Abstract
ColE1-like plasmids constitute the most popular vectors for recombinant protein expression. ColE1 plasmid replication is tightly controlled by an antisense RNA mechanism that is highly dynamic, tuning plasmid metabolic burden to the physiological state of the host. Plasmid homeostasis is upset upon induction of recombinant protein expression because of non-physiological levels of expression and because of the frequently biased amino acid composition of recombinant proteins. Disregulation of plasmid replication is the main cause of collapse of plasmid-based expression systems because of a simultaneous increase in the metabolic burden (due to increased average copy number) and in the probability of generation of plasmid-free cells (due to increased copy number variation). Interference between regulatory elements of co-resident plasmids causes comparable effects on plasmid stability (plasmid incompatibility). Modulating plasmid copy number for recombinant gene expression aims at achieving a high gene dosage while preserving the stability of the expression system. Here I present strategies targeting plasmid replication for optimizing recombinant gene expression. Specifically, I review approaches aimed at modulating the antisense regulatory system (as well as their implications for plasmid incompatibility) and innovative strategies involving modulation of host factors, of R-loop formation, and of the timing of recombinant gene expression.
Keywords: mutagenesis, fermentation, R-loops, RNA degradation, supercoiling, D-loops, plasmid instability, ColE1, dimer catastrophe, plasmid incompatibility
Introduction
Plasmids are DNA entities that replicate autonomously, i.e. independently from the chromosome. Plasmids are used as vectors for the production of recombinant proteins or genes for research and commercial purposes. There are many types of plasmids (reviewed in [1]). ColE1-like plasmids are the most popular ones because they are maintained at high copy number in the cells. This increases the yield of DNA and/or helps raise the level of expression of the desired recombinant product. Popular ColE1 plasmid vectors, with convenient selectable markers and multiple cloning sites include pACYC [2, 3], pUC18 and 19 [3], pBluescript SK−/+ and KS−/+ [4], and pBR322 and derivatives [5].
Encoding a recombinant gene in a plasmid facilitates genetic engineering ex-vivo, recovery of the DNA, and shuttling the gene into different strains or hosts. On the other hand, the high metabolic burden caused by expression of a recombinant gene often causes plasmid instability and runaway plasmid replication. These problems affect expression systems on an industrial scale (for which yield is critical) and are magnified in fed batch fermentation, i.e. in cultures that are prolonged through the addition of nutrients coupled to biomass. Therefore, a significant amount of research has been dedicated to controlling plasmid copy number to improve the yield of DNA and protein for research and biotechnological applications. The present article reviews how ColE1 plasmid replication is regulated normally and how replication is modulated to optimize recombinant gene expression. This review focuses exclusively on ColE1-like plasmids, which are the most popular medium- and high-copy vectors, although some strategies can also be extrapolated to other types of plasmids.
Col E1 Plasmid replication
Replication of ColE1 is under the control of a 550 bp sequence known as the plasmid origin of replication or ori. Selzer et al. noticed a striking conservation between the ori sequences of ColE1, pMB1, p15A, RSF1030, and CloDF13 and proposed that they may have comparable mechanisms of replication [6]. This was later confirmed, and these plasmids are known as ColE1 family plasmids (reviewed in [1]).
The plasmid ori encodes an RNA pre-primer that forms a stable hybrid with a 30-nucleotide stretch of the DNA template strand. This stable hybrid, known as R-loop, is processed by RNAseH to create a 3’ -OH end. Extension of this end by DNA polymerase I initiates leader strand DNA synthesis. The elongating leader strand separates the double-stranded DNA template, creating a structure known as a D-loop and exposing a primosome assembly signal (n’-pas). This signal is bound by PriA, PriB and DnaT to form a complex that initiates replication fork assembly, which includes the replicative polymerase Pol III. Pol III completes the replication of the plasmid. Right positioning of the 3’ –OH end of the pre-primer is critical for RNA processing [7] and for Pol I extension [8].
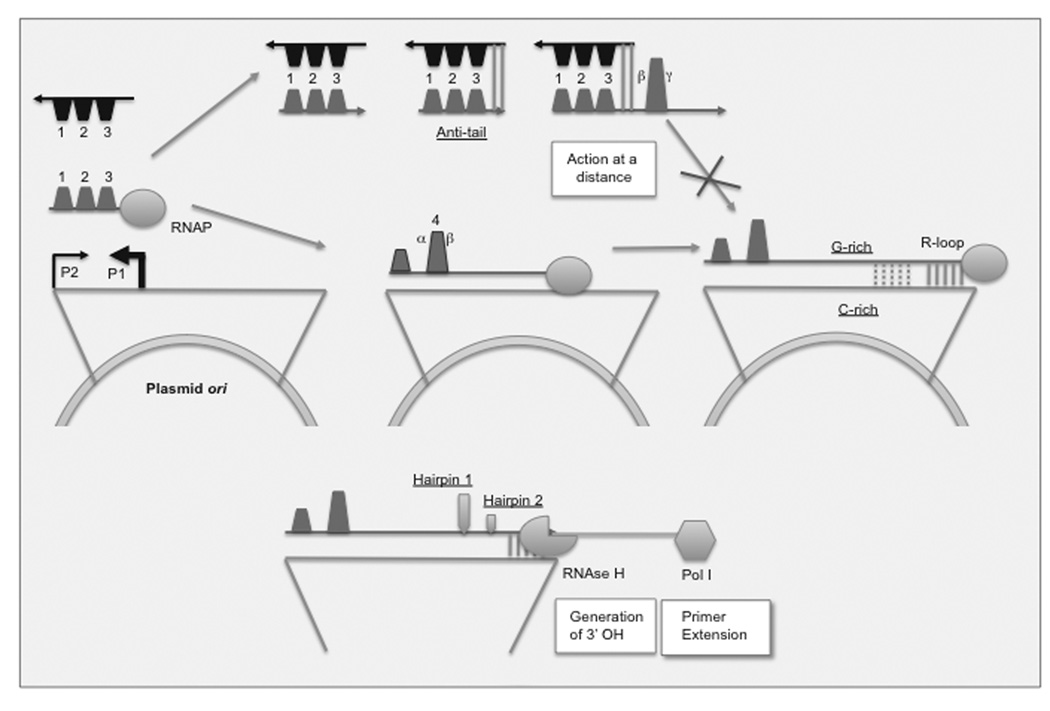
Replication is regulated by transcription of a 108 nt-long antisense RNA encoded by the plasmid (known as RNA I), which is transcribed from antisense promoter P1 (reviewed in [9, 10]) [Fig. (1)]. The pre-primer (known as RNA II) is transcribed from a sense promoter, P2. The antisense promoter is much stronger, resulting in a 100-fold excess of inhibitor relative to pre-primer. This constitutes a negative feedback loop, as the levels of inhibitor are proportional to the number of plasmids. The result is a specific copy number/per cell for a given set of conditions. Plasmid copy number regulation, however, is very dynamic and responds to metabolic fluctuations and to the presence of additional plasmids in the host. The short half-life of RNA I (only 2 minutes during exponential growth) facilitates a rapid response to environmental input. The antisense regulatory system is dispensable for plasmid replication but the ensuing sustained increase in plasmid copy number (known as runaway plasmid replication) may be lethal for the host. The 5’ end of the ori (up to position −386) can be deleted, which makes plasmid copy number proportional to P2 transcription [11].
Fig. (1). Overview of ColE1 plasmid homeostasis.
ColE1 plasmid replication is controlled by a ~550bp sequence element known as plasmid origin of replication (ori).
Plasmid replication: transcription of ori sequence off of the P2 promoter generates a primer RNA that primes leader-strand synthesis. An early (~100 bp) transcript forms three stem-loops with 6 or 7 unpaired bases at the end (SL1, 2, 3). Continued transcription leads to be formation of a new, larger stem-loop (SL4) through the interaction of two complementary sequence segments, α and β. This stem-loop is necessary for stable RNA-DNA hybrid (R-loop) formation. The formation of this hybrid is guided by a G-rich stretch of the primer that hybridizes with the complementary C-rich sequence on the template. The primer is processed by RNAse H to form a 3’-OH end and extended by DNA polymerase I. R-loop formation is the only absolute requirement for plasmid replication initiation.
Antisense regulation: Plasmid copy number is regulated by an antisense RNA whose expression is under the control of the P1 promoter. This inhibitor RNA forms three stem-loops, which are complementary to SL1,2,3 of the pre-primer. The interaction between the tips of these loops forms a transient “kissing complex” and then a permanent hybrid, a process that is nucleated by the antitail portion of the transcript at the 5’ end. This hybrid favors de interaction of the β segment of sequence with another stretch of sequence (γ), preventing the formation of SL4 and therefore of R-loop formation. The levels of antisense transcript are proportional to the number of plasmids, creating a negative feedback loop that keeps plasmid copy number constant for a given set of conditions.
RNAP=RNA polymerase
One unique feature about ColE1 antisense regulation is that the sequence of the inhibitor RNA overlaps with the 5’ end sequence of its target, the pre-primer RNA. Thus, the inhibitor is complementary to its target and hybridizes with it. The inhibitor-target interaction is determined by the formation of three stem-loops that leave 6 to 7 unpaired residues at the tip. These loops, known as stem-loops 1, 2 and 3 (SL1, SL2 and SL3) form in both target and inhibitor RNAs and mediate their initial interaction. Next, the 5’ end of the inhibitor (known as antitail) nucleates the hybridization between the two RNAs to form an RNAI-RNAII duplex. The formation of an RNA-RNA duplex is stabilized by a plasmid-borne protein, Rop [12]. Rom (the gene encoding Rop) has been removed in most vectors, which raises plasmid copy number by 3 to 5-fold [13]
Transcription of the pre-primer past the initial 200 nucleotides leads to the formation of a new loop (SL4), through the interaction of two sequence domains, α and β [Fig. (1)]. Once SL4 is formed, the pre-primer transcript can no longer bind the antisense inhibitor. Thus, although the inhibitor RNA is present in vast excess, it has a short window of action because it is dependent on the kinetics of folding of its target.
Hybridization between the pre-primer RNA and its inhibitor favors subsequent hybridization of β with another sequence domain further downstream in the pre-primer sequence (γ) and this βγ hybrid is incompetent for R-loop formation. This explains the action at a distance of the inhibitor RNA, preventing R-loop formation at the 3’ end of the pre-primer RNA upon hybrid formation at its 5’ end.
