Abstract
A morphologically distinct isolate of Cladosporium sphaerospermum from a North American patent collection, referenced as Cladosporium lignicola in the patent, was examined. Generic affinity was confirmed by scanning electron microscopic examination of conidiogenous loci and conidial hila. Species identity as C. sphaerospermum was indicated by DNA sequence data derived from actin and translation elongation factor 1-α genes, and the internal transcribed spacer region. The isolate broadens the morphological limits of C. sphaerospermum by production of obclavate, occasionally transversely septate conidia with subrostrate conidiogenous apices (‘alternarioid’ conidia), and by production of conidia larger than those in prior standard descriptions. Type material of C. lignicola was re-examined and compared with the North American fungus, from which it is morphologically distinct. The decision to reduce C. lignicola to synonymy under C. herbarum was confirmed.
Keywords: actin, Cladosporium, Davidiella, EF-1α, ITS, morphology, wood-inhabiting
INTRODUCTION
Cladosporium is one of the largest genera of hyphomycetes, with more than 772 names (Braun et al. 2003, Dugan et al. 2004). Cladosporium species are common, widespread fungi, including endophytic, fungicolous, human pathogenic, phytopathogenic and saprobic species (Crous et al. 2007b). Saprobic species occur on various senescing and dead leaves and stems of herbaceous and woody plants, are secondary invaders of necrotic leaf spots, and are frequently isolated from air, soil, foodstuffs, paint, textiles and other organic matter (Riesen & Sieber 1985, Brown et al. 1998, El-Morsy 2000). Furthermore, some Cladosporium species, such as C. bruhnei, are common contaminants in clinical laboratories and cause allergic lung mycoses (de Hoog et al. 2000, Schubert et al. 2007b).
Because the genus Cladosporium is very heterogeneous, David (1997) attempted to circumscribe Cladosporium based on scanning electron microscopic examinations of the scar and hilum structure in Cladosporium and Heterosporium. In so doing, David (1997) demonstrated that the structures of the conidiogenous loci and hila in Heterosporium fully agree with those of Cladosporium, proving that Heterosporium was a synonym of Cladosporium. Furthermore, he introduced the term ‘coronate’ for the Cladosporium scar type, which is characterised by having a central convex part (dome), surrounded by a raised periclinal rim (Schubert et al. 2007b, Zalar et al. 2007). Several workers have employed DNA sequence data to prove that Cladosporium s.str. is a sister clade to Mycosphaerella s.str. (Braun et al. 2003, Crous et al. 2007a, b, Schubert et al. 2007a, b), having teleomorphs in Davidiella. Schoch et al. (2006) employed DNA sequence data of four loci (SSU nrDNA, LSU nrDNA, EF-1α, RPB2), revealing species of Davidiella to cluster in a separate family (Davidiellaceae) from species of Mycosphaerella (Mycosphaerellaceae), with both families residing in the Capnodiales (Dothideomycetes).
A cladosporioid hyphomycete, deposited in the patent collection of the United States Department of Agriculture Northern Regional Research Lab (now National Centre for Agricultural Utilization Research) as NRRL 8131 (= ATCC 38493), was referenced in U.S. Patent 4.086.268 as Cladosporium lignicolum (sic, without author). The patent does not state the original substratum, but wood is a logical inference from the specific epithet. Ho et al. (1999) described NRRL 8131 from culture, and provided several photomicrographs illustrating the fungus, including 0–1 transversely septate, obclavate conidia with subrostrate apices (‘alternarioid’ conidia in their description). A comparison of the latter description and illustrations with the original diagnosis of C. lignicola revealed obvious discrepancies and called into question the correctness of the identification of the NRRL strain. To help ascertain the identity of NRRL 8131, type material of C. lignicola has been re-examined and compared with the North American strain.
We were originally attracted to re-examination of NRRL 8131 not only by the discrepancies referenced above, but by the unique conidial morphology: the ‘alternarioid’ conidia resembled the beaked conidia of Alternaria spp. (Simmons 2007). Scanning electron microscopy (SEM) examination of conidiogenous loci and conidial hila in the NRRL strain of ‘C. lignicola’ confirmed generic affinity, and sequence analyses of the actin, translation elongation factor 1-α and the internal transcribed spacer region (ACT, EF, ITS) were used to assign the strain to species.
MATERIALS AND METHODS
Materials examined
Several specimens deposited at PRM (Herbarium, Department of Mycology, National Museum, Prague, Czech Republic) under C. lignicola were examined: on rotten wood, Czechia, near Prague (PRM 155424, holotype!); on bark, Bohemia, castle Kač ina at Nové Dovory, near Kutná Hora, 1856, J. Peyl (PRM 657824); Germany, Saxony, ‘ad lignam putrida agri Dresdensis’, Rabenhorst, Fungi Eur. 1271 (PRM 657821); ex herb. ‘Verein der Naturfreunde in Reichenberg’ (PRM 657823). These specimens were compared to the type material of C. herbarum (CBS-H 19853), with which C. lignicola has been considered synonymous in the past (Hughes 1958). Furthermore, they were also compared to materials of the U.S. patent strain NRRL 8131 = ATCC 38493 = CBS 117728 = CPC 12098, which had previously been identified as C. lignicola.
