Abstract
Purpose
To determine the growth potential of p63-positive cell clusters maintained in human limbal epithelial sheets.
Methods
Intact human limbal epithelial sheets were isolated from corneoscleral rims and cultured with or without being rendered into single cells by trypsinization on either plastic or 3T3 fibroblast feeder layers. Clonal growth on 3T3 fibroblast feeder layers was compared between monolayers in sheets or single cells and between areas with or without laser-microdissected, p63-enriched cell clusters. Immunostaining, immunoblot analysis, and reverse transcription-polymerase chain reaction of such differentiation markers as keratin (K)−3 and −12 and such progenitor markers as p63, ABCG2, and MDR-1 were also compared.
Results
Clusters of small p63-positive cells were enriched in limbal palisades of dispase-isolated epithelial sheets immediately or after brief cultivation on plastic in SHEM. Clonal growth of p63-rich cell clusters was higher than areas without clusters (P < 0.001). Clonal growth of epithelial monolayers derived from sheets was also higher than that derived from single cells (P < 0.01). Although expression of ABCG2 and MDR-1 transcripts was similar, cells from epithelial sheets expressed higher protein levels of p63 and lower protein levels of K3 and −12 than single cells, whether they were cultured on plastic or as 3T3 fibroblast feeder layers.
Conclusions
Limbal palisades contain clusters of p63-rich progenitor cells. Maintenance of such cluster architecture during ex vivo expansion yields higher growth potential than being dispersed into single cells.
Several studies have substantiated the notion that corneal epithelial stem cells (SCs) reside in the limbal region between the cornea and the conjunctiva.1,2 This unique anatomic enrichment at the limbus allows one to gain easy access to these adult somatic SCs,3 which have the smallest cell size4 and a longer cell cycle,5 do not express K3/K121,6 and connexin 43,7 but preferentially express p63,8 Bcrp1/ABCG2,9 or N-cadherin.10
Limbal epithelial SCs play a critical role in maintaining a healthy corneal surface. Several ocular surface diseases manifest limbal SC deficiency11 when they are either destroyed or their underlying stroma is dysfunctional. Thus, one may naturally speculate that the function of limbal SCs is regulated in part by their microenvironmental niche (for review, see Ref. 12). One way of maintaining such a supportive relationship in vitro is to expand single limbal progenitor cells on growth-arrested 3T3 fibroblast feeder.13–15 The SC-containing limbal epithelium has a higher clonogenic potential when cultured on 3T3 fibroblasts feeder layers.16,17 Such ex vivo–expanded limbal epithelial cells have successfully been transplanted to treat patients with total limbal SC deficiency.18,19
We have reported a method of isolating an intact and viable limbal epithelial sheet.20 Therefore, we questioned whether maintenance of limbal SCs within the sheet is useful in promoting their growth potential during cultivation. Herein, we demonstrate that unique p63-positive cell clusters were enriched in the limbal palisades of Vogt and exhibited a higher clonal growth potential and that limbal sheet cultures generated cells with a higher growth potential and less differentiation than those of single-cell cultures. The significance of these findings is further discussed with respect to tissue engineering methods of generating surgical grafts containing human limbal epithelial progenitor cells for treating limbal SC deficiency.
Materials and Methods
Extraction reagent (TRIzol) was purchased from Invitrogen-Gibco (Grand Island, NY). Dispase II was from Roche (Indianapolis, IN). A cell viability assay (Live/Dead Assay) was from Molecular Probes (Eugene, OR). Monoclonal antibody to keratin (K)−3 (AE5) was purchased from ICN Biomedicals (Aurora, OH). A rabbit polyclonal antibody to human keratin (K)−12 was kindly provided by Winston Kao (University of Cincinnati School of Medicine).21 A monoclonal antibody against p63 (4A4) was from Dako (Carpinteria, CA). Horseradish peroxidase–conjugated goat anti-rabbit IgG antibody was from Bio-Rad (Hercules, CA). The following reagents and chemicals including a monoclonal antibody to β-actin, fluorescein isothiocyanate (FITC)–conjugated goat anti-mouse and goat anti-rat antibodies, mouse-derived epidermal growth factor, cholera toxin, insulin-transferrin-sodium selenite (ITS) medium supplement and sorbitol were purchased from Sigma-Aldrich (St. Louis, MO). Chemiluminescence reagent (Western Lightning) was purchased from Perkin Elmer Life Science (Boston, MA).
Isolation of Human Limbal Epithelial Sheets
Human corneoscleral rims from donors, younger than 60 years and less than 4 days after harvesting, were obtained from the Florida Lions Eye bank and was managed in accordance with the Declaration of Helsinki for research involving human tissue. The method of isolating limbal epithelial sheets followed the method previously reported.20 In short, fresh corneoscleral tissue was cut into four similar segments in a 60-mm culture dish containing HBSS, and the remaining iris and endothelial cells were rubbed off. Each segment was digested with 15 mg/mL Dispase II in SHEM with 100 mM sorbitol at 4°C for 18 hours and separated from the stroma of each segment. SHEM was made of an equal volume of HEPES-buffered DMEM and Ham's F12 containing bicarbonate, 0.5% dimethyl sulfoxide, 2 ng/mL mouse-derived EGF, 5 μg/mL insulin, 5 μg/mL transferrin, 5 ng/mL sodium selenite, 0.5 μg/mL hydrocortisone, 30 ng/mL cholera toxin, 5% FBS, 50 μg/mL gentamicin, and 1.25 μg/mL amphotericin B. Such limbal epithelial sheets were either directly cultured or rendered into single cells by 0.25% trypsin and 1 mM EDTA for 5 minutes before being cultured. Approximately 5 × 104 singe cells or a sheet per well (six-well culture dish) were seeded on plastic with or without mitomycin C-inactivated 3T3 fibroblast feeder layers in the SHEM.
