Abstract
Three new species of Leohumicola (anamorphic Leotiomycetes) are described using morphological characters and phylogenetic analyses of DNA barcodes. Leohumicola levissima and L. atra were isolated from soils collected after forest fires in Crater Lake National Park, United States. Leohumicola incrustata was isolated from burned fynbos from the Cape of Good Hope Nature Reserve, South Africa. The three species exhibit characteristic Leohumicola morphology but are morphologically distinct based on conidial characters. Two DNA barcode regions, the Internal Transcribed Spacer (ITS) nuclear rDNA region and the cytochrome oxidase subunit I (Cox1) mitochondrial gene, were sequenced. Single-gene parsimony, dual-gene parsimony and dual-gene Bayesian inference phylogenetic analyses support L. levissima, L. atra, L. incrustata as distinct phylogenetic species. Both ITS and Cox1 barcodes are effective for the molecular identification of Leohumicola species.
Keywords: aleurioconidium, chlamydospore, cytochrome oxidase subunit I (Cox1), internal transcribed spacer (ITS), Leotiomycetes
INTRODUCTION
The hyphomycete genus Leohumicola was described for four species (L. verrucosa, L. minima, L. terminalis, and L. lenta) by Hambleton et al. (2005). Leohumicola species produce two-celled aleurioconidia, with a round to ellipsoidal, dark-brown terminal cell with slightly thickened walls, and a basal cell that is either cupulate or cylindrical, and hyaline to pale brown. Globose to ellipsoidal, intercalary or terminal chlamydospores are produced by all known species. Conidium ontogeny is usually monoblastic, with sympodial extension of the conidiogenous cells sometimes occurring. Secession is rhexolytic, with the remnants of the empty basal cell remaining attached to the terminal cell. Most Leohumicola strains grow slowly and sporulate sparsely or not at all; they must be grown on various media to stimulate conidial production.
Hambleton et al. (2005) noted that many internal transcribed spacer (ITS) sequences of unidentified soil or root-associated fungi in GenBank belonged to the Leohumicola clade, but did not correspond with the species they described. Several of the known species of Leohumicola were associated with burnt ecosystems, especially commercial blueberry cultivation, and were isolated by heat treatment of soil suspensions or from surface-sterilised roots of ericaceous host plants. This association with the plant family Ericaceae led us to obtain soil samples from a burned area of fynbos in the Cape Floristic Region (Cape of Good Hope Nature Reserve, South Africa), a hotspot of biodiversity for this plant family (Cowling & Richardson 1995). We also obtained soil from a part of Crater Lake National Park, United States, recently affected by forest fires.
The original study of Leohumicola supplemented morphological information with phylogenetic analysis of the ITS. Here, we add analyses of cytochrome oxidase subunit I (Cox1) mitochondrial gene sequences. The ITS is a widely accepted DNA marker for identifying fungi. Cox1 is the DNA barcode gene that has been tested most extensively in the animal kingdom, with a 648-bp region in the 5′ end usually providing species-level resolution (e.g. Ward et al. 2005, Hajibabaei et al. 2006). The Cox1 barcode region was somewhat effective in identifying species of Penicillium (Seifert et al. 2007) but has otherwise been little explored in fungi. This study assesses the utility of both ITS and Cox1 as DNA barcodes for the identification of Leohumicola species.
MATERIALS AND METHODS
Isolation, observation and preservation of cultures
Soil samples from South Africa were collected at Olifantsbos, Cape of Good Hope Nature Reserve, with the kind assistance of Dr Karin Jacobs and her students from the Department of Microbiology, University of Stellenbosch. The sampled region was a fynbos with abundant Erica and Protea species, which had been burned one or two years previously. Samples of surface and rhizosphere soil were collected in sterile 15 mL BD Falcon conical bottom tubes and mailed to Ottawa. Soil samples from Crater Lake National Park, Montana, USA, were collected by Matt Trappe in a region with visible signs of forest fire damage. Approximately 50 g samples were collected into zip lock bags and mailed to Ottawa. All samples were kept at room temperature until processing.
Fungi were isolated using a method modified from Jackson et al. (1995). After using a layer of cheesecloth to remove large particles from the soil sample, 2–4 mL of fine dry soil were placed into 50 mL BD Falcon conical bottom tubes. Sterile 0.1 % (w/v) peptone broth was added to the 25 mL mark, then the tube was vortexed for 30 s. The tube was submerged in a 75 °C water bath until the suspension reached this temperature (~ 15 min), then incubated for a further 30 min with manual shaking every 5 min to ensure even temperature distribution. The tube was cooled at room temperature for 30 min. A well homogenised 1 mL aliquot of the suspension was mixed with 100 mL of molten and cooled (50 °C) half-strength potato-dextrose agar (PDA (Difco), BD, Sparks, Maryland, USA) with 40 mg/L chloramphenicol to inhibit bacterial growth. The mixture was dispensed in polystyrene Petri dishes (~ 20 mL per dish) and then left to solidify. Petri dishes were sealed with Parafilm and incubated upright at 25 °C under ambient light conditions.
Isolation plates were checked for Leohumicola colonies every 1–2 d for 2 wk. Petri dishes with no visible growth after 2 wk were incubated for up to 3 mo before disposal. Leohumicola colonies were recognised by their slow growth combined with the release of yellow pigments into the medium. Putative Leohumicola colonies were transferred to new full-strength PDA plates, then incubated as above for 3 wk at room temperature (22–25 °C) before performing morphological studies and DNA extraction.
For morphological studies, suspected Leohumicola colonies were grown on oatmeal agar (OA, Samson et al. 2004), corn meal agar with dextrose (CMA (Difco), BD, Sparks, Maryland, USA) and PDA. To induce sporulation, PDA was inoculated with a suspension of macerated mycelium as described by Hambleton et al. (2005). Cultures were checked for aleurioconidia and chlamydospores monthly. Cultures that did not sporulate after 3 mo were transferred to potato-carrot agar (PCA, Samson et al. 2004), incubated at 25 °C under ambient light conditions for 2 mo, and then rechecked for conidia. Measurements and photographs were taken from material mounted in 85 % lactic acid using an Evolution MP digital microscope camera on an Olympus BX50 compound microscope with differential interference contrast (DIC) optics and captured using ImagePro 6.0 (Media Cybernetics, Bethesda, Maryland, USA). Colony photographs were taken after 2 wk and 2 mo. Some microphotographs and colony photographs were digitally retouched using Adobe Photoshop CS2 for aesthetic reasons, to reduce background clutter and to remove unwanted reflections, as noted in the figure legends. Colony colours were assessed using Kornerup & Wanscher (1978).
Twenty-three strains selected to represent the genetic diversity of the isolated species were deposited in the Canadian Collection of Fungal Cultures (DAOM), Ottawa, Canada (Table 1). Additional strains are deposited in the Seifert Lab collection at Agriculture and Agri-Food Canada.
Table 1.
