Abstract
PRKAR1A inactivation leads to dysregulated cAMP signaling and Carney complex (CNC) in humans, a syndrome associated with skin, endocrine and other tumors. The CNC phenotype is not easily explained by the ubiquitous cAMP signaling defect; furthermore, Prkar1a+/− mice did not develop skin and other CNC tumors. To identify whether a Prkar1a defect is truly a generic but weak tumorigenic signal that depends on tissue-specific or other factors, we investigated Prkar1a+/− mice when bred within the Rb1+/− or Trp53+/− backgrounds, or treated with a two-step skin carcinogenesis protocol. Prkar1a+/− Trp53+/− mice developed more sarcomas than Trp53+/− mice (P < 0.05) and Prkar1a+/− Rb1+/− mice grew more (and larger) pituitary and thyroid tumors than Rb1+/− mice. All mice with double heterozygosity had significantly reduced life-spans compared with their single-heterozygous counterparts. Prkar1a+/− mice also developed more papillomas than wild-type animals. A whole-genome transcriptome profiling of tumors produced by all three models identified Wnt signaling as the main pathway activated by abnormal cAMP signaling, along with cell cycle abnormalities; all changes were confirmed by qRT–PCR array and immunohistochemistry. siRNA down-regulation of Ctnnb1, E2f1 or Cdk4 inhibited proliferation of human adrenal cells bearing a PRKAR1A-inactivating mutation and Prkar1a+/− mouse embryonic fibroblasts and arrested both cell lines at the G0/G1 phase of the cell cycle. In conclusion, Prkar1a haploinsufficiency is a relatively weak tumorigenic signal that can act synergistically with other tumor suppressor gene defects or chemicals to induce tumors, mostly through Wnt-signaling activation and cell cycle dysregulation, consistent with studies in human neoplasms carrying PRKAR1A defects.
INTRODUCTION
Carney complex (CNC) is a multiple neoplasia syndrome that is inherited in an autosomal dominant manner and is characterized by several types of skin tumors and pigmented lesions, myxomas, schwannomas and endocrine neoplasms (1,2). Inactivating mutations of the PRKAR1A gene coding for the 1-α regulatory (RIα) subunit of protein kinase A (PKA) are responsible for the disease in most CNC patients (3,4). PRKAR1A mutations lead to a net increase in PKA activity and, hence, enhance cAMP signaling (3). Accordingly, Prkar1a haploinsufficiency in mice led to the development of tumors arising in cAMP-responsive tissues, such as the bone, Schwann and thyroid follicular cells (5).
However, Prkar1a+/− mice did not develop some of the most frequent CNC tumors, such as skin and heart myxomas, and pituitary adenomas; in addition, all mouse tumors were observed relatively late, typically after the first 6–9 months of life (5). Thus, tumor formation in Prkar1a+/− mice was not as efficient as in other mouse models of human syndromes leading to multiple neoplasias. One mitigating factor was the degree of increase in cAMP signaling that was observed in mouse tissues (5): generally, Prkar1a+/− mouse tissues did not demonstrate the same level of increased cAMP responsiveness of total PKA signaling seen in human CNC tissues. When a different mouse model was created that led to significantly higher Prkar1a down-regulation and consequently augmented cAMP signaling, more tissues were involved, life-span was decreased and the overall phenotype was more severe (6,7). Accordingly, tissue-specific complete Prkar1a deficiency that led to significant upregulation of PKA signaling in mouse pituitary and heart led to adenomas and myxomas, respectively (8,9).
The Prkar1a mouse model studies not only pointed to the significance of increased cAMP and/or PKA signaling for tumor formation but also suggested that additional factors may be necessary for Prkar1a haploinsufficiency to cause these tumors. Indeed, in vitro studies showed that dysregulation of cyclins and E2F1 were key changes in the process of immortalization of two cell lines, respectively: Prkar1a−/− mouse embryonic fibroblasts (MEFs) (10) and transformed human adrenal PRKAR1A-haploinsufficient cells (11).
Given the role of the p53 (the product of the Trp53 gene) and retinoblastoma (Rb, the product of the Rb1 gene) genes in controlling the cell cycle and the association of Rb with E2F1, we asked the following question: would the tumorigenic properties of Prkar1a haploinsufficiency be augmented or mimic more accurately the CNC phenotype in the background of Trp53 or Rb1 haploinsufficiency? Mouse models such as those for neurofibromatosis type 1 and Peutz-Jeghers syndrome, diseases that are similar to CNC, failed to reproduce exactly or with the same intensity the respective human phenotypes until the mouse gene was knocked out in the Trp53+/− background (12–14), even in the absence of any known involvement of p53 in the human tumors associated with these diseases. In addition, Trp53+/− mice developed bone tumors and bone is a tissue that is frequently affected by Prkar1a haploinsufficiency in mice (5). Rb1+/− mice developed intermediate lobe pituitary adenomas and thyroid C-cell hyperplasia or adenomas at high frequency (15) and mice deficient in both p53 and Rb develop primarily endocrine tumors (16).
And what about skin lesions in CNC and this disease's mouse models? Patients with CNC develop skin pigmented lesions, myxomas, collagenomas and fibromas (1,2); all these lesions, including the melanocytic ones, are always benign (17). As stated earlier, Prkar1a haploinsufficiency or down-regulation by an antisense transcript in mice did not lead to any skin tumors (5–7). There are several transgenic mouse models of skin tumors, but almost all are designed to address questions related to human cutaneous malignancies. A model that produces mostly benign, non-inflammatory, skin lesions (at least in the early phases) is the one that is the result of a two-stage protocol, in which tumor initiation on mouse skin is accomplished through a single topic application of 7,12-dimethylbenz(a)anthracene (DMBA) that causes an irreversible activation of the Hras oncogene in keratinocytes (18,19). Tumor promotion takes place when the initiated cells are expanded due to repeated applications of 12-O-tetradecanoylphorbol-13-acetate (TPA), a proliferation signal that promotes the formation of papillomas (20). Activation of the Hras signaling pathway has been postulated to be implicated in human PRKAR1A haploinsufficiency-induced proliferation (21,22).
Thus, we subjected the skin of Prkar1a+/− mice to DMBA and TPA treatment, an experiment that produced (like the other two described earlier) impressive results: in all three experiments, Prkar1a haploinsufficiency led to a greater number of tumors, and larger and more aggressive lesions. Interestingly, Prkar1a haploinsufficiency did not affect the pattern (the location or histology) of the tumors that these three very different mouse models developed; it only increased their number and/or size.
We then investigated what made these lesions different from those that developed without Prkar1a haploinsufficiency by studying their transcriptome. Beyond expected changes in cell cycle genes, Wnt signaling alterations were shared among all three experiments, despite their very different backgrounds. This finding, along with some recent data from human studies, e.g. the identification of secondary, somatic b-catenin (CTNNB1) mutations in PRKAR1A-haploinsufficient tumors (23,24) point to the central role of Wnt signaling abnormalities in mediating tumorigenesis initiated by Prkar1a haploinsufficiency.