Cell Viability Assay
A cell viability assay (Live/Dead Assay; Molecular Probes) was performed to assess the viability of isolated epithelial sheets according to the manufacturer's instruction. Briefly, after the sheets were isolated as described earlier, the culture medium was removed, and the cells were washed twice with HBSS and incubated in the dark for 40 minutes with 0.5 mL of 2 mM calcein-AM and 4 mM ethidium homodimer in PBS. After they were washed with PBS, green-fluorescent cells (live) and red-fluorescent cells (dead) were examined by epifluorescence microscope (Te-2000u Eclipse; Nikon, Tokyo, Japan).
Epithelial Growth, Laser Micro Dissection, and Clonal Cultures
The size equivalent to one eighth of the entire dispase-isolated limbal epithelial sheets as described earlier was further subdivided into two portions. One portion was directly seeded as a sheet on plastic, whereas the other was treated with trypsin/EDTA, and seeded as single cells on plastic. The epithelial growth of three separate cultures was measured by digitizing phase-contrast micrographs taken every 3 days during a period of 27 days using Image J (developed by Wayne Ras-band, National Institutes of Health, Bethesda, MD; available at http://rsb.info.nih.gov/ij/index.html.) To demonstrate the growth potential, 10 limbal epithelial clusters, each 1 mm in diameter, were removed by laser microdissection (LMD6000; Leica, Tokyo, Japan) from human limbal sheets cultured for 48 hours in SHEM on plastic. As a control, 10 similar sizes were also obtained from areas without clusters. Single cells obtained from the above two cultures were treated with trypsin/EDTA for 5 minutes and seeded on mitomycin C-treated 3T3 fibroblast feeder layers at a density of 40 cells/cm2 for 2 weeks. Resultant clonal growth was assessed by rhodamine B or crystal violet staining, and colony forming efficiency (CFE) was determined by counting the percentage of those clones consisting of more than 100 cells per clone subdivided by seeded cells.
Reverse Transcription–Polymerase Chain Reaction
Total RNA was extracted (Trizol; Invitrogen-Gibco) from confluent cultured epithelial cells and the corneal epithelium scraped from corneal buttons as a positive control. Total RNA equivalent to 1 × 105 cultured cells or one corneal button was subjected to RT-PCR based on a protocol recommended by Promega (Madison, WI). The final concentration of RT reaction was 10 mM Tris-HCl (pH 9.0 at 25°C), 5 mM MgCl2, 50 mM KCl, 0.1% Triton X-100, 1 mM each dNTP, 1 U/μL recombinant RNasin ribonucleases inhibitor, 15 units AMV reverse transcriptase, 0.5 μg oligo(dT)15 primer, and total RNA in a total volume of 20 μL The reaction was kept at 42°C for 60 minutes. One tenth of the RT sample was used for subsequent PCR, with the final concentration of the PCR reaction being 10 mM Tris-HCl (pH 8.3 at 25°C), 50 mM KCl, 1.5 mM Mg(OAc)2, 25 pM of each primer shown in Table 1, and 1.25 units of Taq DNA polymerase in a total volume of 50 μL. The PCR mixture was first denatured at 94°C for 5 minutes, then amplified for 30 cycles (94°C, 1 minute; 60°C, 1 minute; and 72°C, 1 minute) with a programmable thermal controller (PTC-100; MJ Research Inc., Watertown, MA). After amplification, 15 μL of each PCR product and 3 μL of 6× loading buffer were mixed and electrophoresed on a 1.5% agarose gel in 0.5× Tris-boric acid-EDTA (TBE) containing 0.5 μg/mL ethidium bromide. The gels were photographed, scanned, and quantitated by densitometry (Photoshop software; Adobe Systems, San Jose, CA).
Table 1.
