction of luciferase expression by MC (data not shown), suggesting that AHRE at −491 by itself was not sufficient for induction of CYP1A1 promoter activation but probably required the presence of as yet unidentified regulatory elements between −521 and −710. That transfection of pGL3-323 or pGL3-192 into HepG2 cells did not result in significant augmentation of luciferase expression after MC treatment was consistent with the lack of functional AHREs in these regions (Kress et al., 1998).
Our finding that transfection of pGL3-827 or pGL3-710 into HepG2 cells, followed by MC treatment did not result in persistent induction of luciferase expression for up to 96 h (Table 1) supported the hypothesis that AHREs located at −886, −974, and −1047, but not −491, from the start site contributed to the sustained CYP1A1 induction by MC. These observations were consistent with the hypothesis that specific AHREs contribute to the phenomenon of persistent CYP1A1 induction.
To ascertain whether sustained transcriptional activation of CYP1A1 gene by MC was due to persistent activation of the MC-AHR-ARNT complex in the nucleus, we conducted EMSA experiments using nuclear extracts of cells treated with MC at selected time points. The clear band shift upon incubation of nuclear extracts with [γ-32P]ATP-labeled oligonucleotides containing AHREs after 6 to 24 h of MC exposure suggested that the initial induction of CYP1A1 by MC was mediated by AHR-mediated mechanisms, consistent with studies of other investigators (Nebert et al., 2004; Fujii-Kuriyama and Mimura, 2005; Nukaya et al., 2009). The fact that the MC-AHR-ARNT binding was not detectable at 48 to 96 h after treatment with MC indicated that sustained CYP1A1 induction by MC for up to 96 h was mediated by AHR-independent mechanisms. This conclusion was further supported by ChIP experiments, which showed binding of MC-AHR-ARNT complex to the AHREs on the CYP1A1 promoter from 6 to 48 h but not at 72 h. It cannot be ruled out, however, that the initial binding of the MC-AHR-ARNT to the enhancer at the 12- to 24-h time point triggers a long-term induction response.
Our experiments with radiometric TLC or LC-MS/MS showing an intracellular level of MC at 24 h that represented only approximately 0.2% of the concentration that was used to treat the cells indicated that MC was rapidly metabolized in HepG2 cells. Our results showing very little parent compound in the media 12 h after MC treatment lends further credence to this idea and were consistent with the studies of Swedenborg et al. (2008). Thus, our findings indicate that the induction of CYP1A1 by MC at 72 h and beyond was independent of the presence of the parent compound, as we reported previously in the in vivo rat model (Moorthy, 2000).
We tested the possibility that MC metabolites formed in vivo (Kinoshita et al., 1980) or in vitro (Gangarosa and Stoming, 1983) may contribute to the sustained induction of CYP1A1. Our finding that, with the exception of 2-OH-MC, which caused a modest, albeit significant, induction of CYP1A1, none of the metabolites elicited a significant induction of CYP1A1 suggested that persistent CYP1A1 induction by MC was mediated by mechanisms other than classic induction of CYP1A1 by an MC metabolite via the AHR pathway.
It is interesting to note that 2-OH-MC is a carcinogenic metabolite and has DNA binding activity (Osborne et al., 1986). It is possible that 2-OH-MC binds to the CYP1A1 promoter covalently and activates gene transcription by as yet identified mechanisms. Our observations that approximately 78% of cells were viable 96 h after MC treatment suggested that prolonged induction of CYP1A1 by MC was not due to cell toxicity.
In a previous study (Moorthy, 2002), we reported that some MC-DNA adducts are sequence-specific, being specifically targeted to regions containing the AHREs. This conclusion was based on inhibition of PCR amplification of a 360-bp region (−1252 to −892) in the genomic DNA of MC-treated animals but not control animals. The 360-bp regions contain at least five AHREs at −1248, −1192, −1103, −1047, and −974, out of which −1047 and 974 are functional (Fig. 5). In fact, the plasmids we used in the transfection experiments had each of these AHREs. Furthermore, the oligonucleotides we used for EMSAs made up the core AHRE sequence (5′-TNGCGTG-3′), and the primers we used for the ChIP assays amplified the regions −784 to −1156 (Hestermann and Brown, 2003), which also encompassed the functional AHREs.