Source of isolates and GenBank accession numbers of 52 strains used in this study.
| Source |
GenBank accession |
||||||
|---|---|---|---|---|---|---|---|
| Species | Culture | Country | State/Province | Region | Associated plants or substrate | ITS | Cox1 |
| Leohumicola atra | DAOM239515T | United States | Oregon | Crater Lake National Park | Goodyera oblongifolia | EU678386 | EU678425 |
| DAOM239535 | United States | Oregon | Crater Lake National Park | Goodyera oblongifolia | EU678411 | EU678450 | |
| Leohumicola incrustata | DAOM239498T | South Africa | Cape Province | Cape of Good Hope N.R. | Erica | EU678398 | EU678437 |
| DAOM239500 | South Africa | Cape Province | Cape of Good Hope N.R. | Erica | EU678409 | EU678448 | |
| DAOM239501 | South Africa | Cape Province | Cape of Good Hope N.R. | Erica | EU678399 | EU678438 | |
| DAOM239502 | South Africa | Cape Province | Cape of Good Hope N.R. | Erica | EU678404 | EU678443 | |
| DAOM239503 | South Africa | Cape Province | Cape of Good Hope N.R. | Erica | EU678406 | EU678445 | |
| DAOM239504 | South Africa | Cape Province | Cape of Good Hope N.R. | Erica | EU678402 | EU678441 | |
| DAOM239505 | South Africa | Cape Province | Cape of Good Hope N.R. | Erica | EU678403 | EU678442 | |
| DAOM239517 | South Africa | Cape Province | Cape of Good Hope N.R. | Protea tree | EU678397 | EU678436 | |
| HNLHM91 | South Africa | Cape Province | Cape of Good Hope N.R. | Erica | EU678400 | EU678439 | |
| HNLHM92 | South Africa | Cape Province | Cape of Good Hope N.R. | Erica | EU678401 | EU678440 | |
| HNLHM101 | South Africa | Cape Province | Cape of Good Hope N.R. | Erica | EU678405 | EU678444 | |
| HNLHM103 | South Africa | Cape Province | Cape of Good Hope N.R. | Erica | EU678407 | EU678446 | |
| HNLHM107 | South Africa | Cape Province | Cape of Good Hope N.R. | Erica | EU678408 | EU678447 | |
| Leohumicola lenta | DAOM231149T* | Canada | Manitoba | tallgrass prairie | AY706328 | EU678465 | |
| Leohumicola levissima | DAOM239506T | United States | Oregon | Crater Lake National Park | Abies concolor | EU678382 | EU678421 |
| DAOM239507 | United States | Oregon | Crater Lake National Park | Abies concolor | EU678384 | EU678423 | |
| DAOM239508 | United States | Oregon | Crater Lake National Park | Abies concolor | EU678385 | EU678424 | |
| DAOM239509 | United States | Oregon | Crater Lake National Park | Abies concolor | EU678389 | EU678428 | |
| DAOM239510 | United States | Oregon | Crater Lake National Park | Abies concolor | EU678394 | EU678433 | |
| DAOM239511 | United States | Oregon | Crater Lake National Park | Goodyera oblongifolia | EU678396 | EU678435 | |
| DAOM239512 | United States | Oregon | Crater Lake National Park | Ceanothus velatinus | EU678388 | EU678427 | |
| DAOM239513 | United States | Oregon | Crater Lake National Park | Abies concolor | EU678395 | EU678434 | |
| DAOM239514 | United States | Oregon | Crater Lake National Park | Abies concolor | EU678392 | EU678431 | |
| HNLHM2 | United States | Oregon | Crater Lake National Park | Abies concolor | EU678380 | EU678419 | |
| HNLHM2B | United States | Oregon | Crater Lake National Park | Abies concolor | EU678381 | EU678420 | |
| HNLHM4 | United States | Oregon | Crater Lake National Park | Abies concolor | EU678383 | EU678422 | |
| HNLHM21 | United States | Oregon | Crater Lake National Park | Ceanothus velatinus | EU678387 | EU678426 | |
| HNLHM41 | United States | Oregon | Crater Lake National Park | Abies concolor | EU678390 | EU678429 | |
| HNLHM42 | United States | Oregon | Crater Lake National Park | Abies concolor | EU678391 | EU678430 | |
| HNLHM47 | United States | Oregon | Crater Lake National Park | Abies concolor | EU678393 | EU678432 | |
| HNLHM144 | United States | Oregon | Crater Lake National Park | Goodyera oblongifolia | EU678412 | EU678451 | |
| HNLHM145 | United States | Oregon | Crater Lake National Park | Goodyera oblongifolia | EU678413 | EU678452 | |
| HNLHM152 | United States | Oregon | Crater Lake National Park | Goodyera oblongifolia | EU678414 | EU678453 | |
| Leohumicola minima | DAOM232587T* | Chile | Valdivia | volcanic ash | AY706329 | EU678466 | |
| Leohumicola sp. | DAOM230084* | Australia | N.S.W. | Eucalyptus | AY706331 | EU678455 | |
| DAOM231148* | Canada | Manitoba | tallgrass prairie | AY706330 | EU678461 | ||
| DAOM239499 | South Africa | Cape Province | Cape of Good Hope N.R. | Lobelia | EU678417 | EU678469 | |
| DAOM239516 | Canada | Ontario | Stittsville | forest soil | EU678416 | EU678468 | |
| Leohumicola terminalis | DAOM231145T* | Canada | Nova Scotia | Acer saccharum | AY706327 | EU678464 | |
| Leohumicola verrucosa | DAOM226889T* | Canada | Nova Scotia | blueberries | AY706320 | EU678462 | |
| DAOM231141* | Canada | Alberta | Pinus banksiana, Populus tremuloides | AY706321 | EU678457 | ||
| DAOM231142* | Canada | Nova Scotia | blueberries | AY706322 | EU678458 | ||
| DAOM231143* | Canada | Nova Scotia | Pinus spp. | AY706323 | EU678463 | ||
| DAOM231144* | Canada | Nova Scotia | Pinus strobes | AY706324 | EU678459 | ||
| DAOM231147* | Canada | Alberta | Pinus forest | AY706325 | EU678460 | ||
| DAOM230085* | Puerto Rico | not recorded | AY706326 | EU678456 | |||
| DAOM239497 | Canada | Ontario | Stittsville | forest soil | EU678379 | EU678418 | |
| DAOM239536 | Canada | Ontario | Stittsville | forest soil | EU678415 | EU678454 | |
| HNLHM117 | Canada | Ontario | Stittsville | forest soil | EU678410 | EU678449 | |
| Myxotrichium deflexum | UAMH6365** | Canada | Ontario | Toronto | soil | AF062814 | EU678467 |
* Leohumicola strains previously studied by Hambleton et al. (2005). The ITS GenBank accession numbers from Hambleton et al.’s study are listed here for completeness. Cox1 DNA barcodes of Leohumicola species described by Hambleton et al. (2005) were generated in this study.
** Myxotrichium deflexum strain UAMH6365 is from Hambleton et al. (1998).
Abbreviations: DAOM = Canadian Collection of Fungal Cultures, Ottawa, Canada; HNLHM = Seifert Lab Leohumicola Collection, T = ex-type strain; N.R. = Nature Reserve; N.S.W. = New South Wales.
DNA extraction, PCR, sequencing and sequence editing
DNA extractions were performed using UltraClean Microbial DNA Isolation Kits (MO BIO Laboratories Inc., Carlsbad, California, USA) from mycelia scraped from PDA colonies using a sterile scalpel. DNA concentration and quality were determined by Nanodrop ND-1000 spectrometer (Thermo Scientific, Wilmington, Delaware, USA) and preparations were diluted to 1–5 ng/μL of DNA template.