DNA isolation and phylogenetic analysis
Fungal colonies were established on agar plates, and genomic DNA was isolated as described in Gams et al. (2007). Partial gene sequences were determined as described by Crous et al. (2006) for actin (ACT), translation elongation factor 1-α (EF), and part of the nuclear rDNA operon spanning the 3′ end of the 18S rRNA gene, the first internal transcribed spacer, the 5.8S rRNA gene, the second internal transcribed spacer and the 5′ end of the 28S rRNA gene (ITS). The nucleotide sequences were generated using both forward and reverse PCR primers to ensure good quality sequences over the entire length of the amplicon. Sequence data obtained from Schubert et al. (2007b) and Zalar et al. (2007) were used as reference data for the alignments (Table 1). Subsequent sequence alignment and phylogenetic analysis followed the methods of Crous et al. (2006). Gaps longer than 10 bases were coded as single events for the phylogenetic analyses; the remaining gaps were treated as new character states. Sequence data were deposited in GenBank (Table 1) and the alignment and tree in TreeBASE (www.treebase.org).
Table 1.
Cladosporium isolates used for sequence analysis.
| Anamorph | Teleomorph | Accession number1 | Host | Country | Collector | Source | GenBank numbers2 (ITS, EF, ACT) |
|---|---|---|---|---|---|---|---|
| C. antarcticum | – | CBS 690.92 | Caloplaca regalis | Antarctica | C. Möller | Schubert et al. 2007 | EF679334, EF679405, EF679484 |
| C. bruhnei | Davidiella allicina | CBS 157.82 | Quercus robur | Belgium | – | Schubert et al. 2007 | EF679336, EF679407, EF679486 |
| CBS 161.55 | Man, sputum | The Netherlands | – | Schubert et al. 2007 | EF679338, EF679409, EF679488 | ||
| CBS 121624; CPC 12211 | Hordeum vulgare | Belgium | J.Z. Groenewald | Schubert et al. 2007 | EF679350, EF679425, EF679502 | ||
| C. cladosporioides complex | – | CBS 673.69 | Air | The Netherlands | – | Schubert et al. 2007 | EF679353, EF679428, EF679505 |
| Davidiella sp. | CBS 109082 | Silene maritima | United Kingdom | A. Aptroot | Schubert et al. 2007 | EF679354, EF679429, EF679506 | |
| – | CPC 11606 | Musa sp. | India | M. Arzanlou | Schubert et al. 2007 | EF679355, EF679430, EF679507 | |
| C. herbaroides | – | CBS 121626; CPC 12052; EXF-1733 | Hypersaline water from salterns | Israel | P. Zalar | Schubert et al. 2007 | EF679357, EF679432, EF679509 |
| C. herbarum | Davidiella tassiana | CBS 121621; CPC 12177 | Hordeum vulgare | The Netherlands | – | Schubert et al. 2007 | EF679363, EF679440, EF679516 |
| CPC 12181 | Hordeum vulgare | The Netherlands | – | Schubert et al. 2007 | EF679367, EF679444, EF679520 | ||
| CPC 12183 | Hordeum vulgare | The Netherlands | – | Schubert et al. 2007 | EF679368, EF679445, EF679521 | ||
| C. iridis | Davidiella macrospora | CBS 107.20 | Iris sp. | – | – | Schubert et al. 2007 | EF679369, EF679446, EF679522 |
| CBS 138.40 | Iris sp. | The Netherlands | – | Schubert et al. 2007 | EF679370, EF679447, EF679523 | ||
| C. macrocarpum | Davidiella macrocarpa | CBS 299.67 | Triticum aestivum | Turkey | – | Schubert et al. 2007 | EF679372, EF679450, EF679526 |
| CBS 121811; CPC 12755 | Spinacia oleracea | USA | – | Schubert et al. 2007 | EF679376, EF679454, EF679530 | ||
| CPC 12756 | Spinacia oleracea | USA | – | Schubert et al. 2007 | EF679377, EF679455, EF679531 | ||
| C. ossifragi | – | CBS 842.91 | Narthecium ossifragum | Norway | M. di Menna | Schubert et al. 2007 | EF679381, EF679459, EF679535 |
| CBS 843.91 | Narthecium ossifragum | Norway | M. di Menna | Schubert et al. 2007 | EF679382, EF679460, EF679536 | ||
| C. pseudiridis | – | CBS 116463; ICMP 15579 | Iris sp. | New Zealand | C.F. Hill | Schubert et al. 2007 | EF679383, EF679461, EF679537 |