RESULTS
Prkar1a haploinsufficiency increased tumor development in different tissue-specific tumor models
Both the p53 and Rb1 proteins mediate arrest of the cell cycle at late G1. Rb1 functions through sequestration of members of the E2f transcription factor family and prevention of entry into the S phase (25), whereas p53 acts through activation of p21, which directly inhibits the G1 cyclin-dependent kinases that mediate cell cycle progression (26,27). Crosses were made between Trp53+/− mice and Prkar1a+/− mice in the mixed C57BL/6J genetic background and were monitored for subsequent tumor incidence and spectrum. Prkar1a+/− Trp53+/− mice exhibited significantly decreased survival relative to Trp53+/− and Prkar1a+/− mice (P < 0.0001) (Fig. 1A). By 1 year of age, Prkar1a+/− mice bred into the Trp53+/− background developed significantly more sarcomas than Trp53+/− mice (P < 0.001). Prkar1a+/− Trp53+/− mice (n = 30) developed 14 osteosarcomas and 11 splenic hemangiosarcomas, whereas Trp53+/− mice (n = 14) developed 2 splenic hemangiosarcomas, 1 cranial ostema and 1 fibrosarcoma (Fig. 2A). Thyroid neoplasms were also identified in Prkar1a+/− Trp53+/− mice (three follicular thyroid adenomas and one medullary thyroid carcinoma). Interestingly, a medullary thyroid carcinoma emerged from a Prkar1a+/− Trp53+/− mouse.
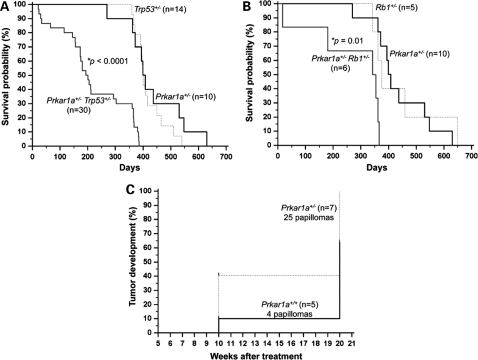
Figure 1.
Prkar1a haploinsufficiency reduced survival in Trp53+/− and RB1+/− mice and increased papilloma development during a two-step skin carcinogenesis protocol. (A) Prkar1a+/− Trp53+/− mice exhibited significantly decreased survival relative to Trp53+/− and Prkar1a+/− mice (P < 0.0001). (B) A reduced survival was also observed in Prkar1a+/− Rb1+/− mice when compared with Rb1+/− and Prkar1a+/− mice (P= 0.01). (C) Prkar1a+/− mice were more susceptible to papilloma formation than WT mice during a 20-week treatment protocol with 7,12-dimethylbenz(a)anthracene (DMBA) and 12-O-tetradecanoylphorbol-13-acetate (TPA) (P =0.004). *P-values were calculated by the log rank test.
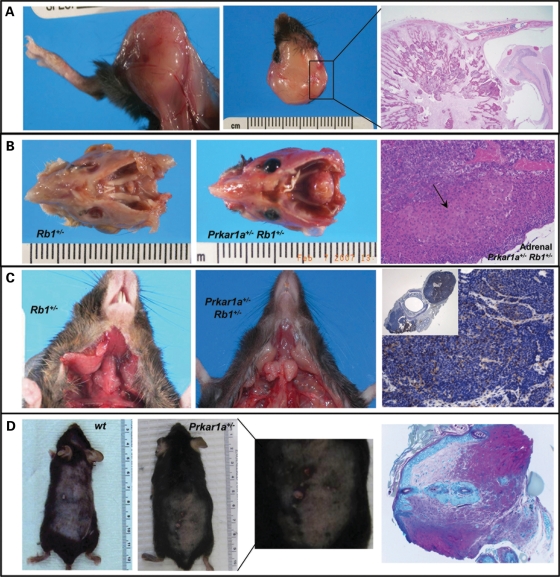
Figure 2.
Prkar1a haploinsufficiency increased tumor development in different tissue-specific tumor models. (A) Gross pathology (hind leg and cranium) and histology (H&E staining 20x) of sarcomas found in Prkar1a+/− Trp53+/− mice. At 1 year, Prkar1a+/− mice bred into the Trp53+/− background developed more sarcomas than Trp53+/− mice. (B) Pituitary tumors were more frequent and larger in Prkar1a+/− Rb1+/− mice than in Rb1+/− mice. A single moderately sized adenoma of the adrenal cortex (arrow) was observed in a Prkar1a+/− Rb1+/− mouse (H&E staining 20x). (C) Thyroid tumors had also a larger size and were more frequently bilateral in Prkar1a+/− Rb1+/− mice than in Rb1+/− mice. Thyroid tumors were positive for calcitonin staining in both Rb1+/− and Prkar1a+/− Rb1+/− groups. (D) Prkar1a+/− mice developed more papillomas than WT mice after the application of a skin carcinogenesis protocol. Papilloma histology (H&E staining 20x).
With the exception of few mild histologic abnormalities (data not shown), Prkar1a+/− mice did not develop any pituitary lesions, consistent with previously published data (5,9,28). Rb1+/− mice developed pituitary and thyroid tumors, as described previously (15). Rb1+/− Prkar1a+/− mice exhibited overall reduced survival when compared with Rb1+/− and Prkar1a+/− mice (P= 0.01) (Fig. 1B). Prkar1a+/− Rb1+/− mice also had a greater number of pituitary tumors and medullary thyroid carcinomas than Rb1+/− mice, and all lesions were larger in the former. Pars intermedia adenomas were identified in 50% of the Prkar1a+/− Rb1+/− mice, whereas only 17% in the Rb1+/− mice (Fig. 2B). Medullary thyroid carcinoma was diagnosed in all but one of the Prkar1a+/− Rb1+/− mice, significantly more than the Rb1+/− mice. In addition, medullary thyroid carcinomas were bilateral in most Prkar1a+/− Rb1+/− mice (data not shown) (Fig. 2C).
Finally, Prkar1a+/− and control mice [wild-type (WT)] from the same litter were treated with the DMBA/TPA carcinogenesis protocol (18–20). It should be noted that different mouse strains have variable susceptibility to this protocol and C57BL/6J mice (of the same background that Prkar1a+/− and WT mice were in this experiment) are relatively resistant to skin tumor induction (29). Prkar1a+/− mice developed more papillomas than WT mice during the 20-week-long treatment protocol (P = 0.004) (Fig. 1C). Prkar1a+/− mice (n = 7) treated with DMBA plus TPA developed a total of 25 papillomas after 20 weeks, whereas only 4 papillomas were observed in WT treated animals (n = 5) (Fig. 2D). Prkar1a+/− and WT mice treated with the vehicle (i.e. acetone) did not develop any skin tumors.
Whole-genome transcriptome profiling
Tumors from the three mouse models were used for microarray analysis using the Illumina Beadarrays® system. All genes were displayed in the heat maps constructed by processing the data using unsupervised hierarchical clustering (Supplementary Material, Fig. S1) (30). The functional analysis of the whole-genome transcriptome profiling was performed using the DAVID Bioinformatic Resources 2008 (NIAID, NIH) (31,32). Multiple pathways are differentially regulated among tumors from single heterozygous and double heterozygous mice, as well as between papillomas from the Prkar1a+/− and WT mice (Table 1 and Supplementary Material, Table S1). The complete data are available at the National Center for Biotechnology Information's Gene Expression Omnibus (geo@ncbi.nlm.nih.gov) and are accessible through accession number GSE19576.
Table 1.