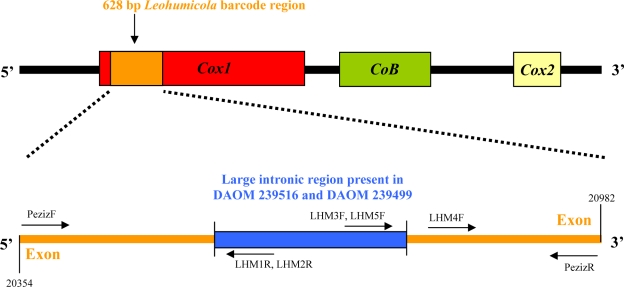
The ITS and Cox1 regions were amplified and sequenced using the primers ITS5 and ITS4 (White et al. 1990) and newly designed primers for the Cox1 of the Pezizomycotina, PezizF (5′-TCAGGRTTAYTAGGWACAGCATTT-3′) and PezizR (5′-ACCTCAGGRTGYCCGAAGAAT-3′) (S. Gilmore, pers. comm.). Primers ITS1, ITS2, and ITS3 (White et al. 1990) were sometimes used as internal sequencing primers when the DNA sequence quality obtained from ITS5 and ITS4 was inadequate. Primer binding sites for Cox1 are illustrated in Fig. 1.
Fig. 1.
Primer binding sites for the 628 bp Leohumicola Cox1 barcode region. Reference positions based on the mitochondrial genomic sequence of Saccharomyces cerevisiae.
For the PCR master mix, 0.1 mM dNTP’s, 0.08 μM forward primer, 0.08 μM reverse primer, 1X Titanium Taq buffer (Clontech, Mountain View, California, USA), 0.5X Titanium Taq enzyme (Clontech, Mountain View, California, USA), and 1.00 μL of DNA template (1–5 ng/μL) were mixed in sterile HPLC water totalling 10 μL per reaction. The PCR reaction was run in a Mastercycler epgradient S thermal cycler (Eppendorf, Mississauga, Ontario, Canada). The following profile was used to amplify ITS: 95 °C for 3 min (initial denaturation), then 40 cycles at 95 °C for 45 s (denaturation), 60 °C for 45 s (annealing), 72 °C for 1.5 min (extension), then 72 °C for 8 min (final extension). The following parameters were used to amplify Cox1: 95 °C for 3 min (initial denaturation), then 40 cycles at 95 °C for 1 min (denaturation), 51 °C for 1 min (annealing), 72 °C for 1.5 min (extension), then 72 °C for 8 min (final extension).
For sufficient amplification of Cox1 for Leohumicola sp. DAOM 239516, a touchdown PCR was performed. The Cox1 touchdown profile was the same as the profile described above except that the annealing temperature started at 54 °C (5 cycles), then changed to 51 °C (5 cycles), then to 49 °C (5 cycles), then finally to 46 °C (35 cycles), for a total of 50 cycles. For sufficient Cox1 amplification from Myxotrichum deflexum UAMH 6365, a step-up PCR was performed. The Cox1 step-up profile differed with the annealing temperature initially at 46 °C (10 cycles), then 49 °C (10 cycles), then finally 51 °C (30 cycles), for a total of 50 cycles. PCR products were separated by electrophoresis on a 1 % agarose gel, stained with ethidium bromide and visualised under UV light.
Both forward and reverse strands were sequenced using Big Dye Terminator (Applied Biosystems, Foster City, California, USA) in 10 μL reactions with the same protocol described by de Cock & Levesque (2004). The following profile was used for the sequencing reaction of ITS: 95 °C for 3 min, then for 40 cycles at 95 °C for 30 s, 50 °C for 15 s, 60 °C for 2 min. For Cox1, the sequencing reaction profile was 95 °C for 3 min, then 40 cycles at 95 °C for 30 s, 51 °C for 15 s, 60 °C for 4 min.
Contigs were assembled and edited using SeqMan II v7.0 from DNA Star (www.dnastar.com/). To confirm that the newly designed PCR primers amplified the expected gene, BLAST analyses were performed with our putative Cox1 sequences to verify that they were homologous with other fungal Cox1 sequences. All sequences are deposited in GenBank and the Barcode of Life Database (BOLD, www.barcodinglife.org) (see Table 1).
Sequence alignment and phylogenetic analyses
Internal Transcribed Spacer (ITS) and Cox1 sequences were aligned using MAFFT v6 (Katoh et al. 2005). A few minor adjustments were made to the ITS alignment using Se-Al v. 1.0 (Rambaut 1996). No manual adjustments were required for the Cox1 alignment, which had no indels. Alignments are deposited in TreeBASE (www.treebase.org/treebase/), study accession no. S2134. To test whether the ITS and Cox1 data sets contained congruent phylogenetic signals and could be combined for analysis, a partition homogeneity test (Farris et al. 1994) was performed using PAUP 4.0 (Swofford 2002) using a heuristic search with 1 000 replicates, TBR branch swapping, unordered and unweighted characters and gaps treated as missing.
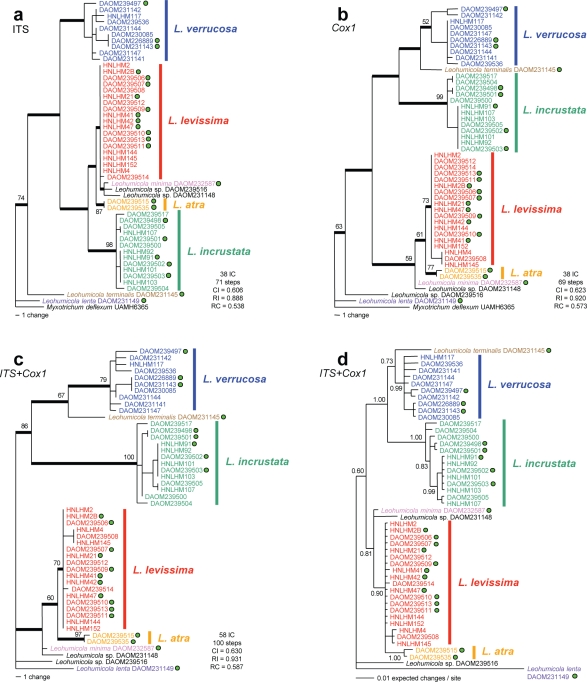
Parsimony analyses of ITS alone (Fig. 4a), Cox1 alone (Fig. 4b), and for both genes combined (Fig. 4c), were performed using heuristic searches in PAUP 4.0 (Swofford 2002), with uninformative characters excluded. Bootstrap analyses (1 000 replicates) were undertaken using full heuristic searches for the two single-gene parsimony analyses, and using fast-stepwise addition for the dual-gene analysis. For all parsimony analyses, parsimony tree scores were calculated and the 70 % consensus tree was computed.
Fig. 4.
ITS and Cox1 phylogenetic analyses. a–c. Single most parsimonious trees based on heuristic analysis. Thick lines indicate branch topology retained in the strict consensus of the MPTs. Green dots represent strains that produced aleurioconidia and chlamydospores. Bootstrap support values above 50 % from 1 000 replicates of a full heuristic search for a and b, and from 1 000 replicates of a fast-step wise search for c are shown; d. Bayesian inference consensus tree based on 20 002 trees from a gene partition analysis of the combined ITS and Cox1 data set. The K80+G and HKY+I models were used for the ITS partition and Cox1 partition, respectively. Abbreviations: IC = informative characters, CI = consistency index, RI = retention index, RC = rescaled consistency index.
For Bayesian analysis, MrModeltest v. 2.2.6 (Nylander 2004) was used to select the most appropriate models of sequence evolution for data sets that contained Leohumicola species only, according to the Akaike information criterion (AIC) (Akaike 1974). The HKY+I model (Hasegawa et al. 1985) and the K80+G (Kimura 1980) model were selected for Cox1 and ITS, respectively. Bayesian inference was performed with MrBayes v. 3.1.2 (Ronquist & Huelsenbeck 2003) using the dual-gene data set with two designated partitions (ITS and Cox1), the appropriate model of sequence of evolution applied to each partition, and L. lenta DAOM 231149 set as outgroup (see below). Two independent MCMC runs were performed simultaneously. Each MCMC ran for 2.0 × 106 generations, sampling every 100 generations, for a total of 20 001 trees. Acceptable convergence was attained after 1.0 × 106 generations, and the first 10 000 trees were discarded as burn-in. The 10 001 trees from each independent MCMC (total 20 002 trees) were combined into one consensus tree with 50 % majority rule consensus (Fig. 4d).