RT-PCR Primer Sequences
| Primer | Sequence 5′-3′ | cDNA Location | Size of PCR Product (bp) | GenBank Accession Number |
|---|---|---|---|---|
| Cytokeratin 3 | 572 | NM057088 | ||
| Sense | CAGAGATCGAGGGTGTCAAGAAG | (1289–1311) | ||
| Antisense | AGGACTGGGAGAACTTGATGCTG | (1861–1839) | ||
| Cytokeratin 12 | 451 | NM000223 | ||
| Sense | GAGCTCCAAAGCTTCCGGGTGGGC | (832–854) | ||
| Antisense | CATTAGTTCTTCAATTTCCTGAAC | (1506–1483) | ||
| ABCG2 | 399 | NM004827.2 | ||
| Sense | GGGTTCTCTTCTTCCTGACGACC | (1776–1798) | ||
| Antisense | TGGTTGTGAGATTGACCAACAGACC | (2174–2150) | ||
| MDR-1 | 484 | NM000927.3 | ||
| Sense | CAGAAACAACGCATTGCCATAGCTC | (3956–3980) | ||
| Antisense | TGATGATGTCTCTCACTCTGTTCC | (4439–4416) | ||
| GAPDH | 573 | BC029618 | ||
| Sense | GAAGGTGAAGGTCGGAGTCAACG | (60–82) | ||
| Antisense | GCGGCCATCACGCCACAGTTTC | (654–633) |
Immunostaining
On confluence, cells were fixed with ice-cold methanol for 15 minutes and then permeabilized with 0.1% Triton X-100 in PBS for 10 minutes. After the reaction was blocked with 3% BSA in PBS for 30 minutes and the cells were rinsed twice for 5 minutes with PBS, they were incubated with a monoclonal antibody against K3, a cornea-specific differentiation marker,1 and with a monoclonal antibody to p63, a putative marker for limbal epithelial progenitors.8 After three PBS washes, each for 5 minutes, the cells were incubated with FITC-conjugated goat anti-rabbit and goat anti-rat IgG (1:100) and IgM (1:100), respectively. After three additional PBS washes, they were mounted with anti-fade mounting medium (Vectashield; Vector Laboratories, Burlingame, CA) and photographed with the epifluorescence microscope.
Immunoblot Analysis
On confluence, cells were rinsed twice with ice-cold PBS at 4°C, and treated by RIPA buffer consisting of 50 mM Tris-HCl, 150 mM NaCl, 1% NP-40, 0.5% deoxycholic acid and 0.1% SDS, supplemented with proteinase inhibitors (2 μg/mL aprotinin, 2 μg/mL leupeptin, 1 μg/mL pepstatin A, and 0.5 mM phenylmethyl sulfonyl fluoride), phosphatase inhibitors (10 mM sodium fluoride, 1 mM sodium orthovanadate), and 1 mM dithiothreitol. The lysate was mixed at 4°C for 30 minutes and then centrifuged at 12,000 rpm at 4°C for 5 minutes. The supernatant was aliquoted and stored at −80°C. The protein concentration was determined by a BCA protein assay (Pierce, Rockford, IL). Equal amounts of proteins (18 μg) in total cell extracts were separated by 10% SDS-PAGE and transferred to nitrocellulose membranes. Nonspecific binding sites on the membrane were blocked with 5% skim milk in TTBS (Tris-buffered saline-Tween 20 containing 20 mM Tris-HCl [pH 7.6], 137 mM NaCl, and 0.1% Tween 20 [vol/vol]) for 1 hour. The membranes were then incubated overnight with antibodies to K3 (1:1000), K12 (1:300), p63 (1:100), and β-actin (1:10,000) in TTBS in 5% skim milk at 4°C. After they were washed with TBST three times, the membranes were incubated with horseradish peroxidase-linked secondary antibody (1:10,000) in TTBS for 1 hour, followed by three washes in TTBS, and the immunoreactive bands were visualized by chemiluminescence reagent (Western Lightning; Pierce). The intensity of the bands was quantitated by densitometry (Photoshop; Adobe Systems).
Statistical Analysis
The significance of the differences between groups was determined by Student's t-test. P < 0.05 was considered statistically significant.
Results
Enrichment of p63-Positive Cell Clusters in Human Limbal Palisades
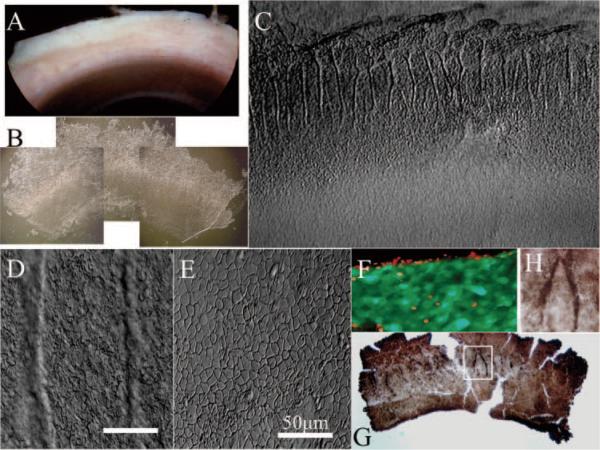
Previous studies based on human limbal cross sections obtained radial8 or tangential21 to the limbus have demonstrated that limbal basal cells preferentially express p63. We wondered whether such p63-positive cells were preferentially distributed in the limbal palisades, an anatomic structure thought to contain limbal epithelial SCs. Immediately after corneal transplantation, we specifically chose those corneo-limbo-scleral tissues that carried heavy pigmentation for ease of identifying the limbal palisades (Fig. 1A). Using a previously reported overnight dispase digestion method,20 we isolated an intact limbal epithelial sheet (Fig. 1B). Under higher magnification, such isolated sheets placed on plastic still preserved undulated folds of pigmented limbal palisades where cells were smaller (Figs. 1C, 1D). In contrast, cells in the corneal region were flat and larger (Figs. 1C, 1E). As previously reported,20 we verified that cells of such isolated limbal sheets were viable as demonstrated by cell viability assay (Live/Dead assay; Molecular Probes; Fig. 1F). Immunostaining showed that p63-positive cells were enriched in limbal palisades (Figs. 1G, 1H).
Figure 1.