That DNA adducts were formed in a sequence-specific manner in the CYP1A1 promoter region was further confirmed by in vitro experiments (Moorthy et al., 2007). We observed that upon incubation of liver microsomes from wild-type mice with MC, NADPH, and plasmids carrying CYP1A1 promoter, some MC-DNA adducts were targeted to the CYP1A1 promoter region (Moorthy et al., 2007). The adducts were dramatically decreased when incubations were conducted using microsomes from Cyp1a2-null mice, suggesting that CYP1A2 catalyzed the formation of an MC metabolite that upon complexing with AHR may have translocated to the nucleus, wherein it covalently bound to AHREs on the CYP1A1 gene promoter and maintained CYP1A1 gene expression in vivo (Moorthy, 2002, 2008; Moorthy et al., 2007). In fact, our major finding was that CYP1A2 plays a key role in the sequence-specific formation of DNA adducts on the CYP1A1 promoter that may in turn modulate gene expression. However, it is not clear as to why the adducted plasmids produced in vitro did not up-regulate CYP1A1 gene expression (Moorthy et al., 2007). It is conceivable that in vivo, the level of adduct formation and/or repair may be tightly coupled to transcriptional activation, leading to sustained induction. It is also possible that DNA adducts in vivo were formed in the regions containing negative regulatory elements (Hines et al., 1988), a phenomenon that may have contributed to long-term CYP1A1 induction. Alternatively, MC-DNA adducts may have formed in the regions of the CYP1A1 promoter that bind chromatin remodeling factors such as histone deacetylase I and/or DNA methyltransferase I, which silence CYP1A1 gene expression in the noninduced state (Schnekenburger et al., 2007). These adducts may have prevented the binding of histone deacetylase I and/or DNA methyltransferase to the CYP1A1 promoter, resulting in continued activation of CYP1A1 transcription.
Irrespective of the mechanism of sustained induction of CYP1A1 by MC, this phenomenon may have important implications for human carcinogenesis by PAHs, as CYP1A1 appears to play a critical role in MC-mediated pulmonary carcinogenesis, based on evidence that very few lung tumors are formed in Cyp1a1-null mice (Moorthy et al., 2009).
In conclusion, the results of this study suggest that persistent induction of CYP1A1 by MC in HepG2 cells was mediated by sustained transcriptional activation of the CYP1A1 promoter. The results also support that hypothesis that MC elicits long-term induction of CYP1A1 by mechanisms other than persistence of the parent carcinogen, probably via by AHR-independent mechanisms. Further studies are warranted to unravel the molecular mechanisms of persistent CYP1A1 induction by MC, in relation to carcinogenesis, and these studies should help in the development of rationale studies for the prevention and/or treatment of carcinogenesis induced by environmental chemicals.
Acknowledgments
We thank Dr. Anil Jaiswal for providing the plasmids containing human CYP1A1 promoter; Dr. Paul Thomas for providing monoclonal antibodies against CYP1A1/1A2; Dr. Thevananther Sundararajah for help with the EMSA experiments; and Danielle Gregory for help with the artwork.
This work was supported in part by the National Institutes of Health National Institute of Environmental Health Sciences [Grant R01-ES009132-08/08S1]; and the National Institutes of Health National Heart, Blood, and Lung Institute [Grants R01-HL087174, R01-HL070921].
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
doi:10.1124/jpet.109.162222.