| C. ramotenellum | – | CBS 121628; CPC 12043; EXF-454 | Hypersaline water from salterns | Slovenia | P. Zalar | Schubert et al. 2007 | EF679384, EF679462, EF679538 |
| CPC 12047; EXF-967 | Air conditioning system | Slovenia | P. Zalar | Schubert et al. 2007 | EF679385, EF679463, EF679539 | ||
| C. sinuosum | – | CBS 121629; CPC 11839; ICMP 15819 | Fuchsia excorticata | New Zealand | A. Blouin | Schubert et al. 2007 | EF679386, EF679464, EF679540 |
| Cladosporium sp. | – | CBS 300.96 | Soil along coral reef coast | Papua New Guinea | A. Aptroot | Zalar et al. 2007 | DQ780352, EU570259, EF101385 |
| C. sphaerospermum | – | CBS 109.14; ATCC 36950 | Carya illinoensis leaf scale | USA | – | Zalar et al. 2007 | DQ780350, EU570260, EF101384 |
| CBS 193.54; ATCC 11289; IMI 49637 | Human nails | The Netherlands | G.A. de Vries | Zalar et al. 2007 | DQ780343, EU570261, EU570269 | ||
| CBS 102045; EXF-2524; MZKI B1066 | Hypersaline water | Spain | P. Zalar | Zalar et al. 2007 | DQ780351, EU570262, EF101378 | ||
| CPC 11822 | Phyllactinia guttata on Corylus sp. | USA | D. Glawe | This study | EU570254, EU570263, EU570270 | ||
| CPC 12476 | Ambrosia artemisiifolia | Germany | J. Nitzsche | This study | EU570255, EU570264, EU570271 | ||
| CPC 13368 | Phaseolus lunatus | Germany | N. Ale-Agha | This study | EU570256, EU570265, EU570272 | ||
| CPC 13995; CAMS 000750 | Thatch | South Africa | – | This study | EU570257, EU570266, EU570273 | ||
| CPC 14016; MRC 10263 | Triticum aestivum | South Africa | – | This study | EU570258, EU570267, EU570274 | ||
| CBS 117728; ATCC 38493; CPC 12098; NRRC 8131 | – | – | – | This study | AF393709, EU570268, EU570275 | ||
| C. spinulosum | – | CBS 102044 | Hypersaline water from salterns | Slovenia | S. Soujak | Schubert et al. 2007 | EF679387, EF679465, EF679541 |
| CBS 119907; CPC 12040; EXF-334 | Hypersaline water from salterns | Slovenia | P. Zalar | Schubert et al. 2007 | EF679388, EF679466, EF679542 | ||
| C. subinflatum | – | CBS 121630; CPC 12041; EXF-343 | Hypersaline water from salterns | Slovenia | P. Zalar | Schubert et al. 2007 | EF679389, EF679467, EF679543 |
| C. subtilissimum | – | CBS 113753 | Bing cherry fruits | USA | – | Schubert et al. 2007 | EF679396, EF679474, EF679550 |
| CBS 113754 | Grape berry | USA | – | Schubert et al. 2007 | EF679397, EF679475, EF679551 | ||
| CPC 12044; EXF-462 | Hypersaline water from salterns | Slovenia | P. Zalar | Schubert et al. 2007 | EF679398, EF679476, EF679552 | ||
| C. tenellum | – | CBS 121634; CPC 12053; EXF-1735 | Hypersaline water from salterns | Israel | P. Zalar | Schubert et al. 2007 | EF679401, EF679479, EF679555 |
| CPC 11813 | Phyllactinia sp. on Corylus sp. | USA | D. Glawe | Schubert et al. 2007 | EF679399, EF679477, EF679553 | ||
| C. variabile | Davidiella variabile | CBS 121636; CPC 12751 | Spinacia oleracea | USA | – | Schubert et al. 2007 | EF679402, EF679480, EF679556 |
| CPC 12753 | Spinacia oleracea | USA | – | Schubert et al. 2007 | EF679403, EF679481, EF679557 |
1 ATCC: American Type Culture Collection, Virginia, USA; CAMS: SERA’s Centre for Applied Mycological Studies, Forestry and Agricultural Biotechnology Institute, University of Pretoria, Pretoria, South Africa; CBS: Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands; CPC: Culture collection of Pedro Crous, housed at CBS; EXF: Extremophilic Fungi Culture Collection of the Department of Biology, Biotechnical Faculty, University of Ljubljana, Slovenia; ICMP: International Collection of Micro-organisms from Plants, Landcare Research, Private Bag 92170, Auckland, New Zealand; IMI: International Mycological Institute, CABI-Bioscience, Egham, Bakeham Lane, U.K.; MRC: Culture collection of PROMEC, Medical Research Council, Cape Town, South Africa; MZKI: Culture Collection of the National Institute of Chemistry, Ljubljana, Slovenia; NRRC: Agricultural Research Culture Collection, Peoria, Illinois, USA.