Functional analysis of whole-genome transcriptome profiling in sarcomas, pituitary and thyroid tumors and papillomas
| Pathways | Genes (n) | Enrichment (%) | P-valuea |
|---|---|---|---|
| Sarcomas from Prkar1a+/−Trp53+/−versus Trp53+/− mice | |||
| Cell cycle | 21 | 21 | 0.008 |
| p53 pathway | 26 | 26 | 0.01 |
| mTOR signaling | 28 | 28 | 0.01 |
| Pituitary tumors from Prkar1a+/−Rb1+/− mice versus Rb1+/− mice | |||
| Cell cycle | 93 | 50 | 0.0001 |
| Wnt signaling | 96 | 16 | 0.02 |
| p53 pathway | 49 | 33 | 0.003 |
| mTOR signaling | 41 | 38 | 0.002 |
| Thyroid tumors from Prkar1a+/−Rb1+/− mice versus Rb1+/− mice | |||
| Cell cycle | 79 | 18 | 0.01 |
| Wnt signaling | 103 | 15 | 0.01 |
| p53 pathway | 51 | 28 | 0.005 |
| Papillomas from Prkar1a+/− mice versus WT mice | |||
| Cell cycle | 60 | 40 | 0.001 |
| Wnt signaling | 69 | 20 | 0.04 |
| p53 pathway | 36 | 39 | 0.01 |
| Oxidative phosphorylation | 84 | 63 | 0.00001 |
aArray functional analysis was performed using DAVID Bioinformatics Resources 2008, NIAID/NIH (http://david.abcc.ncifcrf.gov/home.jsp).
Cell cycle, apoptosis and mTOR-dependent pathways were enriched in Prkar1a+/− Trp53+/− sarcomas when compared with Trp53+/− sarcomas (P < 0.05). Pituitary and thyroid tumors from Prkar1a+/− Rb1+/− mice showed an increase in Wnt signaling, cell cycle and apoptosis pathways in relation to Rb1+/− tumors (P < 0.05). Genes related to Wnt signaling, cell cycle, apoptosis and oxidative phosphorylation pathways were over-expressed in Prkar1a+/− papillomas compared with WT papillomas (P < 0.05).
Confirmatory studies on cell cycle and Wnt signaling genes
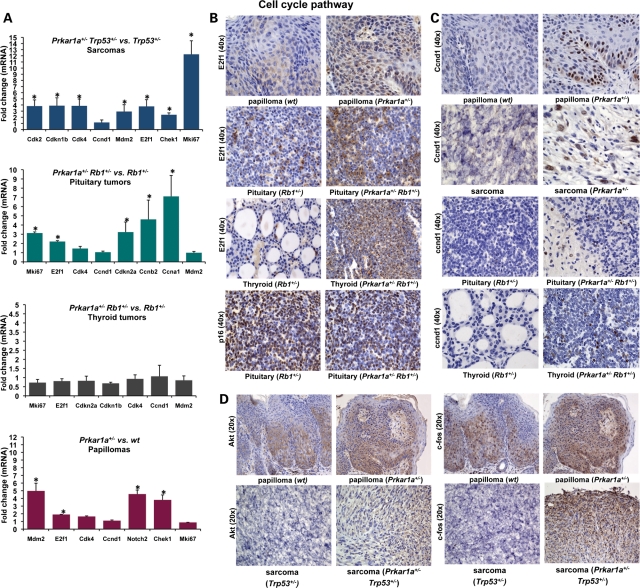
A qRT–PCR array carrying 84 genes of the cell cycle was carried out to confirm the microarray data (Fig. 3A). Key genes (E2f1, Cdk2, Cdk4, Cdknb1, Mdmd2, Chek1 and Mki67) were overexpressed in all Prkar1a+/− Trp53+/− sarcomas when compared with Trp53+/− sarcomas (P < 0.05). Five out of 84 genes (E2f1, Cdkn2a, Ccnb2, Ccna1 and Mki67) were significantly more expressed in pituitary tumors from the Prkar1a+/− Rb1+/− mice, compared with the Rb1+/− mice (P < 0.05). Expression levels of E2f1, Mdm2, Notch2 and Chek1 genes were also higher in Prkar1a+/− papillomas compared with papillomas from WT mice (P < 0.05). A strong staining for E2f1 was demonstrated in Prkar1a+/− papillomas and pituitary and thyroid tumors from Prkar1a+/− Rb1+/− mice (Fig. 3B). Cyclin D1 over-expression at the protein level was also detected in Prkar1a+/− papillomas, Prkar1a+/− Trp53+/− sarcomas, and pituitary and thyroid tumors from Prkar1a+/− Rb1+/− mice (Fig. 3C). Known downstream targets of PKA (Akt and c-fos) were over-expressed in tumors from both the double heterozygous animals and in Prkar1a+/− papillomas (Fig. 3D).
Figure 3.
Cell cycle pathway expression in sarcomas, papillomas, pituitary and thyroid tumors. (A) A qRT–PCR array that included 84 genes involved in the cell cycle demonstrated that key genes were over-expressed in pituitary tumors and sarcomas from double heterozygous mice, as well as in papillomas from Prkar1a+/− mice. (B) Strong staining for E2f1 in Prkar1a+/− papillomas and pituitary and thyroid tumors from Prkar1a+/− Rb1+/− mice. p16 staining was similar in pituitary tumors from Rb1+/− and Prkar1a+/− Rb1+/− mice. (C) Cyclin D1 over-expression at protein level in Prkar1a+/− papillomas, Prkar1a+/− Trp53+/− sarcomas and pituitary and thyroid tumors from Prkar1a+/− Rb1+/− mice−. (D) Downstream targets of PKA (Akt and c-fos) were over-expressed in double heterozygous tumors and in Prkar1a+/− papillomas. *P < 0.05.
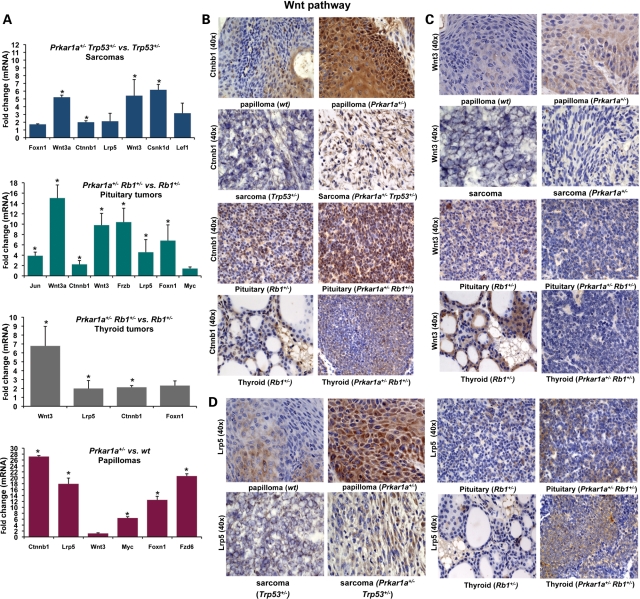
The Wnt signaling pathway was over-expressed in Prkar1a+/− sarcomas, pituitary and thyroid tumors, and papillomas, from the three experiments, respectively. The qRT–PCR array included 84 genes from this pathway (Fig. 4A). The Ctnnb1, Wnt3, Wnt3a and Csnk1d genes were over-expressed in Prkar1a+/−Trp53+/− sarcomas, compared with Trp53+/− sarcomas (P < 0.05). Seven out of 84 genes (Ctnnb1, Wnt3, Wnt3a, Lrp5, Jun, Frzb and Foxn1) were significantly more expressed in pituitary tumors from Prkar1a+/− Rb1+/− mice, compared with Rb1+/− mice (P < 0.05). Expression levels of Ctnnb1, Wnt3, Lrp5 genes were also higher in Prkar1a+/− Rb1+/− thyroid tumors (P < 0.05). In Prkar1a+/− papillomas, five genes (Ctnnb1, Lrp5, Myc, Foxn1 and Fzd6) of the Wnt signaling pathway were found to be over-expressed (P < 0.05). A strong Ctnnb1 protein expression was detected in Prkar1a+/− papillomas, Prkar1a+/− Trp53+/− sarcomas and Prkar1a+/− Rb1+/− pituitary tumors (Fig. 4B). Wnt3 immunoreactivity was clearly higher in Prkar1a+/− papillomas than in WT papillomas (Fig. 4C). The Lrp5 protein was over-expressed in several of the tumors from the two double heterozygote mouse models and in the Prkar1a+/− papillomas (Fig. 4D).
