Abstract
Species of Mycosphaerella and their related anamorphs represent potentially serious foliar pathogens of Eucalyptus. The fungi treated in the present study were isolated from symptomatic Eucalyptus leaves collected in Thailand during June–October 2007. Species were initially identified based on morphological and cultural characteristics. Identifications were confirmed using comparisons of DNA sequence data of the internal transcribed spacers (ITS1, 5.8S nrDNA, ITS2) and the 28S nrDNA (LSU) regions. To help distinguish species of Pseudocercospora, the dataset was expanded by generating partial sequences of the translation elongation factor 1-α and actin genes. By integrating the morphological and molecular datasets, five new taxa were distinguished, namely Mycosphaerella irregulari, M. pseudomarksii, M. quasiparkii, Penidiella eucalypti and Pseudocercospora chiangmaiensis, while M. vietnamensis represents a new record for Thailand.
Keywords: Eucalyptus, Mycosphaerella, Mycosphaerella leaf disease, Penidiella, Pseudocercospora, taxonomy
INTRODUCTION
Species of Eucalyptus (Myrtaceae) are hosts to a wide range of fungal pathogens (Sankaran et al. 1995, Crous et al. 2004a, 2006b, 2007a, Summerell et al. 2006). Eucalyptus spp. are commonly planted as exotics in commercial plantations for fuel wood, timber, and the paper and pulp industries in various tropical and subtropical regions (Ball 1995). Eucalypts have been cultivated extensively because of their fast growth rates and high adaptability to different soil types and climates (Turnbull 2000). In 1995, the total Eucalyptus plantation area in South-East Asia already exceeded 2 million ha (Old et al. 2003) and this number has continually increased over the years. In Thailand alone, a total of 443 000 ha Eucalyptus plantations was established by 2005 (Barney 2005). In spite of these huge areas already afforested to Eucalyptus, this crop is still increasingly being planted in Thailand and other Asian countries to meet rising global timber demand.
Plant pathogenic microfungi associated with Eucalyptus spp. can substantially decrease timber yield (Park et al. 2000, Old et al. 2003). In particular, species of Mycosphaerella (Mycosphaerellaceae) and Teratosphaeria (Teratosphaeriaceae) have proven to be serious pathogens of Eucalyptus, causing severe leaf spot formation, defoliation, shoot die-back and stem cankers (Crous 1998, Maxwell et al. 2003, Crous et al. 2004a, 2006a, c, 2007b, Hunter et al. 2004, 2006b, Jackson et al. 2005, Cortinas et al. 2006, Carnegie 2007). Nonetheless, information about the diversity of Mycosphaerella and related anamorph species on Eucalyptus from Thailand is generally lacking, and presently only five species have been reported, namely Pseudocercospora basiramifera and Ps. flavomargi-nata (Crous 1998, Hunter et al. 2006a), Mycosphaerella heimii, M. konae and M. thailandica (Crous et al. 2007b). Species of this pathogen complex reported from Eucalyptus in Asia include Dissoconium aciculare, Kirramyces destructans, K. eucalypti, M. crystallina, M. eucalyptorum, M. gracilis, M. heimioides, M. marksii, M. obscuris, M. parkii, M. robusta, M. stramenticola, M. sumatrensis, M. verrocosiafricana, M. vietnamensis, M. yunannensis, Ps. deglupta, Ps. eucalyptorum, Ps. fatouae, Ps. paraguayensis, Ps. robusta, Septoria eucalyptorum, S. xenoparkii, Teratosphaeria fimbriata, T. gamsii, T. suberosa and T. suttonii (Crous & Alfenas 1995, Crous & Wingfield 1997, Hunter et al. 2004, 2006a, Crous 1998, Crous & Braun 2003, Crous et al. 2004a, 2006c, 2007c, Burgess et al. 2007). Many Eucalyptus leaf pathogens originally described as related species of Mycosphaerella (i.e. M. cryptica, M. gamsii, M. pseudocryptica, M. pseudosuberosa and M. suttonii) have been re-classified into the genus Teratosphaeria (Teratosphaeriaceae) after substantial taxonomic revisions based on novel morphological characters integrated with their DNA phylogeny obtained by using the 28S nrRNA gene (Crous et al. 2007c, 2008).
DNA sequencing of the ITS nrDNA gene has in the past proven highly effective to distinguish among species of Mycosphaerella (Crous et al. 2000, 2006a, b, 2007b, Cortinas et al. 2006). The main objective of the present study was to identify species of Mycosphaerella and related anamorphs associated with Eucalyptus leaves collected from plantations in Thailand, and to resolve their taxonomy and DNA phylogeny.
MATERIAL AND METHODS
Isolates
Symptomatic Eucalyptus leaves were collected at various locations in Thailand (Table 1). Lesions with ascomata were removed, soaked in distilled water for 2 h, and then placed in the bottom of the Petri dish lids, with the top half of the dish containing 2 % malt extract agar (MEA; Oxoid, Gams et al. 2007). Dishes were incubated at room temperature in the dark. After 24 h ascospore germination patterns from discharged ascospores on MEA were examined under a compound microscope. Single ascospore cultures were established on fresh MEA dishes as described by Crous (1998). Symptomatic leaves were also incubated in moist chambers (Petri dishes containing moist filter paper). Leaves were inspected daily for microfungi, and single conidial colonies of hyphomycetes and coelomycetes established on MEA (Crous 2002). Cultures were plated onto fresh MEA, oatmeal agar (OA; Gams et al. 2007) and pine needle agar (Slippers et al. 2006), and subsequently incubated at 25 °C in dark, to promote sporulation. Reference strains are maintained in the culture collection of the Centraalbureau voor Schimmelcultures (CBS), Utrecht, the Netherlands and BCC, BIOTEC, Thailand (Table 1).
Table 1.
Isolates of Mycosphaerella and related anamorphs used for DNA analysis and morphological studies.
| Species | Accession number1 | Host | Location | GenBank number |
|||
|---|---|---|---|---|---|---|---|
| ITS | LSU | EF | ACT | ||||
| Mycosphaerella heimii | CPC 15428 | Eucalyptus sp. | Burirum, Thailand | EU882123 | – | – | – |
| CPC 15429 | Eucalyptus sp. | Burirum, Thailand | EU882122 | EU882141 | – | – | |
| CPC 15430 | Eucalyptus sp. | Vientein, Laos | EU882121 | EU882140 | – | – | |
| Mycosphaerella irregulari | CPC 15408, CBS 123242 | Eucalyptus sp. | Udonthani, Thailand | EU882110 | – | – | – |
| CPC 15431 | Eucalyptus sp. | Udonthani, Thailand | EU882111 | – | – | – | |
| CPC 15432 | Eucalyptus sp. | Udonthani, Thailand | EU882112 | – | – | – | |
| Mycosphaerella pseudomarksii | CPC 15410, CBS 123241 | Eucalyptus sp. | Chiang Mai, Thailand | EU882113 | EU882137 | – | – |
| CPC 15435 | Eucalyptus sp. | Chiang Mai, Thailand | EU882114 | – | – | – | |
| CPC 15436 | Eucalyptus sp. | Chiang Mai, Thailand | EU882115 | – | – | – | |
| Mycosphaerella quasiparkii | CPC 15409, CBS 123243 | Eucalyptus sp. | Burirum, Thailand | EU882125 | EU882143 | – | – |
| CPC 15433 | Eucalyptus sp. | Burirum, Thailand | EU882126 | – | – | – | |
| CPC 15434 | Eucalyptus sp. | Burirum, Thailand | EU882127 | – | – | – | |
| Mycosphaerella sp. | CPC 15446 | E. camaldulensis | Burirum, Thailand | EU882108 | EU882136 | – | – |
| CPC 15447 | E. camaldulensis | Burirum, Thailand | EU882109 | – | – | – | |
| CPC 15448 | E. camaldulensis | Ubonratchathani, Thailand | EU882124 | EU882142 | – | – | |
| Mycosphaerella thailandica | CPC 15437 | Eucalyptus sp. | Vientein, Laos | EU882120 | – | – | – |
| CPC 15438 | Eucalyptus sp. | Vientein, Laos | EU882119 | – | – | – | |
| CPC 15439 | E. camaldulensis | Mahasarakam, Thailand | EU882118 | EU882139 | – | – | |
| CPC 15440 | E. camaldulensis | Ubonratchathani, Thailand | EU882117 | EU882138 | – | – | |
| CPC 15441 | Eucalyptus sp. | Burirum, Thailand | EU882116 | – | – | – | |
| Mycosphaerella vietnamensis | CPC 15442 | Eucalyptus sp. | Vientein, Laos | EU882104 | – | – | – |
| CPC 15443 | Eucalyptus sp. | Burirum, Thailand | EU882107 | EU882135 | – | – | |
| CPC 15444 | Eucalyptus sp. | Vientein, Laos | EU882106 | EU882134 | – | – | |
| CPC 15445 | E. camaldulensis | Udonthani, Thailand | EU882105 | – | – | – | |
| Penidiella eucalypti | CPC 15411, CBS 123246 | Eucalyptus sp. | Mahasarakam, Thailand | EU882131 | EU882145 | – | – |
| CBS 123245 | E. camaldulensis | Burirum, Thailand | EU882132 | – | – | – | |
| CPC 15449 | E. camaldulensis | Burirum, Thailand | EU882133 | EU882146 | – | – | |
| Pseudocercospora chiangmaiensis | CPC 15412, CBS 123244 | E. camaldulensis | Chiang Mai, Thailand | EU882128 | EU882144 | EU882147 | EU882150 |
| CPC 15450 | E. camaldulensis | Chiang Mai, Thailand | EU882129 | – | EU882148 | EU882151 | |
| CPC 15451 | E. camaldulensis | Chiang Mai, Thailand | EU882130 | – | EU882149 | EU882152 | |
