Abstract
Abs facilitate humoral immunity via the classical mechanisms of opsonization, complement activation, Ab-dependent cellular cytotoxicity, and toxin/viral neutralization. There is also evidence that some Abs mediate direct antimicrobial effects. For example, Ab binding to the polysaccharide capsule of the human pathogenic fungus Cryptococcus neoformans promotes opsonization but also inhibits polysaccharide release and biofilm formation. To investigate whether Ab binding affects C. neoformans directly, we analyzed fungal gene expression after binding of protective and nonprotective mAbs. The 2 IgM Abs and 1 IgG1 Ab tested each induced different changes in gene expression. The protective IgG1 mAb upregulated genes encoding proteins involved in fatty acid synthesis, the protective IgM mAb downregulated genes encoding proteins required for protein translation, and the nonprotective IgM mAb had modest effects on gene expression. Differences in gene expression correlated with mAb binding to different locations of the capsule. Of the 3 Abs tested, the protective IgG1 mAb bound to C. neoformans closest to the cell wall, produced specific differences in the pattern of phosphorylated proteins, caused changes in lipid metabolism, and resulted in increased susceptibility to the antifungal drug amphotericin B. These results suggest what we believe to be a new mode of action for Ab-mediated immunity and raise the possibility that immunoglobulins mediate cross talk between microbes and hosts through their effects on microbial metabolism.
Introduction
Current views of Ab function posit that specific immunoglobulins mediate protection against microbes by promoting phagocytosis, activating complement, neutralizing toxins and viruses, and potentiating Ab-dependent cellular toxicity. Hence, humoral immunity is thought to mediate protection largely by enhancing the ability of other components of the immune system. In contrast, the concept that microbial metabolism is directly affected by immunoglobulins is not part of current immunological thought.
Recently, several mAbs have been shown to mediate direct antimicrobial activity through mechanisms that are yet to be fully elucidated. For example, specific IgM is microbicidal for Borrelia burgdorferi (1); Abs to fungal cell wall components, such as melanin (2, 3) and glucosylceramide (4), are fungistatic; and a genetically recombinant mAb against cell wall HSP90 of Candida albicans is fungicidal against different species of fungi in vitro (5) and increases the fungicidal effect of amphotericin B in clinical trials (6). For encapsulated organisms like Streptococcus pneumoniae and Cryptococcus neoformans, the binding of specific Ab to the capsule can produce the phenomenon of capsular reaction (“quellung” reaction), whereby the capsule becomes visible through differential interference contrast microscopy, as a consequence of refractive index change caused by the addition of protein (7). However, the capsular reaction occurs in the extracellular space and to date has not been associated with changes in microbial metabolism. For C. neoformans, Ab binding to the polysaccharide capsule can prevent subsequent release of capsular polysaccharide in vitro (8). Furthermore, Ab binding can inhibit C. neoformans biofilm formation by a mechanism that probably also involves interference with polysaccharide release (9). These observations have raised the question of whether Ab binding affects fungal metabolism directly.
We investigated this question by comparing gene expression in the presence of 3 capsule-binding mAbs that differ in isotype and protective efficacy in animal models of cryptococcosis (10). Binding of the 3 mAbs resulted in different gene expression profiles. The protective IgM mAb had a direct effect on microbial metabolic rate, while binding of the IgG1 mAb increased susceptibility to the antifungal amphotericin B, changed the pattern of phosphorylated proteins in total cell lysate, and was associated with differences in lipid metabolism. These results imply that specific Abs can affect microbial gene expression, thus opening a new area for investigation in the potential interactions of the humoral immune response and microbes.
Results
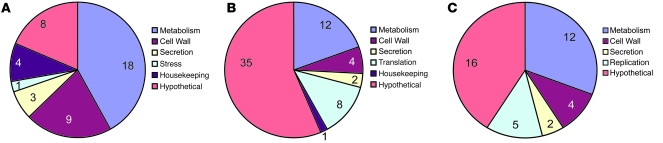
Three capsule-binding mAbs were used in this study, 2 IgM (12A1 and 13F1) and 1 IgG1 (18B7), together with 2 isotype-matched control mAbs, MOPC (IgG1) and TEPC (IgM), which do not bind to the capsular polysaccharide (10). The IgG1 mAb 18B7 is protective and was used in a human trial of passive therapy for cryptococcosis (11), whereas the IgM mAbs differ in both epitope specificity and protective efficacy (10). At mAb concentrations comparable to those measured in serum during passive Ab experiments in animals (12) and humans (11), we measured different microbial responses to each mAb. IgG1 mAb 18B7 binding to C. neoformans strain H99 was associated with the upregulation or downregulation of 43 different genes, relative to cells incubated with a near-saturating concentration of isotype-matched control mAb MOPC. These genes were mostly related to metabolism and cell wall synthesis (Figure 1A and Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI38322DS1). In particular, both the α and β subunits of the fatty-acid synthase and acetyl-CoA carboxylase, the 3 enzymes that catalyze fatty acid synthesis, were upregulated. Real-time RT-PCR confirmed expression changes for 79% of the 14 genes tested for mAb 18B7 binding to H99 (Supplemental Table 2). In contrast, IgM mAb 12A1 binding to H99 was associated with the upregulation or downregulation of 62 genes associated with metabolism, secretion, and translation, relative to H99 incubated with the control IgM mAb TEPC (Figure 1B and Supplemental Table 3). Of the 62 genes showing expression changes upon mAb 12A1 binding, 8 genes associated with ribosome biogenesis and protein translation were downregulated, suggesting that translation was repressed upon mAb binding. Real-time RT-PCR confirmed expression changes in 67% of the 21 genes tested for mAb 12A1 binding to H99 (Supplemental Table 4). When IgM mAb 13F1 bound to H99, 39 miscellaneous genes were upregulated and downregulated, compared with the isotype matched control mAb TEPC (Figure 1C and Supplemental Table 5). However, in contrast to the other 2 mAbs, real-time RT-PCR confirmed expression changes in only 29% of the 14 genes tested, suggesting that many of the microarray expression changes for IgM mAb 13F1 binding to H99 may be false positives from weak signals (Supplemental Table 6). Thus, the microbial response differed depending on the specific mAb, with the relative magnitude of the effects being mAb 18B7 > mAb 12A1 > mAb 13F1.