Figure 4.
Wnt pathway expression in sarcomas, papillomas, pituitary and thyroid tumors. (A) Wnt signaling pathway genes were over-expressed in Prkar1a+/− sarcomas, pituitary and thyroid tumors, papillomas from all three experiments, as demonstrated by a qRT–PCR array study that included 84 genes from this pathway. (B) Strong staining for Ctnnb1 in Prkar1a+/− papillomas, Prkar1a+/− Trp53+/− sarcomas and Prkar1a+/− Rb1+/− pituitary tumors. (C) Wnt3 immunoreactivity was higher in Prkar1a+/− papillomas than in WT papillomas. (D) Lrp5 protein was over-expressed in Prkar1a+/− papillomas and double heterozygous tumors. *P < 0.05.
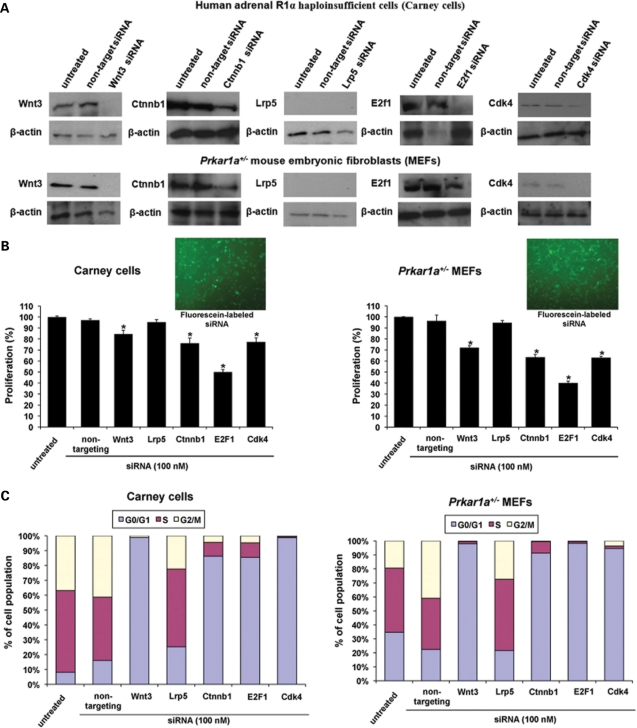
siRNA inhibition of Wnt3, Ctnnb1, E2f1 and Cdk4 decreased proliferation of R1A haploinsufficient human and mouse cells
The Wnt3, Ctnnb1, E2f1 and Cdk4 genes that were significantly altered in all three experiments were selected as targets for confirmation by siRNA studies. A human adrenal cell line bearing a PRKAR1A-inactivating mutation (Carney cells) and Prkar1a+/− MEFs were used for these experiments; a fifth gene that was significantly altered in all three experiments, Lrp5, was not expressed in both cell lines and siRNA treatment for Lrp5 did not have any effect in cell proliferation (to be excluded) (Fig. 5A). On the other hand, siRNA inhibition of Wnt3, Ctnnb1, E2f1 or Cdk4 led to significant decrease in cell proliferation of Carney cells and MEFs (P < 0.05) (Fig. 5B). For example, E2f1 siRNA treatment reduced proliferation by 50 ± 2.3% and 59.9 ± 1.6% when compared with untreated Carney cells and MEFs, respectively.
Figure 5.
siRNA disruption of Wnt3, Ctnnb1, E2f1 or Cdk4 inhibited proliferation of human adrenal cells bearing a PRKAR1A-inactivating mutation (Carney cells) and Prkar1a+/− MEFs. (A) The siRNA effects on the target genes were demonstrated by western blot in Carney cells and Prkar1a+/− MEFs. (B) The treatment with siRNA for Wnt3, Ctnnb1, E2f1 and Cdk4 led to significant decrease in cell proliferation of both cell lines; intra-cellular localization of siRNA in Carney cells and MEFs after transfection with fluorescein-labeled siRNA. (C) siRNA disruption of Wnt3, Ctnnb1, E2f1 and Cdk4 arrested Carney cells and MEFs at the G0/G1 phase of the cell cycle. *P < 0.05.
Flow-cytometric analysis was performed to assess cell cycle distribution of the whole cell population (Fig. 5C). Cell cycle analysis of synchronized Carney cells and MEFs treated with siRNA for Wnt3, Ctnnb1, E2f1 and Cdk4 indicated that both cell lines significantly accumulated in the G0/G1 phase of the cell cycle compared with untreated or non-target siRNA treated cells (P < 0.05).
DISCUSSION
The presented data showed that Prkar1a+/− Trp53+/− and Prkar1a+/− Rb1+/− mice developed more sarcomas and endocrine tumors, respectively, and Prkar1a haploinsufficiency increased susceptibility to skin papillomas induced by DMBA/TPA; Prkar1a haploinsufficiency also reduced overall survival in the Trp53+/− and Rb1+/− backgrounds. This study is to our knowledge the first indication that Prkar1a haploinsufficiency in mice may act as a generic, synergistic, albeit weak, tumorigenic signal. Prkar1a haploinsufficiency appears to be a ‘generic’ tumorigenic signal in the sense that it increased lesions in all three mouse models that were tested, despite the fact that the Trp53+/−, Rb1+/− and DMBA/TPA-induced tumors are quite different in terms of pathways and tissues that are affected. Prkar1a haploinsufficiency appears to also have a synergistic and not an additive effect when combined with Trp53+/− and Rb1+/− defects: the signature lesions of Prkar1a+/− mice, tail and thyroid follicular tumors, and schwannomas (5) were conspicuously not increased in frequency or different in pathology in the Prkar1a+/− Trp53+/− and Prkar1a+/− Rb1+/− mice.
It is also noteworthy, and perhaps indicative of its weakness as a tumorigenic signal, that Prkar1a haploinsufficiency did not alter in any way the tissues or types of cells that are affected by Trp53+/− and Rb1+/− defects or by DMBA/TPA application, despite the almost ubiquitous Prkar1a expression. We selected these mouse models because of the cAMP-responsiveness of the main types of affected cells (sarcomas, thyroid C-cells and pituitary intermediate lobe) or due to the uniqueness of the phenotype and its similarity to CNC (skin papillomas). In all cases, Prkar1a haploinsufficiency simply augmented the previously described phenotype for the respective mouse model, without causing new tumors in other cAMP-responsive (or other) tissues.