1 CBS: Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands; CPC: Culture collection of Pedro Crous, housed at CBS.
DNA isolation, amplification and analyses
Genomic DNA was extracted from mycelia of fungal colonies cultivated on MEA using the UltraCleanTM Microbial DNA Isolation Kit (Mo Bio Laboratories, Inc., Solana Beach, CA, USA). The Primers V9G (de Hoog & Gerrits van den Ende 1998) and LR5 (Vilgalys & Hester 1990) were used to amplify part of the nuclear rDNA operon spanning the 3′ end of the 18S rRNA gene (SSU), the first internal transcribed spacer (ITS1), the 5.8S rRNA gene, the second ITS region (ITS2) and the 5′ end of the 28S rRNA gene (LSU). The primers ITS4 (White et al. 1990) and LR0R (Rehner & Samuels 1994) were used as internal sequence primers to ensure good quality sequences over the entire length of the amplicon. To help resolve species of Pseudocercospora, the ITS region was supplemented with sequences of the translation elongation factor 1-α gene (EF-1α) using the primers EF1-728F and EF1-986R (Carbone & Kohn 1999) and the actin gene (ACT) using the primers ACT-512F and ACT-783R (Carbone & Kohn 1999). The PCR amplifications were performed on a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA, USA) in a total volume of 12.5 μL solution containing 10–20 ng of template DNA, 1 × PCR buffer, 2.5 mM MgCl2, 15 pmol for each primer, 60 μM of each dNTP and 0.75 U Taq DNA polymerase (Bioline GmbH, Luckenwalde, Germany). PCR amplification conditions were set as follows: an initial denaturation temperature of 94 °C for 5 min, followed by 40 cycles of denaturation temperature of 94 °C for 45 s, primer annealing at 48 °C for 30 s, primer extension at 72 °C for 90 s and a final extension step at 72 °C for 7 min. The primer annealing temperature for EF-1α and ACT was at 55 °C. The resulting fragments were sequenced using the PCR primers together with a BigDye Terminator Cycle Sequencing Kit v. 3.1 (Applied Biosystems, Foster City, CA) and analysed on an ABI Prism 3100 DNA Sequencer (Perkin-Elmer, Norwalk, CN).
The generated sequences were compared with other fungal DNA sequences from NCBI’s GenBank sequence database using a blast search; sequences with high similarity were added to the alignments. The additional GenBank sequences were manually aligned using Sequence Alignment Editor v. 2.0a11 (Rambaut 2002). The phylogenetic analyses of the aligned sequence data were performed using PAUP (Phylogenetic Analysis Using Parsimony) v. 4.0b10 (Swofford 2003) and consisted of neighbour-joining analyses with the uncorrected (‘p’), the Kimura 2-parameter and the HKY85 substitution models. Alignment gaps were treated as missing data and all characters were unordered and of equal weight. Any ties were broken randomly when encountered. For parsimony analyses, alignment gaps were treated as a fifth character state and all characters were unordered and of equal weight. Maximum parsimony analysis was performed using the heuristic search option with 100 random simple taxa additions and tree bisection and reconstruction (TBR) as the branch-swapping algorithm. Branches of zero length were collapsed and all multiple, equally parsimonious trees were saved. The robustness of the trees obtained was evaluated by 1 000 bootstrap replications (Hillis & Bull 1993). Tree length (TL), consistency index (CI), retention index (RI) and rescaled consistency index (RC) were calculated and the resulting trees were printed with TreeView v. 1.6.6 (Page 1996). New sequences were lodged in GenBank and the alignments and phylogenetic trees in TreeBASE (www.treebase.org).
Morphology
Preparations from cultured fungal colonies were mounted on glass slides with clear lactic acid for microscopic examination. Sections of ascomata were made by hand for examination purposes. Measurements of all taxonomically relevant parameters were made at 1 000 × magnification, with 30 measurements per structure where possible. Colony colours on MEA (surface and reverse) were determined using the colour charts of Rayner (1970) after 15 d at 25 °C in the dark.
RESULTS
Phylogenetic analysis
Approximately 1 700 bases, spanning the ITS and LSU regions, were obtained for isolates listed in Table 1. These two regions were analysed separately; ITS to determine species level relationships and LSU for the generic placement. Approximately 300 and 220 bases were determined for EF-1α and ACT, respectively, and these were concatenated with the corresponding ITS sequences for a combined analysis of the Pseudocercospora clade.
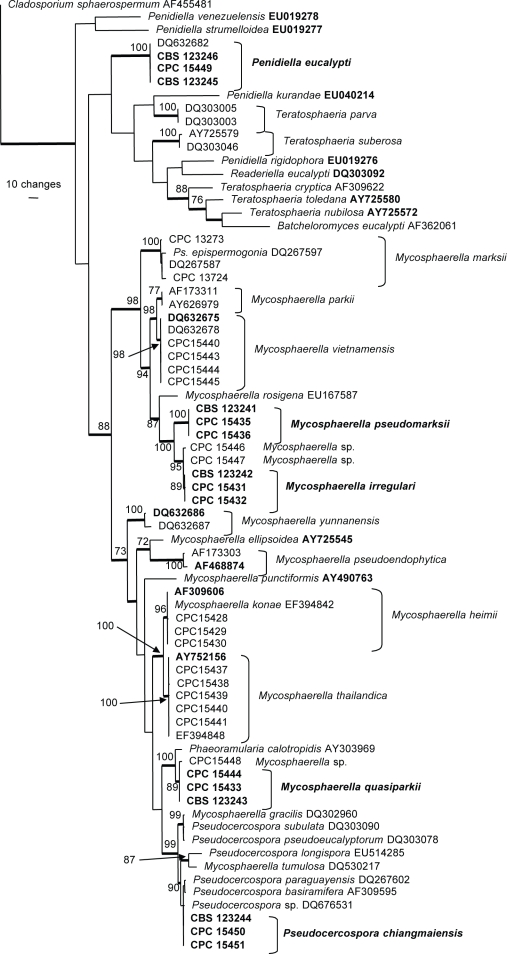
The manually adjusted ITS alignment contained 73 taxa (including the outgroup sequence) and, of the 533 characters used in the phylogenetic analysis, 228 were parsimony-informative, 59 were variable and parsimony-uninformative and 246 were constant. Neighbour-joining analysis using the three substitution models on the sequence data yielded trees with similar topology and bootstrap values; 1 100 equally most parsimonious trees were obtained from the heuristic search, one of which is shown in Fig. 1 (TL = 1386, CI = 0.444, RI = 0.797, RC = 0.354). The phylogenetic tree derived from the ITS region (Fig. 1) showed that some of the isolates belong to known species, whereas others appeared to be new to science.