Myxotrichum deflexum was initially chosen as an outgroup to root all analyses, based on its position as near neighbour to the Leohumicola clade in the 18S analyses by Hambleton et al. (2005). We could not obtain satisfactory sequences for the Cox1 of two other potential outgroups, Myxotrichum arcticum UAMH 9243 and Scytalidium lignicola DAOM 231160. Therefore, M. deflexum was used as the outgroup for the single-gene parsimony analyses. In the Bayesian analysis, the branch connecting M. deflexum to the ingroup was too long, obscuring the phylogenetic structure of the ingroup. Therefore, L. lenta was used to root the tree for the Bayesian and dual-gene parsimony analyses because of its basal position in the single-gene parsimony analyses.
Two Leohumicola isolates (DAOM 239499, 239516) had an intron in the Cox1 region. Internal sequencing primers LHM1R (5′-GGCGTTCTTAGTTCTCCATTTAGT-3′), LHM5F (5′-TTAAGTGGGGTACAAAGTCA-3′) and LHM4F (5′-GGTATAGAAAATGGAGCAGGTA-3′) were designed and used to sequence the poorly resolved region at the ends of the exonic region for Leohumicola sp. DAOM 239499. Similarly internal sequencing primers LHM2R (5′-GGCGTTCTTAGTTTTCCATT-3′), LHM3F (5′-CCGCCTAGTTTATTATTATTTTTA-3′) and LHM4F were used for the intron of Leohumicola sp. DAOM 239516.
RESULTS
By heat treating soil, three distinct species exhibiting characteristic Leohumicola morphology were isolated. On PDA, colonies grew slowly, and initially were yellow and slowly maturing to grey or olive colours, with brown or olive soluble pigments released into the surrounding media (Fig. 2, 3). Characteristic Leohumicola aleurioconidia and chlamydospores were observed for several strains after 3 mo on PDA and CMA, although some strains did not sporulate even after 6 mo in culture. Leohumicola incrustata DAOM 239517 only sporulated on PCA. The morphological descriptions of the new species L. atra, L. incrustata, and L. levissima are presented in the taxonomy section, and the characters of all known species are summarized in Table 2.
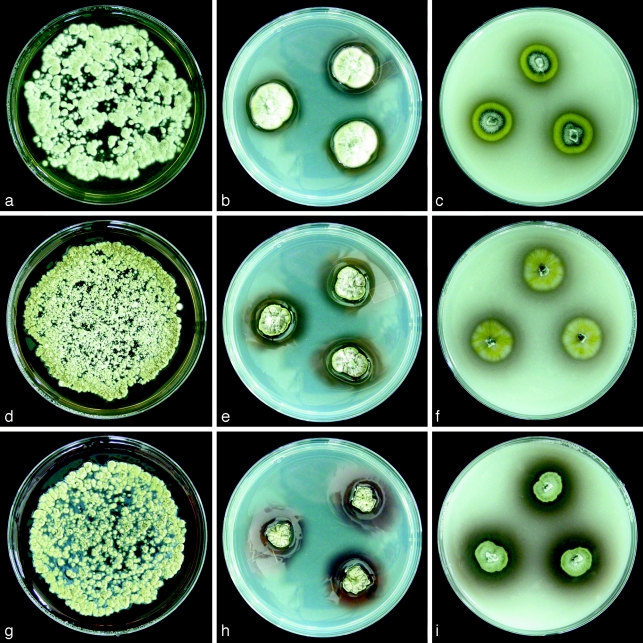
Fig. 2.
Three Leohumicola species growing from macerated inocula on PDA (left column), as three point inocula on PDA (middle column) and OA (right column), after 2 wk at room temperature. a–c. L. levissima HNLHM2; d–f. L. incrustata DAOM 239498; g–i. L. atra DAOM 239515. Reflections on the shiny agar surface from the middle column pictures were removed digitally. — Petri dish diam = 9 cm.
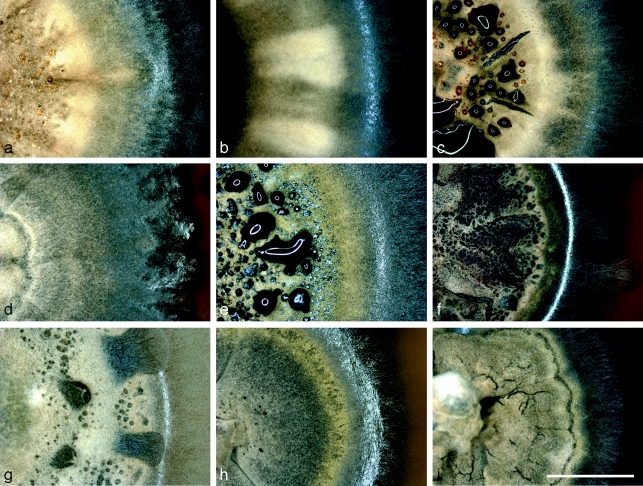
Fig. 3.
Leohumicola colonies on PDA after 2 mo incubation. a. L. levissima HNLHM2B; b. L. levissima DAOM 239511; c. L. levissima DAOM 239512 with dark brown exudates; d. L. atra DAOM 239515; e. L. incrustata DAOM 239500 with black exudates; f. L. incrustata DAOM 239498; g. L. incrustata HNLHM91 with dried clear exudates; h. L. incrustata DAOM 239517; i. Leohumicola sp. DAOM 239516. — Scale bar = 5 mm.
Table 2.
Comparative summary of colony and microscopic characters of all currently known Leohumicola species.
| Species | Colony characters after 2 wk |
Aleurioconidia characteristics (PDA and CMA) | ||||||
|---|---|---|---|---|---|---|---|---|
| PDA | OA | |||||||
| diam (mm) | mycelial colour | soluble pigments | diam (mm) | position | terminal cell (μm) | wall | basal cell (μm) | |
| Leohumicola atra | 10 | olive | greyish red | 13–15 | lateral and terminal | 4.5–5.5 × 4.0–5.5 | smooth dark brown | 2.5–4.5 × 2.5–3.5 |
| Leohumicola incrustata | 12–18 | pastel yellow, greyish yellow, grey | olive-brown, dark yellow | 15–20 | lateral and terminal | 4.0–5.5 × 4.0–5.0 | verrucose and slimy | 2.5–4.5 × 2.0–3.0 |
| Leohumicola lenta* | 1 | olive | minimal | 3–4 | lateral and terminal | 7.0–10 × 6.5–8.5 | smooth | 4.0–11.0 × 2.0–5.0 |
| Leohumicola levissima | 15–20 | olive, grey | minimal olive | 12–17 | lateral and terminal | 4.5–6.0 × 4.0–5.5 | smooth | 1.5–4.0 × 2.5–3.5 |
| Leohumicola minima* | 20–22 | grey, olive grey, brownish grey | olive-grey, brownish grey | 18 | lateral and terminal | 4.5–8.0 × 3.5–5.0 | smooth or partly verrucose | 2.0–5.5 × 2.0–3.0 |
| Leohumicola terminalis* | 10 | greyish yellow | olive | 11–12 | terminal only | 5.0–7.5 × 5.0–8.5 | slightly rough | 3.0–8.5 × 2.5–4.5 |
| Leohumicola verrucosa* | 12–18 | greyish yellow, olive yellow, grey | red, reddish brown, olive brown | 15–19 | lateral and terminal | 4.0–5.5 × 4.0–5.5 | verrucose (usually) | 2.0–4.5 × 2.5–3.0 |
* Leohumicola species previously studied by Hambleton et al. (2005).