R1α haploinsufficiency in human lymphocytes and mouse models causes an increase in total cAMP-stimulated kinase activity and enhances MAPK activity (5,22,33,34). Complete R1α deficiency causes constitutive PKA activation and immortalization of MEFs through up-regulation of cyclin D1; this increase occurred independently of other pathways known to increase cyclin D1 levels (10). The involvement of cyclin D1 in mediating Prkar1a haploinsufficiency signals was confirmed in our studies, along with that of other genes of the cell cycle, that were significantly altered not only in the Rb1+/− and Trp53+/− backgrounds (where comparison was made with the single heterozygotes, Rb1+/− and Trp53+/−, respectively) but also in the DMBA/TPA-induced tumors where no primary cell cycle gene was altered genetically.
Our previous studies of a human cell line harboring a PRKAR1A-inactivating mutation showed that the cell cycle factor E2F1 mediates proliferative effects of defective R1α (11). E2F1 is a downstream effector of Rb and has a pivotal role in controlling cell cycle progression (35,36). Loss of E2f1 in mice reduces the frequency of pituitary and thyroid tumors and lengthens the lifespan of Rb1+/− E2f1−/− mice (37). In the present study, siRNA disruption of E2f1 inhibited proliferation of Carney cells and Prkar1a+/− MEFs and arrested both cell lines at the G0/G1 phase of the cell cycle, preventing entry into and progression through S phase.
The implication of E2F1 in mediating R1α haploinsufficiency signals provides a link to the observed synergy of Prkar1a-haploinsufficiency in the Trp53+/− background and the observed activation of cyclin D1: recent studies suggested that transcriptional activation and repression of E2fs are linked to the p53 tumor suppressor gene (38–40). Specifically, targeted disruption of activator E2fs (E2F1, E2F2 and E2F3) led to p53 activation and sequential induction of p53 target genes, including CDKN1A (38).
The Wnt signaling pathway regulates a wide range of functions such as cell growth and differentiation (41). The functional analysis of array data revealed a significant enrichment of Wnt signaling pathway in Prkar1a+/− Trp53+/− sarcomas, Prkar1a+/− Rb1+/− pituitary and thyroid tumors, as well as in Prkar1a+/− papillomas. Ctnnb1, Wnt3 and Lrp5 over-expression was confirmed by qRT–PCR and immunostaining. In addition, the treatment of PRKAR1A-haploinsufficient cells (Carney cells) and Prkar1a+/− MEFs with siRNAs for Wnt3 and Ctnnb1 blocked the transition to the S phase in both cell lines. cAMP signaling via PKA and its target transcription factor CREB are required for Wnt-directed myogenic gene expression (42). It was previously demonstrated that glycogen synthase kinase (GSK)-3β is a downstream target of cAMP/PKA signaling in steroidogenic cells (43). Interestingly, expression studies in massive macronodular adrenocortical disease, a disorder associated with excess cAMP signaling (44) and primary pigmented nodular adrenocortical disease (PPNAD), a condition mostly associated with PRKAR1A-inactivating mutations (4), indicated the over-expression of genes that regulate or are part of the Wnt signaling pathway such as WISP2, glycogen synthase kinase-3β (GSK3B) and β-catenin (CTNNB1) (45,46). In addition, somatic mutations of the CTNNB1 gene have been found in adrenal tumors from patients with PPNAD, Carney complex and germline PRKAR1A-inactivating mutations (23,24). Recently, a microRNA profile analysis showed that cAMP and/or PKA via microRNA regulation affects the Wnt signaling pathway in both PPNAD (47) and MMAD (48). These studies along with the findings presented here suggest that the Wnt pathway is a primary mediator of tumorigenesis related to increased cAMP and/or PKA signaling and R1α haploinsufficiency, almost regardless of the cellular context in both human and mouse tissues.
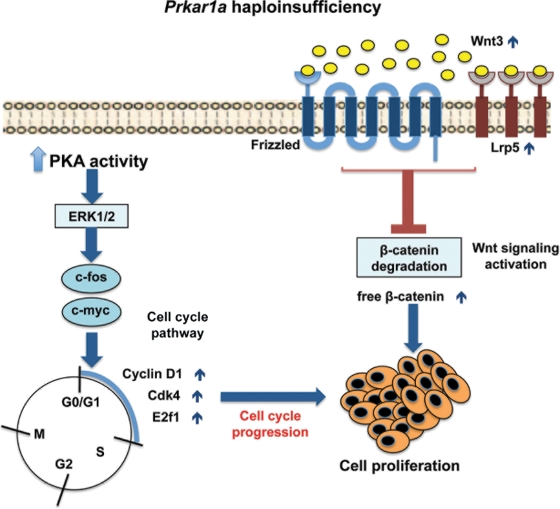
We conclude that Prkar1a haploinsufficiency can act weakly but synergistically with other molecular defects in inducing tumor formation (Fig. 6). Dysregulation of cell cycle genes such as cyclin D1 and E2F1 and abnormal Wnt signaling are essential in these effects.
Figure 6.
Cross-talk between PKA and the Wnt and cell cycle genes in Prkar1a+/− tumors. Prkar1a haploinsufficiency is associated with an increase in the expression of Wnt3, Lrp5 and β-catenin leading to Wnt signaling activation. Additionally, R1α haploinsufficiency in human lymphocytes and mouse models causes an increase in total cAMP-stimulated kinase activity and enhances MAPK activity, which promotes activation of the transcription factors c-myc and c-fos. R1α deficiency also promotes activation of cyclin D1, Cdk4 and E2f1, which facilitates cell cycle progression. PKA, protein kinase A; PRKAR1A, 1-α regulatory (RIα) subunit of PKA.
MATERIALS AND METHODS
Animal procedures
To exclude possible effects of the different genetic backgrounds on molecular signature of tumors, all mice were from a (CD-1 × C57BL/6) F1 hybrid background. Prkar1a+/− mice carrying a deletion of exon 2 have been previously described (5). Trp53+/− mice (C57BL/6; Trp53tm1Tyj) were purchased from Jackson Laboratories (JAX mice and Services, Bar Harbor, ME, USA) and were genotyped as described elsewhere (49). Rb1+/− mice (C57BL/6; Rb1tm1Tyj) were purchased from Jackson Laboratories and were genotyped as previously described (50). Prkar1a+/− mice were crossed with Trp53+/− or Rb+/− mice and the phenotype analyzed at 1 year of age. In the two-step carcinogenesis protocol, WT and Prkar1a+/− mice (both on C57BL/6 background) were treated with a single topical application of 7,12-dimethylbenz(a)anthracene (DMBA) and repeated applications of 12-O-tetradecanoylphorbol-13-acetate (TPA) once per week for 20 weeks (51). All animal work in this study was carried out in accordance with Institutional Laboratory Animal Care and Use Committee guidelines under animals protocol 06-033 (at the NIH, Bethesda, MD, USA).
Microarray analysis and quantitative real-time PCR
Total RNA was extracted from tumor samples using the TRIZOL reagent method. Preparation of cRNA from total RNA, hybridization in Sentrix MouseRef-8 Expression BeadChips, scanning and image analysis was done as previously described (52). Preliminary analysis of Illumina data was performed using Illumina BeadStudio software (Illumina, San Diego, CA, USA), which returns the trimmed mean average intensity for each single gene probe type (non-normalized). Any gene consistently with a P detection value above 0.05 for all samples was eliminated from further analysis. This background filter resulted in the removal of ∼40% of all the genes on the Illumina array. Z-transformation for normalization was performed for each Illumina sample/array (53). Microarray data are in compliance with the Minimal Information About a Microarray Experiment (MIAME) format. The raw and normalized array data have been deposited in National Center for Biotechnology Information's Gene Expression Omnibus http://www.ncbi.nlm.nih.gov/geo and are accessible through Gene Expression Omnibus Series accession number GSE19576. Heatmaps were made using Java Treeview (54).