2 ACT: partial actin gene; EF: partial elongation factor 1-α gene; ITS: internal transcribed spacer region.
Morphology
Strain NRRL 8131 (= CBS 117728) was grown on 2 % potatodextrose agar (PDA), synthetic nutrient-poor agar (SNA), and 2 % malt extract agar (MEA) (Gams et al. 2007), and incubated under continuous near-ultraviolet light at 25 °C to promote sporulation or in the dark for assessing colony characters (Schubert et al. 2007b, Zalar et al. 2007), and suspensions of conidia were preserved in glycerol for long term storage at −80 °C and in liquid nitrogen. Subcultures on MEA plates were used for scanning electron microscopy, and SNA and MEA slide cultures or plates for light microscopy. Microscopic observations were made from colonies cultivated for 7 d under continuous near-ultraviolet light at 25 °C on SNA and MEA. Preparations were mounted in Shear’s solution (Gams et al. 2007) or 85 % lactic acid. To study conidial development and branching patterns, squares of transparent adhesive tape (Titan Ultra Clear Tape, Conglom Inc., Toronto, Canada) were placed on conidiophores growing in the zone between the colony margin and 2 cm inwards, and mounted between two drops of Shear’s solution under a glass cover slip. Conidial terminology follows that of Schubert et al. (2007b). Colonies were cultivated on PDA, SNA and MEA for 14 d at 25 °C in the dark, after which the surface and reverse colours were rated using the charts of Rayner (1970). Linear growth was determined on MEA and SNA plates by inoculating three plates of each medium and incubating them for 14 d at 25 °C.
SEM was conducted by mounting small (2–3 mm3) sections of MEA agar with fungal growth onto SEM stubs, subjecting the mounts to osmium tetroxide vapours for 2 h, and gold-coating the specimens with a Technics Hummer V sputter coater. Specimens were observed and photographed with a Hitachi S-570 scanning electron microscope.
RESULTS
DNA phylogeny
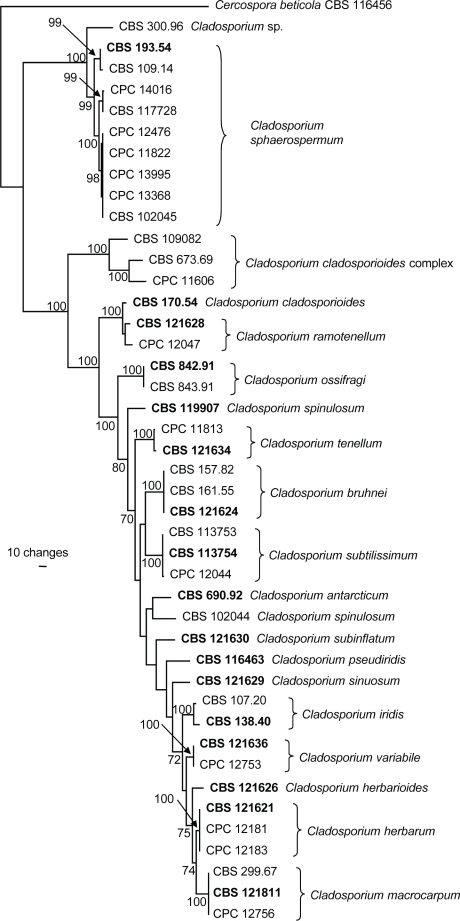
The manually adjusted concatenated alignment contained 44 sequences (including the outgroup sequence) and the three loci were represented by a total of 1094 characters including alignment gaps which were used in the analysis (Table 2). The result of the partition homogeneity test (P = 0.685) indicated that the loci were congruent and the sequence data could therefore be analysed as a concatenated alignment. A single most parsimonious tree (TL = 1253 steps; CI = 0.663; RI = 0.871; RC = 0.578), which is shown in Fig. 1, was obtained from the parsimony analysis of the combined genes. Phylogenetic analyses using the concatenated alignment as well as the individual loci (data not shown) all conclusively demonstrated that CBS 117728 clustered with C. sphaerospermum in accordance with combined ITS, ACT and EF sequences, and that it was the sister taxon of MRC 10263 (= CPC 14016) (Fig. 1). Neighbour-joining analysis using three substitution models (uncorrected ‘p’, Kimura 2-parameter and HKY85) on the sequence data yielded trees with identical topologies (data not shown).
Table 2.
Statistical parameters describing the sequence alignments of three different loci and the combined alignment.
| Parameter | ITS1 | ACT1 | EF1 | Combined |
|---|---|---|---|---|
| Number of alignment positions | 495 | 219 | 380 | 1094 |
| Number of parsimony informative characters | 37 | 99 | 177 | 313 |
| Number of variable and parsimonyuninformative characters | 100 | 30 | 54 | 184 |
| Number of constant characters | 358 | 90 | 149 | 597 |
1 ACT: partial actin gene; EF: partial elongation factor 1-αgene; ITS: internal transcribed spacer region.
Fig. 1.
