Abstract
Triplex forming oligonucleotides (TFOs) modified with C5-alkynyl functionalized LNA (locked nucleic acid) monomers display extraordinary thermal affinity toward double stranded DNA targets, excellent discrimination of Hoogsteen-mismatched targets, and high stability against 3′-exonucleases.
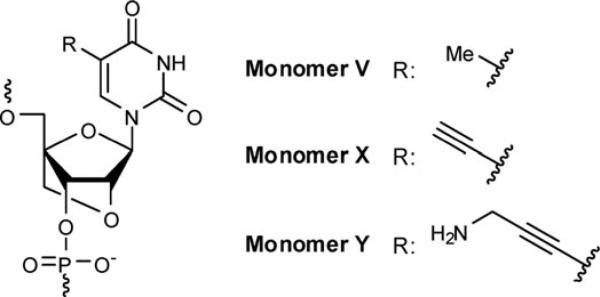
Substantial efforts have been invested to develop probes for use in the antigene technology, i.e., sequence-specific targeting of double stranded DNA (dsDNA).1 The efforts are motivated by the prospect of developing fundamental research tools and new types of therapeutic and diagnostic agents.2,3 (TC)-motif triplex forming oligonucleotides (TFOs), which bind to polypurine regions in dsDNA targets via Hoogsteen base pairs in the major groove, are the most widely studied DNA-targeting agents,1 although promising alternatives are emerging.4 A drawback of the TFO-approach is the instability of triplexes at physiological pH due to electrostatic repulsion between the three strands and insufficient N3-protonation of cytosines resulting in weak Hoogsteen base pairing. Chemical modification of TFOs is therefore essential to increase triplex stability as well as to protect TFOs from enzymatic degradation.1 Three classes of nucleotide building blocks have found particular use as TFO-modifications: (a) ribonucleotides and O2′-alkylated analogs hereof as they favorably preorganize the TFO strand for hybridization to dsDNA targets,5 (b) conformationally restricted nucleotides with RNA-like furanose puckers such as locked nucleic acid (LNA, monomer V, Fig. 1)6 or analogs thereof7 in what is an extension of the TFO preorganization concept, and (c) C5-alkynyl functionalized pyrimidine DNA monomers, as they promote enhanced base stacking interactions in triplexes.8 Interestingly, building blocks that combine C5-alkynyl functionalization and RNA-character induce greater triplex stabilization than the respective C5-alkynyl DNA and RNA TFO monomers by themselves.9 In light of this, we hypothesized that C5-alkynyl functionalized LNA monomers X and Y (Fig. 1) would synergistically integrate beneficial properties from two compound classes, i.e., (1) high thermal affinity toward dsDNA and partial resistance toward nucleases (LNA-component), and (2) resistance toward nucleases due to the presence of steric shields along with favorable contributions to binding affinity (C5-substituent).
Fig. 1.
Structures of LNA and novel C5-alkynyl functionalized LNA monomers.
The corresponding phosphoramidites10 of the C5-ethynyl and C5-propargylamine functionalized LNA monomers X and Y and the commercially available LNA thymine phosphoramidite were incorporated one, three or six times into a 15-mer (TC)-motif DNA TFO sequence (Table 1) that has previously been used to evaluate hybridization properties of LNA,7d ENA7d and 2′,4′-BNANC7e TFOs. TFOs were synthesized on automated DNA synthesizers (0.2 mmol scale) using standard conditions except for extended coupling (15 min, using 4,5-dicyanoimidazole as activator) and oxidation times (45 s).† This resulted in stepwise coupling yields of >98% for the LNA monomers. 5-Methyldeoxycytidine rather than deoxycytidine monomers were used to ameliorate the normal pH dependence of (TC)-motif TFOs.11‡ Following workup and purification, the composition and purityz (>90%) of all synthesized TFOs were verified by MALDI-TOF MS (Table S2)† and HPLC, respectively.†
Table 1.