Fig. 1.
One of 1 100 equally most parsimonious trees obtained from a heuristic search with 100 random taxon additions of the ITS sequence alignment using PAUP v. 4.0b10. The scale bar shows 10 changes and bootstrap support values higher than 70 % from 1 000 replicates are shown at the nodes. Thickened lines indicate the strict consensus branches and ex-type sequences are printed in bold face. The tree was rooted to Cladosporium sphaerospermum (GenBank accession AF455481).
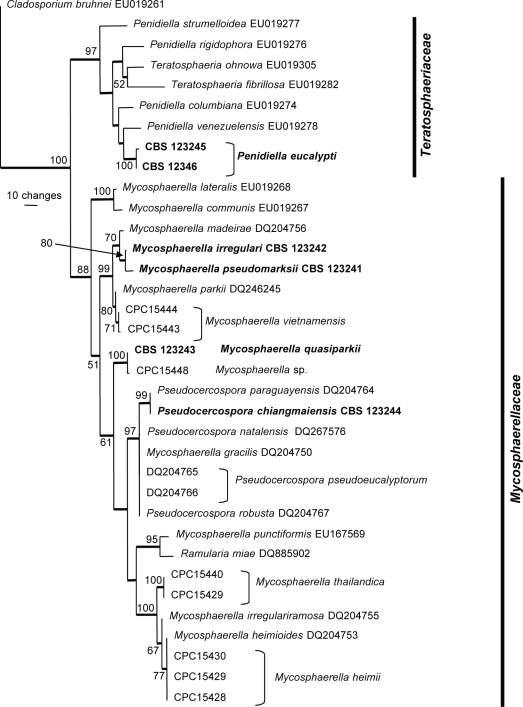
The manually adjusted LSU alignment contained 35 taxa (including the outgroup sequence) and, of the 797 characters used in the phylogenetic analysis, 121 were parsimony-informative, 54 were variable and parsimony-uninformative and 622 were constant. Neighbour-joining analysis using the three substitution models on the sequence data yielded trees with similar topology and bootstrap values. Two equally most parsimonious trees were obtained from the heuristic search, one of which is shown in Fig. 2 (TL = 407, CI = 0.570, RI = 0.805, RC = 0.459). The phylogenetic tree of the LSU region (Fig. 2) showed two distinct groups of fungal isolates; the first was the Teratosphaeriaceae clade (97 % bootstrap support), whereas the second was the Mycosphaerellaceae clade (88 % bootstrap support).
Fig. 2.
One of two equally most parsimonious trees obtained from a heuristic search with 100 random taxon additions of the LSU sequence alignment using PAUP v. 4.0b10. The scale bar shows 10 changes and bootstrap support values from 1 000 replicates are shown at the nodes. Thickened lines indicate the strict consensus branches and ex-type sequences are printed in bold face. The tree was rooted to Cladosporium bruhnei (GenBank accession EU019261).
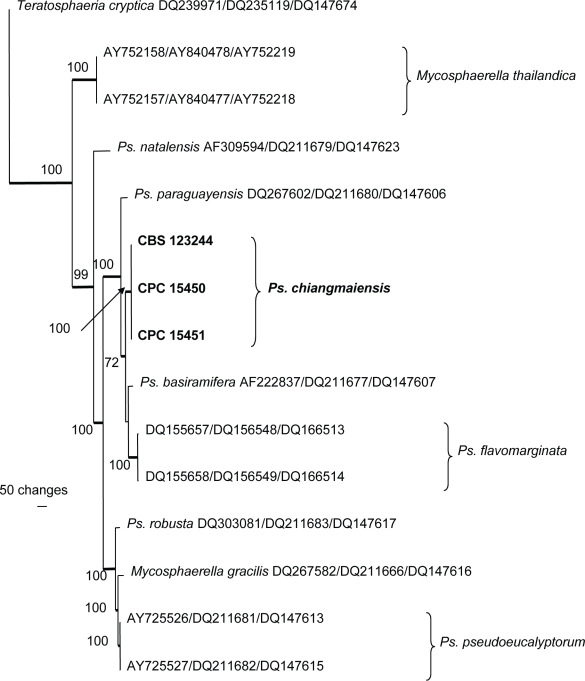
The manually adjusted combined (ITS, EF-1α and ACT) alignment contained 16 taxa (including the outgroup sequence) and, of the 1 012 characters used in the phylogenetic analysis, 339 were parsimony-informative, 139 were variable and parsimony-uninformative and 534 were constant. Neighbour-joining analysis using the three substitution models on the sequence data yielded trees with similar topology and bootstrap values. A single most parsimonious tree was obtained from the heuristic search and is shown in Fig. 3 (TL = 876, CI = 0.822, RI = 0.852, RC = 0.700). The two trees differed only in their placement of Ps. basiramifera; in the one tree it is the closest sister of Ps. chiangmaiensis and in the other it is the sister of Ps. flavomarginata.
Fig. 3.
Single most parsimonious trees obtained from a heuristic search with 100 random taxon additions of the combined ITS, EF and ACT sequence alignment using PAUP v. 4.0b10. The scale bar shows 100 changes and bootstrap support values from 1 000 replicates are shown at the nodes. The tree was rooted to Teratosphaeria cryptica (GenBank accessions DQ239971, DQ235119 and DQ147674, respectively). Ps. = Pseudocercospora.
Taxonomy
Several taxonomic novelties, namely three Mycosphaerella, one Pseudocercospora and one Penidiella species, were found. These species do not match any species presently described from these genera, or any linked to sequences available in GenBank, and are thus described as new below.
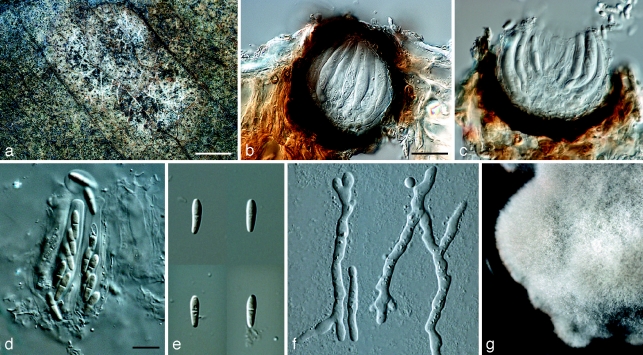
Mycosphaerella irregulari Cheewangkoon, K.D. Hyde & Crous, sp. nov. — MycoBank MB507001; Fig. 4, 5
Fig. 4.
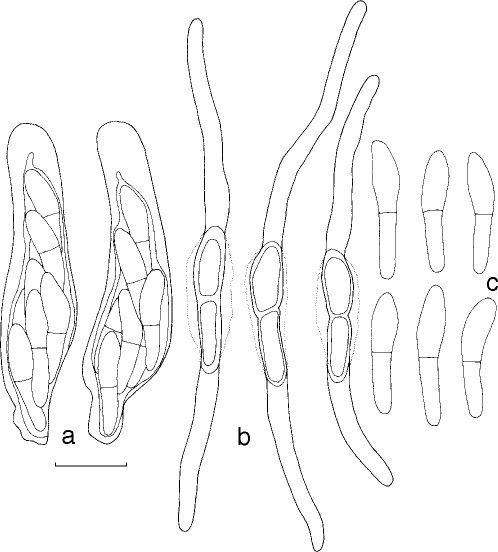
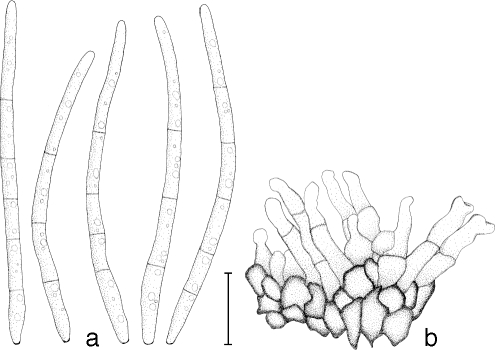
Mycosphaerella irregulari. a. Asci; b. germinating ascospores; c. ascospores. — Scale bar = 10 μm.