The single most parsimonious tree obtained from a heuristic search with 100 random taxon additions of the combined sequence alignment (ITS, ACT, and EF). The scale bar shows ten changes, and bootstrap support values from 1 000 replicates are shown at the nodes. The tree was rooted to sequences of Cercospora beticola strain CBS 116456 (GenBank accession numbers AY840527, AY840458, AY840494, respectively).
Taxonomy
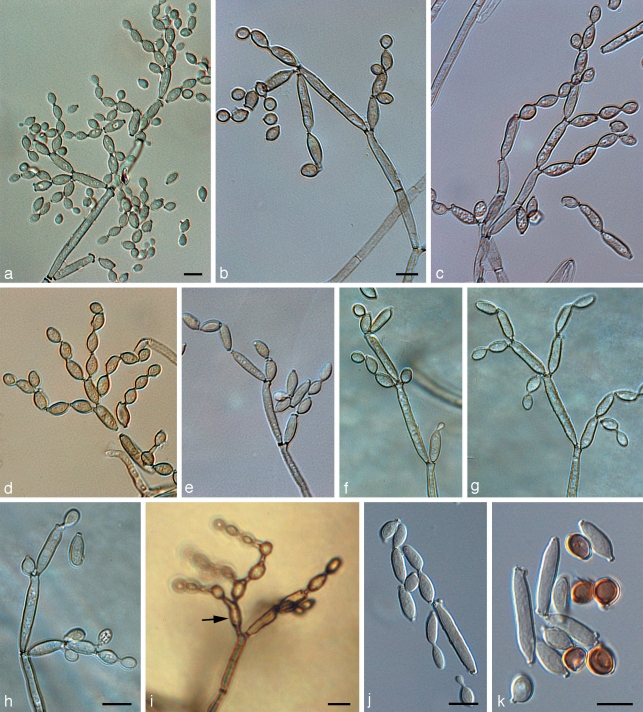
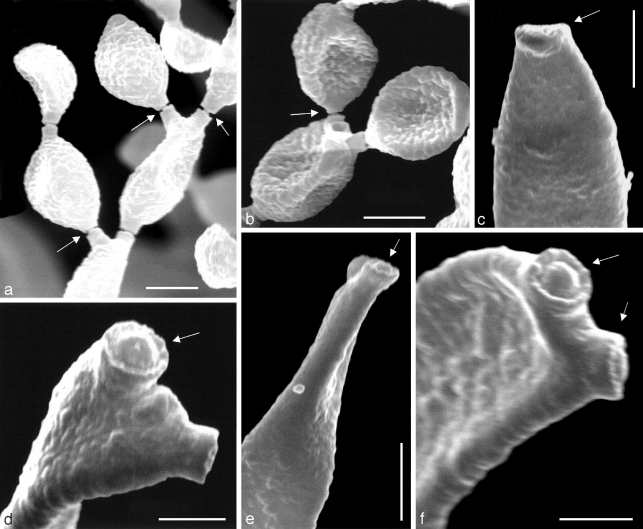
In PRM 657823, only a trimmatostroma-like hyphomycete was found. The three other specimens, including the type material (PRM 155424 holotype, PRM 657824, and PRM 657821) exclusively contained C. herbarum, well-characterised by having nodulose conidiophores and verrucose conidia (Schubert et al. 2007b). The type of C. lignicola was also examined by Hughes (1958), who reduced it to synonymy with C. herbarum, a treatment that was confirmed during the course of the present study. The examination of NRRL 8131 ‘C. lignicola’ clearly showed that this fungus is quite distinct from the true C. lignicola (= C. herbarum) by having non-nodulose conidiophores and almost smooth to verruculose, often short rostrate conidia (Fig. 2, 3). The general habit of this fungus is obviously cladosporioid, including conspicuous, somewhat protuberant conidiogenous loci. The periclinal rim is evident, but the central dome is rather low and not very conspicuous when viewed by light microscopy (Fig. 2). SEM studies confirmed that scar structure conforms to the coronate Cladosporium type (Fig. 3).
Fig. 2.
Light micrographs of Cladosporium sphaerospermum NRRL 8131. a–h. Conidiophores at various stages of development, showing their characteristic branching patterns, ramoconidia, secondary ramoconidia, intercalary conidia, and small, terminal conidia (all on SNA); i. conidiophore with alternarioid secondary ramoconium (arrow), formed on MEA; j, k. secondary ramoconidia and intercalary conidia (note older intercalary conidia, which become dark brown and globose). — Scale bars = 10 μm.
Fig. 3.
Scanning electron micrographs of Cladosporium sphaerospermum NRRL 8131. a, b. Branching chains of conidia, showing conidiogenous loci with disjunctors (arrows); c. apex of conidiophore with conidiogenous scar in profile (arrow); d. two conidiogenous loci at apex of a secondary ramoconidium, the upper (arrow) clearly coronate; e. two conidiogenous loci at apex of a conidiophore, the one facing the viewer is clearly coronate (arrow); f. two conidiogenous loci (arrows) at apex of a secondary ramoconidium are coronate. — Scale bars: a–c = 2.5 μm, d = 1 μm, e = 5 μm, f = 1.25 μm.