Figure 1. Categories and numbers of genes changed upon mAb binding to H99.
Pie charts of the categories of genes upregulated or downregulated upon (A) mAb 18B7, (B) 12A1, or (C) 13F1 binding to H99. Numbers indicate the number of genes in each category.
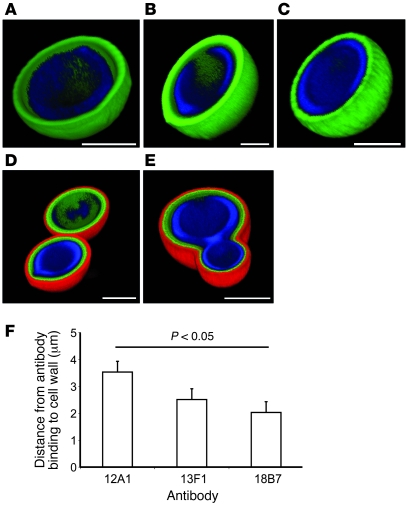
The 3 mAbs were then used in immunofluorescence experiments to establish their binding locations. To avoid potential uncertainties associated with secondary Ab detection, each mAb was conjugated to Alexa Fluor 488. The IgG1 mAb 18B7 bound to H99 in a compact shell-like pattern (Figure 2A). The IgM mAb 12A1 binding to H99 produced a similar binding pattern to that of mAb 18B7 (Figure 2B). However, when the IgM mAb 13F1 bound to H99, the binding pattern was diffuse and punctate throughout the capsule (Figure 2C). Double staining with IgG1 mAb 18B7 conjugated to Alexa Fluor 488 and unlabeled IgM mAbs, 12A1 or 13F1, detected with a TRITC-labeled secondary Ab, revealed that mAb 18B7 bound closer to the cell wall than either IgM (Figure 2, D and E). When the fluorescence profiles of the 3 mAbs directly conjugated to Alexa Fluor 488 were analyzed, the IgG1 mAb 18B7 bound significantly closer to the cell wall than the IgM mAb 12A1 (P < 0.05), with the IgM mAb 13F1 manifesting an intermediate binding pattern, between mAbs 18B7 and 12A1 (Figure 2F). This implies that there are differences in the spatial location of the epitopes recognized by each mAb or in the depth that they can penetrate the capsule.
Figure 2. The 3 Abs differ in their location of binding on the C. neoformans capsule.
(A–E) Three-dimensional reconstructions of confocal fluorescence images of mAbs binding H99. (A) mAb 18B7 conjugated to Alexa Fluor 488 (green) binding H99. (B) mAb 12A1 conjugated to Alexa Fluor 488 (green) binding H99. (C) mAb 13F1 conjugated to Alexa Fluor 488 (green) binding H99. (D) mAb 18B7 conjugated to Alexa Fluor 488 (green) and unlabeled mAb 12A1 detected with anti-IgM–TRITC (red) binding H99. (E) mAb 18B7 conjugated to Alexa Fluor 488 (green) and unlabeled mAb 13F1 detected with anti-IgM–TRITC (red) binding H99. In all images, the cell wall is stained with calcofluor white (blue). Scale bar: 5 μm (A–E). (F) Distance in microns between mAb binding and the cell wall (measured from the fluorescence profiles in ImageJ). ANOVA was used to test for differences in the distance to the cell wall.
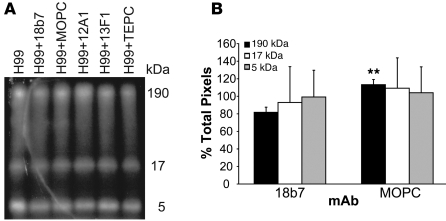
One mechanism for Ab-mediated changes in gene expression could be activation of signal transduction cascades, by virtue of Ab-triggered structural changes to the cell wall. Since signal transduction is often mediated by protein phosphorylation changes, we analyzed cryptococcal lysates for changes in the pattern of phosphorylated proteins, using 2 different methods. Pulse-labeling of C. neoformans strain H99 with α–32P-labeled ATP, followed by binding by the protective IgG1 mAb 18B7, resulted in differences in the pattern of phosphorylated proteins in total cell lysate, relative to the pattern observed after adding the IgG1 control mAb MOPC (Figure 3A). Specifically, the first of the 3 bands seen in each lane had significantly lower intensity in the H99 and mAb 18B7 condition, compared with the same bands in the H99 and mAb MOPC condition (P < 0.03; Figure 3B). The putative proteins that were dephosphorylated following mAb 18B7 binding had a molecular mass of around 190 kDa. There were no differences in phosphorylation seen for binding of the IgM mAbs, 12A1 or 13F1, relative to binding by the IgM control mAb TEPC (data not shown), suggesting that for these mAbs the mechanisms responsible for gene expression changes are different. This suggests that the IgG1 mAb 18B7 is inducing gene expression through changes in phosphorylation.