- PAH
- polycyclic aromatic hydrocarbon
- MC
- 3-methylcholanthrene
- P450
- cytochrome P450
- TCDD
- 2,3,7,8-tetrachlorodibenzo-p-dioxin
- Ah
- aryl hydrocarbon
- AHR
- aryl hydrocarbon receptor
- ARNT
- aryl hydrocarbon receptor nuclear translocator
- AHRE
- aryl hydrocarbon response element
- DMSO
- dimethyl sulfoxide
- TLC
- thin-layer chromatography
- 2-OH-MC
- 2-hydroxy-3-MC
- EROD
- ethoxyresorufin O-de-ethylase
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide)
- LC-MS/MS
- liquid chromatography-tandem mass spectrometry
- MS/MS
- tandem mass spectrometry
- MTBE
- methyl tertiary butyl ether
- PBS
- phosphate-buffered saline
- RT
- reverse transcriptase
- PCR
- polymerase chain reaction
- bp
- base pair(s)
- EMSA
- electrophoretic mobility-shift assay
- ChIP
- chromatin immunoprecipitation
- ANOVA
- analyses of variance.
References
- Bhakta KY, Jiang W, Couroucli XI, Fazili IS, Muthiah K, Moorthy B. (2008) Regulation of cytochrome P4501A1 expression by hyperoxia in human lung cell lines: implications for hyperoxic lung injury. Toxicol Appl Pharmacol 233:169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boobis AR, Nebert DW, Felton JS. (1977) Comparison of beta-naphthoflavone and 3-methylcholanthrene as inducers of hepatic cytochrome(s) P-448 and aryl hydrocarbon (benzo[a]pyrene) hydroxylase activity. Mol Pharmacol 13:259–268 [PubMed] [Google Scholar]
- Bresnick E, Brosseau M, Levin W, Reik L, Ryan DE, Thomas PE. (1981) Administration of 3-methylcholanthrene to rats increases the specific hybridizable mRNA coding for cytochrome P-450c. Proc Natl Acad Sci U S A 78:4083–4087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conney AH. (1986) Induction of microsomal cytochrome P-450 enzymes: the first Bernard B. Brodie lecture at Pennsylvania State University. Life Sci 39:2493–2518 [DOI] [PubMed] [Google Scholar]
- Coumoul X, Diry M, Robillot C, Barouki R. (2001) Differential regulation of cytochrome P450 1A1 and 1B1 by a combination of dioxin and pesticides in the breast tumor cell line MCF-7. Cancer Res 61:3942–3948 [PubMed] [Google Scholar]
- DePierre JW, Lundqvist G, Ernster L. (1982) Return of drug-metabolizing systems to control levels after induction with 3-methylcholanthrene. Acta Chem Scand B 36:497–498 [DOI] [PubMed] [Google Scholar]
- Fujii-Kuriyama Y, Mimura J. (2005) Molecular mechanisms of AhR functions in the regulation of cytochrome P450 genes. Biochem Biophys Res Commun 338:311–317 [DOI] [PubMed] [Google Scholar]
- Gangarosa MA, Stoming TA. (1983) The metabolism of 3-methylcholanthrene by liver and lung microsomes: effect of enzyme inducing agents. Cancer Lett 20:323–331 [DOI] [PubMed] [Google Scholar]
- Guengerich FP. (1988) Roles of cytochrome P-450 enzymes in chemical carcinogenesis and cancer chemotherapy. Cancer Res 48:2946–2954 [PubMed] [Google Scholar]
- Harvey RG. (1982) Polycyclic hydrocarbons and cancer. Am Sci 70:386–393 [PubMed] [Google Scholar]
- Hemminki K. (1990) Environmental carcinogens, in Handbook of Experimental Pharmacology (Cooper CS, Grover PL. eds) pp 33–61, Springer-Verlag, New York: [Google Scholar]
- Hestermann EV, Brown M. (2003) Agonist and chemopreventative ligands induce differential transcriptional cofactor recruitment by aryl hydrocarbon receptor. Mol Cell Biol 23:7920–7925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines RN, Mathis JM, Jacob CS. (1988) Identification of multiple regulatory elements on the human cytochrome P450IA1 gene. Carcinogenesis 9:1599–1605 [DOI] [PubMed] [Google Scholar]