Fig. 5.
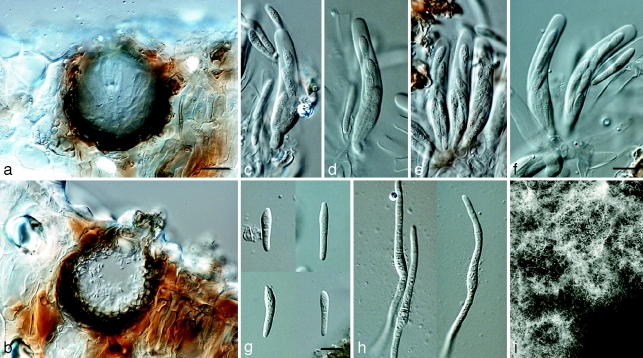
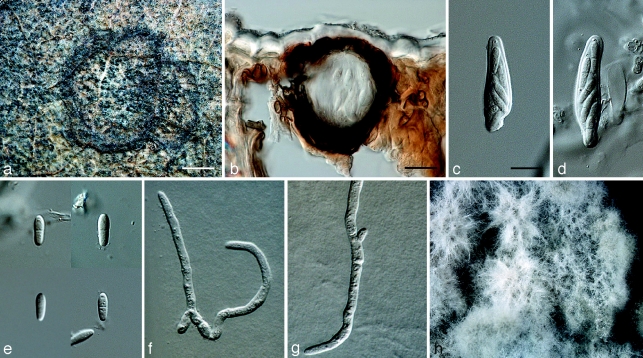
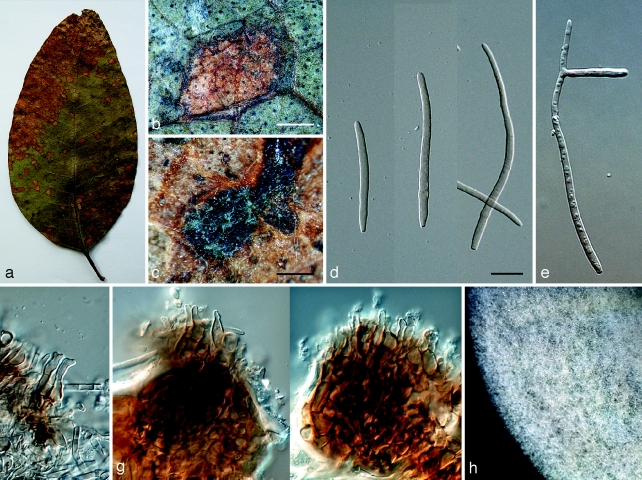
Mycosphaerella irregulari. a. Leaf spot; b, c. sections through ascomata; d. asci; e. ascospores; f. germinating ascospores; g. colony on MEA. — Scale bars: a = 1 mm; b, c = 20 μm; d–f = 10 μm.
Anamorph. Unknown.
Mycosphaerellae tasmaniensis similis, sed ascosporis cum tubis germinalibus irregularister latis.
Etymology. Named after the irregular width of its ascospore germ tubes.
Leaf spots amphigenous, subcircular to oval, pale brown with grey centres, 5–12 mm diam, surrounded by a thin, medium brown margin. Mycelium external, smooth, septate, branch, medium brown, (2–)2.5–3(−4) μm wide hyphae. Ascomata epiphyllous, black, subepidermal to erumpent, ovoid to subglobose, 45–83 × 45–78 μm; apical ostiole 20–25 μm wide; wall thick (6.5–)7–10(−12) μm, consisting of 2–3 layers of medium brown textura angularis. Asci aparaphysate, fasciculate, subsessile, subcylindrical to narrowly obovoid, straight to slightly curved, 8-spored, (25–)35–40(−45) × (6–)7–8(−10) μm. Ascospores bi- to tri-seriate, overlapping, hyaline, guttulate, thin-walled, straight to slightly curved, fusoid-ellipsoidal with obtuse ends, widest just above the septum, or in the middle of the apical cell, medianly 1-septate or slightly longer in the basal cell, slightly constricted at septum, tapering toward both ends, but with more prominent taper towards lower end, at times with a mucous-like coating, (8–)9–11(−13) × 2.5–3(−3.5) μm.
Ascospore germination — Germinating from both ends, remaining hyaline; germ tubes grow parallel to the long axis of the spore, with lateral branches parallel or perpendicular to the long axis of the spore; germination tubes irregular in width, constrict at the median septum of the spore, becoming (11–)13–15(−16.5) × 4–5(−5.5) μm, slightly distorting (Type I; Crous 1998).
Cultural characteristics — Colonies reach 15 mm diam on MEA after 15 d at 25 °C in the dark; circular, convex, with a slightly undulate, smooth margin, and medium aerial mycelium; pale greenish grey to pale olivaceous-grey (surface); olivaceous-black (reverse).
Specimen examined. Thailand, Udonthani, on living leaves of Eucalyptus sp., July 2007, R. Cheewangkoon, holotype CBS H-20135, cultures ex-type CBS 123242 = CPC 15408, CPC 15431, CPC 15432.
Notes — The ascospore morphology of M. irregulari is similar to that of M. tasmaniensis (Crous et al. 1998), M. flexuosa (Crous 1998), M. ellipsoidea (Crous & Wingfield 1996) and M. heimii (Crous & Swart 1995). However, M. irregulari can be distinguished from these species by its irregular germ tubes and germination pattern. Phylogenetically, it is also not closely related to any of the species cited above, but clusters near to M. pseudomarksii (100 % bootstrap, Fig. 1), which has a distinct morphology.
Mycosphaerella pseudomarksii Cheewangkoon, K.D. Hyde & Crous, sp. nov. — MycoBank MB507003; Fig. 6, 7
Fig. 6.
Mycosphaerella pseudomarksii. a. Vertical section through an ascoma; b. vertical section through a spermatogonium; c–f. asci; g. ascospores; h. germinating ascospores; i. colonies on MEA. — Scale bars: a, b = 10 μm; c–h = 10 μm.
Fig. 7.
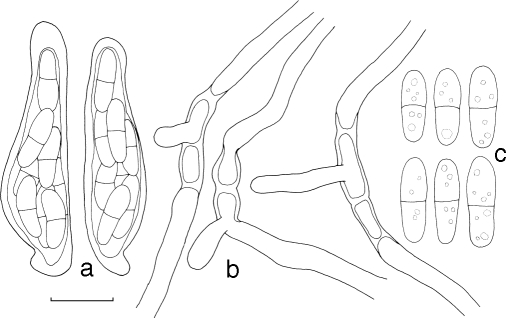
Mycosphaerella pseudomarksii. a. Asci; b. germinating ascospores; c. ascospores. — Scale bar = 10 μm.
Mycosphaerellae marksii similis, sed ascosporis majoribus, (12–)14–17(−18.5) × (2.5–)3(−3.5) μm.
Etymology. Named after Mycosphaerella marksii to which it is morphologically similar.
Leaf spots not observed. Ascomata amphigenous in apparently healthy tissue (endophyte?), occurring on greenish brown part of the leaf after incubation in moist chambers for 2 d, black, subepidermal to erumpent, globose to subglobose, 42–60 × 45–80 μm; apical ostiole 20–35 μm wide; wall consisting of 2–3 layers of medium brown textura angularis. Asci aparaphysate, fasciculate, bitunicate, subsessile, subcylindrical to narrowly ovoid, slightly curved, 8-spored, (40–)42–45(−48) × (6.5–)7–8(−8.5) μm. Ascospores bi- to tri-seriate overlapping, hyaline, guttulate, thin-walled, straight to slightly curved, fusoid with obtuse ends, widest in the middle of the asymmetrical apical cell, medianly 1-septate or with slightly longer basal cell; tapering toward both ends, but with more prominent taper towards lower end, (12–)14–17(−18.5) × (2.5–)3(−3.5) μm. Spermatogonia well developed, amphigenous, dark brown, subepidermal to erumpent, globose to subglobose, up to 90 μm diam. Spermatia hyaline, smooth, rod-shaped, with obtuse ends, (3.5–)4–5(−5.5) × 1.5–2 μm.