Figure 3. mAb binding to C. neoformans is associated with changes in protein phosphorylation.
(A) Autoradiogram of phosphorylated proteins in cell lysates with and without mAb. (B) Graph of the total pixels for each band in cell lysates of H99 and mAb 18B7 and H99 and mAb MOPC, normalized to H99 alone. The labeling experiment was done twice. The autoradiogram shown is representative of both experiments. ANOVA was used to test for differences in protein phosphorylation between mAbs 18B7 and MOPC. **P < 0.03.
To confirm this finding and possibly identify the dephosphorylated proteins, the 190-kDa band was excised and analyzed for differences in phosphorylation with mass spectroscopy. Six different candidate proteins were identified that showed differences in phosphorylation between cells bound with the protective IgG1 mAb 18B7 and cells bound by the isotype-matched control mAb MOPC (data not shown). The only one that matched the expected electrophoretic size was the clathrin heavy chain, which is known to be dephosphorylated during endocytosis (13).
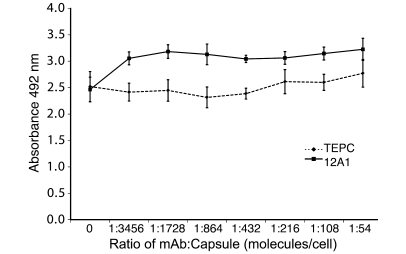
To determine whether Ab binding had direct effects on microbial metabolism, we measured the C. neoformans metabolic rate after incubation with these mAbs. When H99 was incubated with increasing amounts of the IgM mAb 12A1, there was a significant increase in the fungal metabolic rate for most concentrations tested, as measured by the 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino)carbonyl]-2H-tetrazolium hydroxide (XTT) reduction assay, relative to cells incubated with the control IgM mAb TEPC (P < 0.05; Figure 4). This colorimetric test measures the activity of several dehydrogenases present in active mitochondria (14). There was no change in the metabolic rate of H99 incubated with the IgG1 mAb 18B7 or the IgM mAb 13F1, compared with H99 incubated with the control IgG1 mAb MOPC or the control IgM mAb TEPC (data not shown). This observation implies that specific mAb binding induces changes in microbial metabolism.
Figure 4. mAb 12A1 binding can affect the C. neoformans metabolic rate compared with binding of the irrelevant control mAb TEPC, as measured by XTT reduction.
The XTT experiment was done 2 times with 6 wells per concentration per experiment. Data shown is for 1 experiment that is representative of both experiments. ANOVA was used to test for differences in C. neoformans metabolic rate. All mAb/capsule ratios are significantly different except the lowest (P < 0.05).
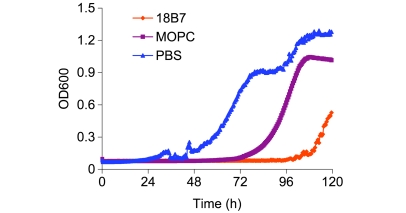
We next evaluated whether the binding of these mAbs to C. neoformans affected susceptibility to amphotericin B, one of the common antifungals used against C. neoformans, using minimum inhibitory concentration (MIC) and growth curves. The IgM mAbs 12A1 and 13F1 had no measurable effect (data not shown); however, when H99 cells were incubated with mAb 18B7 at half-saturating mAb concentrations, followed by incubation with the antifungal drug amphotericin B (2 μg/ml), MIC data showed a 2- to 4-fold increase in death of H99, with the addition of mAb 18B7 relative to mAb control (data not shown). To better evaluate these small differences, we improved the assay by using an automated growth curve machine. When H99 cells were incubated with half-saturating concentrations of mAb 18B7 and 0.125 μg/ml amphotericin B (2 times less than in the MIC in this experiment), the presence of mAb 18B7 led to a delay of approximately 36 hours in the onset of growth (Figure 5). Hence, mAb binding increased the susceptibility of C. neoformans to an important antifungal drug.
Figure 5. Growth curves in presence of IgG1 mAb 18B7 and amphotericin B.
C. neoformans cells were grown in the presence of near-saturating concentrations of mAb 18B7, mAb MOPC, or no Ab at all and 0.125 μg/ml amphotericin B (2 times less than in the MIC). The presence of mAb 18B7 led to a delay of approximately 36 hours in the onset of growth.
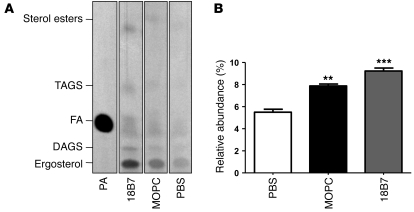
Given that binding of mAb 18B7 to C. neoformans induced upregulation of the fatty acid synthesis pathway, we used radioactively labeled acetate and thin layer chromatography (TLC) to evaluate the neutral lipid profile of cells, following incubation with the mAb (Figure 6A). A significant increase in fatty acids was measured in cells treated with mAb 18B7 relative to cells treated with the IgG1 control mAb MOPC (P < 0.05) or no IgG at all (P < 0.01) (Figure 6B). These observations confirm that binding of H99 by the IgG1 mAb 18B7 results in changes in lipid metabolism.
Figure 6. Neutral lipid composition of C. neoformans in the presence of mAb 18B7, MOPC, or PBS.