Tm-values and association rate constants (kon) for triplexes formed by C5-alkynyl functionalized LNA, conventional LNA or DNA TFOs with dsDNA targeta
|
Tm [ΔTm/mod]/°C |
kon/M–1 s–1 |
|||||||
|---|---|---|---|---|---|---|---|---|
| ON | Seq. 5′ → 3′ | B = | V | X | Y | V | X | Y |
| B1 | TTT TTmCTBT mCTmCTmCT | 40.0 [+11.0] | 42.5 [+13.5] | 42.5 [+13.5] | 3800 | 4400 | ND | |
| B2 | TTT TBmC TTBmCTmC BmCT | 48.5 [ +6.5] | 54.0 [+8.3] | 57.0 [+9.3] | 3600 | 4000 | 3700 | |
| B3 | TTT TBmC TBT mCBmC TmCT | 53.5 [+8.2] | 59.5 [+10.2] | 60.0 [+10.3] | 3900 | 4100 | 3900 | |
| B4 | TTB TBmC BTBmCBmC BmCT | 61.0 [+5.3] | 67.5 [+6.4] | 74.5 [+7.6] | 4300 | 4400 | 5500 | |
Tm's were determined as the first derivative of differential thermal denaturation curves (A260vs. T)6c† recorded in a pH 7.0 phosphate buffer solution containing 140 mM KCl using 1.0 μM of each strand. Tm-values are averages of at least two independent measurements within 1 °C. kon's were measured in pH 7.2 HEPES buffer.† kon's are averages of at least three independent measurements. Data for DNA reference TFO T1 (B = T): Tm = 29.0 °C, kon = 4400 M–1 s–1. Structures of monomers are shown in Fig. 1. mC denotes 5-methyldeoxycytidine monomers. dsDNA target sequence (TFO binding region underlined): 5′-GCT AAA AAG AAA GAG AGA TCG-3′, 3′-CGA TTT TTC TTT CTC TCT ACG-5′.
ND = not determined.
Binding to a central target region in a 21-mer dsDNA was first characterized by non-denaturing PAGE using equimolar quantities of TFOs and dsDNA target in a pH 7.2 HEPES buffer containing Mg2+. Conventional LNA and C5-ethynyl functionalized LNA TFOs form stable triplexes with dsDNA targets under these conditions (V- and X-series, Fig. S1)† as a single band of lower mobility than the corresponding dsDNA target was observed in all instances. Triplex formation between C5-propargylamine functionalized LNA TFOs and dsDNA target is incomplete as a band (~20–40% intensity) of identical mobility as the corresponding dsDNA target is observed in addition to the triplex band (Y-series, Fig. S2).† Triplex formation was, however, driven toward completion using a two-fold excess of C5-propargylamine functionalized LNA TFOs (Fig. S2).†
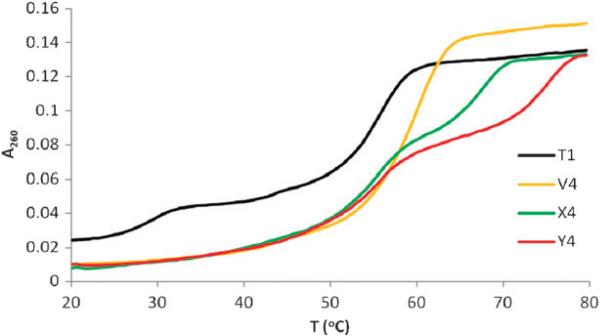
After qualitatively establishing triplex formation, the thermal affinity of C5-functionalized LNA TFOs toward the dsDNA target was determined by UV thermal denaturation experiments at pH 7.0 in the absence of Mg2+-ions and compared relative to reference DNA and LNA TFOs (Table 1). The thermal denaturation curves of the studied triplexes either exhibit (a) biphasic profiles when thermal denaturation temperatures (Tm's) of triplexes differed substantially from the Tm of the dsDNA target (Tm = 56.0 °C), or (b) monophasic profiles when denaturation curves for triplexes and dsDNA targets overlapped substantially (Fig. 2, Fig. S3 and S4).† Similar observations in identical sequence contexts have been made with other LNA analogs.7b,d To facilitate determination of triplex Tm's, differential thermal denaturation curves6c were obtained by ‘subtracting’ the denaturation curve of the dsDNA target from the ‘raw’ triplex denaturation curve (Fig. S3).†
Fig. 2.
Representative thermal denaturation profiles of triplexes between reference DNA T1 or TFOs from the B4-series, and complementary dsDNA target. For sequences and conditions see Table 1.
Singly modified C5-alkynyl functionalized LNA TFOs exhibit dramatically increased thermal affinity toward dsDNA targets relative to reference DNA TFO T1 (ΔTm ≈ +13.5 °C, B1-series, Table 1). Additional modification of TFOs with X/Y monomers results in progressively more stabilized triplexes although less pronounced ΔTm/mod-values are observed (e.g., compare data of B1-, B2- and B4-series, Table 1). C5-Propargylamine functionalized LNAs (Y-series) stabilize triplexes slightly more efficiently than the corresponding C5-ethynyl functionalized LNAs (X-series), which likely results from reduced electrostatic repulsion between the triplex strands due to partial protonation of the propargylamino substituent, or from improved π-stacking with neighboring nucleobases.8b Consequentially, C5-propargylamine functionalized LNA TFO Y4 containing six Y monomers as next-nearest neighbours forms exceedingly stable triplexes with dsDNA targets (Tm = 74.5 °C, Table 1).