Enrichment of p63-positive cells in limbal palisades. From one quarter of corneo-limbo-scleral tissue left after corneal transplantation (A), an intact limbal epithelial sheet was isolated by overnight dispase digestion (B). When one fourth of an isolated sheet (i.e., one sixteenth of the entire limbal sheet) was put on a plastic dish (C), pigmented limbal palisades noted before digestion (A) were preserved and exhibited undulating folds in the limbal location (B, C), where cells were small (D). In contrast, cells in the peripheral corneal region were flat and larger (C, E). Cells of such isolated limbal sheets were viable, as revealed by green fluorescence in a cell-viability assay, whereas some cells in the cut edge were dead, as shown by red fluorescence (F). Immunostaining showed that p63-positive cells were enriched in limbal palisades (G, enlarged inset in H). Bars, 50 μm.
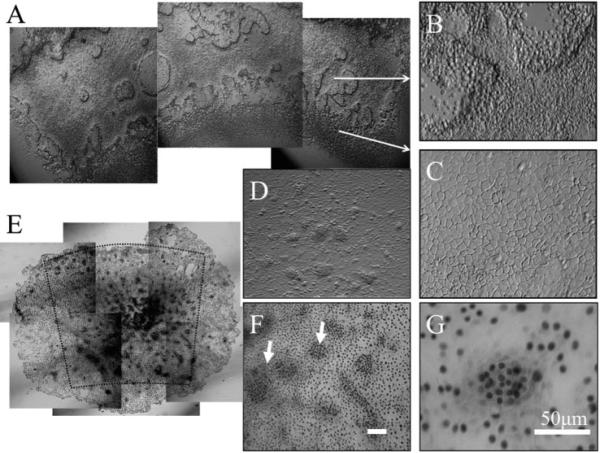
To verify that the aforementioned pattern was generated by basal cells, dispase-isolated human limbal epithelial sheets were cultured on plastic in SHEM for 48 hours so that most superficial and suprabasal cells were detached, resulting in a monolayer with areas of multicellular clusters (Fig. 2A). These cellular clusters were distributed mainly in the limbal region corresponding to palisades (Fig. 2B). In contrast, cells in the peripheral corneal location were larger and did not form clusters (Fig. 2C). When the cultures were extended for another 48 hours, these clusters became more organized into dense cellular aggregates, presumably because of proliferation (Figs. 2D, 2E). Immunostaining showed that these clusters contained small cells with strong p63 nuclear staining (Figs. 2F, 2G). Collectively, these results indicated that human limbal palisades preferentially contained clusters of p63-positive cells.
Figure 2.
Enrichment of p63-positive cell clusters in limbal palisades. When dispase-isolated human limbal epithelial sheets were cultured in SHEM on plastic for 48 hours, most superficial and suprabasal cells detached, resulting in a monolayer with areas of multicellular clusters (A). These cellular clusters were distributed mainly in the limbal region corresponding to the palisade (B). In contrast, cells in the peripheral corneal region were larger without clusters (C). When cultures were extended for another 48 hours, the clusters became more organized into dense cellular aggregates (D). Immunostaining showed that these clusters contained small cells with strong p63 nuclear staining (E–G; arrows in F). Bars, 50 μm.
Higher Clonal Growth by p63-Rich Cell Clusters
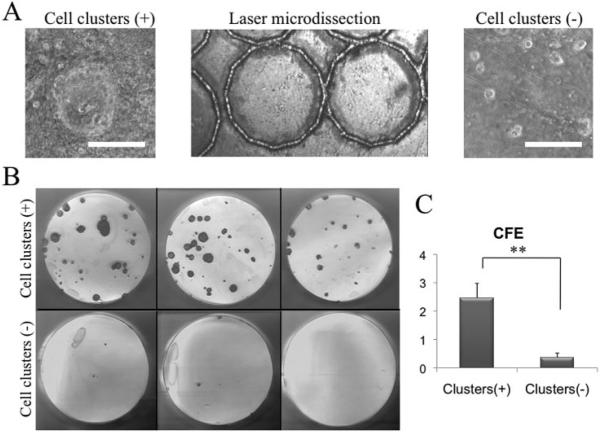
To determine whether areas with p63-positive clusters contained progenitors more so than areas without, we removed 10 areas with and 10 areas without such clusters, each 1 mm in diameter, by laser microdissection (Fig. 3A). After being rendered into single cells, the same number of cells from each group was seeded on 3T3 fibroblast feeder layers. After 14 days of cultivation, the clonal growth-formed areas containing p63-positive clusters had a significantly higher CFE than that from areas without (Figs. 3B, 3C; P < 0.001).
Figure 3.
Higher clonal growth by p63-positive cell clusters. When dispase-isolated human limbal sheets were cultured on plastic in SHEM for 48 hours (Fig. 2), the cells were collected from areas with (+) and without (−) p63+ clusters by a laser-assisted microdissection microscope (A) and rendered into single cells by trypsin/EDTA. After being seeded on 3T3 fibroblast feeder layers for 14 days, cells derived from p63+ cluster areas exhibited more vivid clonal growth revealed by rhodamine B staining (B) and a significantly higher CFE than those from the noncluster areas (C, **P < 0.01). Bars, 50 μm.