Ascospore germination — Germinating from both ends, with a thin, mucous-like coat visible surrounding ascospores on agar; germ tube growing parallel to the long axis of the spore, regular in width, remaining hyaline, not distorting or becoming constricted at septum (Type B; Crous 1998).
Cultural characteristics — Colonies reach 15 mm diam on MEA after 15 d at 25 °C in the dark; subcircular, convex, with even margin, slightly folded, with sparse aerial mycelium; pale olivaceous-grey to olivaceous-grey (surface); olivaceous-black (reverse).
Specimen examined. Thailand, Chiang Mai, Mae Tang, on living leaves of Eucalyptus sp., June 2007, R. Cheewangkoon, holotype CBS H-20134, cultures ex-type CBS 123241 = CPC 15410, CPC 15435, CPC 15436.
Notes — Mycosphaerella pseudomarksii was most similar to M. marksii (Carnegie & Keane 1994) based on its asymmetrical apical ascospore cells and ascospore germination patterns. However, germinating ascospores of M. pseudomarksii have a visible mucilaginous sheath, which was not observed in germinating ascospores of M. marksii. Furthermore, ascospores of M. pseudomarksii were slightly longer and wider (12–)14–17(−18.5) × (2.5–)3(−3.5) μm than those of M. marksii (11–)12–14(−16) × 2–2.5(−3) μm. Crous & Wingfield (1996) observed considerable variation in ascospore dimensions of several collections of M. marksii (12.5–22.5 × 2.5–5 μm) commenting that this may represent a species complex. Phylogenetically the two species are also distinct (Fig. 1).
Mycosphaerella quasiparkii Cheewangkoon, K.D. Hyde & Crous, sp. nov. — MycoBank MB507002; Fig. 8, 9
Fig. 8.
Mycosphaerella quasiparkii. a. Leaf spot; b. section through an ascoma; c, d. asci; e. ascospores; f, g. germinating ascospores; h. colony on MEA. — Scale bars: a = 1 mm; b, = 20 μm; c–g = 10 μm.
Fig. 9.
Mycosphaerella quasiparkii. a. Asci; b. germinating ascospores; c. ascospores. — Scale bar = 10 μm.
Anamorph. Unknown.
Mycosphaerellae parkii similis, sed ascosporis ellipsoideis et coloniis bubalinis in agaro MEA.
Etymology. Named after its similarity to Mycosphaerella parkii.
Leaf spots amphigenous, round to irregular, separate, becoming confluent, 5–15 mm diam, medium brown on adaxial surface, pale brown on abaxial surface, surrounded by a raised border, which is dark brown on the adaxial surface and paler brown on the abaxial surface. Ascomata epiphyllous, black, subepidermal to erumpent, subglobose, 40–60 × 40–55 μm; apical ostiole 10–15 μm wide; wall consisting of 3–4 layers of medium to dark brown textura angularis. Asci aparaphysate, fasciculate, bitunicate, subsessile, broadly ellipsoid to obclavate, straight to slightly curved, 8-spored, (30–)45–50(−57) × (7–)8.5–9(−9.5) μm. Ascospores bi- to multiseriate, overlapping, hyaline, guttulate, thin-walled, straight to slightly curved, ellipsoidal to obovoid with obtuse ends, widest in the middle of the apical cell, medianly 1-septate, not constricted at the septum, tapering toward both ends, with a thin mucilaginous sheath, (9–)10–11(−12.5) × (2.5–)3–3.5(−4.5) μm.
Ascospore germination — Germinating with more than one germ tube per cell. Initial germ tubes originating from polar ends, growing parallel to the long axis of the spore, with perpendicular germ tubes developing later; ascospores remain hyaline, but become constricted at the median septum, and distorting, (2.8–)3.5–4(−4.5) μm wide (Type D; Crous 1998).
Cultural characteristics — Colonies reach 27 mm diam on MEA after 15 d at 25 °C in the dark; circular, low convex, with entire edge and sparse aerial mycelium; buff (surface); vinaceous-buff (reverse).
Specimen examined. Thailand, Burirum, on living leaves of Eucalyptus sp., July 2007, P. Suwannawong, holotype CBS H-20132, cultures ex-type CBS 123243 = CPC 15409, CPC 15433, CPC 15434.
Notes — Mycosphaerella quasiparkii is morphologically similar to M. parkii (Crous et al. 1993, 2006c) with which it also shares the same ascospore germination pattern. However, M. quasiparkii has more ellipsoid ascospores and paler colonies on MEA. Phylogenetically, M. quasiparkii clusters close to Phaeoramularia calotropidis (AY303969, Fig. 1).
Penidiella eucalypti Cheewangkoon, K.D. Hyde & Crous, sp. nov. — MycoBank MB507004; Fig. 10, 11
Fig. 10.
Penidiella eucalypti. a. Colony on MEA; b–h. catenulate conidia; i. conidiogenous cell and primary ramoconidia; j. primary ramoconidium (left) and secondary ramoconidium (right); k. conidia with mucilaginous sheath. — Scale bars: b = 60 μm; c, d = 80 μm; e = 30 μm; f, g = 20 μm; h, i = 15 μm; j, k = 10 μm.
Fig. 11.
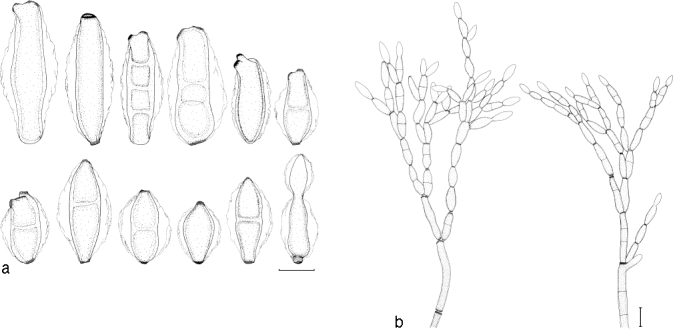
Penidiella eucalypti. a. Conidia; b. conidial branching system. — Scale bars = 10 μm.
Teleomorph. Unknown.
Differt a omnibus speciebus Penidiellae conidiis mucosis in vitro (MEA) formantibus.
Etymology. Named after its host genus, Eucalyptus.
Leaf spots not observed. Mycelium consisting of branched, septate, smooth to slightly verruculose, pale to medium brown, (2.5–)3–4(−5) μm wide hyphae. Conidiophores macronematous, occasionally micronematous, arising from superficial mycelium, solitary, erect, straight to slightly curved, branched laterally or not, medium to dark brown, slightly thick-walled, wall ≤ 1 μm wide, smooth to finely verruculose, (30–)150–200(−220) × (4–)4.5–5.5(−6.5) μm, wider at the base, (5.5–)6–7(−8.5) μm, (0–)4–6(−8) septate. Conidiogenous cells terminal, cylindrical to subcylindrical, tapering to a flattened apical region, smooth to finely verruculose, medium brown, (10–)13–17(−25) × (3–)4–5(−6) μm; scars thickened, somewhat darkened, 2–2.5 μm wide. Ramoconidia: primary ramoconidia subcylindrical or obovoid, (0–)1(−3)-septate, base truncate, with (1–)2–3(−4) apical hila, pale olivaceous to pale brown, smooth to finely verruculose, wall < 1 μm wide, (25–)35–40(−48) × (3.5–)4–5(−6.5) μm; scars thickened and slightly darkened, 2–2.5 μm wide; giving rise to chains of up to 12 conidia; secondary ramoconidia obovoid, 0–1-septate, some constricted at septum, base truncate, with 2–3(−4) apical hila, pale olivaceous, smooth, (13–)15–20(−28) × (4.5–)5–6(−7.5) μm; intercalary conidia obovoid, 0–1(−2)-septate, some constricted at septa, base truncate, with 2–3 apical hila, pale olivaceous, smooth, (10–)12–15(−18) × (3.5–)4–5(−5.5) μm. Conidia in branched acropetal chains, broadly fusiform to obovoid, 0–1-septate, pale olivaceous, paler towards the apex, (9–)12–15(−19) × (4–)5–6(−7) μm; terminal conidia ovoid, 0-septate, pale olivaceous to hyaline, paler towards the apex, base truncate, conidia have a wing-like mucilaginous sheath in culture, extending up to 5 μm wide on each side, tapering towards the polar ends.