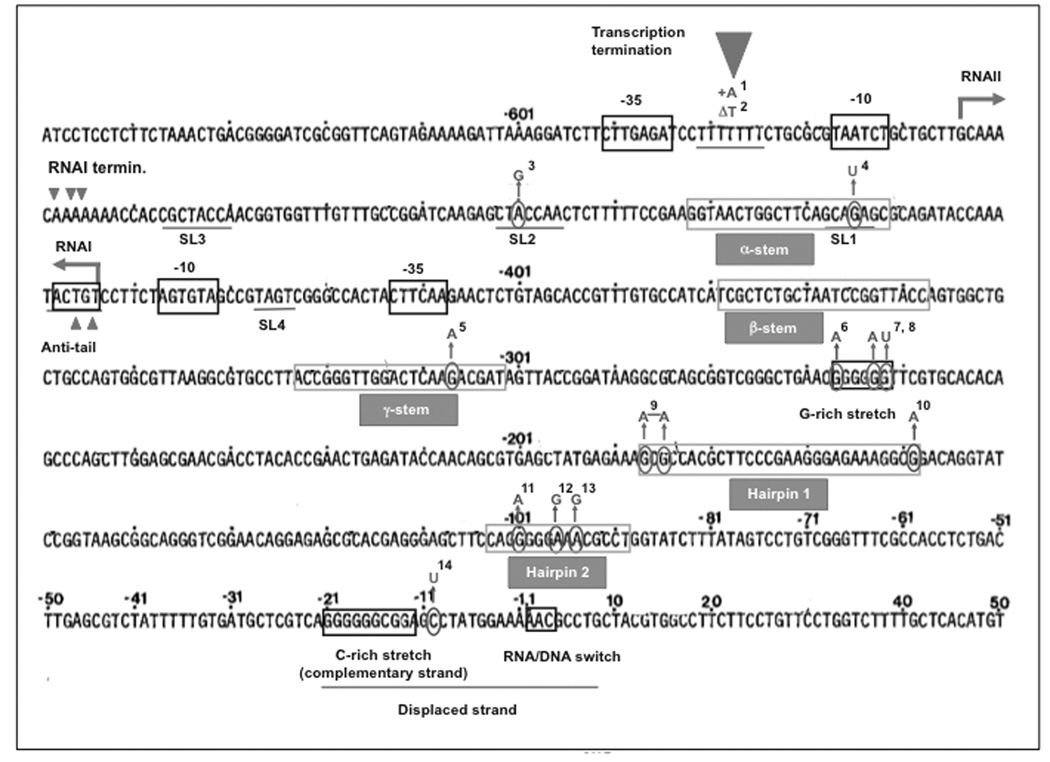
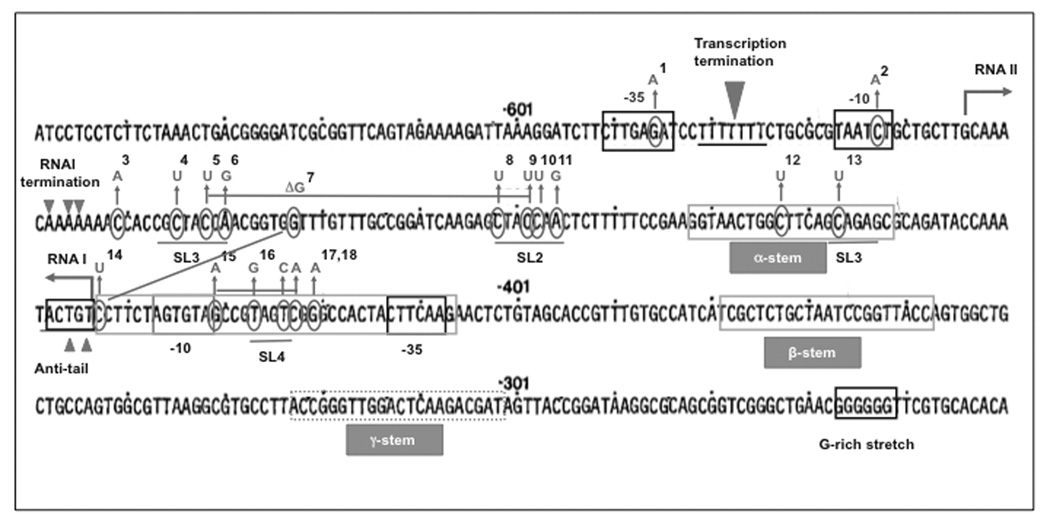
Additional functional sequence domains have been identified through loss of function mutations and their suppressors [Fig. (1), (2) and Table (1)]. These include an A T-rich motif between the −35 and −10 boxes important for promoter activity, a G-rich patch in the primer RNA and a C-rich patch on the template DNA whose interaction assists in R-loop formation [7], and two hairpin structures toward the 3’ of the pre-primer. The first hairpin is important for pre-primer processing and extension [7, 14], whereas the second one plays a role in R-loop formation [15]. Representative loss-of-function mutants are presented in Fig. (2). The phenotypes associated with these mutants and additional second-site suppressors are listed in Table (1).
Fig. (2). Functional determinants of ColE1 plasmid origins of replication and examples of loss-of-function mutations.
The sequence of the ColE1 ori is presented as originally described [84]. Functional elements are highlighted as boxes and secondary structures are underlined. Of these, three elements are conserved, suggesting common functionality between ColE1, pMB1, p15A, RSF1030, and CloDF13: 1) stem-loops 1, 2, and 3, although the unpaired sequences at the tip show divergence (for antisense regulation) [6]; 2) the potential to expose an anti-tail (for the formation of a stable RNAI/RNAII complex) [53]; 3) G-rich and C-rich stretches (whose interaction is likely involved in preventing the release of the nascent transcript) [9]. The RNA primer hybridizes with a ~30 nucleotide stretch around the DNA/RNA switch. Beyond 7 nucleotides downstream of the RNA/DNA switch, sequence homology within ColE1 family plasmids is lost, except for positions +110 and +170 [6], which contains a putative binding site for the PriA [85]. PriA is normally recruited to restart replication at sites of replication fork collapse (reviewed in [86]). It seems that plasmids have co-opted this mechanism of DNA repair to complete the replication of their own genomes (reviewed in [85]). In agreement with their relative lack of conservation, sequences downstream of position +14 are dispensable [87]. Mutations that decrease the efficiency of replication initiation in ColE1 or in the closely related pMB1 ori are indicated and numbered. Mutations isolated together in combination are joined with a line. The reference key for loss of function mutations is presented separately in Table (1), which lists phenotype, references and second-site suppressors for each mutant.
Table (1).
Representative loss-of-function mutants of ColE1 family plasmids and corresponding suppressors.
| # | Mutation name |
Ref | Mechanism | Suppressors | Ref |
|---|---|---|---|---|---|
| 1 | pri10, cer7 | [7,14] [14] |
Inactivates P2 PolA essential in rnhA strains [16] |
her (RNAseH- deficient) |
[14] |
| 2 | cer17 | [14] |
Her (RNAseH- deficient) |
[14] | |
| 3 | svir 11 | [44] | |||
| 4 | pri7 | [15] | Favors β, γ conformation |
spr71 , spr77 , spr76 (stem of loop 3, restore stem IV formation) |
|
| 5 | pri1 | [7] | Preventing R-loop formation PolA essential in rnhA strains [16] |
||
| 6 | pri2 | [7] | |||
| 7 | pri3 | [7] | |||
| 8 | pri4 | [7] |
spr42 spr41 (enhance Pri3) |
[7] | |
| 9 | pri6 | [7] | Preventing primer extension. Insensitive to RNAseH and polA inactivation[16] |
spr61 Also addition of excess Pol I |
|
| 10 | cer6 | [80,14] | Conformational change that breaks a hairpin structure; makes the primer hypersensitive to RNAseH |
rev16 her (RNAseH- deficient) herB (increased DNA polymerase I activity) |
[14 , 80 , 81] |
| 11 | pri9 | [82] | Preventing R-loop formation (destabilizes hairpin) |
||
| 12 | cer8 | [82] | |||
| 13 | cer114 | [82] | Defective in RNAseH-dependent and independent modes; Conformational change |
rev20 herC |
[82] |
| 14 | pri5 | [7] | RNAseH cleavage sites shifter 5 nt upstream Prevents primer extension Insensitive to RNAseH and polA inactivation[16] |
Mutations with a negative effect on plasmid copy number, their names, phenotype and probable mechanism of action are listed. The number in column 1 identifies the mutation presented in Fig. (1). Pri= Priming-defective. Cer=ColE1 Replication defective; host factor suppressors are called her, for Host factor affecting colE1 Replication. svir=suppressor of virulence.
Overall, the final copy number of a ColE1 family plasmid is determined by the relative strengths of promoters P2 and P1 and by the efficiency of inhibitor-target interaction. Single nucleotide changes in the area of RNA I/RNA II overlap alter both the effector (RNAI) and the target (RNA II), maintaining complementarity while potentially altering the affinity of the interaction. This built-in tolerance to point mutations greatly facilitates the mutational fine-tuning of the system.
Finally, ColE1 replication is very versatile. It occurs at some frequency in the absence of RNAseH (by extension of unprocessed pre-primers), [16] and even in the absence of both RNAseH and Pol I (by PriA-dependent fork assembly at the D-loop) (reviewed in [9]). The one element that is shared between all modes of ColE1 plasmid replication is the requirement for R-loop formation. We’ll see that the field is moving from modulating the antisense regulatory system to trying to modulate host factors, particularly R-loop formation. While the later approach is more challenging, it is potentially more robust to metabolic alterations caused by recombinant gene expression.
Role of host factors
I have already mentioned the direct role of some host proteins for ColE1 plasmid replication. These include: RNA polymerase (synthesis of the RNA pre-primer and of its antisense inhibitor), RNAseH (pre-primer processing), DNA polymerase I (leader-strand synthesis), and PriA-dependent fork assembly (completing plasmid replication). Three additional processes have a significant impact on plasmid homeostasis: modulation of R-loop formation, control of RNA I decay, and resolution of dimers and multimers generated by recombination. Each of these processes involves several additional host factors.
E. coli maintains a global level of negative supercoiling within a 15% range. According to the “twin domain model”, local supercoiling is caused by the movement of an RNA or DNA polymerase along the template, which (assisted by DNA gyrase) causes the DNA to rotate around its helical axis creating negative supercoiling in its wake [17]. Local supercoiling immediately behind the RNA polymerase during transcription leads to R-loop formation, which causes transcription blocks and inhibits elongation [18,19]. Topoisomerase I is primarily responsible for suppressing R-loop formation during transcription elongation to allow gene expression, although it is partially redundant with Topoisomerase IV. RNA hydrolysis by RNAseH, and unwinding by RecG also suppress R-loop formation. Coupling transcription to translation and factor-dependent transcription termination are additional mechanisms preventing the hybridization of nascent transcripts with DNA in E. coli (reviewed in [19]). Thus, uncoupling transcription from translation (by nutrition, temperature shifts or by chloramphenicol treatment) favors R-loop formation [20]. These treatments increase plasmid copy number, presumably by titration of R-loop suppressing factors [20]. Conversely, overexpression of RecG causes a dramatic fall in ColE1 plasmid copy number [21]. The effects of altering the two other major suppressors of R-loop formation, namely Topoisomerase I (encoded by topA) and RnaseH (encoded by rnhA) are more complicated. TopA cells exhibit R-loop formation in a plasmid but, RNAse H appears to be sufficient to suppress unscheduled R-loop formation in these cells. RnaseH inactivation on the other hand leads to unscheduled R-loop formation (and unscheduled replication fork assembly). In spite of such promiscuous R-loop formation, rnhA strains show decreased ColE1 plasmid copy number because RNAseH is also required for processing the RNA pre-primer [16].