(A) Autoradiography of TLC of neutral lipids. H99 cells were grown for 36 hours, and 6 × 107 cells were incubated with 12 mCi 14C-acetate, together with saturating concentrations of the indicated mAbs for 2 hours at 37°C. Total lipids were extracted and loaded onto a silica TLC plate. Lanes were loaded with equal amounts of total lipids, measured by scintillation counting. The positions of triacylglycerols (TAGS), diacylglycerols (DAGS), fatty acids (FA), sterols esters, and ergosterol are indicated. The left lane shows a standard with 100 nCi of 14C-palmitic acid (PA). (B) The pixel intensity of the band corresponding to fatty acids was measured and compared with the total pixel intensity of every lane. Mean and SD from 3 replicates are shown. **P < 0.05, ***P < 0.001.
Discussion
Despite increasing evidence that many specific Abs can interfere with the growth and metabolism of microbes directly (15), little is known about the microbial response to Ab binding. For the human pathogenic fungus C. neoformans, Ab-mediated protection is associated with interference of polysaccharide release and biofilm formation (8, 9). For example, the IgG1 mAb 18B7 and IgM mAb 12A1 each inhibit biofilm formation and polysaccharide release, while the IgM mAb 13F1 has no effect on either phenomenon (9). Since these are direct immunoglobulin effects on microbial physiologic functions that are not related to the host immune system, we investigated whether binding by these mAbs to C. neoformans strain H99 had a direct effect on fungal metabolism.
When H99 was bound by the mAb 18B7, a variety of genes involved in metabolism, secretion, and fatty acid synthesis were upregulated. We focused on the genes involved in fatty acid synthesis, because it appeared that an entire pathway was upregulated in unison, but note that mAb 18B7 binding also induced other metabolic and secretory changes that may have important consequences for fungal metabolism. When H99 was bound by the mAb 12A1, a variety of protein translation genes were downregulated. In contrast, when H99 was bound by mAb 13F1, various genes involved in metabolism, secretion, and cell wall synthesis were upregulated or downregulated, but only a small fraction of these were validated with real-time RT-PCR. This result implies both quantitative and qualitative changes in the gene expression profile response depending on the mAb. We note that these effects occurred at mAb concentrations that are similar to concentrations used to measure phagocytosis in vitro. Using immunoglobulin and glucuronoxylomannan (GXM) molecular masses, we calculated that these effects occurred at immunoglobulin/GXM molecular ratios of 1:14 for IgG1 and 1:269 for IgM (or ~1:54 considering that IgM is pentameric) (16). When we evaluated the dose response at lower concentrations of IgG1 mAb, the number of genes that had altered expression changed in a dose-dependent manner, such that there were no gene changes at immunoglobulin/GXM molar ratios of less than 1:500 (data not shown). One explanation for the relatively high mAb concentrations required for this effect is that the cryptococcal capsule can sequester large amounts of Ab (17), and consequently, the effects are only observed at near-saturating concentrations, which are nonetheless comparable to those used in vitro and in animal experiments (12).
Immunofluorescence revealed that the IgG1 mAb 18B7 bound significantly closer to the cell wall than the IgM mAbs 12A1 or 13F1, possibly due to increased penetration given its smaller size. Furthermore, the IgM mAbs 12A1 and 13F1 produced qualitatively different immunofluorescent patterns (18). The observation that each of the mAbs bound to a different area of the capsule suggests that the differences in gene expression profiles may be related to mAb binding at distinct areas, where the mAb epitope is located.
The observation that mAb 18B7 binding to the capsule was associated with significant differences in dephosphorylation is a bit surprising. However, dephosphorylation has been implicated in at least one signal transduction cascade in plants (19) as well as in a raft-mediated signaling pathway in astrocytes (20), suggesting that it is possible that this mechanism is involved in signal transduction in C. neoformans. Thus, the finding of differences in phosphorylation in association with mAb binding is consistent with changes in gene expression through phosphorylation-related signaling.
When H99 was incubated with mAb 12A1, there was an increase in metabolic rate that was not observed upon incubation with either mAb 18B7 or mAb 13F1. This observation, in addition to the microarray data, is consistent with upregulation of many genes associated with metabolism. When H99 was incubated with mAb 18B7, there was a 2- to 4-fold increase in susceptibility to the antifungal amphotericin B, as measured with MIC. The MIC test revealed that the concentration of amphotericin B necessary to kill C. neoformans was lower in the presence of mAb. Given that amphotericin B mediates its antifungal action by binding to and inhibiting the synthesis of membrane sterols, the observation of mAb 18B7 binding increasing cellular susceptibility to this drug is consistent with observation that this mAb affects lipid metabolism. A similar effect has been seen before in C. neoformans biofilms (21) but not planktonic cells. Interestingly, when near-saturating concentrations of mAb 18B7 are used, there seemed to be a protective effect against amphotericin B. Since mAb binding crosslinks the capsule and reduces its permeability (22) and amphotericin B is a large molecule, saturating concentrations of the mAb may protect the cell from the effects of the antifungal drug by reducing accessibility.
In considering the question of whether a 2- to 4-fold increase in antifungal susceptibility has biological relevance, we note that small differences between groups have been shown to be biologically significant in other studies. For example, a 2-fold increase in intracellular adenylyl cyclase can have profound biological consequences (23), and a 1.4-fold increase in Cyp3a expression levels has a significant biological impact on drug metabolism in SPF mice (24). Thus, while the magnitude of the change in antifungal susceptibility is numerically small, it was consistently reproduced. Hence, we believe that it has the potential to be biologically relevant, and we note that capsule binding Ab has been shown to enhance amphotericin B efficacy in murine studies (25, 26). Additionally, the presence of the mAb 18B7 and amphotericin B led to a delay in the onset of growth by approximately 36 hours. In the grand scheme of fungal growth in the host this is biologically meaningful, in that addition of both mAb 18B7 and amphotericin B would allow the host immune response a 36-hour head start, which could mean the difference between clearance and control of the fungal infection.