The internal transcribed spacer (ITS) nrDNA sequences were 462 bp long for all strains of L. levissima; 463 bp for most L. incrustata strains (except DAOM 239503, HNLHM91, HNLHM103, 464 bp, and DAOM 239500 462 bp) and 462 bp for both L. atra strains. ITS sequences of the isolated strains closely matched reference Leohumicola sequences in GenBank using BLAST searches. The barcode region of the cytochrome oxidase subunit I (Cox1) mitochondrial gene was 628 bp for all strains, except DAOM 239499 and DAOM 239516, which had an intron making the PCR product roughly 1500 bp. There are currently few fungal Cox1 reference sequences in GenBank, but BLAST results of Cox1 sequences from our strains matched fungal sequences for that gene at a maximum of 87 % DNA sequence identity. The sequences generated for Leohumicola and the outgroup Myxotrichum deflexum are currently the only Leotiomycetes with Cox1 DNA barcode sequences.
Parsimony analyses of ITS and Cox1 alignments (Fig. 4a, b) revealed that all previously recognized and newly discovered Leohumicola species form monophyletic groups in strict consensus trees, with weak bootstrap support for some clades. In the ITS analysis, L. atra (bootstrap support 87 %) and L. incrustata (98 %) form well-supported clades. Leohumicola levissima is paraphyletic with L. minima and two unidentified strains, but is resolved as a monophyletic group in the Cox1 tree. In the Cox1 analysis, the three new species are monophyletic, supported by bootstrap values of 73 % for L. levissima, 77 % for L. atra, and 99 % for L. incrustata. The monophyly of the strains of the type species of the genus L. verrucosa is not strongly supported by bootstrapping. The topology of the ITS and Cox1 trees present differing sister group relationships. In the Cox1 analysis, L. terminalis is sister to L. verrucosa whereas it is sister to L. lenta in the ITS analysis. Furthermore, L. incrustata is sister to the L. atra/L. minima clade in the ITS analysis, but most closely related to the L. verrucosa/L. terminalis clade in the Cox1 analysis.
The partition homogeneity test confirmed that the ITS and Cox1 data sets could be combined (P = 0.50). A dual-gene parsimony analysis (Fig. 4c) and a dual-gene Bayesian inference (Fig. 4d) were performed using L. lenta to root the tree.
In the dual-gene parsimony analysis, all three new species form monophyletic groups supported by strict consensus tree topology and bootstrap values of 70 % for L. levissima, 97 % for L. atra, and 100 % for L. incrustata. In the Bayesian analysis (Fig. 4d), L. atra and L. incrustata samples form monophyletic clusters both with a branch support value of 1.00. However, L. levissima is paraphyletic with L. atra in the Bayesian analysis. Leohumicola verrucosa is more strongly supported as a clade in both dual-gene analyses (79 % in parsimony and 0.99 in Bayesian) than in the single-gene analyses. The topology of the dual-gene parsimony and Bayesian analyses are identical and most similar to the Cox1 parsimony analysis (Fig. 4b).
As reviewed by Seifert et al. (2007), introns are a frequently reported problem in fungal Cox1 genes. Most of our strains of Leohumicola amplified easily but three strains were problematic. In DAOM 230084, chromatograms contained an ambiguous stretch with double peaks from position 574 to 590 of the Cox1 amplicon; consequently, we removed this strain from all phylogenetic analyses. Both Leohumicola sp. DAOM 239516 and Leohumicola sp. DAOM 239499 had large introns (about 875 bp) in the target region of the Cox1 gene. When these introns were removed and the sequences were translated, the amino acid alignment revealed conserved protein sequences in relation to the Cox1 of most other Leohumicola strains. ITS parsimony analyses placed Leohumicola sp. DAOM 239499 (a sterile strain from South Africa) in the L. incrustata clade (data not shown), which corresponds with its colony characters. However, in a Cox1 parsimony analysis with the intron removed, this strain formed a monophyletic clade with another intron-containing strain, Leohumicola sp. DAOM 239516 (from Ontario, Canada). The phylogenetic conflicts between ITS and Cox1 were unexpected and thus strains DAOM 239499 and 239516 remain unidentified. Strain DAOM 239499 was removed from all analyses presented here to increase phylogenetic congruency, and allow our ITS and Cox1 data to be combined for dual-gene analysis.
Taxonomy
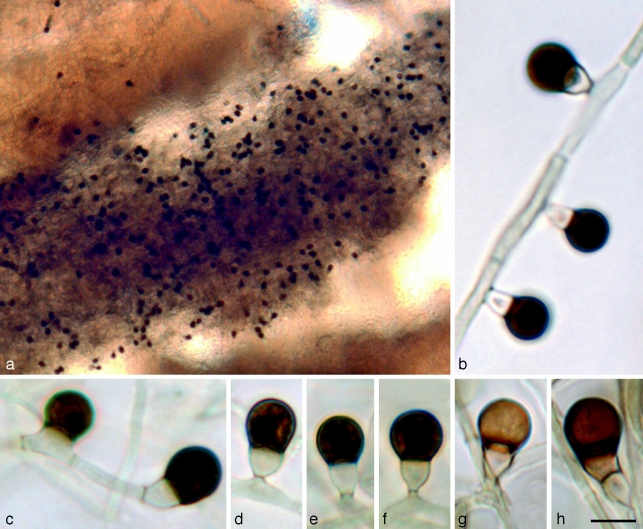
Leohumicola atra Nguyen & Seifert, sp. nov. — MycoBank MB512015; Fig. 5
Fig. 5.
Leohumicola atra. a. Sporulating part of colony on PDA; b. aleurioconidial development; c. single-celled aleurioconidium (left) and terminal aleurioconidium (right); d–f. mature aleurioconidia; g, h. lighter coloured aleurioconidia. All panels = DAOM 239515. — Scale bars: a = 50 μm, b–h = 5 μm.
Conidia lateralia vel modice terminalia, cellula terminali 4.5–5.5 ×4.0–5.5 μm, (sub)globosa, atra, laevi; cellula basilari 2.5–4.5 ×2.5–3.5 μm, crateriformis, obconica vel cylindrica. Coloniae in agaro PDA dicto ca. 10 diam post 14 dies.
Holotypus. Cultura ex solo isolata, exsiccata in herbario DAOM 239515, viva ex-typo CCFC.
Etymology. Named after the dark-brown or nearly black colour of the terminal conidial cells (atra Lat. = dark).