RNAI is degraded by the combined action of endonucleases and exonucleases and is regulated by polyadenylation. Polyadenylation is catalyzed by a poly(A)polymerase encoded by pcnB gene. Addition of a poly(A) tail to the inhibitor RNAI reduces its half-life and its affinity for its target RNA II [22]. RNAse E cleaves RNA I at a single site. This intermediate is further degraded by the poly(A)-dependent activity of polynucleotide phosphorylase, RNAse II exonuclease and RNAse III endonuclease (reviewed in [23]).
Homologous recombination between plasmids generates dimers and higher multimers at low frequency. This decreases the number of independently segregating units, increasing plasmid copy number variability in individual cells. With time in culture, multimer representation gradually increases, possibly because the presence of more than one origin of replication confers a selective advantage. The net result is a substantial decrease in plasmid stability. ColE1-like plasmids encode a 250bp region (the cer site [24]) that resolves multimers through site-specific recombination mediated by host proteins [25]. Cer sites, however were inadvertently removed during the construction of ColE1-based vectors as they are far from the plasmid ori and not essential for plasmid replication or maintenance. Strains used as hosts for recombinant gene expression are typically deficient in recA function, which mediates initiation of homologous recombination and therefore lack significant plasmid multimerization over time.
Disregulation of ColE1 plasmid replication
Cultures tend to lose plasmids unless they are under selection (reviewed in [26]). On a laboratory scale, sthis problem can be easily solved using antibiotic or auxotrophic selections. On an industrial scale, however, auxotrophic selections are not desirable because defined media decrease biomass and product yields. The use of antibiotics is not practical on an industrial scale either for two reasons, namely the amount of antibiotics involved (3 kg to achieve the standard 100 mg/l concentration of ampicillin for a 30m3 fermentation) and the need to subsequently purify the antibiotic away from the product. Genetic complementation strategies can sometimes be used as alternative positive selections, but achieving maximal plasmid stability remains a major goal in biotechnology (reviewed in [27]).
Plasmid instability
Plasmid instability is determined by two major variables, namely the frequency of plasmid loss and the relative selective advantage of plasmid-free cells. Plasmid loss is stochastic, and depends on the average copy number per cell and on the copy number variability in individual cells. For multicopy plasmids, given their high average copy number, the standard deviation is the most important of the two variables (reviewed in [28]). Thus, removing one component of the replication regulatory system (Rop) results increased plasmid instability despite a 5-fold increase in plasmid copy number [13]. We’ll see below that conditions that alter plasmid replication regulatory circuitry tend to increase copy number variability, facilitating plasmid loss.
The selective advantage of plasmid-free cells is proportional to the metabolic burden caused by recombinant gene expression. This burden has two components: one that is caused by plasmid replication within the cell and one that is caused by expression of recombinant genes outside their natural context.
The burden of plasmid maintenance is proportional to the size of the plasmid. Against initial expectations, the metabolic cost of plasmid maintenance is not simply due to ATP consumption. Instead it appears to be caused by increased glucose uptake, possibly mediated by cAMP-CRP induction and upregulation of the phosphotransferase (pet) operon. This carbon overflow causes an accumulation of inhibitory intermediate metabolites and byproducts that is detrimental to the cell [29]. In general, though, the metabolic cost of plasmid maintenance to the host is limited [26].
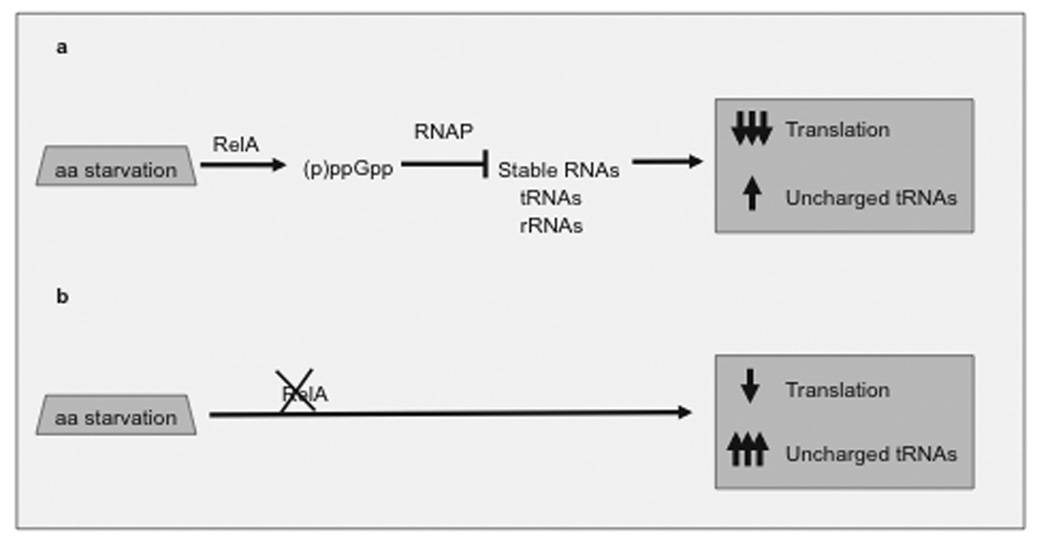
The metabolic burden of recombinant plasmid maintenance can be largely attributed to expression of recombinant genes outside their natural context. Expression of high levels of recombinant proteins upsets the balance of tRNAs in the cell, which is proportional to the amino acid composition and relative abundance of E. coli proteins [30]. This creates a situation akin to amino acid starvation. Increased levels of uncharged tRNAs cause idling reactions at the A-site of the 50S ribosome, which activate a ribosome-bound protein, RelA. Rel A synthesizes an unusual guanosine nucleotide, (p)ppGpp, which binds to the RNA polymerase, suppressing synthesis of stable RNA (tRNA and rRNA) synthesis. This so-called stringent response (reviewed in [31, 32]) limits expression of foreign recombinant proteins [Fig. (3a)].
Fig. (3). Metabolic response to amino acid starvation.
a. Stringent response. Amino acid depletion causes ribosome idling, which is sensed by a ribosomal protein, ppGpp-synthetase I (encoded by relA). Rel A activation produces an alarmon, (p)ppGpp, which interacts with RNA Polymerase. Conformational changes in the RNA polymerase lead to changes in promoter specificity, reducing the synthesis of stable RNAs and increasing expression of biosynthetic pathways (reviewed in [32]). This leads to a marked suppression in protein synthesis. b Relaxed response. RelA strains, which are defective in ppGpp synthetase I, maintain low levels of (p)ppGpp under conditions of amino acid starvation. This maintains the level of protein synthesis to continue, but generates high levels of uncharged tRNAs, which eventually also have an impact on levels of translation.
To avoid the stringent response, bacterial strains used for recombinant gene expression are typically deficient in relA function [Fig. (3b)]. In these strains, however, imbalanced amino acid consumption alters plasmid homeostasis. Disregulated replication raises the average copy number and increases the variation of the plasmid copy number in individual cells. This in turn amplifies the metabolic stress caused by recombinant expression, leading to plasmid instability (reviewed in [27]). Therefore, the increase in plasmid copy number associated with induction of recombinant gene expression has been recognized as one of the main factors causing the collapse of the expression system and is being intensively investigated (reviewed in [33, 34]).
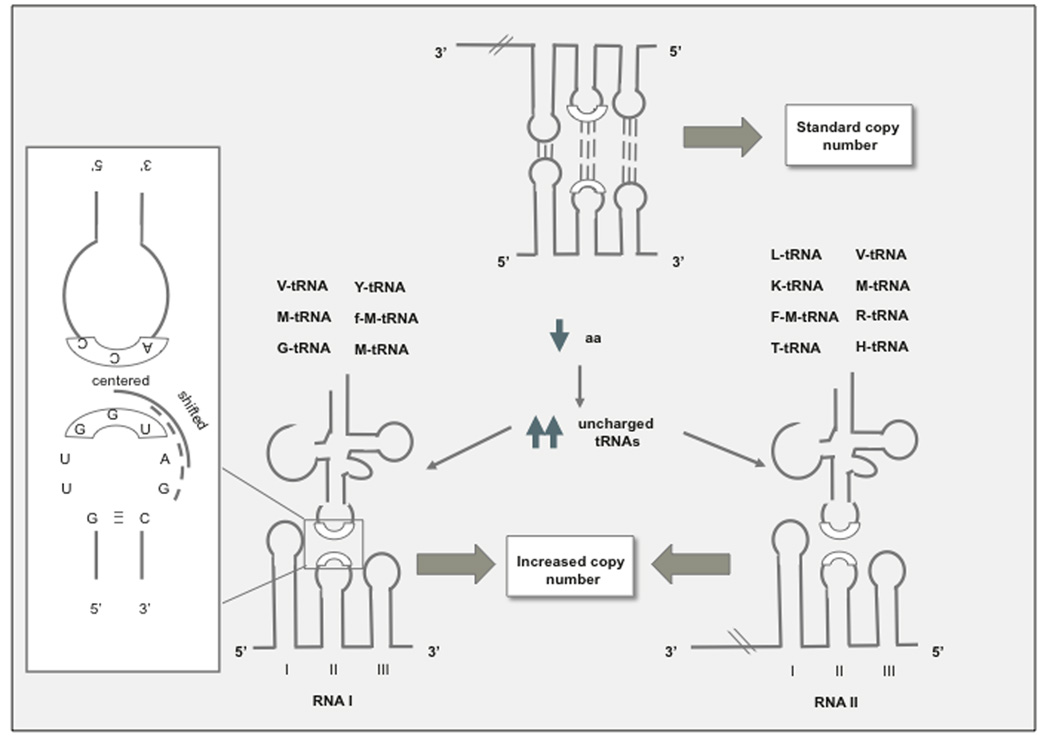
Plasmid disregulation associated with recombinant gene expression appears to be mediated by uncharged tRNAs. The first clue suggesting tRNA involvement was the observation that the outcome of amino acid starvation varies depending on the specific amino acid that is lacking (reviewed in [35]). Two models have been reported to explain the link between amino acid starvation and increased plasmid copy number in relA strains [Fig. (4)]. Yavachev et al. noticed a structural similarity between unpaired residues at the tip of SL1, 2 and 3 and the cloverleaf structure of t-RNAs. In addition to this structural similarity, they found >40% homology between the anticodon loops of eleven tRNAs and RNA I (R, H, L, K, F, T), RNA II (f-Met, Y, G), or both (M, V) [36]. They postulated that competitive hybridization between the tRNA and RNA I or RNA II could interfere with the formation of RNAI/RNAII hybrids [36]. This model predicts that changes in plasmid copy number associated with deprivation of individual amino acids should correlate with the homology between the corresponding anticodon tRNAs and RNA I and II inhibitory loops. To explain the preferential hybridization of uncharged tRNAs, this model invokes rapid titration of charged tRNAs by the replication machinery [Fig. (4a)]. The second model suggests that the 3’-CCA of uncharged tRNAs hybridizes with the GGU…G sequence of RNA I, and that this bond is stabilized because a proton given by the CCA is trapped by an electron hole (GG+) at RNA I loops [37]. This would neatly explain the preferential effect of uncharged tRNAs. To explain the specific effects of individual amino acid deprivation, it points out that since RNA I is the more highly-expressed of the two ori transcripts, it is more sensitive to the levels of abundant uncharged tRNAs. This model predicts that members of the ColE1 family of enzymes that encode the GGU…G motif in the RNAII transcript (which is rate-limiting), starvation for any amino acid should lead to runaway plasmid replication [Fig. (4b)]. Experimental evidence is consistent with both models [35, 37]. In addition, uncharged tRNAala was also shown to cleave RNA I both in vitro and in vivo [38], directly tying plasmid replication to an increase in levels of uncharged tRNAs.