When H99 was incubated with the IgG1 mAb 18B7, we measured changes in lipid metabolism by TLC. The results were consistent with the upregulation of the entire fatty acid synthesis pathway seen in the microarray when H99 was bound by mAb 18B7. Given prior observations that mAb 18B7 inhibits polysaccharide shedding in vitro (8) and biofilm formation (9), together with recent evidence that the major component of the capsule, GXM (27), and other major virulence factors (28), are exported to the cell exterior in lipid vesicles, our data suggest an association between lipid metabolism and capsule production/shedding, which appears to be reflected in the metabolic changes observed. Additionally, it is possible that changes in lipid metabolism are seen because mAb 18B7 binds very close to the cell wall, and this binding may somehow perturb membrane lipids that in turn produce regulatory changes in lipid metabolism (29).
The observation that ergosterol synthesis was increased by the binding of mAb 18B7 may provide an explanation for the changes in antifungal drug susceptibility following specific Ab binding. Given that amphotericin B mediates some of its antifungal effects by binding to cell membrane ergosterol, an increase in ergosterol synthesis would increase the amount of amphotericin B target and thereby enhance fungal cell susceptibility to the polyene antifungal agent.
While coincubation of C. neoformans with specific Ab produced changes, relative to those observed with irrelevant mAb, we also measured changes in gene expression, lipid metabolism, and susceptibility to amphotericin B when fungi were incubated with irrelevant mAb, relative to conditions in which no immunoglobulin was present. This result cannot be attributed to the presence of protein alone, since the effect was observed relative to conditions that contained albumin. Although we do not have a good explanation for this effect, we note that immunoglobulins are antimicrobial proteins that have been described to have numerous properties that could affect microbial metabolism, directly and indirectly. For example, immunoglobulins have been reported to have catalytic activities that result in the generation of oxygen-derived radicals (30, 31), which may have influenced fungal metabolism through oxidative stress. Furthermore, peptides derived from immunoglobulins can have antimicrobial effects (32), and it is conceivable that low-grade degradation of Abs by fungal proteases results in peptide products that affect fungal metabolism. Immunoglobulins can chelate metals (33), and metal scavenging by the irrelevant Ab could have affected trace concentrations in solution, producing nonspecific metabolic changes. Hence, immunoglobulins caused specific and nonspecific changes in fungal metabolism, and it is conceivable that both contribute to their antimicrobial effects.
In summary, our results establish that Ab binding to a microbe can affect microbial metabolism. mAb binding to C. neoformans was associated with changes in gene expression, protein phosphorylation, lipid metabolism, metabolic rate, and susceptibility to antifungal agents. Ab-mediated changes in gene expression involved genes implicated in lipid metabolism, while biosynthetic labeling studies documented changes in lipid profiles. Given that amphotericin B damages membranes, we note that there is a reassuring internal consistency in our findings of mAb-mediated changes in lipid metabolism, gene expression, lipid biosynthesis, and susceptibility to this drug. From a more global perspective on host-microbe interactions, the results imply a connection between a product of the adaptive immune system and microbial responses, which highlight new possibilities for the ability of immune responses to modulate microbial metabolism and engage in cross talk with microbes through production of specific Abs.
Methods
Strains.
C. neoformans strain H99 (serotype A) was grown from frozen stock in yeast peptone dextrose (YPD) broth for 36–42 hours (mid-log phase) at 37°C and then washed 3 times and resuspended in PBS. Ab was added to 4 × 107 cells slowly to avoid aggregation.
Abs.
Abs used were mAbs 18B7 (IgG1), 12A1 (IgM), and 13F1 (IgM) and the control mouse IgG1, MOPC, and mouse IgM, TEPC183. Control mAbs do not bind C. neoformans. mAb concentrations used were normalized to the total number of cells used in the experiment, based on the ratio of mAb molecules to GXM molecules in the capsule (34).
Microarray.
The cells and mAbs were incubated at 37°C for 1 hour, and RNA was extracted (RNAeasy Kit; Qiagen) and genomic DNA was removed (Message Clean Kit; GenHunter). Multiple different pools of RNA were analyzed at Washington University in St. Louis (St. Louis, Missouri), using the C. neoformans JEC21 genomic microarray, which was developed by the Cryptococcus Community Microarray Consortium with financial support from individual researchers and the Burroughs Wellcome Fund (The Genome Center at Washington University; http://genome.wustl.edu/services/microarray/cryptococcus_neoformans). The array includes 7,775 probes in duplicate. For the mAb 18B7 microarray, 6 different pools of RNA were used, in which H99 and mAb 18B7 were compared with H99 and mAb MOPC. For the IgM microarrays, 3 different pools of RNA were used, in which H99 and mAb 12A1 and mAb 13F1 were compared with H99 and mAb TEPC. Each comparison was done with a Cy3-Cy5 dye swap between the RNA pools. The consistency of the dye swap was measured by correlating the average mean between channel 1 and 2. The r2 value was 0.78, suggesting that the dye swap was successful and the effect of the dye was relatively insignificant. The gene expression data were averaged across the RNA pools and analyzed (GeneSpring 7.2; Agilent), and the data were filtered for genes with more than 2-fold change and P values of less than 0.05. The Benjamini and Hochberg false discovery rate for multiple testing corrections was then done for all genes showing changes in gene expression.
Real-time RT-PCR.