Importantly, the observed stabilization of triplexes by C5-alkynyl LNA TFOs compares favourably with the corresponding parent LNA TFOs (V-series) as additional increases in Tm-values of between +1.1 °C and +2.8 °C per modification are observed (compare Tm-data for X-, Y-and V-series, Table 1). In fact, direct comparison with previously reported hybridization data for ENA7d and 2′,4′-BNANC7e TFOs in the same sequence, strongly suggests that C5-alkynyl LNA TFOs stabilize triplexes more efficiently than these ‘gold standard’ TFO building blocks. Thus, incorporation of C5-alkynyl functionalized LNA monomers into DNA TFOs is a simple,† yet powerful approach to improve thermal affinity of TFOs toward dsDNA targets.
To evaluate the influence of C5-alkynyl functionalized LNA monomers on association kinetics of triplex formation, the association rate constant kon was determined for TFOs by fitting a second order rate equation to the A260 decay curve13 obtained by mixing pre-annealed dsDNA target with equimolar quantities of TFOs (Fig. S6).† Generally little variation in the kon-values between DNA, LNA or C5-functionalized LNA TFOs was observed (~4000 M–1 s–1, Table 1), which verifies that LNA-type TFOs primarily influence triplex dissociation kinetics.6a As an exception hereto, densely modified C5-propargylamine functionalized LNA Y4 exhibits faster triplex association kinetics which most likely is linked to the presence of partially positively charged ‘patches’ of the C5-propargylamine substituent.9b
Increased affinity of TFOs toward dsDNA targets should preferably be accompanied by concomitant improvements in mismatch discrimination to avoid off-targets effects.5b Accordingly, the ability of singly modified C5-alkynyl LNA TFOs (B1-series) to discriminate dsDNA targets with a single Hoogsteen base pair mismatch opposite of the modification was evaluated in a thermal denaturation buffer containing 10 mM Mg2+ (see footnote Table 2), which is known to accelerate and stabilize triplex formation.14§,†
Table 2.
Tm-values of matched and mismatched triplexes in the presence of Mg2+-ionsa
|
Tm[ΔTm]/°C |
||||
|---|---|---|---|---|
| ON | A : T (match) | T : A | C : G | G : C |
| T1 | 37.0 | 10.0 [–27.0] | 19.5 [–17.5] | 13.5 [–23.5] |
| V1 | 49.0 | 10.0 [–39.0] | 28.5 [–20.5] | 24.0 [–25.0] |
| X1 | 51.5 | 10.0 [–41.5] | 24.5 [–27.0] | 27.5 [–24.0] |
| Y1 | 50.0 | 10.0 [–40.0] | 26.5 [–23.5] | 27.5 [–22.5] |
Tm-values (ΔTm = change in Tm-value relative to matched triplex). Conditions and TFO-sequences (B1-series) as described in Table 1 except for addition of 10 mM MgCl2. dsDNA targets = 5′-GCT AAA AAG AMA GAG AGA TCG-3′ :3′-CGA TTT TTC TM′T CTC TCT ACG-5′, where M : M′ = A : T, T : A, C : G and G : C, respectively.
As anticipated, less stable triplexes are generally formed with mismatched dsDNA targets. Introduction of C5-functionalized LNA monomers into TFOs greatly improves discrimination of Hoogsteen TA- and CG-mismatches compared to reference TFOs T1 and V1, while discrimination of GC-mismatches is largely unaffected (Table 2).
Consequentially, the most demanding mismatched dsDNA targets are more efficiently discriminated by C5-functionalized LNA TFOs (ΔTm = –24.0 °C and –22.5 °C for X1 and Y1, respectively) than by reference TFOs (ΔTm = –17.5 °C and –20.5 °C, for T1 and V1, respectively). This data demonstrates that C5-alkynyl functionalized LNAs exhibit increased thermal affinity and specificity relative to parent LNAs.