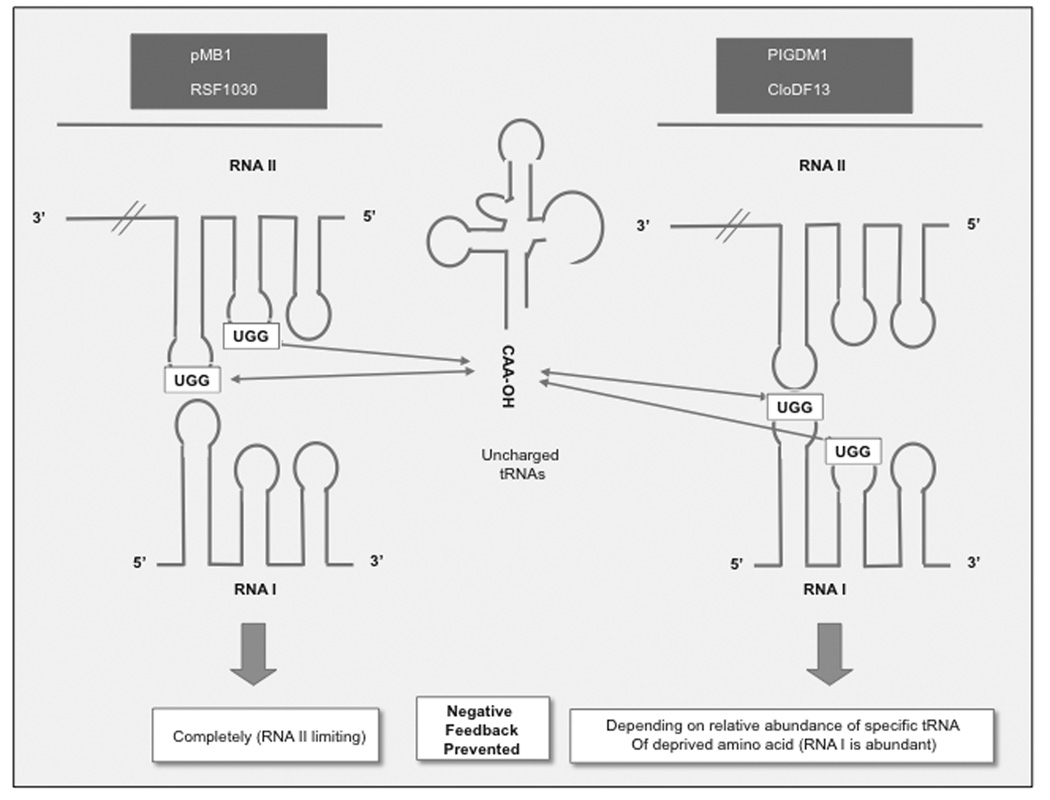
Fig. (4). Models of disregulation of plasmid replication caused by accumulation of uncharged tRNAs in relA stains.
a Formation of codon-anticodon complexes with tRNA. A structural similarity and >40% sequence homology was noticed between SL1, 2, and 3 of RNA I, II or both and the cloverleaf structure of t-RNAs. Yavachev et al. postulated that competitive hybridization between the anticodon loop of tRNA and the corresponding anticodon-like loops of RNA I or RNA II could interfere with the formation of RNAI/RNAII hybrids [36]. This model predicts that changes in plasmid copy number with deprivation of individual amino acids should correlate with the homology between the corresponding tRNAs and loops in RNA I and II. Note that each unpaired loop provides three different options for hybridization with tRNA anticodon sequences. A 7-nt loop is shown in the inset as an example: centered (boxed), shifted (continuous line) and very shifted (broken line). Based on solvent exposure, centered interactions are assumed to be the preferred ones, while the very shifted ones would be the least preferred ones. b. CAA-OH tRNA hybridization with UGG sequence of RNAI or RNA II. 3’-CAA of uncharged tRNAs hybridizes with the GGU motif of either RNA I or RNAII (depending on the specific sequence of each ori), and this bond is stabilized because a proton given by the CCA is trapped by an electron hole (GG+) at RNA I, RNA II loops [37]. The specific effects of individual amino acid deprivation are explained because in ColE1 plasmids, the GGU…G sequence is encoded in RNA I, which is the more abundant of the two ori transcripts and therefore sensitive to the levels of uncharged tRNA, which in turn corresponds to the relative abundance of different amino acids in E. coli proteins. This model predicts that starvation for any amino acid should lead to runaway plasmid replication in the case of PIGDM1, CloDF13 and other members of the ColE1/pMB1 family of enzymes that encode the GGU…G in the RNAII transcript. Note that both models propose a role of tRNAs in regulation of replication initiation. It has been proposed that tRNAs originated from RNA replication primers not unlike RNA II, as clover leaf-like structures frequently prime replication of viral and plasmid genomes (note the clover leaf-like structure of RNA II) [88].
A different interpretation is based on the observation that in bacteria translation is coupled to transcription, and that translation suppresses R-loop formation by transcripts emerging from the RNA polymerase during transcription [18, 20]. According to this model, amino acid starvation uncouples translation from transcription, facilitating unscheduled R-loop formation. This would titrate out factors that destabilize R-loop formation in ColE1 plasmids, leading to runaway plasmid replication [20]. Besides neatly explaining the role of amino acid starvation, this model explains two other instances of increased plasmid copy number: inhibition of translation by chloramphenicol treatment [39], and inhibition of factor-dependent transcriptional termination by rho or nusG mutations [40]. However, this model doesn’t account for the differential effect of individual amino acid deprivation. Most likely plasmid disregulation associated with recombinant gene expression is caused by a combination both mechanisms, namely modulation of antisense RNA inhibition by uncharged tRNAs and titration of suppressors of R-loop formation.
Incompatibility
Incompatibility is the inability of two plasmids to coexist stably in a population of growing bacteria without external selective pressure. It can be understood as a form of disregulation. In this case, the disregulation is caused by the introduction of one or more elements belonging to a second plasmid that regulate replication initiation or partitioning of the resident plasmid in trans. In turn, this second plasmid can be susceptible to inhibition by shared regulatory elements of the resident plasmid. Thus, contralateral regulation between plasmids interferes with the autologous (ipsilateral) regulation of each plasmid.
As in the case of plasmid instability, incompatibility is associated with increased plasmid copy number variation (reviewed in [28]). If contralateral inhibition is comparable between the two plasmids (symmetrical), either of them is eliminated stochastically. Asymmetrical contralateral regulation, on the other hand, preferentially eliminates one of the plasmids. If two plasmids are refractory to contralateral regulation by each other, they belong to different incompatibility groups. This means that plasmids belonging to each of these groups can stably coexist in the same cell even if they share regulatory elements. For example, ColE1 is compatible with pA15 and CloDF13 and RSF1030, but not with pMB1. We’ll see below that compatibility within ColE1 family plasmids can be modulated by mutagenesis of the antisense RNA regulatory element.
In sum, plasmid copy number regulation is very dynamic, tuning plasmid metabolic burden to the physiological state of the host. This delicate balance is upset upon induction of recombinant protein expression, as proteins are being expressed at non-physiological levels, often have biased amino acid compositions, and may aggregate or exhibit toxicity (reviewed in [26]). Because of a simultaneous increase in the metabolic burden (due to increased average copy number) and in the probability of generation of plasmid-free cells (due to increased variation), disregulation of plasmid replication has a serious negative impact on recombinant expression systems.
Engineering ColE1 plasmid copy number
Modulating plasmid copy number for optimal recombinant gene expression is a delicate balancing act involving increasing gene dosage while maintaining a stable copy number. Approaches used to modulate plasmid replication for recombinant gene expression will be discussed below. One group of methods primarily aims at raising plasmid copy number as a way of increasing gene dosage. This in turn causes the disregulatory effects just discussed. We’ll also discuss a second class of methods whose primary target is minimizing plasmid metabolic burden.
These methods modulate plasmid regulation through mutations in the ori, through mutations in the host strain or through alterations in culture conditions. Fig. (5) below gives examples of mutations in ColE1 or the closely related pMB1 origins of replication that increase plasmid copy number. Table (2) lists their references and phenotype. The Rop protein, which stabilizes the binding of inhibitor RNA I to its target will not be discussed because it is absent in most ColE1-like plasmids used for recombinant protein expression.
Fig. (5). Examples of gain-of-function mutations.
Mutations that increase plasmid copy number in ColE1 or in the closely related pMB1 ori are indicated and numbered on the sequence of ColE1 originally described in [84]. Mutations isolated together in combination are joined with a line. The reference key for the mutations is presented separately in Table (2), which lists phenotype and references for each mutant.
Table (2).