RNA was made from H99 incubated with near-saturating concentrations of all mAbs, as above. As an additional control, RNA was made from H99 cells incubated with boiled mAbs (18B7, 12A1, and 13F1). cDNA was made from 2 pools of RNA (Quantitech Reverse Transcription Kit; Qiagen), and real-time RT-PCR was done using SYBR Green (Applied Biosystems), cDNA, and primer (see Supplemental Table 7) in an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). Each cDNA was done in quadruplicate and normalized with actin, and the fold change was determined (35). Fold change for each mAb and the boiled mAb control was relative to the control mAb. Real-time RT-PCR was repeated twice.
Immunofluorescence microscopy.
The mAbs 18B7, 12A1, and 13F1 were conjugated to Alexa Fluor 488 (Invitrogen), and immunofluorescent microscopy was done as described (36). For reconstruction in ImageJ software ( http://rsbweb.nih.gov/ij/), the color palettes used were nonlinear. ANOVA was used to test for differences in the distance of mAb binding to the cell wall.
Phosphorylated protein analysis.
Approximately 6 × 107 H99 cells were incubated with 100 μCi/ml 32P-labeled ATP for 30 minutes at 37°C. Radioactive cells were then incubated with mAb for 1 hour at 37°C. Cells were collected in PBS with inhibitors of serine, cysteine, and metalloproteases, and cell lysates were generated by homogenization in a bead beater 10 times with 30-second bursts. The cell lysates were concentrated through a 10-kDa molecular cutoff filter, and the protein concentration measured (37). Ten milligrams of total protein were electrophoresed on a 10%–20% polyacrylamide gel, which was then dried and exposed to radiographic film for 24 hours. The film was scanned, and the lanes were digitized and quantified using Un-Scan-It software (v6.3, Silk Scientific). This experiment was repeated twice.
Mass spectroscopy.
Approximately 4 × 107 H99 cells were incubated with near-saturating concentrations of mAb, and cell lysates were generated as above. Ten milligrams of total protein were electrophoresed on a 10%–20% polyacrylamide gel, and then the gel was stained for 1 hour with Gelcode Blue (Pierce). The 190-kDa band was carefully excised from the gel, stored in 100 μl dH20, and shipped overnight to NextGen Sciences Inc., where it was treated with trypsin and analyzed for differences in phosphorylation modifications with mass spectroscopy (GeLCMS).
XTT assay.
H99 cells were grown to mid-log phase at 37°C, washed, and diluted in PBS and 1% BSA. The XTT assay was done as in ref. 21, with mAb concentrations of (6 wells each) 0, 5.2, 10.4, 20.8, 41.6, 83.25, 166, and 333 μg/ml, and 5 × 106 H99 cells in each well. The mAb concentrations were correlated with the following ratios of mAb/capsule (6 wells each): 0; 1:3,456; 1:1,728; 1:864; 1:432; 1:216; 1:108; and 1:54. Plates were incubated at 37°C for 20 hours, and XTT and Menadione were added. The absorbance was measured at 492 nm after incubation for 4 hours at 37°C. Each assay was repeated twice. The data for mAb 12A1 at the highest concentration consist of 5 data points (instead of 6), as an outlier was omitted.
Antifungal drug susceptibility.
For determination of the MIC, H99 cells were grown as described above and washed once with 2X minimal media. Approximately 1 × 104 cells were incubated without mAb or with half- or near-saturating concentrations of mAbs 18B7 or MOPC for 1 hour at 37°C. Two-fold concentrations of amphotericin B (1, 0.5, 0.25, 0.125, 0.063, 0.031, and 0 μg/ml) were prepared in water, added to opsonized H99 cells, and incubated at 37°C. At 36 hours and 5 days, the 96-well plate was photographed to document the lowest amphotericin B concentration capable of restricting the growth of the H99 cells. The MIC experiment was repeated 3 times.
For the growth curve assay, H99 cells grown as above were added to appropriate plates for incubation in a Bioscreen C automated microbiology growth curve system (Growth Curves USA). Each well contained in a final volume of 200 μl minimal medium: 1 × 104 H99 cells; mAbs 18B7 or MOPC at 166 μg/ml; amphotericin B in 2-fold dilutions, starting at 4 μg/ml and ending at 16 ng/ml. Each Ab/drug concentration combination was done in triplicate in the plate, and the growth was carried on for 5 days with optical density readings every 30 minutes.
Lipid metabolism methods.
H99 cells were grown from frozen stock in YPD for 36 hours at 37°C. Cells were harvested, washed in PBS, suspended in 1% BSA in PBS at 3 × 107 cells/ml, and incubated with gentle agitation with 6 μCi/ml of [1-14C]-acetate at 37°C, with or without near-saturating concentrations of mAbs 18B7 or MOPC. After 2 hours, cells were harvested and suspended in 1 ml of PBS. Neutral lipids were extracted using the Bligh-Dyer method (38). Briefly, 3.75 ml of chloroform/methanol (1:2) were added to 1 ml of PBS-washed cells with 0.5 mm glass beads, and the cells vortexed for 15 minutes. Next, 1.25 ml of chloroform was added, and the cells were vortexed for 1 minute. One milliliter of water was finally added, and the cells were vortexed for 1 minute before centrifugation at 110 g for 10 minutes. The lower phase was collected and dried. The lipid extract was redissolved in a small volume of chloroform/methanol (2:1). Neutral lipids were separated in a silica TLC plate, using petroleum ether (boiling point, 60°–80°C) diethyl ether-acetic acid (90:15:1, vol/vol/vol) as the solvent system. The plate was exposed for 5 days on a BioMax radiographic film (Kodak). The identity of the fatty acid band was determined by comparison with a radioactive palmitic acid standard (39). To determine differences in fatty acids synthesis, the pixel intensity of the band corresponding to fatty acids was measured using the software multigauge v2.3 (Fujifilm) and expressed relative to the accumulated pixel intensity of every lane. Mean and SD are representative of 3 independent experiments.