- Jiang W, Wang L, Zhang W, Coffee R, Fazili IS, Moorthy B. (2009) Persistent induction of cytochrome P450 (CYP)1A enzymes by 3-methylcholanthrene in vivo in mice is mediated by sustained transcriptional activation of the corresponding promoters. Biochem Biophys Res Commun 390:1419–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Welty SE, Couroucli XI, Barrios R, Kondraganti SR, Muthiah K, Yu L, Avery SE, Moorthy B. (2004) Disruption of the Ah receptor gene alters the susceptibility of mice to oxygen-mediated regulation of pulmonary and hepatic cytochromes P4501A expression and exacerbates hyperoxic lung injury. J Pharmacol Exp Ther 310:512–519 [DOI] [PubMed] [Google Scholar]
- Kawajiri K, Nakachi K, Imai K, Hayashi S, Watanabe J. (1991) Individual differences in lung cancer susceptibility in relation to polymorphisms of P-4501A1 gene and cigarette dose, in Xenobiotics and Cancer (Ernstner L, Gelboin HV, Sugimura T. eds) pp 55–61, Japan Society, Tokyo, Japan and Taylor: & Francis, London, UK: [PubMed] [Google Scholar]
- Kinoshita K, Hashimoto K, Takahashi G, Yasuhira K. (1980) Gas chromatography-mass spectrometric analysis of 3-methyl-cholanthrene metabolism in vivo. Gann 71:181–189 [PubMed] [Google Scholar]
- Kondraganti SR, Jiang W, Moorthy B. (2002) Differential regulation of expression of hepatic and pulmonary cytochrome P4501A enzymes by 3-methylcholanthrene in mice lacking the CYP1A2 gene. J Pharmacol Exp Ther 303:945–951 [DOI] [PubMed] [Google Scholar]
- Kondraganti SR, Jiang W, Jaiswal AK, Moorthy B. (2008) Persistent induction of hepatic and pulmonary phase II enzymes by 3-methylcholanthrene in rats. Toxicol Sci 102:337–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress S, Reichert J, Schwarz M. (1998) Functional analysis of the human cytochrome P4501A1 (CYP1A1) gene enhancer. Eur J Biochem 258:803–812 [DOI] [PubMed] [Google Scholar]
- Lekas P, Tin KL, Lee C, Prokipcak RD. (2000) The human cytochrome P450 1A1 mRNA is rapidly degraded in HepG2 cells. Arch Biochem Biophys 384:311–318 [DOI] [PubMed] [Google Scholar]
- Masaki R, Matsuura S, Tashiro Y. (1984) A biochemical and electron microscopic study of changes in the content of cytochrome P-450 in rat livers after cessation of treatment with phenobarbital, β-napthoflavone or 3-methylcholanthrene. Cell Struct Funct 9:53–66 [DOI] [PubMed] [Google Scholar]
- Moorthy B. (2000) Persistent expression of 3-methylcholanthrene-inducible cytochromes P4501A in rat hepatic and extrahepatic tissues. J Pharmacol Exp Ther 294:313–322 [PubMed] [Google Scholar]
- Moorthy B. (2002) 3-Methylcholanthrene-inducible hepatic DNA adducts: a mechanistic hypothesis linking sequence-specific DNA adducts to sustained cytochrome P4501A1 induction by 3-methylcholanthrene. Redox Rep 7:9–13 [DOI] [PubMed] [Google Scholar]
- Moorthy B. (2008) The CYP1A subfamily, in Cytochromes P450. Role in Drug Metabolism and Toxicity of Drugs and other Xenobiotics (Ioannides C. ed) pp 97–135, RSC Publishing, Cambridge, UK: [Google Scholar]