Representative gain of function mutants of ColE1 family plasmids
| # | Mutation Name |
Ref/Patent | Phenotype | Mechanism |
|---|---|---|---|---|
| 1 |
svir017 (cop1 in CloDF13) |
[41, 42] | Increased PCN | Increased P2 expression (stabilizes secondary structure in promoter) |
| 2 | [83] | Increased PCN | Increased P2 expression | |
| 3 | pOPIΔ6 (same in pBR322) |
[54, 55] | Increased PCN (200–300 copies) |
Read-through transcription of RNA I, altering its secondary structure |
| 4 | inc4 | [45] | Increased PCN(3×) | |
| 5 | inc1 | [45] | Increased PCN (2×) | |
| 6 | inc9 | [45] | Increased PCN (1.5×) | SL3 disruption |
| 7 | Patent | [48] | Increased PCN | |
| 8 | svir 3,115 | [44] | Runaway plasmid replication |
|
| 9 |
svir 2,7; inc1 inc2 pWT11cop |
[44, 45] | Increased Increased (7×) New compatibility group |
|
| 10 | svir12 | [44] | Increased | |
| 11 | svir102 | [44] | ||
| 12 | svir19 | |||
| 13 | inc3 | [45] | Increased (1.5×) Altered compatibility |
Decreased inhibition by RNAI due to weaker interaction to homologous residues in unpaired loop |
| 14 |
svir002 (C to A in pUC) |
[41, 51, 65] | Increased (Temperature-dependent) |
Stabilizes α, β; conformation less sensitive to inhibition |
| 15 | pEW2705 | [64, 52] | Te- dependent PCN 30 °C normal 42 °C runaway (host affected) Growth phase-dependent @ 37 °C Exp: 2–4× Sat: 20× |
Stabilizes α, β; conformation less sensitive to inhibition because antitail is hidden in a stem |
| 16 | pMM7 | [64, 52, 53] | ~ pEW2705 (less Ts?) |
|
| 17 | pMM4 | [64, 52, 53] | ~ pEW2705 (less Ts?) |
|
| 18 | pMM1 | [64, 52] | ~ pEW2705 |
Mutations increasing plasmid copy number, their names, phenotype and probable mechanism of action are listed. The number in column 1 identifies the mutation presented in Fig. (5). inc=incompatibility mutants, allow plasmids pNT7 and pMB9 to be stably inherited in the same cell. svir= suppressors of virulence. cop=copy number. PCN=plasmid copy number
Strategies aimed at raising plasmid copy number
Plasmid copy number can be raised through increased expression of P2 (which is normally weak). svir017 increases the strength of this promoter by stabilizing a secondary structure in the −35 box [41, 42]. It can also be increased by inactivating antisense regulation. As mentioned above, RNA I regulation is dispensable. Removing RNA I completely from ColE1 plasmids, however, leads to lethal runaway plasmid replication. This can be solved by reducing the levels of pre-primer expression. P1 (the promoter for the inhibitor RNA I transcript) was deleted and P2 (the promoter for priming RNA II transcript) was replaced with an IPTG-inducible promoter, creating an inducible system of plasmid replication [11]. In this case, lethal runaway plasmid replication is avoided by keeping the levels of RNAII pre-primer low. Alternatively, RNAI can be only partially inactivated, leaving sufficient inhibitory activity to reach a new regulatory state with a stable, elevated copy number. This concept was patented early on [43] and proved extremely successful.
Partial RNA I inactivation can be achieved in different ways:
1. Introducing mutations in the area of RNA I/RNA II overlap that decrease the affinity of RNA I/RNA II interaction
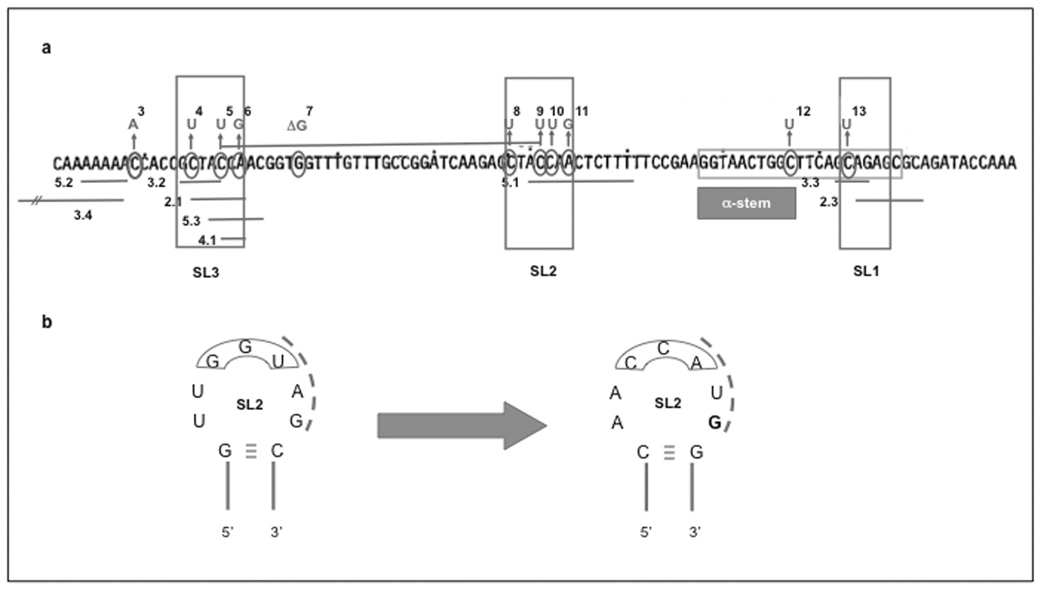
As mentioned above, point mutations will preserve the complementarity of both RNAs, but may alter the affinity of the interaction. In some cases mutations alter the energetics of loop-loop interaction, as A-U bonds have a lower free energy content than G-C bonds. Examples of C→U mutations in inhibitory loops include inc4, and inc1 in SL3; svir2,7 and svir12 in SL2; and inc3 in SL1[44, 45] [Fig. (5), (6a), and Table (2)]. Alterations in the energetics of loop-loop interaction have been exhaustively explored by Grabherr et al. with the goal of tuning plasmid copy number to the metabolic burden of specific recombinant expression systems [46]. Given that these same residues are known to mediate incompatibility and responses to amino-acid starvation, changes in the unpaired sequence of SL1, 2, 3 have the potential added advantage of being less responsive to metabolic alterations [46, 47].
Fig. (6). Complex proprietary mutants.
a Deletion mutants in SL1, 2 and 3 [49, 50]. Deleted sections as reported in [50] (and patented [49]) are represented as lines below the sequence, with their identifiers next to them. For comparison, point mutants that increase copy number are also presented as numbers above the sequence. The legend to the numbers can be found in Table (2). b Inverted homology to tRNAs. Seven bases of SL2 (all but one G, in bold) were replaced by their complement bases, thereby inverting tRNA homologies without changing the complementarity between inhibitor and target RNA or any structural features of the stem-loop. A construct bearing the mutant ori and superoxide dismutase showed no signs of disregulation after induction of the recombinant protein in batch fermentation culture.
2. Mutations causing structural alterations
These can be found in the loop (possibly svir11, an A→G mutation in SL2) [44] or in the stem (possibly a G deletion affecting SL3) [48] [Fig. (5) and Table (2)]. Altering RNA I secondary structure has been thoroughly exploited by Smider et al. They selected mutants exhibiting increased plasmid copy number from large libraries of ori insertions and deletions [49]. They report that the vast majority of mutations mapped to the area of RNAI/RNA II overlap, and that, while most indels were small (10 nt in length) some large ones were tolerated [49]. Nine small deletions reported in a subsequent publication affect specifically SL1, 2 and 3, consistent with the distribution of partially-inactivating RNA I point mutations [Fig. (6a)] [50]. It seems that large deletions should lead to runaway plasmid replication; maybe the rare the ones identified in these screens decrease the efficiency of RNA II primer as well as abolishing antisense RNA regulation, thus preventing lethality.
3. Mutations that stabilize the αβ conformation of the pre-primer
These mutations are just downstream of RNA I/RNA II overlap. With one exception, svir002 [41, 51] these mutations correspond to unpaired residues of SL4 [64, 52, 53]. They all show a temperature-dependent increase in copy number that is sensitive to increased RNA I inhibition. In the case of SL4 mutations, at 37 °C the increase in copy number is also growth phase-dependent, with an additional 5–10× increase in saturated cultures [52]. One proprietary double mutant exhibiting a substantial increase in copy number probably combines structural alterations in SL3 (ΔG at position −525) with stabilization of the αβ conformation [48].
4. Mutations that allow read-through transcription of RNAI
This longer transcript is less effective as an inhibitor, probably due to alterations in its secondary structure. An example is pOPIΔ6, which is immediately adjacent to the polyA track that marks the end of RNA I transcription [54, 55].
Implications for incompatibility
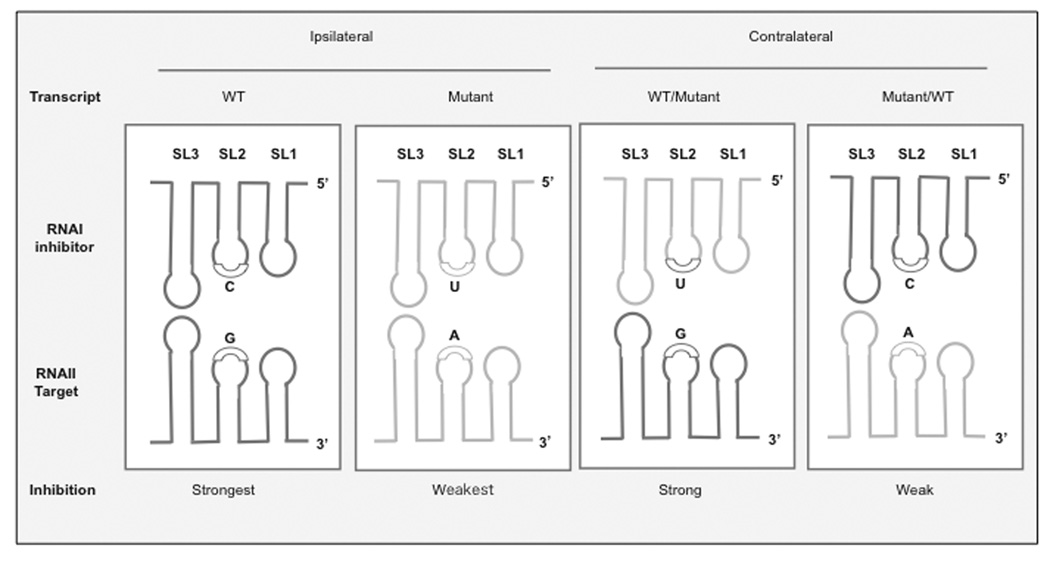
As discussed above, plasmid incompatibility can be considered a form of disregulation due to contralateral inhibition between two coexisting plasmids. Since this inhibition is mediated by RNA I, altering RNA I activity typically has a dual effect on copy number and on the compatibility properties of the plasmid. Thus, screens for mutations conferring compatibility to incompatible plasmids identified mutants with increased copy number (inc mutants) [45] and vice-versa (svir mutants) [44]. The mutations identified in these screens mapped to the loops of SL1, 2 and 3, altering the affinity of the inhibitor RNA I for its target RNA II. These mutations typically create disparities between ipsilateral and contralateral inhibitor-target interactions because these mutations rarely affect inhibitor and target function equally. Fig. (7) shows the effect of G to A transition on ipsilateral and contralateral interactions as an example. When asymmetrical contralateral effects occur, the plasmid showing stronger contralateral inhibition will preferentially be maintained.
Fig. (7). Effect of mutations affecting SL1, 2 ,or 3 interactions on contralateral regulation.
A G→ A point mutation is presented as an example. If the mutant plasmid coexists with the wild-type plasmid, two inhibitor RNA I species are present. The corresponding contralateral interactions are: WT RNA I (encoding C) is opposite an A, a weak interaction, and mutant RNA I (encoding a U) opposite a G, a less-destabilizing interaction. This asymmetrical effect leads to the preferential elimination of the plasmid that is more efficiently repressed, the wild-type plasmid in this case.
Deletions affecting SL1, 2 or 3 loops also frequently have a dual effect on plasmid copy number and on compatibility, but in this case the two effects appear to be separable, as deletions in SL3 had little effect on compatibility while increasing plasmid copy number [49, 50].
