Abstract
Evaluation of transcriptional changes in the striatum may be an effective approach to understanding the natural history of changes in expression contributing to the pathogenesis of Huntington disease (HD). We have performed genome-wide expression profiling of the YAC128 transgenic mouse model of HD at 12 and 24 months of age using two platforms in parallel: Affymetrix and Illumina. The data from these two powerful platforms were integrated to create a combined rank list, thereby revealing the identity of additional genes that proved to be differentially expressed between YAC128 and control mice. Using this approach, we identified 13 genes to be differentially expressed between YAC128 and controls which were validated by quantitative real-time PCR in independent cohorts of animals. In addition, we analyzed additional time points relevant to disease pathology: 3, 6 and 9 months of age. Here we present data showing the evolution of changes in the expression of selected genes: Wt1, Pcdh20 and Actn2 RNA levels change as early as 3 months of age, whereas Gsg1l, Sfmbt2, Acy3, Polr2a and Ppp1r9a RNA expression levels are affected later, at 12 and 24 months of age. We also analyzed the expression of these 13 genes in human HD and control brain, thereby revealing changes in SLC45A3, PCDH20, ACTN2, DDAH1 and PPP1R9A RNA expression. Further study of these genes may unravel novel pathways contributing to HD pathogenesis. DDBJ/EMBL/GenBank accession no: GSE19677
INTRODUCTION
Huntington disease (HD) is a neurological disorder characterized by early selective neuronal cell death in striatum and cortex caused by mutations in the human HTT gene (1). Expansion of the CAG tract in exon 1 of the HTT gene leads to an abnormally long polyglutamine tract in huntingtin, a protein ubiquitously expressed throughout the body and brain. Neuropathological features are observed in the neostriatum, globus pallidus and cerebral cortex with a selective loss of the large pyramidal neurons in cortical layers III, IV and VI (2,3).
Evidence derived from cellular and animal models of HD has identified a number of potential mechanisms by which polyglutamine-expanded huntingtin protein results in neuronal cell death. These include disruption of the axonal transport, mitochondrial dysfunction, imbalance in calcium homeostasis and excitotoxicity, altered proteolysis, impairment of the ubiquitin proteosome system and transcriptional alterations (4,5). Specific changes in mRNA expression have been described in human postmortem brains and in cellular and mouse models of HD. Down-regulation of dopamine D1 and D2 receptors, enkephalin, substance P (6–8) and several striatal-enriched genes have been found altered in postmortem caudate samples from HD cases using quantitative real-time PCR analysis (qPCR) (9). Microarray studies performed on human HD brain samples (grade 0–2) have demonstrated that the caudate nucleus manifests the greatest number and magnitude of transcriptional changes, followed by motor cortex and then cerebellum, with no significant change detectable in prefrontal association cortex (10).
There is concordance of striatal-enriched genes differentially expressed in HD mouse models with gene expression changes identified in human microarray studies (11). For example, several of the genes showing the greatest magnitude of changes, such as the cannabinoid CB1 receptor, dopamine D1 receptor and enkephalin genes, have also been identified as differentially expressed in expression profiling studies of transgenic mice (10–12). Also, a considerable number of Ca2+-binding and Ca2+, K+ and Na+ channel mRNAs are down-regulated in humans in concordance with previous observations in the R6/2 transgenic mouse model (13–15).
The YAC128 mouse model of HD expresses full-length human mutant huntingtin with 128 glutamines from a yeast artificial chromosome (YAC) (16,17). This mouse model expresses full-length huntingtin under the endogenous human huntingtin promoter and spans both up- and downstream regulatory regions. YAC128 mice display disease progression over time with both neuropathological and behavioral deficits similar to human disease. Initially, YAC128 mice exhibit behavioral changes with a hyperkinetic phenotype and rotarod deficits at 3 months of age (18). YAC128 mice display several neuropathological changes characteristic of HD with a significant decrease in brain weight and striatal volume observed at 8 months (19), and a selective neuronal loss in striatum and cortex at 12 months of age (18). The fact that YAC128 transgenic mice display early onset cognitive and motor deficits with selective neuropathology recapitulating key features of human disease makes this mouse model useful for studies into disease pathogenesis.