Higher Clonal Growth Potential by Limbal Epithelial Sheets
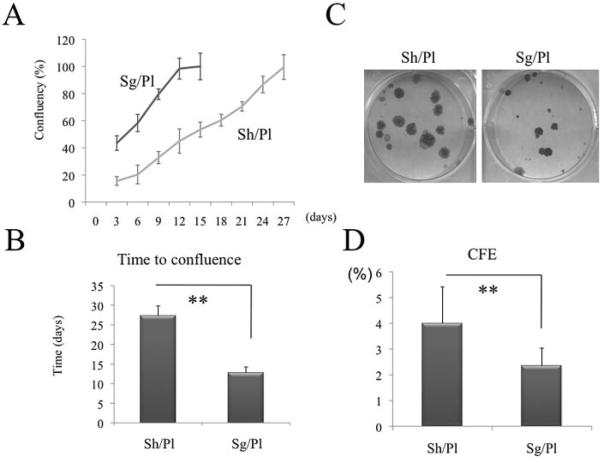
We thus wondered whether maintenance of the aforementioned p63-positive clusters by limbal sheet cultures may retain a higher growth potential. To test this hypothesis, we cultivated isolated limbal epithelial sheets either directly or as single cells after being treated by trypsin/EDTA. The latter treatment retained high viability in the range of 81% to 92% (n = 10). We confirmed that the viability of cells treated with dispase alone was not different from that treated with dispase followed by trypsin-EDTA based on trypan blue dye exclusion (n = 3, 66.5% ± 9.9%, 67.6% ± 21.1%, respectively). Just after the enzymatic treatment, the growth potential between the two was not significantly different as judged by the CFE counted from 3T3 fibroblast feeder layers (n = 3, 8.4% ± 6.5%, 7.8% ± 1.7%, respectively). Within 24 hours after seeding, more than 50% of epithelial cells attached on plastic surface and contained both small and large cells. To facilitate attachment of the epithelial sheets, we applied a minimal amount of medium sufficient to cover the sheet for 1 or 2 days. As expected, epithelial growth measured by digitizing phase-contrast micrographs in single cells on plastic (Sg/Pl) was significantly greater and faster than that in limbal sheets on plastic (Sh/Pl) when plotted as a percentage of confluence (Figs. 4A, 4B; P < 0.01). Afterward, the same number of single cells obtained from the two cultures was seeded on 3T3 fibroblast feeder layers. The resultant clones of Sh/Pl were larger than Sg/Pl (Fig. 4C).
Figure 4.
Comparison of clonal growth potential between sheets and single cells. Dispase-isolated limbal epithelial sheets were cultivated Sh/Pl or Sg/Pl after trypsinization for 27 days. As expected, epithelial growth measured by digitizing phase-contrast micrographs and plotted as the percentage of confluence in Sg/Pl was significantly greater (A) and faster (B, P < 0.01) than that in Sh/Pl. Afterward, the same number of single cells obtained from the above two cultures was seeded on 3T3 fibroblast feeder layers. The resultant clonal growth by Sh/Pl was more vivid (C) and had a significantly higher CFE (D) than that by Sg/Pl (**P < 0.01).
Expression of Differentiation and Progenitor Markers
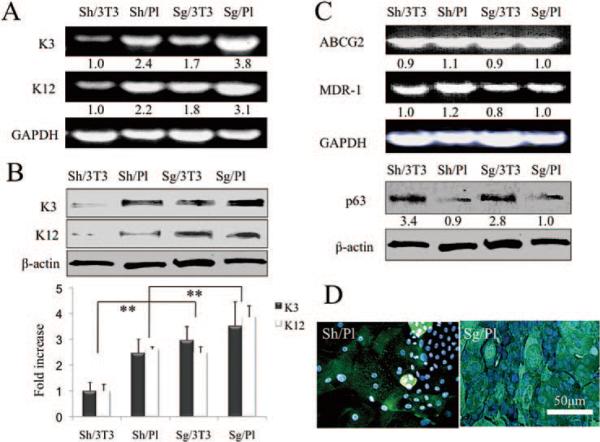
To determine the differentiation status of the cells, we measured the expression of K3 and -12 transcripts by RT-PCR and their proteins by Western blot analysis. The same amounts of limbal sheets and single cells were seeded on plastic or 3T3 fibroblast feeder layers (3T3). On confluence at different times (Fig. 4B), we isolated epithelial cells, their total RNAs and protein extracts were prepared. Before that, 3T3 fibroblasts were much easier to remove by trypsin/EDTA than were the epithelial cells. With GAPDH used as a loading control and the expression of K3 transcript treated as 1 for Sh/3T3 cultures, the expression of Sh/Pl, Sg/3T3, and Sg/Pl increased by 2.7-, 2.3-, and 4.5-fold, respectively. Similarly, with the expression of K12 transcript by Sh/3T3 cultures treated as 1, the expression in Sh/Pl, Sg/3T3, and Sg/Pl cultures was found to increase by 1.8-, 1.8-, and 2.3-fold, respectively (Fig. 5A). With β-actin as a loading control and the expression of K3 protein by Sh/3T3 treated as 1, the expression of Sh/Pl, Sg/3T3, and Sg/Pl were found to increase by 2.5 ± 0.5-, 3.0 ± 0.5-, and 3.5 ± 0.9-fold (mean ± SD, n = 3), respectively. Similarly, with the expression of K12 protein by Sh/3T3 treated as 1, the expression of Sh/Pl, Sg/3T3, and Sg/Pl cultures increased by 2.6 ± 0.1-, 2.5 ± 0.2-, and 3.9 ± 0.4-fold, respectively (n = 3; Fig. 5B, **P < 0.01). Collectively, these results indicate that expression of both corneal differentiation markers was the lowest in sheet cultures expanded on 3T3 fibroblast feeder layers (Sh/3T3) when compared with the other three culturing conditions.