The functional analysis of the whole-genome transcriptome profiling was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatic Resources 2008 (NIAID, NIH, http://david.abcc.ncifcrf.gov/home.jsp) (31,32). The lists of genes (induced or repressed) were submitted to the DAVID database (http://david. abcc.ncifcrf.gov), which clusterizes genes according to a series of common keywords. The proportion of each keyword in the list is compared with the one in the whole genome, making it possible to compute P-values and enrichment scores (geometric mean of the inverse log of each P-value). The detailed information of gene alterations was systematically reported on KEGG pathways (Supplementary Material, Table S1).
Gene expression data were confirmed by quantitative real-time PCR using specific array plates for cell cycle and Wnt signaling pathways (SABiosciences, Frederick, MD, USA). Relative quantification was performed using the 2−ΔΔCT method (55).
Immunohistochemistry
Tumor tissues were removed from mice and fixed in formalin, processed and paraffin embedded for subsequent H&E staining and immunohistochemistry, as previously described (56). For the immunohistochemical analysis, the following antibodies were used: E2f1 (H-137, Santa Cruz Biotechnology, Santa Cruz, CA, USA), Cncd1 (ab16663, Abcam, Cambridge, MA, USA), p16 (ab54210, Abcam), AKT (H-136, Santa Cruz Biotechnology), c-fos (ab7963-1, Abcam), Wnt3 (ab32249, Abcam), Ctnnb1 (ab6302, Abcam) and Lrp5 (ab38311, Abcam).
siRNA transfections
The human adrenal cells bearing a PRKAR1A-inactivating mutation (Carney cells) and Prkar1a+/− MEFs were previously characterized (10,11). Carney cells and Prkar1a+/− MEFs were transfected with 100 nM ON-TARGETplus Smartpool siRNA (Dharmacon Thermo Scientific, Lafayette, CO, USA) specific to human (WNT3, LRP5, CTNNB1, E2F1 and CDK4) and mouse (WNT3, Wnt3, Ctnnb1, Lrp5, E2f1 and Cdk4), respectively, or ON-TARGETplus non-targeting pool using transfection reagent DharmaFECT1 (Dharmacon) as per manufacturer's instruction.
Immunoblotting
The efficiency of siRNA target gene disruption was confirmed by western blot analysis following standard procedures (57). The following antibodies were used: Wnt3 (ab32249, Abcam), Ctnnb1 (ab6302, Abcam), Lrp5 (ab38311, Abcam), E2f1 (H-137, Santa Cruz Biotechnology), Cdk4 (DCS-35, Santa Cruz Biotechnology) and beta-actin (ab8227, Abcam). Briefly, cells were lysed by homogenization in 20 mm Tris–HCl (pH 7.5), 100 mm NaCl, 5 mm MgCl2, 1% Nonidet P-40, 0.5% sodium deoxycholate and protease inhibitor cocktail I (EMD Biosciences, La Jolla, CA, USA) with subsequent centrifugation at 10 000 rpm for 10 min at 4°C. Equal amounts of protein lysate were subjected to SDS–PAGE, transferred to nitrocellulose membranes and probed with antibodies.
Cell proliferation assay and cell cycle analysis
After 72 h of siRNA transfection, cell proliferation assays were performed on Carney cells and Prkar1a+/− MEFs using the CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega, Madison, WI, USA). Data represent mean ± SE of three separate experiments.
Flow-cytometry analysis was performed to assess cell cycle distribution of the whole cell population. Carney cells and Prkar1a+/− MEFs, synchronized in low-serum (0.5%) medium for 48 h, were released from cell cycle arrest by adding 10% FBS. We measured the cell cycle 48 h after addition of serum. Cells were harvested, fixed with ice-cold 70% ethanol, stained with propidium iodide (10 µg/ml) and ribonuclease A (100 µg/ml) (Sigma, St Louis, MO, USA), and subjected to cell cycle analysis using FACSCalibur (Becton Dickinson, Mountain View, CA, USA). The percentage of aneuploid cells and cell cycle distribution were calculated with ModFit LT cell-cycle analysis software (Verity Software House, Topsham, ME, USA). Data represent mean ± SE of three separate experiments.
Statistical analysis
All statistical analyses were performed with the SPSS 16.0 (SPSS Inc., Chicago, IL, USA). Survival analysis was performed using the log rank test. The X2 test was used to test the number of observed tumors between groups. Continuous data are expressed as mean ± SE. A two-sample t-test was used for statistical analysis of cell proliferation and cell cycle data. A P-value less than 0.05 was considered significant.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by US National Institutes of Health, Eunice Kennedy Shriver National Institute of Child Health and Human Development intramural project Z01-HD-000642-04 (to C.A.S.).
Supplementary Material
REFERENCES
- 1.Carney J.A., Gordon H., Carpenter P.C., Shenoy B.V., Go V.L. The complex of myxomas, spotty pigmentation, and endocrine overactivity. Medicine (Baltimore) 1985;64:270–283. doi: 10.1097/00005792-198507000-00007. [DOI] [PubMed] [Google Scholar]
- 2.Stratakis C.A., Kirschner L.S., Carney J.A. Clinical and molecular features of the Carney complex: diagnostic criteria and recommendations for patient evaluation. J. Clin. Endocrinol. Metab. 2001;86:4041–4046. doi: 10.1210/jcem.86.9.7903. [DOI] [PubMed] [Google Scholar]
- 3.Kirschner L.S., Carney J.A., Pack S.D., Taymans S.E., Giatzakis C., Cho Y.S., Cho-Chung Y.S., Stratakis C.A. Mutations of the gene encoding the protein kinase A type I-alpha regulatory subunit in patients with the Carney complex. Nat. Genet. 2000;26:89–92. doi: 10.1038/79238. [DOI] [PubMed] [Google Scholar]
- 4.Kirschner L.S., Sandrini F., Monbo J., Lin J.P., Carney J.A., Stratakis C.A. Genetic heterogeneity and spectrum of mutations of the PRKAR1A gene in patients with the carney complex. Hum. Mol. Genet. 2000;9:3037–3046. doi: 10.1093/hmg/9.20.3037. [DOI] [PubMed] [Google Scholar]
- 5.Kirschner L.S., Kusewitt D.F., Matyakhina L., Towns W.H., 2nd, Carney J.A., Westphal H., Stratakis C.A. A mouse model for the Carney complex tumor syndrome develops neoplasia in cyclic AMP-responsive tissues. Cancer Res. 2005;65:4506–4514. doi: 10.1158/0008-5472.CAN-05-0580. [DOI] [PubMed] [Google Scholar]
- 6.Griffin K.J., Kirschner L.S., Matyakhina L., Stergiopoulos S., Robinson-White A., Lenherr S., Weinberg F.D., Claflin E., Meoli E., Cho-Chung Y.S., et al. Down-regulation of regulatory subunit type 1A of protein kinase A leads to endocrine and other tumors. Cancer Res. 2004;64:8811–8815. doi: 10.1158/0008-5472.CAN-04-3620. [DOI] [PubMed] [Google Scholar]