Dominant mutations occurring outside the overlap region (e.g. stabilizing RNA II conformations that are insensitive to RNA I inhibition) tend to eliminate the wild-type plasmid, because the mutant plasmid is not sensitive to WT RNA I inhibition, whereas the WT plasmid is.
Two plasmids that are mutually refractory to contralateral regulation belong to two different incompatibility groups. Plasmid compatibility can be generated by point mutations in the region of RNAI/RNAII overlap [44, 45] or by removal of RNA I regulation, although in this case the efficiency of priming by RNA II needs to be controlled to avoid potentially runaway plasmid replication [11]. There is an interest in generating compatible origins for ColE1 plasmids to facilitate co-expression of multiple proteins (for functional proteomics, functional genomics, metabolic engineering, etc…) and for in vivo shuffling of random DNA libraries [56, 57]. Currently, standard ColE1/pMB1 vectors can coexist with plasmids containing the related pA15, CloDF13, RSF1030 origins of replication but when ColE1 family plasmids reside together at least one of them has very low copy. Unrelated vectors such as pSC101 could be used, but again these tend to have lower copy number and lack the cloning and expression features of the more sophisticated ColE1 vectors. A recent report suggested that incompatible plasmids may coexist more stably that previously accepted (to the tune of 50% retention after 5 cycles of overnight growth) [58]. This result is in sharp contrast with earlier studies [44, 45]. It would be of great interest to see whether the way plasmids were introduced into the cell in the more recent report (by infection rather than by transformation) influenced the outcome. This would imply that introducing plasmids through phage infection could substantially improve plasmid compatibility.
Modulating host factors
The mutations described above provide a range of regulatory states leading to increased plasmid copy number. An alternative strategy to optimize plasmid copy number consists of modulating host factors that are directly or indirectly involved in plasmid replication. This greatly facilitates tuning of gene dosage according to the metabolic burden of the construct. Instead of cloning the gene of interest into vectors bearing a range of mutant ori plasmids, the same construct can be transformed into a panel of mutant strains in order to empirically find the optimal copy number. In this case, changes in both directions (i.e. increased and decreased copy number) are of interest. Two recent patents are based on this strategy [59, 60]. Notably, the effects on plasmid copy number are largely specific for ColE1 family plasmids even in the case of treatments expected to have pleiotropic effects on the host [61–63].
1. Culture conditions
Temperature and stress affect plasmid replication. Shocks of all kinds, including heat- and cold-shock, nutritional shifts and growth in stationary phase favor R-loop formation. This effect on R-loop formation could be due to effects on translation (ribosomes are prime stress sensors) and/or to substantial changes in gene transcription profile, which tend to generate excess negative supercoiling [18]. Mutations that stabilize the αβ conformation of the pre-primer further increase plasmid copy number in stationary phase [64]. In addition, high temperature (42 °C) destabilizes the interaction of inhibitor RNA with the pre-primer. Thus, increasing temperature leads to an increase in plasmid copy number, at least in the case of pUC19 [51, 65] and of mutants with stabilized αβ conformations [64].
2. R-loop formation
Unscheduled R-loop formation leads to dramatic increases in ColE1 plasmid copy number, presumably by titrating suppressors of R-loop formation. However, induction of R-loop formation in the host needs to be carefully controlled, as it creates blocks for transcription and is potentially lethal [40]. Strategies for induction of R-loop formation include:
2a. Global translation arrest in de absence of any limitation on transcription. Examples include treatment with low levels of chloramphenicol or spectinomycin [39], letting cultures reach stationary phase [64, 66], and possibly other stressors (reviewed in [18, 20]). Given that these conditions inhibit translation, they can only be used to enhance the production of genes, not of gene products.
2b. Genetically decoupling transcription from translation. Mutants defective in factor-dependent transcription termination such as Rho and NusG fall into this category [40]. A systematic screen for Tn10 mutants exhibiting increased plasmid copy number identified the rho operon again, along with other genes involved in translation and transcription [59, 67]. These additional genes include rpsA (a ribosomal protein that facilitates binding to mRNA), rpoC (β’ subunit of RNA polymerase), orn (an oligoribonuclease that targets small mRNAs), and threonyl-tRNA synthetase [59, 67]. In addition to possibly decoupling transcription from translation, RNA polymerase mutants may modulate R-loop formation through the velocity and efficiency of elongation or through subtle alterations in the positioning of the nascent transcript (reviewed in [19]).
2c Altering the relaxation state of DNA: gyrases and topoisomerases maintain the supercoiling state of plasmid DNA within a narrow margin. In addition, Topoisomerase I appears to suppress the formation of R-loops during transcription elongation. Alterations in topoisomerase function would be expected to alter global R-loop homeostasis in a manner not unlike decoupling translation from transcription. RNAse H, however, appears to be an effective suppressor as topA cells show limited unscheduled R-loop formation. Locally, promoters may influence R-loop balance by modulating supercoiling in the ori region [19].
3. RNA Polymerase promoter specificity
A point mutation or a 41-amino acid deletion located near the 3'-terminal region in the rpoC lead to a 2–10× reduction in plasmid copy number [61]. This appeared to be due to an alteration of the relative specificity of the RNA polymerase for P1 and P2 promoters [61].
4. Rates of RNA I decay
Mutations in pcnB, which is the poly(A)polymerase responsible for RNA I polyadenylation decreased plasmid copy number by increasing the half-life of RNA I and of its active decaying intermediates [22, 62, 67]. Other genes in this pathway that are likely to have an effect on plasmid copy number are: RNase E (responsible for initial cleavage of RNAI) and three enzymes involved in further RNA I degradation, namely polynucleotide phosphorylase, RNAse II exonuclease and RNAse III endonuclease.
5. Primer processing and extension
This involves RNAse H (for processing) and DNA polymerase I (for extension). In general, RNAseH inactivation has a negative effect on ColE1 plasmid copy number, as it is required for pre-primer processing [16]. Some ori mutants, however, are refractory to rnhA inactivation [16] [Table (1)]. Further, inactivation of RNAse H is essential for modes II and III of plasmid replication.
Pol A, the gene coding for DNA polymerase I, is essential for ColE1 replication unless RNAseH is also inactive, in which case plasmid copy number is very low [16]. Ori mutations that stabilize the αβ conformation can be suppressed by point mutations in DNA polymerase I [8]. Suppression is allele-specific, suggesting that Pol I may recognize specific structures on the RNA primer.
6. Replication fork assembly
Mutations that affect replication fork assembly at the n’-pas site would be expected to affect the completion of plasmid replication. Two such mutants have been described, one in PriB [68], and one in DnaC, a core component of replication forks [60, 69]. Both mutants appear to affect specifically the PriA-PriB replication fork assembly pathway and decrease plasmid copy number because of incomplete replication.
Minimizing plasmid metabolic burden
The approaches discussed above primarily control copy number. The goal is tuning plasmid copy number to particular expression systems, maximizing gene dosage while maintaining plasmid stability. An alternative approach consists of addressing the problem of disregulation more directly by either making the plasmids refractory to metabolic input or by uncoupling biomass production from recombinant gene expression.
1. Preventing disregulation
Disregulation of plasmid replication caused by metabolic stress is a self-amplifying process. Therefore, finding ways to preserve plasmid homeostasis in the face of recombinant gene expression would go a long way toward limiting the metabolic burden of recombinant gene expression on host cells. One strategy has been to redesign the unpaired residues at SL1, 2 and 3 with potential to interact with the anticodon loop of tRNAs so as to minimize possible homologies with them ([46], reviewed in [47]). Seven bases of SL2 were replaced by their complement bases, thereby inverting tRNA homologies without changing the complementarity between inhibitor and target RNA or any structural features of the stem-loop, [Fig. (6b)]. The copy number of this proprietary mutant ori did not change after induction of recombinant gene expression in fed-batch cultures [46]. As striking as this result was, it was difficult to interpret because the basal (pre-induction) copy number of the ori mutant was significantly lower than that of its wild-type counterpart [34].
2. Uncoupling biomass production from recombinant gene expression
These strategies aim at controlling the timing of expression in order to promote recombinant protein synthesis. Here I discuss two of these approaches: runaway plasmid replication and the quiescent cell (QC) system.
Runaway plasmid replication consists of increasing plasmid copy number when the desired biomass has been reached, ideally to coincide with induction of recombinant protein expression. The triggers to induce increased plasmid replication are diverse, including inducible promoters and temperature-sensitive mutations. This strategy keeps the metabolic burden of the construct to a minimum during expansion of the culture while allowing a dramatic increase in gene dosage during recombinant protein expression. However, high levels of recombinant production are short-lived using this strategy unless plasmid replication retains some regulatory capacity after induction because of the deleterious effects of runaway plasmid replication [70, 71].
An alternative, proprietary strategy consists of inducing a quiescent state once the recombinant expression system has reached an adequate cell density [72]. In this case, cell growth is arrested through induction of a small regulatory RNA (rcd). This regulatory transcript (whose natural function is to arrest chromosomal replication while cells resolve plasmid multimers) induces nucleoid condensation and wholesale inhibition of chromosomal gene expression but leaves the cell metabolically active. Thus, this induced quiescent state leads to preferential and sustained expression of plasmid-borne proteins [73, 74].
Current and Future Developments
Great progress has been made in our understanding of regulation of ColE1 plasmid replication. This has allowed the design of strategies aimed at optimizing plasmid copy number for recombinant gene expression. The antisense regulatory system of ColE1 plasmids has been intensively modified by elimination [11], through point mutations, [43, 48] or through targeted indels [49]. These modifications, particularly those involving the area of RNAI/RNAII overlap, frequently alter the compatibility properties of the plasmids as well. Increasing the number of compatible ColE1 origins of replication available will facilitate the study of protein complexes and metabolic processes or the generation of genetic diversity by recombination.
Modulating host factors is emerging as an alternative strategy for control of plasmid copy number [20, 59]. This strategy has the advantage of not requiring plasmid engineering but it is not applicable to expression systems that require specific host strains for complementation or for metabolic engineering purposes. The number of gene dosage-modulating strains is likely to increase significantly in the future, through additional genetic screening or by rational design. This will facilitate tuning plasmid replication to individual expression systems. As our understanding of the relationship between supercoiling and plasmid homeostasis improves, it should become possible in the future to fine-tune plasmid copy number by modulating local coiling at the ori, either by controlling the level of transcription in the vicinity of the RNA/DNA switch or by controlling the availability of suppressors of R-loop formation. Also, improvements in our understanding of the cross-talk between transcription and translation machineries and their functional interactions with plasmid replication will likely allow for more sophisticated strategies aimed at preserving plasmid homeostasis in the face of metabolic alterations associated with recombinant gene expression. RNA polymerase could become a key target, as it is emerging as both a sensor and an effector of the stress response. Rifampin resistance, which produces active mutants in the active site of the β subunit facilitates the generation of active mutant libraries. These libraries have already been used to screen for new activities, including mimicking (p)ppGpp binding [75], increasing resistance to DNA damage [76, 77], or preventing constitutive SOS induction in rnhA strains [78]. Rifampin-resistant mutants with effects on plasmid copy number have been described, suggesting a role of RNAP in regulating plasmid copy number [79]. Therefore it is conceivable that RNAP mutants can be isolated favoring plasmid replication initiation or insulating plasmid replication from metabolic input.