Conidiogenous hyphae hyaline, approximately 1–2.5 μm wide, often in fascicles in aerial mycelium (Fig. 5a). Conidiogenous cells reduced to a single denticle 0.5–1.0 μm long (mean ± SE = 0.6 ± 0.1, n = 10) and 1.0–2.0 μm wide (1.5 ± 0.1, n = 10). Conidia initially two-celled, single or side by side in small clusters, or successively produced sympodially from hyphae, either lateral (Fig. 5b, d–f) or terminal on conidiogenous hyphae. Terminal cell 4.5–5.5 ×4.0–5.5 μm (5.0 ± 0.1 ×5.2 ± 0.1, n = 20), globose to subglobose, at first hyaline like the basal cell, becoming dark brown (Fig. 5h) or almost black; conidial walls slightly thick, remaining smooth after 3 mo. Conidial connection to basal cell 2.5–3.5 μm wide, not constricted. Aleurioconidia sometimes single-celled with terminal cell directly attached to the hypha, with no basal cell (Fig. 5c). Basal cell 2.5–4.5 ×2.5–3.5 μm (3.3 ± 0.1 ×3.1 ± 0.1, n = 20), obconical or cupulate, often symmetrical or sometimes asymmetrical or irregular, hyaline or slightly pale brown, paler than the terminal cell. Ratio of lengths of terminal : basal cell 1.5–2.0 (1.6 ± 0.1). Basal cell of conidium rupturing during secession, resulting in a functionally single-celled conidium bearing the remnant of the basal cell. Chlamydospores sparsely produced, intercalary, single, rarely in chains, concolorous with conidial terminal cell, subglobose to ellipsoidal, 3–11 ×4–7 μm, with thin or slightly thickened walls. Vegetative mycelium often with swollen, monilioid, hyaline or subhyaline hyphae 1.5–2 μm wide, septate, with slightly thickened walls.
Colonies on PDA after 2 wk (Fig. 2) under ambient light at room temperature roughly 10 mm diam; entirely olive (3E3) at first becoming grey (3E1) in the centre and olive (3E3) at the margins as the colonies mature, convex, wrinkled with a felty appearance. Exudates produced around the colony centre as black droplets; soluble pigments greyish red (8C4) or faint yellow/orange around the colony and becoming darker with age. Margin entire, slightly gnawed, or smooth. Colony reverse dark-brown (7F8).
Colonies on OA after 2 wk (Fig. 2) under ambient light at room temperature roughly 13–15 mm diam; grey (4E1) in the centre and olive-brown (4E4) at the margins, sometimes with felty white aerial mycelium; soluble pigments brown (6E4) around the colony. Margin entire and smooth. Colony reverse brown (5F5) to olive (3F5).
Specimens examined. USA, Oregon, Crater Lake National Park, 45°56′N 122°08′W, from heated soil, 28 July 2006, M. Trappe, holotype and ex-type strain DAOM 239515. Two strains were isolated from one soil sample (Table 1).
Notes — The terminal cell of the conidia of L. atra becomes much darker brown compared to other Leohumicola species and is nearly black. It remains smooth-walled even after 5 mo incubation.
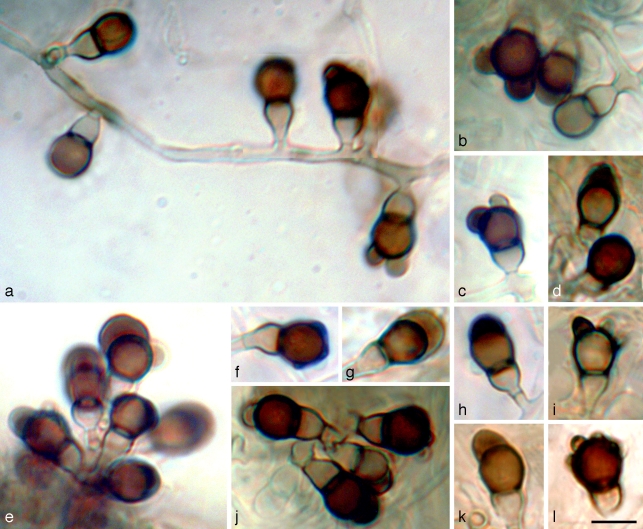
Leohumicola incrustata Nguyen & Seifert, sp. nov. — MycoBank MB512014; Fig. 6
Fig. 6.
Leohumicola incrustata. a, e. Aleurioconidial development; b–d, f–l. terminal cells of aleurioconidia are often incrusted with a brown slime or warts around the apex. Panels a–c, e, j = DAOM 239501; d, f–i, k, l = DAOM239502. The background of panel j was altered digitally for aesthetic reasons. — Scale bar = 5 μm.
Conidia lateralia vel modice terminalia, cellula terminali 4.0–5.5 × 4.0–5.0 μm, (sub)globosa, brunnea, incrustata; cellula basilari 2.5–4.5 × 2.0–3.0 μm, crateriformis vel obconica. Coloniae in agaro PDA dicto 12–18 mm diam post 14 dies.
Holotypus. Cultura ex solo isolata, exsiccata in herbario DAOM 239498, viva ex-typo CCFC.
Etymology. Named after the appearance of the terminal conidial cells which are incrusted with a crust-like slime and warts.
Conidiogenous hyphae hyaline, 1.5–2.0 μm wide, often in fascicles in aerial mycelium. Conidiogenous cells reduced to a single denticle 1.0–3.0 μm long (mean ± SE = 1.4 ± 0.1, n = 15) and 1.5–3.5 μm wide (2.1 ± 0.1, n=15). Conidia initially two-celled, single or side by side in small clusters, or successively produced sympodially (Fig. 6a, e) from hyphae, either lateral or terminal on conidiogenous hyphae. Terminal cell 4.0–5.5 ×4.0–5.0 μm (4.9 ± 0.1 ×4.5 ± 0.1, n = 20), globose to subglobose, at first hyaline like the basal cell, becoming either pale brown to dark brown; conidial walls slightly thick, smooth or slightly verrucose with large warts 0.75–1.5 μm, usually incrusted with a brown coloured slime 1–2 μm thick around the apex (Fig. 6b–d, f–l). Conidial connection to basal cell 2–3 μm wide, not constricted. Basal cell 2.5–4.5 ×2.0–3.0 μm (3.7 ± 0.1 ×2.7 ± 0.1, n = 20), obconical or cupulate, often symmetrical or sometimes asymmetrical or irregular, hyaline to pale brown, paler than the terminal cell. Ratio of lengths of terminal : basal cell 1.0–2.0 (1.5 ± 0.1). Basal cell of conidium rupturing during secession, resulting in a functionally single-celled conidium bearing the remnant of the basal cell. Chlamydospores sparsely produced in submerged mycelium, commonly found in poorly sporulating colonies, intercalary, single, concolorous with conidial terminal cell, subglobose or ellipsoidal or irregularly shaped with a rough and wrinkled appearance, 5–6 ×3–4.5 μm, with slightly thickened walls. Vegetative mycelium often with swollen, monilioid, hyaline or brown coloured hyphae 1.5–3 μm wide, septate, with slightly thickened walls.
Colonies on PDA after 2 wk (Fig. 2) under ambient light at room temperature 12–18 mm diam; pastel yellow (2A4) or greyish yellow (2B3) or grey (2B1) in the centre and olive-yellow (2C8) or white at the margins; sometimes wrinkled, sometimes splitting the agar near the colony centre, with short and felty white aerial mycelium. Exudates produced; around colony centre as small reddish brown droplets, at the colony margins and centre as large clear droplets, or around the colony centre as small dark brown to black droplets; soluble pigments variable ranging from brown (6D8), olive (3E8), and dark yellow (4C8). Margin smooth, entire, sometimes irregular or gnawed. Colony reverse dark-olive (2F6) or dark-brown (5F8).
Colonies on OA after 2 wk (Fig. 2) under ambient light at room temperature roughly 15–20 mm diam; yellowish white (2A2) to pastel-yellow (2A4); soluble pigments sometimes absent and sometimes minimal and purplish white (14A2) around the colony. Margin entire and smooth. Colony reverse pale yellow (3A2) to light yellow (3A5) at the margin and olive (2E3) in the centre.
Specimens examined. South Africa, Western Cape Province, Cape of Good Hope Nature Reserve, 34°20′S 18°27′E, from heated soil, 7 April 2006, K.A. Seifert, holotype and ex-type strain DAOM 239498. Thirteen strains were isolated from four soil samples (Table 1).