Cultural characteristics — Colonies on MEA reaching 20 mm diam after 15 d at 25 °C; margin feathery, colonies erumpent, spreading, with moderate aerial mycelium. Surface grey-olivaceous, reverse olivaceous-black.
Specimens examined. Thailand, Payakpoompisai, Mahasarakam, on leaves of Eucalyptus camaldulensis, July 2007, P. Suwannawong, holotype CBS H-20136, cultures ex-type CBS 123246 = CPC15411, AGI064.1, AGI064.2 (occurring on a lesion in association with Harknessia sp.); Satuk, Burirum, on leaves of Eucalyptus camaldulensis, July 2007, R. Cheewangkoon, cultures CBS 123245, CPC15449 (occurring on a lesion in association with several microfungi).
Notes — Penidiella eucalypti is a typical species of Penidiella by having solitary conidiophores with a branching system consisting of ramoconidia that form secondary ramoconidia and conidia, with slightly thickened, darkened scars (Crous et al. 2007c). Other than being phylogenetically distinct (Fig. 1, 2), P. eucalypti is distinct from most other species of Penidiella by having a prominent branching system which develops on a single terminal conidiogenous cell. Another character that has not previously been reported in the genus is the distinct mucilaginous sheath observed on conidia formed on MEA.
Pseudocercospora chiangmaiensis Cheewangkoon, K.D. Hyde & Crous, sp. nov. — MycoBank MB507005; Fig. 12, 13
Fig. 12.
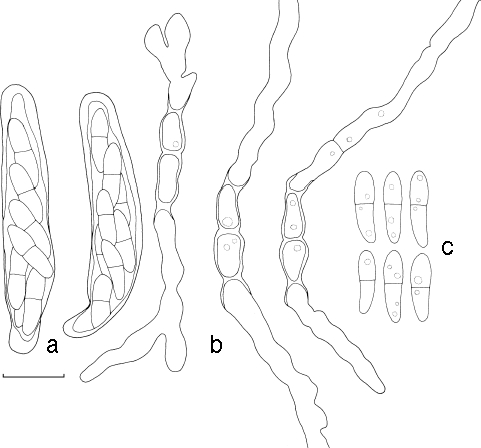
Pseudocercospora chiangmaiensis. a. Leaf spots; b, c. close up of leaf spot; d, e. conidia; f, g. conidiogenous cells; h. colony on MEA. — Scale bars: b = 1 mm; c = 300 μm; d–g = 10 μm.
Fig. 13.
Pseudocercospora chiangmaiensis. a. Conidia; b. conidiophores with conidiogenous cells. — Scale bar = 10 μm.
Teleomorph. Unknown.
Pseudocercosporae basiramiferae similis, sed cellulis conidiogenis terminalibus et intercalaribus et stromatibus bene evolutis.
Etymology. Named after Chiang Mai, the province in Thailand from where it was collected.
Leaf spots amphigenous, subcircular to angular, 2–6 mm diam, pale to medium brown, surrounded by a slightly raised, dark-brown border, becoming confluent with age, leading to leaf blight from the leaf tip. Mycelium predominantly internal, consisting of smooth, septate, branched (1.5–)2–3(−4) μm wide hyphae. Caespituli amphigenous, more prominent on abaxial leaf surface, pale grey on leaves, often on dark brown to black and thickened leaf tissue, 25–50 × 50–100 μm. Stromata immersed, becoming erumpent, medium brown, 25–70 × 30–70 μm wide. Conidiophores reduced to conidiogenous cells or one supporting cell, occasionally arising from upper cells of stroma, subcylindrical, 1–3(−6)-septate with intercalary conidiogenous cells, (13–)20–25(−60) × 3–4(−4.5) μm, situated on the superficial part of the stroma. Conidiogenous cells terminal or intercalary, subcylindrical to obclavate, medium brown, becoming paler toward apex, straight to geniculate-sinuous, tapering to a truncate or bluntly rounded apex, at times subdenticulate, smooth, medium to thick-walled, variable in length, (5–)10–11(−12) × (2–)3(−5) μm, unbranched, proliferating sympodially; conidial scars thickened at the rim, not darkened, inconspicuous. Conidia solitary, subcylindrical to narrowly obclavate, tapering toward the subobtuse apex; base obconic-subtruncate, (2–)3–5(−10)-septate, straight to slightly curved, pale to medium brown, smooth, thin-walled, guttulate, (40–)50–60(−100) × (2–)2.5–3(−3.5) μm (up to 140 μm long in moist chambers); hilum thickened and somewhat darkened at the rim (paracercospora-like), not refractive, at times inconspicuous; microcyclic conidiation observed when incubated in a moist chamber.
Cultural characteristics — Colonies reaching 18 mm diam on MEA after 15 d at 25 °C in the dark; colonies circular, convex, with entire margin and medium aerial mycelium; pale greenish grey (surface), fuscous-black (reverse).
Specimen examined. Thailand, Chiang Mai, Doi Lor, on leaves of Eucalyptus camaldulensis, June 2007, P. Suwannawong, holotype CBS H-20133, cultures ex-type CBS 123244 = CPC 15412, CPC 15450, CPC 15451.
Notes — Of the Pseudocercospora species known from Eucalyptus (Crous 1998, Braun & Dick 2002, Crous et al. 2004a, 2006c, 2007b, Hunter et al. 2006a, Carnegie et al. 2007), Ps. chiangmaiensis is morphologically most similar to Ps. basiramifera (Crous 1998) in conidium morphology (dimensions, shape, microcyclic conidiation and scar thickening along the rim). Pseudocercospora chiangmaiensis is distinct from Ps. basiramifera based on its terminal and intercalary conidiogenous cells in vivo, and by its conidiophores occurring on well-developed stromata. Phylogenetically it is closely related to Ps. basiramifera (Fig. 3) but differs from it with 5 nucleotide positions on ITS, 34 on EF-1α and 12 on ACT.
DISCUSSION
Although numerous species of Mycosphaerella have been associated with Eucalyptus leaf diseases in tropical regions around the world (Crous 1998, Crous et al. 2000, 2006c, 2007b, c, 2008), only a few of these species have been documented from Asia. The number of novel species found in the present study from sampled Eucalyptus leaves collected in Thailand, was thus not totally unexpected. The three new Mycosphaerella spp. (i.e. M. irregulari, M. pseudomarksii and M. quasiparkii) identified here were difficult to distinguish from other species of Mycosphaerella based on their ascospore morphology and germination patterns alone, which are the characters that have been commonly used in the past for taxonomic classification of Mycosphaerella spp. (Park & Keane 1982, Crous 1998). Therefore, the DNA sequencing data generated here proved particularly helpful in distinguishing these species.