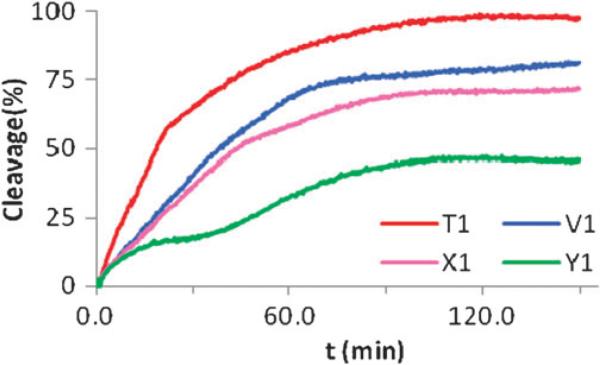
Finally, the resistance of singly modified C5-alkynyl functionalized LNA TFOs (B1-series) against degradation by the 3′-exonuclease snake venom phosphodiesterase (SVPDE) was evaluated, by following the increase in absorbance (hyperchromicity) at 260 nm15 of TFOs in the presence of SVPDE (Fig. 3). As expected, DNA TFO T1 is rapidly degraded (>85% cleavage after 1 h) whereas singly modified LNA V1 confers some protection against SVPDE-mediated degradation (~70% cleavage after 1 h). C5-Ethynyl and, in particular, C5-propargylamine functionalized LNA TFOs are considerably more resistant against nucleolytic degradation (X1 ~60%, Y1 ~35% after 1 h). This suggests that the C5-substituents act as a steric shield, to ensure greater biostability.
Fig. 3.
Exonuclease (SVPDE) degradation of singly modified C5-functionalized LNA and reference TFOs. Nuclease degradation studies performed in Tris-buffer (50 mM Tris-HCl, 10 mM MgCl2, pH9.0) at 37 °C using 2 μM of TFO strands and 0.43 μg of SVPDE (snake venom phosphordiesterase). For sequences of T1–V1–X1–Y1,see Table 1.
To conclude, C5-alkynyl functionalized LNAs exhibit unsurpassed TFO-hybridization properties, which are likely to enable efficient DNA-targeting under physiological conditions. Studies along these lines are ongoing.
We appreciate financial support from Idaho NSF EPSCoR, the BANTech Center at the Univ. of Idaho, a University of Idaho Research Office and Research Council Seed Grant, and National Institutes of Health [grant number P20 RR016448 from the COBRE Program of the National Center for Research Resources]. MEØ acknowledges a scholarship from the College of Graduate Studies. We thank the EBI Murdock Mass Spectrometry center for mass spectrometric analyses.
Supplementary Material
Footnotes
Electronic supplementary information (ESI) available: Synthetic outline of phosphoramidites, protocols for synthesis/purification of TFOs, determination of Tm- and kon-values, and exonuclease studies; MALDI-TOF-MS data of TFOs; thermal denaturation profiles; PAGE experiments; absorption decay profiles; discussion regarding parallel duplex formation. See DOI: 10.1039/b917312a
O4-Triazolyl-dT-CE phosphoramidites12 were incorporated into TFOs, and converted to 5-methyldeoxycytidine monomers during deprotection (32% aq. NH3).
Notes and references
- 1.Duca M, Vekhoff P, Oussedik K, Halby L, Arimondo PB. Nucleic Acids Res. 2008;36:5123. doi: 10.1093/nar/gkn493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rogers FA, Lloyd JA, Glazer PM. Curr. Med. Chem.: Anti-Cancer Agents. 2005;5:319. doi: 10.2174/1568011054222300. [DOI] [PubMed] [Google Scholar]
- 3.Ghosh I, Stains CI, Ooi AT, Segal DJ. Mol. BioSyst. 2006;2:551. doi: 10.1039/b611169f. [DOI] [PubMed] [Google Scholar]
- 4.a Hrdlicka PJ, Kumar TS, Wengel J. Chem. Commun. 2005:4279. doi: 10.1039/b506986f. [DOI] [PubMed] [Google Scholar]; b Ge R, Heinonen JE, Svahn MG, Mohamed AJ, Lundin KE, Smith CIE. FASEB J. 2007;21:1902. doi: 10.1096/fj.06-7225com. [DOI] [PubMed] [Google Scholar]; c Beane R, Gabillet S, Montaillier C, Arar K, Corey DR. Biochemistry. 2008;47:13147. doi: 10.1021/bi801930p. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Ishizuka T, Yoshida J, Yamamoto Y, Sumaoka J, Tedeschi T, Corradini R, Sforza S, Komiyama M. Nucleic Acids Res. 2008;36:1464. doi: 10.1093/nar/gkm1154. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Simon P, Cannata F, Concordet J-P, Giovannangeli C. Biochimie. 2008;90:1109. doi: 10.1016/j.biochi.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 5.a Asensio JL, Carr R, Brown T, Lane AN. J. Am. Chem. Soc. 1999;121:11063. [Google Scholar]; b Alam MR, Majumdar A, Thazhathveetil AK, Liu ST, Liu JL, Puri N, Cuenoud B, Sasaki S, Miller PS, Seidman MM. Biochemistry. 2007;46:10222. doi: 10.1021/bi7003153. [DOI] [PubMed] [Google Scholar]