Figure 5.
Expression of differentiation and progenitor markers. RT-PCR showed that expression of K3 and −12 transcripts (i.e., corneal differentiation markers), was the lowest in sheets expanded on 3T3 fibroblast feeder layers (Sh/3T3) when compared to the rest (A). Western blot analysis also showed that expression of K3 and −12 proteins was the lowest in Sh/3T3 (B, **P < 0.01). Although expression of ABCG2 and MDR-1 transcripts did not show any difference among the four conditions, that of p63 protein by either sheets or single cells was promoted by 3T3 feeder cells (C). Immunostaining for K3 was heterogeneously positive in sheet cultures on plastic, but was uniformly positive in single cell suspension cultures (D).
To determine whether there was any difference in the progenitor status, levels of ABCG2 and MDR1 transcripts measured by RT-PCR were not found to be significantly different (Fig. 5C). However, Western blot analysis showed that p63 protein expression was promoted by 3T3 feeder cells either in a sheet or single cells (Fig. 5C). Immunostaining to K3 was heterogeneously positive in sheet cultures on plastic with small cells being negative, but was uniformly positive in single-cell suspension cultures (Fig. 5D). These results indicate that the sheet cultures preserved more progenitors with less differentiation than did single cells.
Discussion
Previously, nuclear expression of p63 was found to be enriched in limbal basal cells in radial and tangential cross sections.8 Three-dimensional reconstruction of the limbal region has revealed that expression of p63 is enriched in the limbal region along the basal layer of the palisade ridge.22 Using dispase-isolated limbal sheets for two-dimensional flatmount preparations, the present study indicated that p63-positive small cells were distributed in clusters along the palisade (Figs. 1, 2). Such a cluster pattern was distinctively different from the corneal region immediately adjacent to the palisade. Although overall p63 expression was more in the limbal region, cells isolated from this cluster area by laser microdissection exhibited significantly more vivid and higher clonal growth than those isolated from areas without clusters when reseeded on 3T3 fibroblast feeder layers (Fig. 3). Collectively, these results indicate that not all limbal basal cells are SCs. We thus speculate that such a pattern of p63-positive clusters would help us home in on the exact locus of limbal SCs in the future and that selective isolation of such clusters will help us characterize limbal SCs better in the future.
Because clusters of p63-positive small cells were not uniformly distributed in an intact human limbal epithelial sheet, we wondered whether maintenance of the cell– cell interaction in sheets plays any role in preserving their progenitor status. As expected, single cells reached confluence much faster than sheets as the growth for the latter was affected by contact inhibition that was maintained in epithelial sheets (Fig. 4). Nevertheless, our study surprisingly showed that dissociation of such clusters into single cells could affect the overall growth potential. That is, cells of the monolayer derived from the sheet plastic culture exhibited more vivid and higher clonal growth than those from the single cell plastic culture when reseeded on 3T3 fibroblast feeder layers (Fig. 4). Further studies are needed to determine what kind of cell– cell interactions is used in p63-rich clusters and how such interactions may help retain their clonal growth potential.
Without knowing the exact action mechanism at this moment, our study further showed that continuous expansion of both sheets and single cells on plastic resulted in more differentiation as evidenced by notable expression of K3 and −12 and reduction of p63 expression (Fig. 5). Consistent with other reports,23,24 such a deleterious effect could be counteracted by 3T3 fibroblast feeder layers, as revealed by significant reduction of K3 and −12 expression in either sheet or single-cell cultures. Even if seeded on 3T3 fibroblast feeder layers, sheet cultures still expressed the lowest level of K3 and −12 transcripts and proteins (Fig. 5).
Several protocols have been exercised clinically for expanding human limbal epithelial progenitor cells from a small limbal biopsy for treating limbal SC deficiency.18,19,25–28 There are many differences regarding the three key steps (i.e., isolation, expansion, and transplantation on carrier or not) among these protocols. Some used intact28 or denuded human amniotic membrane29 or fibrin30 as a substrate, but some did not.31 Besides this obvious difference in the use of carrier for transplantation, all start with a small limbal biopsy for autologous transplantation and entire limbus from cadaveric donors for allogenic transplantation. However, some use it as an explant for autograft,25 but others isolate limbal epithelial cells from the biopsy using trypsin/EDTA32 or dispase33 for allografts. The results of our study imply that there is an advantage of preserving cell– cell interactions as in sheets. However, if sheets are to be dissociated into single cells, it becomes crucial to be in contact with 3T3 fibroblast feeder layers, either directly18,19,26 or indirectly,29 or other feeder layers derived from amniotic epithelial cells,34 to maintain the progenitor status. Only if we begin to dissect carefully each step of ex vivo expansion protocol will we finally grasp the key essence of effective expansion of limbal epithelial progenitors by maintaining their clonal growth potential (i.e., one salient feature of stemness).