Description — On MEA or SNA, hyphae 1–5 μm wide, sparingly to richly branched (angles between 45 and 90°), sometimes anastomosing, loosely to densely septate, thin-walled, almost smooth to distinctly rough-walled, subhyaline in narrow hyphae to medium dark olivaceous-brown, subcylindrical to irregular in outline by swellings and constrictions at septa. Conidiophores little differentiated, micronematous, barely discernable and distinguishable from ordinary hyphae, becoming macronematous on SNA after 14 d (15–)31–125(−250) × (2.5–)3–4.5(−5.5) μm, predominantly unbranched. Conidiogenous cells integrated, terminal and intercalary, 10–25 × (2.5–)3–4(−5) μm, with a single or up to three conidiogenous loci, 1–1.5 μm diam, coronate, but central dome low and not very conspicuous when viewed by light microscopy. Conidia catenate, in simple or usually branched chains, subglobose, ellipsoid-ovoid, obovoid, broadly fusiform, limoniform, straight to somewhat curved; terminal conidia with a single basal hilum and intercalary conidia with two hila on MEA (3–)4–10(−13) × (2–)3–4(−5) μm, 0(−1)-septate, length becoming shorter towards the apex; on SNA (2.5–)3.5–8(−10.5) × 2.5–4.5(−5) μm (mean 5.9 (std dev 1.4) × 3.4 (std dev 0.4), n = 50), length/width 1.1–3.8 (mean = 1.8); secondary ramoconidia on MEA with 3–5 hila, (6–)10–20(−25) × 3–5 μm; on SNA with 2–4 hila, 8.5–20 × 2.5–4.5 μm (mean 12.5 (std dev 2.7) × 2.9 (std dev 0.4), n = 25), 0–1-septate, sometimes alternarioid, obclavate, subrostrate (the alternarioid ones seldom observed when cultivated on SNA after 7 d, but readily observed on PDA and MEA); ramoconidia (‘true ramoconidia’ sensu Crous et al. 2007b) on SNA (11–)13–34(−43) × (2.5–)3–4 μm; ramoconidia and small terminal conidia in general subhyaline to pale olivaceous or olivaceous-brown, thin-walled, almost smooth to distinctly verruculose, hila conspicuous, 0.75–1.25 μm diam, coronate, often at the end of protuberant, short, terminal projections, 1–2 μm long or even longer in secondary ramoconidia with beak-like ends.
Cultural characteristics — On MEA, colonies attaining 45 mm diam at 25 °C after 14 d; dark olivaceous, powdery, velvety, reverse dark olivaceous-grey. Colonies on PDA attaining 50 mm diam at 25 °C after 14 d, olivaceous-grey in centre, iron-grey in outer region, reverse iron-grey. On SNA, colonies attaining 30 mm diam at 25 °C after 14 d, semi-translucent and olivaceous-grey to iron-grey, with wide translucent margin.
Specimen examined. USA (no additional data known), isolated from wood, CBS-H 20086 (HAL 1846 F), dried culture ex ATCC 38493, cultures ATCC 38493 = CBS 117728 = CPC 12098 = NRRL 8131.
Notes — NRRL 8131 differs from previously known isolates of C. sphaerospermum in having mostly unbranched, micronematous conidiophores, only becoming macronematous with age, and frequently subrostrate, occasionally ‘alternarioid’ conidia. Also, conidia on MEA and SNA exceed in size conidia of other isolates. Conidial length/width (mean 1.8, n = 50) on SNA exceeds that from the standard description (range = 1.1–1.5, Zalar et al. 2007), with 32 % of conidia of NRRL 8131 falling within the range from Zalar et al. (2007).
DISCUSSION
Re-examination of type material of C. lignicola and a putatively North American strain NRRL 8131, originally referred to the latter species, demonstrated clearly that the two fungi are distinct. The type of C. lignicola was formerly examined by Hughes (1958), who reduced it to synonymy with C. herbarum, a treatment confirmed during the course of the present investigations.
The North American strain (NRRL 8131) is C. sphaerospermum, but differs in morphology from previously known isolates of that species. It is easily distinguishable from C. herbarum (including C. lignicola) and all other know species of Cladosporium s.str., by having obclavate, short rostrate, sometimes ‘alternarioid’ conidia. Individual conidia often conformed to the spherical shape generally typical of isolates of C. sphaerospermum, but such conidia of NRRL 8131 could be somewhat larger than the upper limits of 4(−7) × 3.5(−4.5) μm given for C. sphaerospermum in Zalar et al. (2007). Furthermore, the conidiophores are at first consistently micronematous, much later they may become more macronematous, and they are usually unbranched. The conidiophores in other isolates of C. sphaerospermum are often branched in vivo as well as in vitro. However, not only did NRRL 8131 cluster with strains of C. sphaerospermum (Fig. 1), but the neotype of C. sphaerospermum (CBS 193.54) occasionally displayed subrostrate ‘beaks’ on ramoconidia (e.g., fig. 5G in Zalar et al. 2007). Because our sequence data conclusively place NRRL 8131 into C. sphaerospermum (Fig. 1) and because the subrostrate ‘beaks’ could also be located in the neotype, we have refrained from designating the morphologically distinct NRRL 8131 as a new variety.