- 6.a Torigoe H, Hari Y, Sekiguchi M, Obika S, Imanishi T. J. Biol. Chem. 2001;276:2354. doi: 10.1074/jbc.M007783200. [DOI] [PubMed] [Google Scholar]; b Obika S. Chem. Pharm. Bull. 2004;52:1399. doi: 10.1248/cpb.52.1399. [DOI] [PubMed] [Google Scholar]; c Sun B-W, Babu BR, Sørensen MD, Zakrzewska K, Wengel J, Sun J-S. Biochemistry. 2004;43:4160. doi: 10.1021/bi036064e. [DOI] [PubMed] [Google Scholar]; d Brunet E, Alberti P, Perrouault L, Babu R, Wengel J, Giovannangeli C. J. Biol. Chem. 2005;280:20076. doi: 10.1074/jbc.M500021200. [DOI] [PubMed] [Google Scholar]
- 7.a Koizumi M, Morita K, Daigo M, Tsutsumi S, Abe K, Obika S, Imanishi T. Nucleic Acids Res. 2003;31:3267. doi: 10.1093/nar/gkg416. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Kumar N, Nielsen KE, Maiti S, Petersen MJ. J. Am. Chem. Soc. 2006;128:14. doi: 10.1021/ja055483r. [DOI] [PubMed] [Google Scholar]; c Højland T, Kumar S, Babu BR, Umemoto T, Albaek N, Sharma PK, Nielsen P, Wengel J. Org. Biomol. Chem. 2007;5:2375. doi: 10.1039/b706101c. [DOI] [PubMed] [Google Scholar]; d Rahman SMA, Seki S, Obika S, Haitani S, Miyashita K, Imanishi T. Angew. Chem., Int. Ed. 2007;46:4306. doi: 10.1002/anie.200604857. [DOI] [PubMed] [Google Scholar]; e Rahman SMA, Seki S, Obika S, Yoshikawa H, Miyashita K, Imanishi T. J. Am. Chem. Soc. 2008;130:4886. doi: 10.1021/ja710342q. [DOI] [PubMed] [Google Scholar]
- 8.a Lacroix L, Lacoste J, Reddoch JF, Mergny JL, Levy DD, Seidman MM, Matteucci MD, Glazer PM. Biochemistry. 1999;38:1893. doi: 10.1021/bi982290q. [DOI] [PubMed] [Google Scholar]; b Bijapur J, Keppler MD, Bergqvist S, Brown T, Fox KR. Nucleic Acids Res. 1999;27:1802. doi: 10.1093/nar/27.8.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Brazier JA, Shibata T, Townsley J, Taylor BF, Frary E, Williams NH, Williams DM. Nucleic Acids Res. 2005;33:1362. doi: 10.1093/nar/gki254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a Sollogoub M, Darby RAJ, Cuenoud B, Brown T, Fox KR. Biochemistry. 2002;41:7224. doi: 10.1021/bi020164n. [DOI] [PubMed] [Google Scholar]; b Li H, Miller PS, Seidman MM. Org. Biomol. Chem. 2008;6:4212. doi: 10.1039/b810709b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Østergaard ME, Kumar P, Baral B, Raible DJ, Kumar TS, Anderson BA, Guenther DC, Deobald L, Paszczynski AJ, Sharma PK, Hrdlicka PJ. ChemBioChem. doi: 10.1002/cbic.200900500. DOI: 10.1002/cbic.200900500. [DOI] [PubMed] [Google Scholar]
- 11.Lee JS, Woodsworth ML, Latimer LJ, Morgan AR. Nucleic Acids Res. 1984;12:6603. doi: 10.1093/nar/12.16.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ivanov SA, Volkov EM, Oretskaya TS, Muller S. Tetrahedron. 2004;60:9273. [Google Scholar]
- 13.Xodo LE. Eur. J. Biochem. 1995;228:918. doi: 10.1111/j.1432-1033.1995.tb20340.x. [DOI] [PubMed] [Google Scholar]
- 14.Sugimoto N, Wu P, Hara H, Kawamoto Y. Biochemistry. 2001;40:9396. doi: 10.1021/bi010666l. [DOI] [PubMed] [Google Scholar]
- 15.Jørgensen PN, Stein PC, Wengel J. J. Am. Chem. Soc. 1994;116:2231. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.