Statistics.
For the microarray analysis, the data were initially filtered for genes with more than 2-fold change and P values of less than 0.05. The Benjamini and Hochberg false discovery rate for multiple testing corrections was then done for all genes showing changes in gene expression. A P value of less than 0.01 was considered significant. A 1-way ANOVA was used to measure the distance of mAb binding to the cell wall, differences in protein phosphorylation, differences in microbial metabolic rate, and differences in lipid fatty acids. For all of the above tests, except the microarray, a P value of less than 0.05 was considered significant. For Figure 2F, Figure 3B, Figure 4, and Figure 6B, the data shown are mean ± SD.
Supplementary Material
Acknowledgments
We thank Antonio Nakouzi for help with purifying the mAbs 12A1 and 13F1 and for help with the immunofluorescence. We thank Luis Martinez for help with the XTT assays. We also thank Seth Crosby and Mike Heinz at Washington University in St. Louis for their help and analysis of the microarray data. A. Casadevall is supported by the NIH awards AI033142, AI033774, and HL059842-08.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(4):1355–1361. doi:10.1172/JCI38322.
See the related Commentary beginning on page 1039.
References
- 1.Connolly SE, Thanassi DG, Benach JL. Generation of a complement-independent bactericidal IgM against a relapsing fever Borrelia. J Immunol. 2004;172(2):1191–1197. doi: 10.4049/jimmunol.172.2.1191. [DOI] [PubMed] [Google Scholar]
- 2.Alviano DS, et al. Melanin from Fonsecaea pedrosoi induces production of human antifungal antibodies and enhances the antimicrobial efficacy of phagocytes. Infect Immun. 2004;72(1):229–237. doi: 10.1128/IAI.72.1.229-237.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosas AL, Nosanchuk JD, Casadevall A. Passive immunization with melanin-binding monoclonal antibodies prolongs survival of mice with lethal Cryptococcus neoformans infection. Infect Immun. 2001;69(5):3410–3412. doi: 10.1128/IAI.69.5.3410-3412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodrigues ML, et al. Human antibodies against a purified glucosylceramide from Cryptococcus neoformans inhibit cell budding and fungal growth. Infect Immun. 2000;68(12):7049–7060. doi: 10.1128/IAI.68.12.7049-7060.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matthews RC, et al. Preclinical assessment of the efficacy of mycograb, a human recombinant antibody against fungal HSP90. Antimicrob Agents Chemother. 2003;47(7):2208–2216. doi: 10.1128/AAC.47.7.2208-2216.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pachl J, et al. A randomized, blinded, multicenter trial of lipid-associated amphotericin B alone versus in combination with an antibody-based inhibitor of heat shock protein 90 in patients with invasive candidiasis. Clin Infect Dis. 2006;42(10):1404–1413. doi: 10.1086/503428. [DOI] [PubMed] [Google Scholar]
- 7.MacGill TC, MacGill RS, Casadevall A, Kozel TR. Biological correlates of capsular (quellung) reactions of Cryptococcus neoformans. J Immunol. 2000;164(9):4835–4842. doi: 10.4049/jimmunol.164.9.4835. [DOI] [PubMed] [Google Scholar]
- 8.Martinez LR, Moussai D, Casadevall A. Antibody to Cryptococcus neoformans glucuronoxylomannan inhibits the release of capsular antigen. Infect Immun. 2004;72(6):3674–3679. doi: 10.1128/IAI.72.6.3674-3679.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinez LR, Casadevall A. Specific antibody can prevent fungal biofilm formation and this effect correlates with protective efficacy. Infect Immun. 2005;73(10):6350–6362. doi: 10.1128/IAI.73.10.6350-6362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mukherjee J, Nussbaum G, Scharff MD, Casadevall A. Protective and nonprotective monoclonal antibodies to Cryptococcus neoformans originating from one B cell. J Exp Med. 1995;181(1):405–409. doi: 10.1084/jem.181.1.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larsen RA, et al. Phase I evaluation of the safety and pharmacokinetics of murine-derived anticryptococcal antibody 18B7 in subjects with treated cryptococcal meningitis. Antimicrob Agents Chemother. 2005;49(3):952–958. doi: 10.1128/AAC.49.3.952-958.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mukherjee J, Scharff MD, Casadevall A. Variable efficacy of passive antibody administration against diverse Cryptococcus neoformans strains. Infect Immun. 1995;63(9):3353–3359. doi: 10.1128/iai.63.9.3353-3359.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crotzer VL, Mabardy AS, Weiss A, Brodsky FM. T cell receptor engagement leads to phosphorylation of clathrin heavy chain during receptor internalization. J Exp Med. 2004;199(7):981–991. doi: 10.1084/jem.20031105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roehm NW, Rodgers GH, Hatfield SM, Glasebrook AL. An improved colorimetric assay for cell proliferation and viability utilizing the tetrazolium salt XTT. J Immunol Methods. 1991;142(2):257–265. doi: 10.1016/0022-1759(91)90114-U. [DOI] [PubMed] [Google Scholar]