In their treatment of C. sphaerospermum-like species (Zalar et al. 2007) some variation was observed in the ITS sequence data of all members studied, suggesting that they may not present a single monophyletic group, but could belong to a species complex within Cladosporium. Cladosporium sphaerospermum was described by Penzig (1882) from decaying Citrus leaves and branches in Italy. Isolate CBS 193.54, originating from a human nail, was accepted as neotype (de Vries 1952, Zalar et al. 2007). During the course of this study, attempts to generate the EF sequence data of isolate CBS 193.54 (ex-type of C. sphaerospermum) failed due to the presence of numerous double-peaks in the raw sequence, indicative of contaminated DNA or a possible hybrid. However, sequencing this gene for isolates cultured from single spores derived from isolate CBS 193.54 also generated the same results, and therefore led us to discard the idea of contaminated DNA. It was only possible to obtain the correct sequence (when compared to the EF sequences of other C. sphaerospermum isolates) upon cloning the obtained EF amplicon and sequencing the clones. The correct sequence was one of three sequence types obtained from the clones; blastn results of these three sequences all had distant affinity with Cladosporium EF sequences in GenBank. The origin of the problematic EF sequences is difficult to explain. As part of a larger project we have generated more than 400 EF sequences for Cladosporium spp., none of which gave a similar problem. Whether isolate CBS 193.54 contains pseudogene copies of the translation elongation factor 1-α gene, or whether it is a hybrid, remains to be investigated. Numerous strains with similar ITS rDNA sequences have in subsequent years been isolated from hypersaline water or organic substrata including plants and bathroom walls. Zalar et al. (2007) reported that strains of C. sphaerospermum could grow under in vitro conditions at a water activity of up to 0.860 (Hocking et al. 1994), or even 0.815 (Aihara et al. 2002). Therefore, Zalar et al. (2007) considered C. sphaerospermum as halo- or osmotolerant. Although C. sphaerospermum has commonly been isolated from osmotically stressed environments, it is also known from non-stressed niches, though the exact niche of NRRL 8131 (presumably from wood), remains unknown. Cladosporium sphaerospermum is a cosmopolitan species that has been studied from the perspectives of phylogeny, halo-tolerance and general ecology (summarised in Zalar et al. 2007), biodegradative capacities (e.g., Weber et al. 1995, Prenafeta-Boldú et al. 2001, Potin et al. 2004, Nieves-Rivera et al. 2006, Kim et al. 2007), and clinical aspects (summarised in de Hoog et al. 2000 and Zalar et al. 2007).
Acknowledgments
The authors thank Mr Michael J. Adams, Ms Shari L. Lupien (Washington State Univ.), Ms Mieke Starink, Marjan Vermaas and Arien van Iperen (CBS) for technical assistance.
REFERENCES
- Aihara M, Tanaka T, Ohta T, Takatori K. 2002. Effect of temperature and water activity on the growth of Cladosporium sphaerospermum and Cladosporium cladosporioides. Biocontrol Science 7: 193 – 196 [Google Scholar]
- Braun U, Crous PW, Dugan FM, Groenewald JZ, Hoog GS de. 2003. Phylogeny and taxonomy of cladosporium-like hyphomycetes, including Davidiella gen. nov., the teleomorph of Cladosporium s.str. Mycological Progress 2: 3 – 18 [Google Scholar]
- Brown KB, Hyde KD, Guest DI. 1998. Preliminary studies on endophytic fungal communities of Musa acuminata species complex in Hong Kong and Australia. Fungal Diversity 1: 27 – 51 [Google Scholar]
- Crous PW, Braun U, Groenewald JZ. 2007a. Mycosphaerella is polyphyletic. Studies in Mycology 58: 1 – 32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Braun U, Schubert K, Groenewald JZ. 2007b. Delimiting Cladosporium from morphologically similar genera. Studies in Mycology 58: 33 – 56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Slippers B, Wingfield MJ, Rheeder J, Marasas WFO, Phillips AJL, Alves A, Burgess T, Barber P, Groenewald JZ. 2006. Phylogenetic lineages in the Botryosphaeriaceae. Studies in Mycology 55: 235 – 253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David JC. 1997. A contribution to the systematics of Cladosporium. Revision of the fungi previously referred to Heterosporium. Mycological Papers 172: 1 – 157 [Google Scholar]