CONFLICT OF INTEREST AND ACKNOWLEDGEMENTS
This work was supported by K08 award CA116429-04 (NCI) to Manel Camps.
Footnotes
The author has no conflicts of interest that are directly relevant to the contents of this manuscript
REFERENCES
- 1.del Solar G, Giraldo R, Ruiz-Echevarria MJ, Espinosa M, Diaz-Orejas R. Replication and control of circular bacterial plasmids. Microbiol Mol Biol Rev. 1998;62:434–464. doi: 10.1128/mmbr.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang AC, Cohen SN. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 4.Alting-Mees MA, Short JM. pBluescript II: gene mapping vectors. Nucleic Acids Res. 1989;17:9494. doi: 10.1093/nar/17.22.9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balbas P, Bolivar F. Back to basics: pBR322 and protein expression systems in E. coli. Methods Mol Biol. 2004;267:77–90. doi: 10.1385/1-59259-774-2:077. [DOI] [PubMed] [Google Scholar]
- 6.Selzer G, Som T, Itoh T, Tomizawa J. The origin of replication of plasmid p15A and comparative studies on the nucleotide sequences around the origin of related plasmids. Cell. 1983;32:119–129. doi: 10.1016/0092-8674(83)90502-0. [DOI] [PubMed] [Google Scholar]
- 7.Masukata H, Tomizawa J. Effects of point mutations on formation and structure of the RNA primer for ColE1 DNA replication. Cell. 1984;36:513–522. doi: 10.1016/0092-8674(84)90244-7. [DOI] [PubMed] [Google Scholar]
- 8.Yang YL, Polisky B. Suppression of ColE1 high-copy-number mutants by mutations in the polA gene of Escherichia coli. J Bacteriol. 1993;175:428–437. doi: 10.1128/jb.175.2.428-437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cesareni G, Helmer-Citterich M, Castagnoli L. Control of ColE1 plasmid replication by antisense RNA. Trends Genet. 1991;7:230–235. doi: 10.1016/0168-9525(91)90370-6. [DOI] [PubMed] [Google Scholar]
- 10.Polisky B. ColE1 replication control circuitry: sense from antisense. Cell. 1988;55:929–932. doi: 10.1016/0092-8674(88)90235-8. [DOI] [PubMed] [Google Scholar]
- 11.Panayotatos N. DNA replication regulated by the priming promoter. Nucleic Acids Res. 1984;12:2641–2648. doi: 10.1093/nar/12.6.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomizawa J. Control of ColE1 plasmid replication. Interaction of Rom protein with an unstable complex formed by RNA I and RNA II. J Mol Biol. 1990;212:695–708. doi: 10.1016/0022-2836(90)90231-a. [DOI] [PubMed] [Google Scholar]
- 13.Tomizawa J, Som T. Control of ColE1 plasmid replication: enhancement of binding of RNA I to the primer transcript by the Rom protein. Cell. 1984;38:871–878. doi: 10.1016/0092-8674(84)90282-4. [DOI] [PubMed] [Google Scholar]
- 14.Naito S, Kitani T, Ogawa T, Okazaki T, Uchida H. Escherichia coli mutants suppressing replication-defective mutations of the ColE1 plasmid. Proc Natl Acad Sci U S A. 1984;81:550–554. doi: 10.1073/pnas.81.2.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masukata H, Tomizawa J. Control of primer formation for ColE1 plasmid replication: conformational change of the primer transcript. Cell. 1986;44:125–136. doi: 10.1016/0092-8674(86)90491-5. [DOI] [PubMed] [Google Scholar]
- 16.Dasgupta S, Masukata H, Tomizawa J. Multiple mechanisms for initiation of ColE1 DNA replication: DNA synthesis in the presence and absence of ribonuclease H. Cell. 1987;51:1113–1122. doi: 10.1016/0092-8674(87)90597-6. [DOI] [PubMed] [Google Scholar]
- 17.Wu HY, Shyy SH, Wang JC, Liu LF. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988;53:433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- 18.Drolet M. Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation and DNA topology. Mol Microbiol. 2006;59:723–730. doi: 10.1111/j.1365-2958.2005.05006.x. [DOI] [PubMed] [Google Scholar]
- 19.Drolet M, Broccoli S, Rallu F, Hraiky C, Fortin C, Masse E, et al. The problem of hypernegative supercoiling and R-loop formation in transcription. Front Biosci. 2003;8:d210–d221. doi: 10.2741/970. [DOI] [PubMed] [Google Scholar]
- 20.Gowrishankar J, Harinarayanan R. Why is transcription coupled to translation in bacteria? Mol Microbiol. 2004;54:598–603. doi: 10.1111/j.1365-2958.2004.04289.x. [DOI] [PubMed] [Google Scholar]
- 21.Fukuoh A, Iwasaki H, Ishioka K, Shinagawa H. ATP-dependent resolution of R-loops at the ColE1 replication origin by Escherichia coli RecG protein, a Holliday junction-specific helicase. EMBO J. 1997;16:203–209. doi: 10.1093/emboj/16.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu FF, Gaggero C, Cohen SN. Polyadenylation can regulate ColE1 type plasmid copy number independently of any effect on RNAI decay by decreasing the interaction of antisense RNAI with its RNAII target. Plasmid. 2002;48:49–58. doi: 10.1016/s0147-619x(02)00023-9. [DOI] [PubMed] [Google Scholar]
- 23.Jung YH, Lee Y. RNases in ColE1 DNA metabolism. Mol Biol Rep. 1995;22:195–200. doi: 10.1007/BF00988728. [DOI] [PubMed] [Google Scholar]
- 24.Summers DK, Sherratt DJ. Resolution of ColE1 dimers requires a DNA sequence implicated in the three-dimensional organization of the cer site. EMBO J. 1988;7:851–858. doi: 10.1002/j.1460-2075.1988.tb02884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colloms SD, McCulloch R, Grant K, Neilson L, Sherratt DJ. Xer-mediated site-specific recombination in vitro. EMBO J. 1996;15:1172–1181. [PMC free article] [PubMed] [Google Scholar]
- 26.Diaz Ricci JC, Hernandez ME. Plasmid effects on Escherichia coli metabolism. Crit Rev Biotechnol. 2000;20:79–108. doi: 10.1080/07388550008984167. [DOI] [PubMed] [Google Scholar]
- 27.Friehs K. Plasmid copy number and plasmid stability. Adv Biochem Eng Biotechnol. 2004;86:47–82. doi: 10.1007/b12440. [DOI] [PubMed] [Google Scholar]
- 28.Novick RP. Plasmid incompatibility. Microbiol Rev. 1987;51:381–395. doi: 10.1128/mr.51.4.381-395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diaz-Ricci JC, Tsu M, Bailey JE. Influence of expression of the pet operon on intracellular metabolic fluxes of Escherichia coli. Biotechnol Bioeng. 1992;39:59–65. doi: 10.1002/bit.260390110. [DOI] [PubMed] [Google Scholar]
- 30.Pramanik J, Keasling JD. Stoichiometric model of Escherichia coli metabolism: Incorporation of growth-rate dependent biomass composition and mechanistic energy requirements. Biotechnol Bioeng. 1997;56:398–421. doi: 10.1002/(SICI)1097-0290(19971120)56:4<398::AID-BIT6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 31.Chatterji D, Ojha AK. Revisiting the stringent response, ppGpp and starvation signaling. Curr Opin Microbiol. 2001;4:160–165. doi: 10.1016/s1369-5274(00)00182-x. [DOI] [PubMed] [Google Scholar]
- 32.Jain V, Kumar M, Chatterji D. ppGpp: stringent response and survival. J Microbiol. 2006;44:1–10. [PubMed] [Google Scholar]
- 33.Wegrzyn G, Wegrzyn A. Is tRNA only a translation factor or also a regulator of other processes? J Appl Genet. 2008;49:115–122. doi: 10.1007/BF03195257. [DOI] [PubMed] [Google Scholar]
- 34.Grabherr R, Nilsson E, Striedner G, Bayer K. Stabilizing plasmid copy number to improve recombinant protein production. Biotechnol Bioeng. 2002;77:142–147. doi: 10.1002/bit.10104. [DOI] [PubMed] [Google Scholar]
- 35.Wegrzyn G. Replication of plasmids during bacterial response to amino acid starvation. Plasmid. 1999;41:1–16. doi: 10.1006/plas.1998.1377. [DOI] [PubMed] [Google Scholar]
- 36.Yavachev L, Ivanov I. What does the homology between E. coli tRNAs and RNAs controlling ColE1 plasmid replication mean? J Theor Biol. 1988;131:235–241. doi: 10.1016/s0022-5193(88)80240-6. [DOI] [PubMed] [Google Scholar]
- 37.Wang Z, Le G, Shi Y, Wegrzyn G, Wrobel B. A model for regulation of ColE1-like plasmid replication by uncharged tRNAs in amino acid-starved Escherichia coli cells. Plasmid. 2002;47:69–78. doi: 10.1006/plas.2001.1562. [DOI] [PubMed] [Google Scholar]
- 38.Wang Z, Yuan Z, Xiang L, Shao J, Wegrzyn G. tRNA-dependent cleavage of the ColE1 plasmid-encoded RNA I. Microbiology. 2006;152:3467–3476. doi: 10.1099/mic.0.29134-0. [DOI] [PubMed] [Google Scholar]
- 39.Frenkel L, Bremer H. Increased amplification of plasmids pBR322 and pBR327 by low concentrations of chloramphenicol. DNA. 1986;5:539–544. doi: 10.1089/dna.1.1986.5.539. [DOI] [PubMed] [Google Scholar]
- 40.Harinarayanan R, Gowrishankar J. Host factor titration by chromosomal R-loops as a mechanism for runaway plasmid replication in transcription termination-defective mutants of Escherichia coli. J Mol Biol. 2003;332:31–46. doi: 10.1016/s0022-2836(03)00753-8. [DOI] [PubMed] [Google Scholar]
- 41.Castagnoli L, Lacatena RM, Cesareni G. Analysis of dominant copy number mutants of the plasmid pMB1. Nucleic Acids Res. 1985;13:5353–5367. doi: 10.1093/nar/13.14.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stuitje AR, Spelt CE, Veltkamp E, Nijkamp HJ. Identification of mutations affecting replication control of plasmid Clo DF13. Nature. 1981;290:264–267. doi: 10.1038/290264a0. [DOI] [PubMed] [Google Scholar]
- 43.Gelfand DH. US4966840. 1990
- 44.Lacatena RM, Cesareni G. Base pairing of RNA I with its complementary sequence in the primer precursor inhibits ColE1 replication. Nature. 1981;294:623–626. doi: 10.1038/294623a0. [DOI] [PubMed] [Google Scholar]
- 45.Tomizawa J, Itoh T. Plasmid ColE1 incompatibility determined by interaction of RNA I with primer transcript. Proc Natl Acad Sci U S A. 1981;78:6096–6100. doi: 10.1073/pnas.78.10.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bayer K, Grabherr R, Nilsson E, Striedner G. US 680666 B2. 2004