Notes — Large warts or slime production on the terminal cell are the defining characteristics of L. incrustata aleurioconidia. The conidia are most similar to those of L. verrucosa, but in that species the wall ornamentation forms smaller, discrete warts.
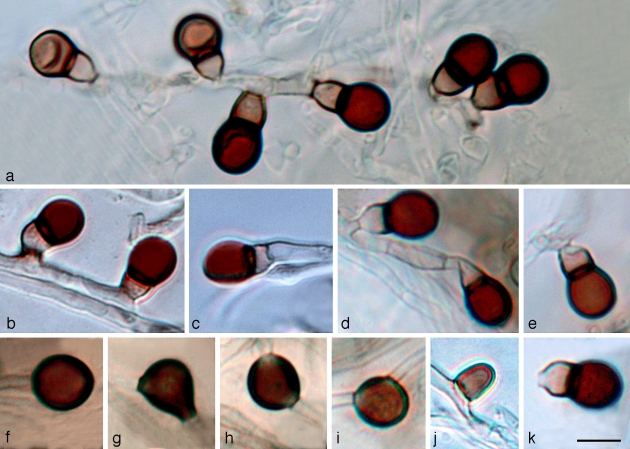
Leohumicola levissima Nguyen & Seifert, sp. nov. — MycoBank MB512013; Fig. 7
Fig. 7.
Leohumicola levissima. a. Aleurioconidial development, with younger aleurioconidia (left) and older aleurioconidia (right); b. laterial aleurioconidia; c. terminal aleurioconidium; d. laterial (left) and terminal (right) aleurioconidia; e. basal cell of aleurioconidium rupturing during secession; f. terminal chlamydospore; g–i. intercalary chlamydospores; j. single-celled aleurioconidium; k. functionally single-celled aleurioconidum bearing the remnant of the basal cell. Panels a, d, e, k = DAOM239511; b, c, j = DAOM 239509; f–i = DAOM 239513. The background of panels a and c were altered digitally for aesthetic reasons. — Scale bar = 5 μm.
Conidia lateralia vel modice terminalia, cellula terminali 4.5–6.0 × 4.0–5.5 μm, (sub)globosa, brunnea, laevi; cellula basilari 1.5–4.0 × 2.5–3.5 μm, crateriformis vel obconica. Coloniae in agaro PDA dicto 15–20 mm diam post 14 dies.
Holotypus. Cultura ex solo isolata, exsiccata in herbario DAOM 239506, viva ex-typo CCFC.
Etymology. Named after the smooth walled and unornamented appearance of the terminal conidial cells (levissima Lat. = smooth).
Conidiogenous hyphae hyaline, 1–2 μm wide, often in fascicles in aerial mycelium. Conidiogenous cells reduced to a single denticle, 0.5–1.5 μm long (mean ± SE = 0.8 ± 0.1, n = 12) and 1.0–3.5 μm wide (2.2 ± 0.2, n = 12). Conidia initially two-celled, single (Fig. 7j) or side by side in small clusters, or successively produced sympodially (Fig. 7a) from hyphae, either lateral (Fig. 7b) or terminal (Fig. 7c) on conidiogenous hyphae. Terminal cell 4.5–6.0 × 4.0–5.5 μm (5.3 ± 0.1 × 4.9 ± 0.1, n = 20), globose to subglobose, at first the same colour as the basal cell, becoming dark brown while still attached; conidial walls slightly thick, remaining smooth after 3 mo. Conidial connection to basal cell 2.5–3.5 μm wide, not constricted. Basal cell 1.5–4.0 × 2.5–3.5 μm (2.6 ± 0.1 × 3.1 ± 0.1, n = 20), obconical or cupulate, often symmetrical or sometimes asymmetrical or irregular, hyaline to pale brown, paler than the terminal cell. Ratio of lengths of terminal : basal cell 1.5–2.5 (1.8 ± 0.1). Basal cell of conidium rupturing during secession (Fig. 7e), resulting in a functionally single-celled conidium bearing the remnant of the basal cell (Fig. 7k). Chlamydospores sparsely produced in submerged mycelium, commonly found in poorly sporulating colonies, intercalary, single, concolorous with conidial terminal cell, subglobose to ellipsoidal, sometimes with irregular constrictions, 5.5–7.5 × 5–6 μm, with thin or slightly thickened walls (Fig. 7f–i). Vegetative mycelium often with swollen, monilioid, hyaline or subhyaline hyphae 1–2 μm wide, septate, with slightly thickened walls.
Colonies on PDA after 2 wk (Fig. 2) under ambient light at room temperature 15–20 mm diam; olive (2E4) or grey (2D1) in the centre and greyish yellow (2C4) or yellowish grey (2C2) at the margins, planar or convex, sometimes wrinkled, sometimes splitting the agar near the colony centre, with low, felty, slightly lanose white aerial mycelium. Soluble pigments not produced after 2 wk. Margin smooth and entire. Colony reverse olive-grey (2F2) to olive (2E4).
Colonies on OA after 2 wk (Fig. 2) under ambient light at room temperature roughly 12–17 mm diam; pale yellow (2A3) to light yellow (2A5); soluble pigments minimal around the colony. Margin entire and smooth. Colony reverse olive (2E5) in the centre and light yellow (3A5) at the edge.
Specimens examined. USA, Oregon, Crater Lake National Park, 45°56′N 122°08′W, from heated soil, 28 July 2006, M. Trappe, holotype and ex-type culture DAOM 239506. Nineteen strains were isolated from three soil samples (Table 1).
Notes — The terminal cell of L. levissima conidia remains smooth even after 3 mo, in contrast to the roughened or encrusted terminal cells of L. verrucosa and L. incrustata. The conidia of L. atra, described above, have similarly smooth terminal cells, but are much darker. The colony morphologies of L. levissima and L. atra are distinct, particularly on OA. On OA, L. levissima is light yellow and L. atra is grey and olive brown (Fig. 2). Also, L. levissima grows faster on PDA than L. atra. Based on the similarity in aleurioconidia and the topology of the parsimony analyses, perhaps these two species share a common ancestor. PDA colonies of L. levissima after 6 wk incubation are less variable than they are after 2 wk. Older colonies eventually produce pale olive, soluble pigments.
DISCUSSION
This study proposes three new species of Leohumicola, namely L. atra, L. incrustata, and L. levissima, using morphological characters and DNA barcoding. The three species exhibit the characteristic two-celled aleurioconidia of the genus, which become functionally single-celled after secession. Brown to olive pigments diffuse into agar media, and colonies grow slowly. The new species can be distinguished by features of the terminal cells of the aleurioconidia. In L. levissima they are smooth and brown, whereas they are smooth and almost black in L. atra, and covered with warts (that sometimes appear slimy rather than composed of cell wall material) in L. incrustata. Chlamydospores, similar in pigmentation to the aleurioconidia, are produced by the three new species and the four previously described species. A revised key to the seven known species of Leohumicola is provided below.
Dense mycelial growth, and abundant soluble pigment production on PDA, renders microscopic observations of Leohumicola conidia difficult. They are more conspicuous on the optically clear CMA medium, where the mycelia are sparser and soluble pigments are reduced; sporulation is more abundant on inoculum blocks originally transferred from PDA. Generally, strains were more likely to produce conidia after 3 mo on CMA than on PDA, OA, or when macerated on PDA. As indicated on the phylogenetic trees, not all strains of L. incrustata and L. levissima sporulated. Of note, Leohumicola sp. DAOM 239516 produced almost exclusively chlamydospores and only one aleurioconidium was seen. This isolate is phylogenetically distinct from other Leohumicola species, but we chose not to describe it here because of the paucity of diagnostic morphological characters in the single culture available.