- Dugan FM, Schubert K, Braun U. 2004. Checklist of Cladosporium names. Schlechtendalia 11: 1 – 103 [Google Scholar]
- El-Morsy EM. 2000. Fungi isolated from the endorhizosphere of halophytic plants from the Red Sea Coast of Egypt. Fungal Diversity 5: 43 – 54 [Google Scholar]
- Gams W, Verkley GJM, Crous PW. 2007. CBS Course in Mycology, 5th ed CBS, Utrecht, Netherlands: [Google Scholar]
- Ho MH-M, Castañeda RF, Dugan FM, Jong SC. 1999. Cladosporium and Cladophialophora in culture: descriptions and an expanded key. Mycotaxon 72: 115 – 157 [Google Scholar]
- Hocking AD, Miscamble BF, Pitt J. 1994. Water relations of Alternaria alternata, Cladosporium cladosporioides, Cladosporium sphaerospermum, Curvularia lunata and Curvularia pallescens. Mycological Research 98: 91 – 94 [Google Scholar]
- Hoog GS de, Guarro J, Gené J, Figueras MJ. 2000. Atlas of Clinical Fungi, 2nd ed CBS, Utrecht, Netherlands, and Universitat Rovira i Virgili, Reus, Spain: [Google Scholar]
- Hughes SJ. 1958. Revisiones hyphomycetum aliquot cum appendice de nominibus rejiciendis. Canadian Journal of Botany 36: 727 – 836 [Google Scholar]
- Kim JJ, Kang S-M, Choi Y-S, Kim G-H. 2007. Microfungi potentially disfiguring CCA-treated wood. International Biodeterioration & Biodegradation 60: 197 – 201 [Google Scholar]
- Nieves-Rivera ÁM, Rodríguez NJ, Dugan FM, Zaidi BR, Williams EH., Jr 2006. Characterization of Cladosporium oxysporum and C. sphaerospermum using polyaromatic hydrocarbons (PAHs) as their sole carbon source in tropical coastal seawater. In: Mendez-Vilas A. (ed.), Modern multidisciplinary applied microbiology: Exploiting microbes and their interactions: 483–487 Wiley-VCH, Weinheim, Germany: [Google Scholar]
- Penzig AJO. 1882. Fungi Agromiculi. Michelia 2: 385 – 503 [Google Scholar]
- Potin O, Veignie E, Rafin C. 2004. Biodegradation of polycyclic aromatic hydrocarbons (PAHs) by Cladosporium sphaerospermum isolated from an aged PAH contaminated soil. FEMS Microbiology Ecology 51: 71 – 78 [DOI] [PubMed] [Google Scholar]
- Prenafeta-Boldú FX, Kuhn A, Luykx DMAM, Anke H, Groenestijn JW van, Bont JAM de. 2001. Isolation and characterization of fungi growing on volatile aromatic hydrocarbons as their sole carbon and energy source. Mycological Research 4: 477 – 484 [Google Scholar]
- Rayner RW. 1970. A Mycological Colour Chart Commonwealth Mycological Institute & British Mycological Society, Kew, Surrey, England: [Google Scholar]
- Riesen T, Sieber T. 1985. Endophytic fungi in winter wheat (Triticum aestivum L.) Swiss Federal Institute of Technology, Zürich: [Google Scholar]
- Schoch C, Shoemaker RA, Seifert KA, Hambleton S, Spatafora JW, Crous PW. 2006. A multigene phylogeny of the Dothideomycetes using four nuclear loci. Mycologia 98: 1041 – 1052 [DOI] [PubMed] [Google Scholar]
- Schubert K, Braun U, Groenewald JZ, Crous PW. 2007a. Cladosporium leaf-blotch and stem rot of Paeonia spp. caused by Dichocladosporium chlorocephalum gen. nov. Studies in Mycology 58: 95 – 104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert K, Groenewald JZ, Braun U, Dijksterhuis J, Starink MS, Hill CF, Zalar P, Hoog GS de, Crous PW. 2007b. Biodiversity in the Cladosporium herbarum complex (Davidiellaceae, Capnodiales), with standardisation of methods for Cladosporium taxonomy and diagnostics. Studies in Mycology 58: 105 – 156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons EG. 2007. Alternaria: An identification manual CBS Biodiversity Centre, Utrecht, Netherlands: [Google Scholar]
- Vries GA de. 1952. Contribution to the knowledge of the genus Cladosporium Link ex Fr Centraalbureau voor Schimmelcultures, Baarn, Netherlands: [Google Scholar]
- Weber FJ, Hage KC, Bont JA de. 1995. Growth of the fungus Cladosporium sphaerospermum with toluene as the sole carbon and energy source. Applied and Environmental Microbiology 61: 3562 – 3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalar P, Hoog GS de, Schroers H-J, Crous PW, Groenewald JZ, Gunde-Cimerman N. 2007. Phylogeny and ecology of the ubiquitous saprobe Cladosporium sphaerospermum, and descriptions of seven new species from hypersaline environments. Studies in Mycology 58: 157 – 183 [DOI] [PMC free article] [PubMed] [Google Scholar]