- 47.Grabherr R, Bayer K. Impact of targeted vector design on Co/E1 plasmid replication. Trends Biotechnol. 2002;20:257–260. doi: 10.1016/s0167-7799(02)01950-9. [DOI] [PubMed] [Google Scholar]
- 48.Devauchelle G, Garnier L, Cerutti M, Valverde V, Masson JM. US005565333A. 1996
- 49.Smider V. US20080057545A1. 2008
- 50.Kim D, Rhee Y, Rhodes D, Sharma V, Sorenson O, Greener A, et al. Directed evolution and identification of control regions of ColE1 plasmid replication origins using only nucleotide deletions. J Mol Biol. 2005;351:763–775. doi: 10.1016/j.jmb.2005.06.051. [DOI] [PubMed] [Google Scholar]
- 51.Miki T, Yasukochi T, Nagatani H, Furuno M, Orita T, Yamada H, et al. Construction of a plasmid vector for the regulatable high level expression of eukaryotic genes in Escherichia coli: an application to overproduction of chicken lysozyme. Protein Eng. 1987;1:327–332. doi: 10.1093/protein/1.4.327. [DOI] [PubMed] [Google Scholar]
- 52.Fitzwater T, Yang YL, Zhang XY, Polisky B. Mutations affecting RNA-DNA hybrid formation of the ColE1 replication primer RNA. Restoration of RNA I sensitivity to a copy-number mutant by second-site mutations. J Mol Biol. 1992;226:997–1008. doi: 10.1016/0022-2836(92)91048-t. [DOI] [PubMed] [Google Scholar]
- 53.Fitzwater T, Zhang XY, Elble R, Polisky B. Conditional high copy number ColE1 mutants: resistance to RNA1 inhibition in vivo and in vitro. EMBO J. 1988;7:3289–3297. doi: 10.1002/j.1460-2075.1988.tb03196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boros I, Posfai G, Venetianer P. High-copy-number derivatives of the plasmid cloning vector pBR322. Gene. 1984;30:257–260. doi: 10.1016/0378-1119(84)90130-6. [DOI] [PubMed] [Google Scholar]
- 55.Muesing M, Tamm J, Shepard HM, Polisky B. A single base-pair alteration is responsible for the DNA overproduction phenotype of a plasmid copy-number mutant. Cell. 1981;24:235–242. doi: 10.1016/0092-8674(81)90519-5. [DOI] [PubMed] [Google Scholar]
- 56.Gomez A, Galic T, Mariet JF, Matic I, Radman M, Petit MA. Creating new genes by plasmid recombination in Escherichia coli and Bacillus subtilis. Appl Environ Microbiol. 2005;71:7607–7609. doi: 10.1128/AEM.71.11.7607-7609.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sblattero D, Bradbury A. Exploiting recombination in single bacteria to make large phage antibody libraries. Nat Biotechnol. 2000;18:75–80. doi: 10.1038/71958. [DOI] [PubMed] [Google Scholar]
- 58.Velappan N, Sblattero D, Chasteen L, Pavlik P, Bradbury AR. Plasmid incompatibility: more compatible than previously thought? Protein Eng Des Sel. 2007;20:309–313. doi: 10.1093/protein/gzm005. [DOI] [PubMed] [Google Scholar]
- 59.Cheng Q, Rouviere PE, Tao L, Suh W. US20040191863A1. 2004
- 60.Gowrishankar J, Harinarayanan R. US007176028B2. 2007
- 61.Ederth J, Isaksson LA, Abdulkarim F. Origin-specific reduction of ColE1 plasmid copy number due to mutations in a distinct region of the Escherichia coli RNA polymerase. Mol Genet Genomics. 2002;267:587–592. doi: 10.1007/s00438-002-0689-y. [DOI] [PubMed] [Google Scholar]
- 62.Lopilato J, Bortner S, Beckwith J. Mutations in a new chromosomal gene of Escherichia coli K-12, pcnB, reduce plasmid copy number of pBR322 and its derivatives. Mol Gen Genet. 1986;205:285–290. doi: 10.1007/BF00430440. [DOI] [PubMed] [Google Scholar]
- 63.Tao L, Jackson RE, Cheng Q. Directed evolution of copy number of a broad host range plasmid for metabolic engineering. Metab Eng. 2005;7:10–17. doi: 10.1016/j.ymben.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 64.Polisky B, Zhang XY, Fitzwater T. Mutations affecting primer RNA interaction with the replication repressor RNA I in plasmid CoIE1: potential RNA folding pathway mutants. EMBO J. 1990;9:295–304. doi: 10.1002/j.1460-2075.1990.tb08108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin-Chao S, Chen WT, Wong TT. High copy number of the pUC plasmid results from a Rom/Rop-suppressible point mutation in RNA II. Mol Microbiol. 1992;6:3385–3393. doi: 10.1111/j.1365-2958.1992.tb02206.x. [DOI] [PubMed] [Google Scholar]
- 66.Hong X, Cadwell GW, Kogoma T. Activation of stable DNA replication in rapidly growing Escherichia coli at the time of entry to stationary phase. Mol Microbiol. 1996;21:953–961. doi: 10.1046/j.1365-2958.1996.591419.x. [DOI] [PubMed] [Google Scholar]
- 67.Tao L, Jackson RE, Rouviere PE, Cheng Q. Isolation of chromosomal mutations that affect carotenoid production in Escherichia coli: mutations alter copy number of ColE1-type plasmids. FEMS Microbiol Lett. 2005;243:227–233. doi: 10.1016/j.femsle.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 68.Berges H, Oreglia J, Joseph-Liauzun E, Fayet O. Isolation and characterization of a priB mutant of Escherichia coli influencing plasmid copy number of delta rop ColE1-type plasmids. J Bacteriol. 1997;179:956–958. doi: 10.1128/jb.179.3.956-958.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Harinarayanan R, Gowrishankar J. A dnaC mutation in Escherichia coli that affects copy number of ColE1-like plasmids and the PriA-PriB (but not Rep-PriC) pathway of chromosomal replication restart. Genetics. 2004;166:1165–1176. doi: 10.1534/genetics.166.3.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bachvarov D, Jay E, Ivanov I. Construction of a Co1E1 plasmid bearing inducible high-copy-number phenotype. Folia Microbiol (Praha) 1990;35:177–182. doi: 10.1007/BF02820482. [DOI] [PubMed] [Google Scholar]
- 71.Togna AP, Shuler ML, Wilson DB. Effects of plasmid copy number and runaway plasmid replication on overproduction and excretion of beta-lactamase from Escherichia coli. Biotechnol Prog. 1993;9:31–39. doi: 10.1021/bp00019a005. [DOI] [PubMed] [Google Scholar]
- 72.Summers DK, Rowe DCD. US6190867. 1999
- 73.Mukherjee KJ, Rowe DC, Watkins NA, Summers DK. Studies of single-chain antibody expression in quiescent Escherichia coli. Appl Environ Microbiol. 2004;70:3005–3012. doi: 10.1128/AEM.70.5.3005-3012.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rudolph CJ, Dhillon P, Moore T, Lloyd RG. Avoiding and resolving conflicts between DNA replication and transcription. DNA Repair (Amst) 2007;6:981–993. doi: 10.1016/j.dnarep.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 75.Trinh V, Langelier MF, Archambault J, Coulombe B. Structural perspective on mutations affecting the function of multisubunit RNA polymerases. Microbiol Mol Biol Rev. 2006;70:12–36. doi: 10.1128/MMBR.70.1.12-36.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McGlynn P, Lloyd RG. Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell. 2000;101:35–45. doi: 10.1016/S0092-8674(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 77.Trautinger BW, Lloyd RG. Modulation of DNA repair by mutations flanking the DNA channel through RNA polymerase. EMBO J. 2002;21:6944–6953. doi: 10.1093/emboj/cdf654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kogoma T. Escherichia coli RNA polymerase mutants that enhance or diminish the SOS response constitutively expressed in the absence of RNase HI activity. J Bacteriol. 1994;176:1521–1523. doi: 10.1128/jb.176.5.1521-1523.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang YL, Polisky B. Allele-specific suppression of ColE1 high-copy-number mutants by a rpoB mutation of Escherichia coli. Plasmid. 1999;41:55–62. doi: 10.1006/plas.1998.1378. [DOI] [PubMed] [Google Scholar]
- 80.Naito S, Uchida H. Initiation of DNA replication in a ColE1-type plasmid: isolation of mutations in the ori region. Proc Natl Acad Sci U S A. 1980;77:6744–6748. doi: 10.1073/pnas.77.11.6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Naito S, Uchida H. RNase H and replication of ColE1 DNA in Escherichia coli. J Bacteriol. 1986;166:143–147. doi: 10.1128/jb.166.1.143-147.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kawakami K, Naito S, Inoue N, Nakamura Y, Ikeda H, Uchida H. Isolation and characterization of herC, a mutation of Escherichia coli affecting maintenance of ColE1. Mol Gen Genet. 1989;219:333–340. doi: 10.1007/BF00259604. [DOI] [PubMed] [Google Scholar]
- 83.Bert AG, Burrows J, Osborne CS, Cockerill PN. Generation of an improved luciferase reporter gene plasmid that employs a novel mechanism for high-copy replication. Plasmid. 2000;44:173–182. doi: 10.1006/plas.2000.1474. [DOI] [PubMed] [Google Scholar]
- 84.Ohmori H, Tomizawa J. Nucleotide sequence of the region required for maintenance of colicin E1 plasmid. Mol Gen Genet. 1979;176:161–170. doi: 10.1007/BF00273210. [DOI] [PubMed] [Google Scholar]
- 85.Masai H, Arai K. Mechanisms of primer RNA synthesis and D-loop/R-loop-dependent DNA replication in Escherichia coli. Biochimie. 1996;78:1109–1117. doi: 10.1016/s0300-9084(97)86737-5. [DOI] [PubMed] [Google Scholar]
- 86.Marians KJ. Mechanisms of replication fork restart in Escherichia coli. Philos Trans R Soc Lond B Biol Sci. 2004;359:71–77. doi: 10.1098/rstb.2003.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Backman K, Betlach M, Boyer HW, Yanofsky S. Genetic and physical studies on the replication of ColE1-type plasmids. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):69–76. doi: 10.1101/sqb.1979.043.01.012. [DOI] [PubMed] [Google Scholar]
- 88.Maizels N, Weiner AM. Phylogeny from function: evidence from the molecular fossil record that tRNA originated in replication, not translation. Proc Natl Acad Sci U S A. 1994;91:6729–6734. doi: 10.1073/pnas.91.15.6729. [DOI] [PMC free article] [PubMed] [Google Scholar]