- 15.Casadevall A, Pirofski LA. New concepts in antibody-mediated immunity. Infect Immun. 2004;72(11):6191–6196. doi: 10.1128/IAI.72.11.6191-6196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bryan RA, Zaragoza O, Zhang T, Ortiz G, Casadevall A, Dadachova E. Radiological studies reveal radial differences in the architecture of the polysaccharide capsule of Cryptococcus neoformans. Eukaryot Cell. 2005;4(2):465–475. doi: 10.1128/EC.4.2.465-475.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rakesh V, et al. Finite-element model of interaction between fungal polysaccharide and monoclonal antibody in the capsule of Cryptococcus neoformans. J Phys Chem B. 2008;112(29):8514–8522. doi: 10.1021/jp8018205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nussbaum G, Cleare W, Casadevall A, Scharff MD, Valadon P. Epitope location in the Cryptococcus neoformans capsule is a determinant of antibody efficacy. J Exp Med. 1997;185(4):685–694. doi: 10.1084/jem.185.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim TW, et al. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Biol. 2009;11(10):1254–1260. doi: 10.1038/ncb1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park SJ, et al. Oxidative stress induces lipid-raft-mediated activation of Src homology 2 domain-containing protein-tyrosine phosphatase 2 in astrocytes. Free Radic Biol Med. 2009;46(12):1694–1702. doi: 10.1016/j.freeradbiomed.2009.03.026. [DOI] [PubMed] [Google Scholar]
- 21.Martinez LR, Christaki E, Casadevall A. Specific antibody to Cryptococcus neoformans glucurunoxylomannan antagonizes antifungal drug action against cryptococcal biofilms in vitro. J Infect Dis. 2006;194(2):261–266. doi: 10.1086/504722. [DOI] [PubMed] [Google Scholar]
- 22.Zaragoza O, Casadevall A. Monoclonal antibodies can affect complement deposition on the capsule of the pathogenic fungus Cryptococcus neoformans by both classical pathway activation and steric hindrance. Cell Microbiol. 2006;8(12):1862–1876. doi: 10.1111/j.1462-5822.2006.00753.x. [DOI] [PubMed] [Google Scholar]
- 23.Pieroni JP, Harry A, Chen J, Jacobowitz O, Magnusson RP, Iyengar R. Distinct characteristics of the basal activities of adenylyl cyclases 2 and 6. J Biol Chem. 1995;270(36):21368–21373. doi: 10.1074/jbc.270.36.21368. [DOI] [PubMed] [Google Scholar]
- 24.Toda T, et al. Intestinal flora induces the expression of Cyp3a in the mouse liver. Xenobiotica. 2009;39(4):323–334. doi: 10.1080/00498250802651984. [DOI] [PubMed] [Google Scholar]
- 25.Mukherjee J, Zuckier LS, Scharff MD, Casadevall A. Therapeutic efficacy of monoclonal antibodies to Cryptococcus neoformans glucuronoxylomannan alone and in combination with amphotericin B. Antimicrob Agents Chemother. 1994;38(3):580–587. doi: 10.1128/aac.38.3.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dromer F, Charreire J. Improved amphotericin B activity by a monoclonal anti-Cryptococcus neoformans antibody: study during murine cryptococcosis and mechanisms of action. J Infect Dis. 1991;163(5):1114–1120. doi: 10.1093/infdis/163.5.1114. [DOI] [PubMed] [Google Scholar]
- 27.Rodrigues ML, et al. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot Cell. 2007;6(1):48–59. doi: 10.1128/EC.00318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodrigues ML, et al. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot Cell. 2008;7(1):58–67. doi: 10.1128/EC.00370-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oliveira DL, et al. Cryptococcus neoformans cryoultramicrotomy and vesicle fractionation reveals an intimate association between membrane lipids and glucuronoxylomannan. Fungal Genet Biol. 2009;46(12):956–963. doi: 10.1016/j.fgb.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang PX, Sanders PW. Immunoglobulin light chains generate hydrogen peroxide. J Am Soc Nephrol. 2007;18(4):1239–1245. doi: 10.1681/ASN.2006111299. [DOI] [PubMed] [Google Scholar]
- 31.Nieva J, Wentworth P., Jr The antibody-catalyzed water oxidation pathway--a new chemical arm to immune defense? Trends Biochem Sci. 2004;29(5):274–278. doi: 10.1016/j.tibs.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 32.Magliani W, Conti S, Cunha RL, Travassos LR, Polonelli L. Antibodies as crypts of antiinfective and antitumor peptides. Curr Med Chem. 2009;16(18):2305–2323. doi: 10.2174/092986709788453104. [DOI] [PubMed] [Google Scholar]
- 33.Lewis RA, Hultquist DE, Baker BL, Falls HF, Gershowitz H, Penner JA. Hypercupremia associated with a monoclonal immunoglobulin. J Lab Clin Med. 1976;88(3):375–388. [PubMed] [Google Scholar]
- 34.Macura N, Zhang T, Casadevall A. Dependence of macrophage phagocytic efficacy on antibody concentration. Infect Immun. 2007;75(4):1904–1915. doi: 10.1128/IAI.01258-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maxson ME, Dadachova E, Casadevall A, Zaragoza O. Radial mass density, charge, and epitope distribution in the Cryptococcus neoformans capsule. Eukaryot Cell. 2007;6(1):95–109. doi: 10.1128/EC.00306-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 38.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37(8):911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 39.Siafakas AR, Wright LC, Sorrell TC, Djordjevic JT. Lipid rafts in Cryptococcus neoformans concentrate the virulence determinants phospholipase B1 and Cu/Zn superoxide dismutase. Eukaryot Cell. 2006;5(3):488–498. doi: 10.1128/EC.5.3.488-498.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.