The results of our phylogenetic analyses are in agreement with the morphological data. Most Leohumicola species are monophyletic, although L. levissima is paraphyletic with L. minima in the ITS parsimony analysis and with L. atra in the dual-gene Bayesian analysis. Bootstrap support is variable for some groups and the sister relationships among species are inconsistent. Although this renders the phylogenetic structure of the genus uncertain, it does not interfere with monophyletic species recognition. Perhaps as additional species are discovered and the species sampling of this genus is more complete, the phylogenetic structure will be clearer.
As DNA barcodes, the ITS and Cox1 loci provide similar sequence variation and reveal similar phylogenetic groupings of Leohumicola species (Table 3). However, Cox1 provides slightly better species resolution, particularly for the minima/levissima/atra clade. Both Cox1 and ITS sequences are suitable DNA barcodes for the currently known Leohumicola species because the mean sequence divergence between species is about 10 times greater than the mean divergence within species.
Table 3.
Comparison of ITS and Cox1 markers for DNA barcoding.
| Marker | No. of isolates analysed | No. of species | Sequence length | Mean infraspecific divergence (%) | Range of means (%) | Mean interspecific divergence (%) | Range of means (%) |
|---|---|---|---|---|---|---|---|
| ITS | 45 | 7 | 462 | 0.35 | 0.04–0.78 | 3.71 | 0.45–8.09 |
| Cox1 | 45 | 7 | 628 | 0.24 | 0.11–0.32 | 2.42 | 0.54–4.99 |
Genetic anomalies in the target region of the Cox1 gene were found in three Leohumicola isolates. Leohumicola sp. DAOM 230084 contained a short ambiguous stretch of double peaks that may indicate multiple copies of the Cox1 gene. Leohumicola sp. DAOM 239499 and DAOM 239516 contained introns. They were partially sequenced from the 5′ and 3′ ends, but additional sequencing primers would be needed to sequence the entire length of the introns.
The search for Leohumicola species has only begun. Hambleton et al. (2005) noted that Leohumicola ITS sequences correspond (with > 95 % identity and BLAST E-value of 0) to many unidentified fungi from environmental studies or plant roots in GenBank. Based on these observations and our discovery of three new species from soil samples from two previously unsampled locations, Leohumicola is likely to be a diverse, wide-spread soil-borne genus. The soil heating procedure should be effective for isolating additional Leohumicola species from other countries.
KEY TO THE SPECIES OF LEOHUMICOLA
-
1.
Colony diam < 5 mm after 2 wk on PDA, terminal cell of conidium 7–10 μm long……………L. lenta
-
1.
Colony diam > 5 mm after 2 wk on PDA, terminal cell of conidium shorter……………2
-
2.
Conidia lateral and terminal……………3
-
2.
Conidia terminal only and usually smooth-walled…………………………L. terminalis
-
3.
Terminal cell of conidium mostly ellipsoidal……………L. minima
-
3.
Terminal cell of conidium globose or subglobose, with a width of about 5 μm diam……………4
-
4.
Terminal cell of conidium smooth after 3 mo incubation……………5
-
4.
Terminal cell of conidium verrucose or encrusted……………6
-
5.
Terminal cell of conidium pale brown……………L. levissima
-
5.
Terminal cell of conidium dark-brown……………L. atra
-
6.
Many small warts occurring on the terminal cell of the conidium……………L. verrucosa
-
6.
Few large warts or slime produced around the apex of the terminal cell……………L. incrustata
Acknowledgments
We would like to thank Dr Sarah Hambleton for discussions on Leohumicola and for her donations of genomic DNA and chromatograms of the previously published Leohumicola species; Matt Trappe (Oregon State University) and Dr Karin Jacobs (University of Stellenbosch) for their assistance with collecting soil samples; Dr Tom Graefenhan for his guidance; Gerry Louis-Seize for his technical help and advice in the lab; Dr Miao Liu and Caroline Lamoureux for interesting project discussions; and the rest of the mycology group at the Eastern Cereal and Oilseed Research Centre. This research was supported through funding to the Canadian Barcode of Life Network and Genome Canada through the Ontario Genomics Institute, NSERC, and other sponsors listed at www.BOLNET.ca.
REFERENCES
- Akaike H. 1974. A new look at the statistical model identification. IEEE Transactions on Automatic Control 19: 716 – 723 [Google Scholar]
- Cock AWAM de, Levesque CA. 2004. New species of Pythium and Phytophthora. Studies in Mycology 50: 481 – 487 [Google Scholar]
- Cowling RM, Richardson DM. 1995. Fynbos: South Africa’s Unique Floral Kingdom Fernwood Press, Cape Town: [Google Scholar]
- Farris JS, Kallersjo M, Kluge AG, Bult C. 1994. Testing the significance of incongruence. Cladistics 10: 315 – 319 [Google Scholar]
- Hajibabaei M, Janzen DH, Burns JM, Hallwachs W, Hebert PDN. 2006. DNA barcodes distinguish species of tropical Lepidoptera. Proceedings of the National Academy of Sciences of the United States of America 103: 968 – 971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambleton S, Egger KN, Currah RS. 1998. The genus Oidiodendron: species delimitation and phylogenetic relationships based on nuclear ribosomal DNA analysis. Mycologia 90: 854 – 869 [Google Scholar]
- Hambleton S, Nickerson NL, Seifert KA. 2005. Leohumicola, a new genus of heat-resistant hyphomycetes. Studies in Mycology 53: 29 – 52 [Google Scholar]
- Hasegawa M, Kishino H, Yano T. 1985. Dating of human-ape splitting by a molecular clock of mitochondrial DNA. Journal of Molecular Evolution 22: 160 – 174 [DOI] [PubMed] [Google Scholar]
- Jackson ED, Hughes TJ, Ells TC, Renderos WE, Bell CR. 1995. The incidence, source and significance of heat resistant moulds in fresh and freeze-processed wild lowbush blueberries. Agriculture and Agri-Food Canada Atlantic Food Horticulture Research Centre Kentville, N.S. Tech. Memo. 9601
- Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Research 33: 511 – 518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16: 111 – 120 [DOI] [PubMed] [Google Scholar]
- Kornerup A, Wanscher JH. 1978. Methuen handbook of colour. Third Ed Eyre Methuen, London, United Kingdom; [Google Scholar]
- Nylander JAA. MrModeltest v. 2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University; 2004. [Google Scholar]
- Rambaut A. 1996. Se–Al: Sequence alignment editor. Version 1.0 Department of Zoology, University of Oxford, Oxford, United Kingdom: [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572 – 1574 [DOI] [PubMed] [Google Scholar]
- Samson RA, Hoekstra ES, Frisvad JC. 2004. Introduction to food- and airborne fungi, 7th ed Centraalbureau voor Schimmelcultures, Utrecht: [Google Scholar]
- Seifert KA, Samson RA, Waard JR de, Houbraken J, Levesque CA, Moncalvo J-M, Louis-Seize G, Hebert PDN. 2007. Prospects for fungus identification using CO1 DNA barcodes, with Penicillium as a test case. Proceedings of the National Academy of Sciences of the United States of America 104: 3901 – 3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. 2002. PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods) Sinauer Associates, Sunderland, MA, USA: [Google Scholar]
- Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN. 2005. DNA barcoding Australia’s fish species. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 360: 1847 – 1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfland DH, Sninsky JJ, White TJ. (eds), PCR protocols: a guide to methods and applications: 315–322 Academic Press, San Diego, USA: [Google Scholar]