Mycosphaerella pseudomarksii has ascospores with asymmetrical apical cells similar to M. marksii, the only difference being ascospore dimensions and the presence of a mucilaginous ascospore sheath in M. pseudomarksii. Ascospores of M. irregulari are fusoid-ellipsoidal, thus being similar as those of M. ellipsoidea, M. flexuosa, M. heimii and M. tasmaniensis. Although these species have similar ascospore dimensions, M. irregulari is characterised by the distinctive irregular width of its ascospore germination tubes. Mycosphaerella quasiparkii has an ascospore morphology and germination pattern almost identical to M. parkii, but the presence of a thin mucous-like layer on ascospores of M. quasiparkii and its buff colonies, distinguish it from M. parkii, which lacks an ascospore sheath and has olivaceous-grey colonies. Pseudocercospora chiangmaiensis, which is also newly described from Eucalyptus, shares some morphological features (conidial dimensions, hilum thickening and microcyclic conidiation) with Ps. basiramifera. It is distinct, however, by having terminal and intercalary conidiogenous cells in vivo and having conidiophores arising from a well-defined stroma. Phylogenetic analyses of the ITS and ACT genes showed limited differences (only 1 nucleotide) between Ps. chiangmaiensis and Ps. assamensis, which Arzanlou et al. (2008) recently described from banana. Morphologically, however, they differ in the basal conidial cell shape and marginal thickening along the hilum. These differences were supported by analyses based on the EF gene, which indicated these two species to differ by 38 nucleotides.
Other than new species of Mycosphaerella and Pseudocercospora, the present study also led to the discovery of a novel species of Penidiella. Penidiella eucalypti is distinct in that it has a distinctive branching system developed chiefly on a single conidiogenous locus, and conidia with a persistent, characteristic mucilaginous sheath in vitro. Results from the phylogenetic analyses also indicated that this species belongs to a clade represented by an undescribed Teratosphaeria sp. (DQ632682) which could represent its teleomorph.
Two strains that were phylogenetically closely related to M. irregulari (95 % bootstrap), namely CPC 15446 and CPC 15447, represent two undescribed species of Mycosphaerella. Mycosphaerella sp. CPC 15446 differs from M. irregulari by having ascospores with more obtuse apical cells and germination tubes that are regular in width. Although insufficient material was available of Mycosphaerella sp. CPC 15447, it occurred in lesions in association with M. thailandica and M. heimii. Another undescribed Mycosphaerella species, CPC 15448, was closely related to M. quasiparkii (89 % bootstrap). More collections are required to resolve their status. In the present study, three known species were also identified from the diseased Eucalyptus leaf samples, namely M. heimii, M. thailandica and M. vietnamensis. Although M. thailandica and M. heimii were previously reported on Eucalyptus in Thailand (Crous et al. 2007b), M. vietnamensis represents a record for this country.
This study has again demonstrated that morphological characters and molecular techniques are complementary, and necessary, to uncover the diversity and geographical range of Mycosphaerella and Teratosphaeria species occurring on Eucalyptus. Although five new fungal species have been identified and one species represents a new record from diseased Eucalyptus leaves from Thailand, it is still unknown whether these species are native or exotic. We expect that more unidentified disease-causing microfungi await discovery in Thailand, because the expanding area of Eucalyptus plantations allow fungal pathogens to cross geographical barriers to infect new hosts (i.e. from exotic Eucalyptus to other native trees) more easily, and also increase the chance of infection by native fungi to the exotic plantations (Slippers et al. 2005). Some examples of introduced pathogens from exotic Eucalyptus are T. cryptica, T. nubilosa (Park & Keane 1982, Wingfield et al. 1995), and T. suttonii (Chipompha 1987, Crous et al. 1989, 1997). These species were described from Australia where Eucalyptus is native, but were found later in other countries where this host has been planted as an exotic. In addition, the study of Mycosphaearella spp. on exotic Acacia in the tropics (Crous et al. 2004b) again revealed examples of host sharing of Mycosphaearella citri on Citrus, Acacia and Musa. More extensive research should thus be carried out to provide information concerning the fungal diversity of exotic Eucalyptus plantations in Thailand and other Asian countries to promote the understanding of the evolution of new pathogens and the movement of fungi between continents.
Acknowledgments
Dr G. Hunter is thanked for providing assistance with various Pseudocercospora spp. examined during this study. We thank Miss Marjan Vermaas for help in preparing the photographic plates and Arien van Iperen for preparing all the fungal cultures for examination.
REFERENCES
- Arzanlou M, Groenewald JZ, Fullerton RA, Abeln ECA, Carlier J, Zapater M-F, Buddenhagen IW, Viljoen A, Crous PW. 2008. Multiple gene genealogies and phenotypic characters differentiate several novel species of Mycosphaerella and related anamorphs on banana. Persoonia 20: 19 – 37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball JB. 1995. Development of Eucalyptus plantations – an overview. In: White K, Ball J, Kashio M. (eds), Proceedings of the Regional Expert Consultation on Eucalyptus Vol. I (Bangkok, Thailand 4–8 October 1993): 15–27 FAO Regional Office for Asia and the Pacific, Bangkok, Thailand: [Google Scholar]
- Barney K. 2005. At the supply edge: Thailand’s forest policies, plantation sector, and commodity export links with China: China and Forest trade in the Asia-Pacific Region: Implications for forests and livelihoods (25 May 2005). Forest Trends and CIFOR, Washington, DC: [Google Scholar]
- Braun U, Dick MA. 2002. Leaf spot diseases of eucalypts in New Zealand caused by Pseudocercospora species. New Zealand Journal of Forestry Science 32: 221 – 234 [Google Scholar]
- Burgess TI, Barber PA, Sufaati S, Xu D, Hardy GEStJ, Dell B. 2007. Mycosphaerella spp. on Eucalyptus in Asia; new species, new host and new records. Fungal Diversity 24: 135 – 157 [Google Scholar]
- Carbone I, Kohn LM. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553 – 556 [Google Scholar]
- Carnegie AJ. 2007. Forest health condition in New South Wales, Australia, 1996–2005. II. Fungal damage recorded in eucalypt plantations during forest health surveys and their management. Australasian Plant Pathology 36: 225 – 239 [Google Scholar]
- Carnegie AJ, Burgess TI, Beilharz V, Wingfield MJ. 2007. New species of Mycosphaerella from Myrtaceae in plantations and native forests in eastern Australia. Mycologia 99: 461 – 474 [DOI] [PubMed] [Google Scholar]
- Carnegie AJ, Keane PJ. 1994. Further Mycosphaerella species associated with leaf diseases of Eucalyptus. Mycological Research 98: 413 – 418 [Google Scholar]
- Chipompha NWS. 1987. Phaeoseptoria eucalypti: A new pathogen of Eucalyptus in Malawi. South African Forestry Journal 142: 10 – 12 [Google Scholar]
- Cortinas MN, Crous PW, Wingfield BD, Wingfield MJ. 2006. Multilocus gene phylogenies and phenotypic characters distinguish two fungi previously identified as Colletogloeopsis zuluensis causing Eucalyptus cankers. Studies in Mycology 55: 133 – 146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW. 1998. Mycosphaerella spp. and their anamorphs associated with leaf spot diseases of Eucalyptus. Mycologia Memoir 21: 1 – 170 [Google Scholar]
- Crous PW. 2002. Taxonomy and pathology of Cylindrocladium (Calonectria) and allied genera APS Press, Minnesota, USA: [Google Scholar]
- Crous PW, Alfenas AC. 1995. Mycosphaerella gracilis and other species of Mycosphaerella associated with leaf spots of Eucalyptus in Indonesia. Mycologia 87: 121 – 126 [Google Scholar]