- 7.Griffin K.J., Kirschner L.S., Matyakhina L., Stergiopoulos S.G., Robinson-White A., Lenherr S.M., Weinberg F.D., Claflin E.S., Batista D., Bourdeau I., et al. A transgenic mouse bearing an antisense construct of regulatory subunit type 1A of protein kinase A develops endocrine and other tumours: comparison with Carney complex and other PRKAR1A induced lesions. J. Med. Genet. 2004;41:923–931. doi: 10.1136/jmg.2004.028043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin Z., Jones G.N., Towns W.H., 2nd, Zhang X., Abel E.D., Binkley P.F., Jarjoura D., Kirschner L.S. Heart-specific ablation of Prkar1a causes failure of heart development and myxomagenesis. Circulation. 2008;117:1414–1422. doi: 10.1161/CIRCULATIONAHA.107.759233. [DOI] [PubMed] [Google Scholar]
- 9.Yin Z., Williams-Simons L., Parlow A.F., Asa S., Kirschner L.S. Pituitary-specific knockout of the Carney complex gene Prkar1a leads to pituitary tumorigenesis. Mol. Endocrinol. 2008;22:380–387. doi: 10.1210/me.2006-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nadella K.S., Kirschner L.S. Disruption of protein kinase a regulation causes immortalization and dysregulation of D-type cyclins. Cancer Res. 2005;65:10307–10315. doi: 10.1158/0008-5472.CAN-05-3183. [DOI] [PubMed] [Google Scholar]
- 11.Nesterova M., Bossis I., Wen F., Horvath A., Matyakhina L., Stratakis C.A. An immortalized human cell line bearing a PRKAR1A-inactivating mutation: effects of overexpression of the wild-type allele and other protein kinase A subunits. J. Clin. Endocrinol. Metab. 2008;93:565–571. doi: 10.1210/jc.2007-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cichowski K., Shih T.S., Schmitt E., Santiago S., Reilly K., McLaughlin M.E., Bronson R.T., Jacks T. Mouse models of tumor development in neurofibromatosis type 1. Science. 1999;286:2172–2176. doi: 10.1126/science.286.5447.2172. [DOI] [PubMed] [Google Scholar]
- 13.Vogel K.S., Klesse L.J., Velasco-Miguel S., Meyers K., Rushing E.J., Parada L.F. Mouse tumor model for neurofibromatosis type 1. Science. 1999;286:2176–2179. doi: 10.1126/science.286.5447.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takeda H., Miyoshi H., Kojima Y., Oshima M., Taketo M.M. Accelerated onsets of gastric hamartomas and hepatic adenomas/carcinomas in Lkb1+/–p53−/− compound mutant mice. Oncogene. 2006;25:1816–1820. doi: 10.1038/sj.onc.1209207. [DOI] [PubMed] [Google Scholar]
- 15.Jacks T., Fazeli A., Schmitt E.M., Bronson R.T., Goodell M.A., Weinberg R.A. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- 16.Harvey M., Vogel H., Lee E.Y., Bradley A., Donehower L.A. Mice deficient in both p53 and Rb develop tumors primarily of endocrine origin. Cancer Res. 1995;55:1146–1151. [PubMed] [Google Scholar]
- 17.Zembowicz A., Knoepp S.M., Bei T., Stergiopoulos S., Eng C., Mihm M.C., Stratakis C.A. Loss of expression of protein kinase a regulatory subunit 1alpha in pigmented epithelioid melanocytoma but not in melanoma or other melanocytic lesions. Am. J. Surg. Pathol. 2007;31:1764–1775. doi: 10.1097/PAS.0b013e318057faa7. [DOI] [PubMed] [Google Scholar]
- 18.Hennings H., Shores R., Wenk M.L., Spangler E.F., Tarone R., Yuspa S.H. Malignant conversion of mouse skin tumours is increased by tumour initiators and unaffected by tumour promoters. Nature. 1983;304:67–69. doi: 10.1038/304067a0. [DOI] [PubMed] [Google Scholar]
- 19.Roop D.R., Lowy D.R., Tambourin P.E., Strickland J., Harper J.R., Balaschak M., Spangler E.F., Yuspa S.H. An activated Harvey ras oncogene produces benign tumours on mouse epidermal tissue. Nature. 1986;323:822–824. doi: 10.1038/323822a0. [DOI] [PubMed] [Google Scholar]
- 20.Hawley-Nelson P., Stanley J.R., Schmidt J., Gullino M., Yuspa S.H. The tumor promoter, 12-O-tetradecanoylphorbol-13-acetate accelerates keratinocyte differentiation and stimulates growth of an unidentified cell type in cultured human epidermis. Exp. Cell Res. 1982;137:155–167. doi: 10.1016/0014-4827(82)90017-9. [DOI] [PubMed] [Google Scholar]
- 21.Robinson-White A., Hundley T.R., Shiferaw M., Bertherat J., Sandrini F., Stratakis C.A. Protein kinase-A activity in PRKAR1A-mutant cells, and regulation of mitogen-activated protein kinases ERK1/2. Hum. Mol. Genet. 2003;12:1475–1484. doi: 10.1093/hmg/ddg160. [DOI] [PubMed] [Google Scholar]
- 22.Robinson-White A.J., Leitner W.W., Aleem E., Kaldis P., Bossis I., Stratakis C.A. PRKAR1A inactivation leads to increased proliferation and decreased apoptosis in human B lymphocytes. Cancer Res. 2006;66:10603–10612. doi: 10.1158/0008-5472.CAN-06-2200. [DOI] [PubMed] [Google Scholar]
- 23.Tadjine M., Lampron A., Ouadi L., Horvath A., Stratakis C.A., Bourdeau I. Detection of somatic beta-catenin mutations in primary pigmented nodular adrenocortical disease (PPNAD) Clin. Endocrinol. (Oxf.) 2008;69:367–373. doi: 10.1111/j.1365-2265.2008.03273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaujoux S., Tissier F., Groussin L., Libe R., Ragazzon B., Launay P., Audebourg A., Dousset B., Bertagna X., Bertherat J. Wnt/beta-catenin and 3′,5'-cyclic adenosine 5'-monophosphate/protein kinase A signaling pathways alterations and somatic beta-catenin gene mutations in the progression of adrenocortical tumors. J. Clin. Endocrinol. Metab. 2008;93:4135–4140. doi: 10.1210/jc.2008-0631. [DOI] [PubMed] [Google Scholar]
- 25.Nevins J.R. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 26.el-Deiry W.S., Tokino T., Velculescu V.E., Levy D.B., Parsons R., Trent J.M., Lin D., Mercer W.E., Kinzler K.W., Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 27.Harper J.W., Adami G.R., Wei N., Keyomarsi K., Elledge S.J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 28.Yin Z., Williams-Simons L., Rawahneh L., Asa S., Kirschner L.S. Development of a pituitary-specific cre line targeted to the Pit-1 lineage. Genesis. 2008;46:37–42. doi: 10.1002/dvg.20362. [DOI] [PubMed] [Google Scholar]
- 29.Sundberg J.P., Sundberg B.A., Beamer W.G. Comparison of chemical carcinogen skin tumor induction efficacy in inbred, mutant, and hybrid strains of mice: morphologic variations of induced tumors and absence of a papillomavirus cocarcinogen. Mol. Carcinog. 1997;20:19–32. doi: 10.1002/(sici)1098-2744(199709)20:1<19::aid-mc4>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 30.Eisen M.B., Spellman P.T., Brown P.O., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang D.W., Sherman B.T., Tan Q., Kir J., Liu D., Bryant D., Guo Y., Stephens R., Baseler M.W., Lane H.C., et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 33.Robinson-White A., Meoli E., Stergiopoulos S., Horvath A., Boikos S., Bossis I., Stratakis C.A. PRKAR1A Mutations and protein kinase A interactions with other signaling pathways in the adrenal cortex. J. Clin. Endocrinol. Metab. 2006;91:2380–2388. doi: 10.1210/jc.2006-0188. [DOI] [PubMed] [Google Scholar]