Acknowledgments
Supported by Grants R01-EY06819 and R01-EY15735 (SCGT) from the National Institutes of Health, National Eye Institute, Bethesda, MD; and Grant-in-Aid H18-Tissue Engineering-Young-002 for Scientific Research from the Ministry of Health and Welfare, Japan.
Footnotes
Disclosure: T. Kawakita, None; S. Shimmura, None; K. Higa, None; E.M. Espana, None; H. He, None; J. Shimazaki, None; K. Tsubota, None; S.C.G. Tseng, None
References
- 1.Schermer A, Galvin S, Sun T-T. Differentiation-related expression of a major 64K corneal keratin in vivo and in culture suggests limbal location of corneal epithelial stem cells. J Cell Biol. 1986;103:49–62. doi: 10.1083/jcb.103.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lavker RM, Tseng SC, Sun TT. Corneal epithelial stem cells at the limbus: looking at some old problems from a new angle. Exp Eye Res. 2004;78:433–446. doi: 10.1016/j.exer.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 3.Lavker RM, Sun T-T. Epidermal stem cells: properties, markers and location. Proc Natl Acad Sci USA. 2000;97:13473–13475. doi: 10.1073/pnas.250380097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romano AC, Espana EM, Yoo SH, Budak MT, Wolosin JM, Tseng SC. Different cell sizes in human limbal and central corneal basal epithelia measured by confocal microscopy and flow cytometry. Invest Ophthalmol Vis Sci. 2003;44:5125–5129. doi: 10.1167/iovs.03-0628. [DOI] [PubMed] [Google Scholar]
- 5.Cotsarelis G, Cheng SZ, Dong G, Sun T-T, Lavker RM. Existence of slow-cycling limbal epithelial basal cells that can be preferentially stimulated to proliferate: implications on epithelial stem cells. Cell. 1989;57:201–209. doi: 10.1016/0092-8674(89)90958-6. [DOI] [PubMed] [Google Scholar]
- 6.Liu C-Y, Zhu G, Converse R, et al. Characterization and chromosomal localization of the cornea-specific murine keratin gene Krt1.12. J Biol Chem. 1994;260:24627–24636. [PubMed] [Google Scholar]
- 7.Matic M, Petrov IN, Chen S, Wang C, Dimitrijevich SD, Wolosin JM. Stem cells of the corneal epithelium lack connexins and metabolite transfer capacity. Differentiation. 1997;61:251–260. doi: 10.1046/j.1432-0436.1997.6140251.x. [DOI] [PubMed] [Google Scholar]
- 8.Pellegrini G, Dellambra E, Golisano O, et al. p63 identifies keratin-ocyte stem cells. Proc Natl Acad Sci USA. 2001;98:3156–3161. doi: 10.1073/pnas.061032098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watanabe K, Nishida K, Yamato M, et al. Human limbal epithelium contains side population cells expressing the ATP-binding cassette transporter ABCG2. FEBS Lett. 2004;565:6–10. doi: 10.1016/j.febslet.2004.03.064. [DOI] [PubMed] [Google Scholar]
- 10.Hayashi R, Yamato M, Sugiyama H, et al. N-cadherin is expressed by putative stem/progenitor cells and melanocytes in the human limbal epithelial stem cell niche. Stem Cells. 2007;25:289–296. doi: 10.1634/stemcells.2006-0167. [DOI] [PubMed] [Google Scholar]
- 11.Puangsricharern V, Tseng SCG. Cytologic evidence of corneal diseases with limbal stem cell deficiency. Ophthalmology. 1995;102:1476–1485. doi: 10.1016/s0161-6420(95)30842-1. [DOI] [PubMed] [Google Scholar]
- 12.Li W, Hayashida Y, Chen YT, Tseng SC. Niche regulation of corneal epithelial stem cells at the limbus. Cell Res. 2007;17:26–36. doi: 10.1038/sj.cr.7310137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rheinwald JG, Green H. Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell. 1975;6:331–337. doi: 10.1016/s0092-8674(75)80001-8. [DOI] [PubMed] [Google Scholar]
- 14.Lindberg K, Brown ME, Chaves HV, Kenyon KR, Rheinwald JG. In vitro preparation of human ocular surface epithelial cells for transplantation. Invest Ophthalmol Vis Sci. 1993;34:2672–2679. [PubMed] [Google Scholar]
- 15.Wei Z-G, Wu R-L, Lavker RM, Sun T-T. In vitro growth and differentiation of rabbit bulbar, fornix, and palpebral conjunctival epithelia: implication on conjunctival epithelial transdifferentiation and stem cells. Invest Ophthalmol Vis Sci. 1993;34:1814–1828. [PubMed] [Google Scholar]
- 16.Lehrer MS, Sun T-T, Lavker RM. Strategies of epithelial repair: modulation of stem cell and transient amplifying cell proliferation. J Cell Sci. 1998;111:2867–2875. doi: 10.1242/jcs.111.19.2867. [DOI] [PubMed] [Google Scholar]