- Crous PW, Aptroot A, Kang JC, Braun U, Wingfield MJ. 2000. The genus Mycosphaerella and its anamorphs. Studies in Mycology 45: 107 – 121 [Google Scholar]
- Crous PW, Braun U. 2003. Mycosphaerella and its anamorphs. 1. Names published in Cercospora and Passalora. CBS Biodiversity Series 1: 1 – 571 [Google Scholar]
- Crous PW, Braun U, Groenewald JZ. 2007c. Mycosphaerella is polyphyletic. Studies in Mycology 58: 1 – 32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Ferreira FA, Sutton BC. 1997. A comparison of the fungal genera Phaeophleospora and Kirramyces (coelomycetes). South African Journal of Botany 63: 111 – 115 [Google Scholar]
- Crous PW, Groenewald JZ, Mansilla JP, Hunter GC, Wingfield MJ. 2004a. Phylogenetic reassessment of Mycosphaerella spp. and their anamorphs occurring on Eucalyptus. Studies in Mycology 50: 195 – 214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Groenewald JZ, Pongpanich K, Himaman W, Arzanlou M, Wingfield MJ. 2004b. Cryptic speciation and host specificity among Mycosphaerella spp. occurring on Australian Acacia species grown as exotics in the tropics. Studies in Mycology 50: 457 – 469 [Google Scholar]
- Crous PW, Knox-Davies PS, Wingfield MJ. 1989. A summary of fungal leaf pathogens of Eucalyptus and the diseases they cause in South Africa. South African Forestry Journal 149: 9 – 16 [Google Scholar]
- Crous PW, Mohammed C, Glen M, Verkley GJM, Groenewald JZ. 2007a. Eucalyptus microfungi known from culture. 3. Eucasphaeria and Sympoventuria genera nova, and new species of Furcaspora, Harknessia, Heteroconium and Phacidiella. Fungal Diversity 25: 19 – 36 [Google Scholar]
- Crous PW, Rong IH, Wood A, Lee S, Glen H, Botha W, Slippers B, Beer WZ de, Wingfield MJ, Hawksworth DL. 2006a. How many species of fungi are there at the tip of Africa? Studies in Mycology 55: 13 – 33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Summerell BA, Carnegie A, Mohammed C, Himaman W, Groenewald JZ. 2007b. Foliicolous Mycosphaerella spp. and their anamorphs on Corymbia and Eucalyptus. Fungal Diversity 26: 143 – 185 [Google Scholar]
- Crous PW, Summerell BA, Mostert L, Groenewald JZ. 2008. Host specificity and speciation of Mycosphaerella and Teratosphaeria species associated with leaf spots of Proteaceae. Persoonia 20: 59 – 86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Swart WJ. 1995. Foliicolous fungi of Eucalyptus spp. from eastern Madagascar: Implications for South Africa. South African Forestry Journal 172: 1 – 5 [Google Scholar]
- Crous PW, Verkley GJM, Groenewald JZ. 2006b. Eucalyptus microfungi known from culture. 1. Cladoriella and Fulvoflamma genera nova, with notes on some other poorly known taxa. Studies in Mycology 55: 53 – 63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ. 1996. Species of Mycosphaerella and their anamorphs associated with leaf blotch disease of Eucalyptus in South Africa. Mycologia 88: 441 – 458 [Google Scholar]
- Crous PW, Wingfield MJ. 1997. New species of Mycosphaerella occurring on Eucalyptus leaves in Indonesia and Africa. Canadian Journal of Botany 75: 781 – 790 [Google Scholar]
- Crous PW, Wingfield MJ, Ferreira FA, Alfenas A. 1993. Mycosphaerella parkii and Phyllosticta eucalyptorum, two new species from Eucalyptus leaves in Brazil. Mycological Research 97: 582 – 584 [Google Scholar]
- Crous PW, Wingfield MJ, Mansilla JP, Alfenas AC, Groenewald JZ. 2006c. Phylogenetic reassessment of Mycosphaerella spp. and their anamorphs occurring on Eucalyptus. II. Studies in Mycology 55: 99 – 131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Mohammed C, Yuan ZQ. 1998. New foliar pathogens of Eucalyptus from Australia and Indonesia. Mycological Research 102: 527 – 532 [Google Scholar]
- Gams W, Verkley GJM, Crous PW. 2007. CBS course of mycology, 5th ed Centraalbureau voor Schimmelcultures, Utrecht, Netherlands: [Google Scholar]
- Hillis DM, Bull JJ. 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology 42: 182 – 192 [Google Scholar]
- Hoog GS de, Gerrits van den Ende AHG. 1998. Molecular diagnostics of clinical strains of filamentous basidiomycetes. Mycoses 41: 183 – 189 [DOI] [PubMed] [Google Scholar]
- Hunter GC, Crous PW, Wingfield BD, Pongpanich K, Wingfield MJ. 2006a. Pseudocercospora flavomarinata sp. nov from Eucalyptus leaves in Thailand. Fungal Diversity 22: 71 – 91 [Google Scholar]
- Hunter GC, Roux J, Wingfield BD, Crous PW. 2004. Mycosphaerella species causing leaf diseases in South African Eucalyptus plantations. Mycological Research 108: 672 – 681 [DOI] [PubMed] [Google Scholar]
- Hunter GC, Wingfield BD, Crous PW, Wingfield MJ. 2006b. A multi-gene phylogeny for species of Mycosphaerella occurring on Eucalyptus leaves. Studies in Mycology 55: 147 – 161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SL, Maxwell A, Dell B, Hardy GEStJ. 2005. New disease records of Mycosphaerella leaf disease from eucalypts in western Australia. Australasian Plant Pathology 34: 423 – 424 [Google Scholar]
- Maxwell A, Dell B, Neumeister-Kemp H, Hardy GEStJ. 2003. Mycosphaerella species associated with Eucalyptus in southwestern Australia – new species, new records and a key. Mycological Research 107: 351 – 359 [DOI] [PubMed] [Google Scholar]
- Old KM, Wingfield MJ, Yuan ZQ. 2003. A manual of diseases in Eucalyptus in South East Asia Centre for International Forestry Research (CIFOR), Bogor, Indonesia: [Google Scholar]
- Page RDM. 1996. TREEVIEW: An application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences 12: 357 – 358 [DOI] [PubMed] [Google Scholar]
- Park RF, Keane PJ. 1982. Leaf diseases of Eucalyptus associated with Mycosphaerella species. Transactions of the British Mycological Society 79: 101 – 115 [Google Scholar]
- Park RF, Keane PJ, Wingfield MJ, Crous PW. 2000. Fungal diseases of eucalypt foliage. In: Keane PJ, Kile GA, Podger FD, Brown BN. (eds), Diseases and pathogens of Eucalypts: 153–239 CSIRO Publishing, Australia: [Google Scholar]
- Rambaut A. 2002. Sequence Alignment Editor. Version 2.0 Department of Zoology, University of Oxford, Oxford, United Kingdom: [Google Scholar]
- Rayner RW. 1970. A mycological colour chart CMI and British Mycological Society, Kew, Surrey, England: [Google Scholar]
- Rehner SA, Samuels GJ. 1994. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycological Research 98: 625 – 634 [Google Scholar]
- Sankaran KV, Sutton BC, Minter DW. 1995. Checklist of fungi recorded on Eucalyptus. Mycological Papers 170: 1 – 376 [Google Scholar]
- Slippers B, Smit WA, Crous PW, Coutinho TA, Wingfield BD, Wingfield MJ. 2006. Taxonomy, phylogeny and identification of Botryosphaeriaceae associated with pome and stone fruit trees in South Africa and other regions of the world. Plant Pathology 56: 128 – 139 [Google Scholar]
- Slippers B, Stenlid J, Wingfield MJ. 2005. Emerging pathogens: fungal host jumps following anthropogenic introduction. Trends in Ecology and Evolution 20: 420 – 421 [DOI] [PubMed] [Google Scholar]
- Summerell BA, Groenewald JZ, Carnegie A, Summerbell RC, Crous PW. 2006. Eucalyptus microfungi known from culture. 2. Alysidiella, Fusculina and Phlogicylindrium genera nova, with notes on some other poorly known taxa. Fungal Diversity 23: 323 – 350 [Google Scholar]
- Swofford DL. 2003. PAUP*. Phylogenetic analysis using parsimony (*and their methods). Version 4. Sinauer Associates, Sunderland, Massachusetts: [Google Scholar]
- Turnbull JW. 2000. Economic and social importance of eucalypts. In: Keane PJ, Kile GA, Podger FD, Brown BN. (eds), Diseases and pathogens of Eucalypts: 1–9 CSIRO Publishing, Australia: [Google Scholar]
- Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238 – 4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee J, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (eds), PCR protocols: a guide to methods and applications: 315–322 Academic Press, San Diego, California, USA: [Google Scholar]
- Wingfield MJ, Crous PW, Peredo HL. 1995. Foliar pathogens of Eucalyptus spp. in Chile. South African Forestry Journal 173: 53 – 57 [Google Scholar]