- 34.Meoli E., Bossis I., Cazabat L., Mavrakis M., Horvath A., Stergiopoulos S., Shiferaw M.L., Fumey G., Perlemoine K., Muchow M., et al. Protein kinase A effects of an expressed PRKAR1A mutation associated with aggressive tumors. Cancer Res. 2008;68:3133–3141. doi: 10.1158/0008-5472.CAN-08-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson D.G., Schwarz J.K., Cress W.D., Nevins J.R. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature. 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- 36.Qin X.Q., Livingston D.M., Kaelin W.G., Jr, Adams P.D. Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc. Natl Acad. Sci. USA. 1994;91:10918–10922. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamasaki L., Bronson R., Williams B.O., Dyson N.J., Harlow E., Jacks T. Loss of E2F-1 reduces tumorigenesis and extends the lifespan of Rb1(+/−)mice. Nat. Genet. 1998;18:360–364. doi: 10.1038/ng0498-360. [DOI] [PubMed] [Google Scholar]
- 38.Timmers C., Sharma N., Opavsky R., Maiti B., Wu L., Wu J., Orringer D., Trikha P., Saavedra H.I., Leone G. E2f1, E2f2, and E2f3 control E2F target expression and cellular proliferation via a p53-dependent negative feedback loop. Mol. Cell. Biol. 2007;27:65–78. doi: 10.1128/MCB.02147-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma N., Timmers C., Trikha P., Saavedra H.I., Obery A., Leone G. Control of the p53-p21CIP1 Axis by E2f1, E2f2, and E2f3 is essential for G1/S progression and cellular transformation. J. Biol. Chem. 2006;281:36124–36131. doi: 10.1074/jbc.M604152200. [DOI] [PubMed] [Google Scholar]
- 40.Wu L., Timmers C., Maiti B., Saavedra H.I., Sang L., Chong G.T., Nuckolls F., Giangrande P., Wright F.A., Field S.J., et al. The E2F1-3 transcription factors are essential for cellular proliferation. Nature. 2001;414:457–462. doi: 10.1038/35106593. [DOI] [PubMed] [Google Scholar]
- 41.Conacci-Sorrell M., Zhurinsky J., Ben-Ze'ev A. The cadherin-catenin adhesion system in signaling and cancer. J. Clin. Invest. 2002;109:987–991. doi: 10.1172/JCI15429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen A.E., Ginty D.D., Fan C.M. Protein kinase A signalling via CREB controls myogenesis induced by Wnt proteins. Nature. 2005;433:317–322. doi: 10.1038/nature03126. [DOI] [PubMed] [Google Scholar]
- 43.Roy L., McDonald C.A., Jiang C., Maroni D., Zeleznik A.J., Wyatt T.A., Hou X., Davis J.S. Convergence of 3',5'-cyclic adenosine 5'-monophosphate/protein kinase A and glycogen synthase kinase-3beta/beta-catenin signaling in corpus luteum progesterone synthesis. Endocrinology. 2009;150:5036–5045. doi: 10.1210/en.2009-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsiao H.P., Kirschner L.S., Bourdeau I., Keil M.F., Boikos S.A., Verma S., Robinson-White A.J., Nesterova M., Lacroix A., Stratakis C.A. Clinical and genetic heterogeneity, overlap with other tumor syndromes, and atypical glucocorticoid hormone secretion in adrenocorticotropin-independent macronodular adrenal hyperplasia compared with other adrenocortical tumors. J. Clin. Endocrinol. Metab. 2009;94:2930–2937. doi: 10.1210/jc.2009-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horvath A., Mathyakina L., Vong Q., Baxendale V., Pang A.L., Chan W.Y., Stratakis C.A. Serial analysis of gene expression in adrenocortical hyperplasia caused by a germline PRKAR1A mutation. J. Clin. Endocrinol. Metab. 2006;91:584–596. doi: 10.1210/jc.2005-1301. [DOI] [PubMed] [Google Scholar]
- 46.Bourdeau I., Antonini S.R., Lacroix A., Kirschner L.S., Matyakhina L., Lorang D., Libutti S.K., Stratakis C.A. Gene array analysis of macronodular adrenal hyperplasia confirms clinical heterogeneity and identifies several candidate genes as molecular mediators. Oncogene. 2004;23:1575–1585. doi: 10.1038/sj.onc.1207277. [DOI] [PubMed] [Google Scholar]
- 47.Iliopoulos D., Bimpaki E.I., Nesterova M., Stratakis C.A. MicroRNA signature of primary pigmented nodular adrenocortical disease: clinical correlations and regulation of Wnt signaling. Cancer Res. 2009;69:3278–3282. doi: 10.1158/0008-5472.CAN-09-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bimpaki E.I., Iliopoulos D., Moraitis A., Stratakis C.A. MicroRNA signature in massive macronodular adrenocortical disease and implications for adrenocortical tumorigenesis. Clin. Endocrinol. (Oxf.) 2009 doi: 10.1111/j.1365-2265.2009.03725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jacks T., Remington L., Williams B.O., Schmitt E.M., Halachmi S., Bronson R.T., Weinberg R.A. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 50.Keramaris E., Ruzhynsky V.A., Callaghan S.M., Wong E., Davis R.J., Flavell R., Slack R.S., Park D.S. Required roles of Bax and JNKs in central and peripheral nervous system death of retinoblastoma-deficient mice. J. Biol. Chem. 2008;283:405–415. doi: 10.1074/jbc.M701552200. [DOI] [PubMed] [Google Scholar]
- 51.Rutberg S.E., Lee E.J., Hansen L.H., Glick A.B., Yuspa S.H. Identification of differentially expressed genes in chemically induced skin tumors. Mol. Carcinog. 1997;20:88–98. [PubMed] [Google Scholar]
- 52.Cheadle C., Nesterova M., Watkins T., Barnes K.C., Hall J.C., Rosen A., Becker K.G., Cho-Chung Y.S. Regulatory subunits of PKA define an axis of cellular proliferation/differentiation in ovarian cancer cells. BMC Med. Genomics. 2008;1:43. doi: 10.1186/1755-8794-1-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheadle C., Vawter M.P., Freed W.J., Becker K.G. Analysis of microarray data using Z score transformation. J. Mol. Diagn. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saldanha A.J. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 55.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 56.Shi S.R., Key M.E., Kalra K.L. Antigen retrieval in formalin-fixed, paraffin-embedded tissues: an enhancement method for immunohistochemical staining based on microwave oven heating of tissue sections. J. Histochem. Cytochem. 1991;39:741–748. doi: 10.1177/39.6.1709656. [DOI] [PubMed] [Google Scholar]
- 57.Neary C.L., Nesterova M., Cho Y.S., Cheadle C., Becker K.G., Cho-Chung Y.S. Protein kinase A isozyme switching: eliciting differential cAMP signaling and tumor reversion. Oncogene. 2004;23:8847–8856. doi: 10.1038/sj.onc.1208165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.