- Moorthy B, Chen S, Li D, Randerath K. (1993) 3-Methylcholanthrene-inducible liver cytochrome(s) P450 in female Sprague-Dawley rats: possible link between P450 turnover and formation of DNA adducts and I-compounds. Carcinogenesis 14:879–886 [DOI] [PubMed] [Google Scholar]
- Moorthy B, Muthiah K, Fazili IS, Kondraganti SR, Wang L, Couroucli XI, Jiang W. (2007) 3-Methylcholanthrene elicits DNA adduct formation in the CYP1A1 promoter region and attenuates reporter gene expression in rat H4IIE cells. Biochem Biophys Res Commun 354:1071–1077 [DOI] [PubMed] [Google Scholar]
- Moorthy B, Wang L, Kondraganti SR, Zhou GD, Jiang W. (2009) Mice lacking the gene for cytochrome P450 1A2 (CYP1A2) display persistent induction of CYP1A1 expression in lung and increased susceptibility to lung tumorigenesis. Proc Am Assoc Cancer Res 50:288 [Google Scholar]
- Nebert DW, Gelboin HV. (1970) The role of ribonucleic acid and protein synthesis in microsomal aryl hydrocarbon hydroxylase induction in cell culture. The independence of transcription and translation. J Biol Chem 245:160–168 [PubMed] [Google Scholar]
- Nebert DW, Dalton TP, Okey AB, Gonzalez FJ. (2004) Role of aryl hydrocarbon receptor-mediated induction of the CYP1 enzymes in environmental toxicity and cancer. J Biol Chem 279:23847–23850 [DOI] [PubMed] [Google Scholar]
- Nukaya M, Moran S, Bradfield CA. (2009) The role of the dioxin-responsive element cluster between the Cyp1a1 and Cyp1a2 loci in aryl hydrocarbon receptor biology. Proc Natl Acad Sci U S A 106:4923–4928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne MR, Brookes P, Lee HM, Harvey RG. (1986) The reaction of a 3-methylcholanthrene diol epoxide with DNA in relation to the binding of 3-methylcholanthrene to the DNA of mammalian cells. Carcinogenesis 7:1345–1350 [DOI] [PubMed] [Google Scholar]
- Pendurthi UR, Okino ST, Tukey RH. (1993) Accumulation of the nuclear dioxin (Ah) receptor and transcriptional activation of the mouse Cyp1a-1 and Cyp1a-2 genes. Arch Biochem Biophys 306:65–69 [DOI] [PubMed] [Google Scholar]
- Poland A, Glover E. (1974) Comparison of 2,3,7,8-tetrachlorodibenzo-p-dioxin, a potent inducer of aryl hydrocarbon hydroxylase, with 3-methylcholanthrene. Mol Pharmacol 10:349–359 [PubMed] [Google Scholar]
- Postlind H, Vu TP, Tukey RH, Quattrochi LC. (1993) Response of human CYP1-luciferase plasmids to 2,3,7,8-tetrachlorodibenzo-p-dioxin and polycyclic aromatic hydrocarbons. Toxicol Appl Pharmacol 118:255–262 [DOI] [PubMed] [Google Scholar]
- Riddick DS, Huang Y, Harper PA, Okey AB. (1994) 2,3,7,8-Tetrachlorodibenzo-p-dioxin versus 3-methylcholanthrene: comparative studies of Ah receptor binding, transformation, and induction of CYP1A1. J Biol Chem 269:12118–12128 [PubMed] [Google Scholar]
- Schnekenburger M, Peng L, Puga A. (2007) HDAC1 bound to the Cyp1a1 promoter blocks histone acetylation associated with Ah receptor-mediated trans-activation. Biochim Biophys Acta 1769:569–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swedenborg E, Rüegg J, Hillenweck A, Rehnmark S, Faulds MH, Zalko D, Pongratz I, Pettersson K. (2008) 3-Methylcholanthrene displays dual effects on estrogen receptor (ER) alpha and ER beta signaling in a cell-type specific fashion. Mol Pharmacol 73:575–586 [DOI] [PubMed] [Google Scholar]
- Xu LC, Sinclair PR, Bresnick E. (1993) Induction of cytochrome P450IA1 and its recombinant construct in H4IIE rat hepatoma cells. Int J Biochem 25:13–21 [DOI] [PubMed] [Google Scholar]