- 17.Pellegrini G, Golisano O, Paterna P, et al. Location and clonal analysis of stem cells and their differentiated progeny in the human ocular surface. J Cell Biol. 1999;145:769–782. doi: 10.1083/jcb.145.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pellegrini G, Traverso CE, Franzi AT, Zingirian M, Cancedda R, De Luca M. Long-term restoration of damaged corneal surface with autologous cultivated corneal epithelium. Lancet. 1997;349:990–993. doi: 10.1016/S0140-6736(96)11188-0. [DOI] [PubMed] [Google Scholar]
- 19.Rama P, Bonini S, Lambiase A, et al. Autologous fibrin-cultured limbal stem cells permanently restore the corneal surface of patients with total limbal stem cell deficiency. Transplantation. 2001;72:1478–1485. doi: 10.1097/00007890-200111150-00002. [DOI] [PubMed] [Google Scholar]
- 20.Espana EM, Romano AC, Kawakita T, Di Pascuale M, Smiddy R, Tseng SC. Novel enzymatic isolation of an entire viable human limbal epithelial sheet. Invest Ophthalmol Vis Sci. 2003;44:4275–4281. doi: 10.1167/iovs.03-0089. [DOI] [PubMed] [Google Scholar]
- 21.Chen WYW, Mui M-M, Kao WW-Y, Liu C-Y, Tseng SCG. Conjunctival epithelial cells do not transdifferentiate in organotypic cultures: expression of K12 keratin is restricted to corneal epithelium. Curr Eye Res. 1994;13:765–778. doi: 10.3109/02713689409047012. [DOI] [PubMed] [Google Scholar]
- 22.Shortt AJ, Secker GA, Munro PM, Khaw PT, Tuft SJ, Daniels JT. Characterization of the limbal epithelial stem cell niche: novel imaging techniques permit in vivo observation and targeted biopsy of limbal epithelial stem cells. Stem Cells. 2007;25:1402–1409. doi: 10.1634/stemcells.2006-0580. [DOI] [PubMed] [Google Scholar]
- 23.Grueterich M, Espana EM, Tseng SC. Modulation of keratin and connexin expression in limbal epithelium expanded on denuded amniotic membrane with and without a 3T3 fibroblast feeder layer. Invest Ophthalmol Vis Sci. 2003;44:4230–4236. doi: 10.1167/iovs.02-0943. [DOI] [PubMed] [Google Scholar]
- 24.Liu S, Li J, Wang C, Tan D, Beuerman R. Human limbal progenitor cell characteristics are maintained in tissue culture. Ann Acad Med Singapore. 2006;35:80–86. [PubMed] [Google Scholar]
- 25.Tsai RJF, Li L-M, Chen J-K. Reconstruction of damaged corneas by transplantation of autologous limbal epithelial cells. N Engl J Med. 2000;343:86–93. doi: 10.1056/NEJM200007133430202. [DOI] [PubMed] [Google Scholar]
- 26.Schwab IR, Reyes M, Isseroff RR. Successful transplantation of bioengineered tissue replacements in patients with ocular surface disease. Cornea. 2000;19:421–426. doi: 10.1097/00003226-200007000-00003. [DOI] [PubMed] [Google Scholar]
- 27.Koizumi N, Inatomi T, Suzuki T, Sotozono C, Kinoshita S. Cultivated corneal epithelial stem cell transplantation in ocular surface disorders. Ophthalmology. 2001;108:1569–1574. doi: 10.1016/s0161-6420(01)00694-7. [DOI] [PubMed] [Google Scholar]
- 28.Grueterich M, Espana EM, Touhami A, Ti SE, Tseng SC. Phenotypic study of a case with successful transplantation of ex vivo expanded human limbal epithelium for unilateral total limbal stem cell deficiency. Ophthalmology. 2002;109:1547–1552. doi: 10.1016/s0161-6420(02)01105-3. [DOI] [PubMed] [Google Scholar]
- 29.Koizumi N, Fullwood NJ, Bairaktaris G, Inatomi T, Kinoshita S, Quantock AJ. Cultivation of corneal epithelial cells on intact and denuded human amniotic membrane. Invest Ophthalmol Vis Sci. 2000;41:2506–2513. [PubMed] [Google Scholar]
- 30.Higa K, Shimmura S, Kato N, et al. Proliferation and differentiation of transplantable rabbit epithelial sheets engineered with or without an amniotic membrane carrier. Invest Ophthalmol Vis Sci. 2007;48:597–604. doi: 10.1167/iovs.06-0664. [DOI] [PubMed] [Google Scholar]
- 31.Nishida K, Yamato M, Hayashida Y, et al. Corneal reconstruction with tissue-engineered cell sheets composed of autologous oral mucosal epithelium. N Engl J Med. 2004;351:1187–1196. doi: 10.1056/NEJMoa040455. [DOI] [PubMed] [Google Scholar]
- 32.Zito-Abbad E, Borderie VM, Baudrimont M, et al. Corneal epithelial cultures generated from organ-cultured limbal tissue: factors influencing epithelial cell growth. Curr Eye Res. 2006;31:391–399. doi: 10.1080/02713680600681228. [DOI] [PubMed] [Google Scholar]
- 33.Ueta M, Kweon MN, Sano Y, et al. Immunosuppressive properties of human amniotic membrane for mixed lymphocyte reaction. Clin Exp Immunol. 2002;129:464–470. doi: 10.1046/j.1365-2249.2002.01945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen YT, Li W, Hayashida Y, et al. Human amniotic epithelial cells as novel feeder layers for promoting ex vivo expansion of limbal epithelial progenitor cells. Stem Cells. 2007;25:1995–2005. doi: 10.1634/stemcells.2006-0677. [DOI] [PMC free article] [PubMed] [Google Scholar]