The majority of genome-wide expression studies performed including studies on different HD mouse models have employed the Affymetrix Gene chip platform when studying gene expression changes. More recent technologies, such as the Illumina Bead array platform, are now also widely used for this kind of analysis. Affymetrix arrays are produced by in situ synthesis of 25-mer oligonucleotides with multiple probes for each gene. The Affymetrix arrays contain one-base mismatch probes as controls for cross-hybridization. In contrast, the Illumina Bead arrays are based on self-assembly of 50-mer-long oligonucleotides to microbeads. Bead arrays contain 30 copies of the same oligonucleotide as an internal technical replication, which Affymetrix arrays lack. We employed both platforms to increase the power of the gene expression profile analysis of the YAC128 and controls with the ultimate goal of studying transcriptional alterations in striatum and identifying genes of importance for disease pathogenesis.
In this study, we present evidence for both early and late transcriptional changes of genes in the striatum that we initially identified by genome-wide expression profiling of the YAC128 mouse model of HD using the Affymetrix and Illumina platforms. As our experiments were designed to analyze both platforms in parallel as a validation tool, we first analyzed the same striatal mRNA separately across both platforms. We identified modest transcriptional changes on the individual platforms. This prompted us to an alternative approach, to combine the results from the Affymetrix and Illumina platforms to create combined rank lists. We show that combining data sets obtained from two powerful independent platforms revealed the identity of additional genes that were differentially expressed between YAC128 and control mice at 12 and 24 months of age. To further validate our results, we analyzed the top 10 genes ranked from the Illumina, Affymetrix and the combined rank lists, respectively, by applying the more sensitive qPCR methodology. For genes validated to be differentially expressed, we also analyzed earlier time points: 3-, 6- and 9-month-old YAC128 mice and controls.
In summary, we have applied an unbiased approach to combine results from two powerful expression array platforms that identified a set of genes that showed a differential regulation between YAC128 and controls. In total, we evaluated expression of 13 genes at time points relevant to disease pathology. This analysis revealed several genes Wt1, Pcdh20 and Actn2 that display transcriptional changes already at 3 months of age, early in the manifestation of the HD phenotype of these mice. The expression of additional genes, Gsg1l, Sfmbt2, Acy3, Polr2a and Ppp1r9a, was found to be altered only at later time points. We examined the expression of these genes in human HD caudate and controls and demonstrated transcriptional changes in SLC45A3, PCDH20, ACTN2, DDAH1 and PPP1R9A, validating the relevance of these findings in mouse to the human disease. Further study of these genes might elucidate novel disease mechanisms of importance in HD.
RESULTS
Genome-wide expression profiling of 24-month-old YAC128 mice and controls using Affymetrix and Illumina platforms
We initially compared the transcriptional changes in the striatum of 24-month-old YAC128 and wild-type littermate control mice. YAC128 mice display a clear progression of disease with age that correlates with the degree of selective neuronal loss and neurodegeneration. There is an established correlation between levels of mutant huntingtin, disease severity and neuronal loss in YAC128 mice (20). We studied the gene expression profiles of the same mRNA using two different platforms: Illumina and Affymetrix (Supplementary Material, Fig. S1). YAC128 mice showed both an up- (light yellow) and down-regulation (dark brown) of genes compared with controls. However, no genes were significantly different on either platform after correction for multiple comparisons (21).
Calculating combined ranks from the Affymetrix and Illumina profiles for 24-month-old YAC128 and controls
Gene expression studies using Affymetrix GeneChips and Illumina Bead arrays have previously been compared side by side with a high agreement for genes that were predicted to be differentially expressed (22). We therefore examined our genome-wide expression data to determine whether genes had similar ranks on both platforms, taking into account that ∼48 000 probe sets are analyzed on the Illumina arrays and 45 000 probe sets are measured on the Affymetrix counterpart. In order to focus on the very best candidate changes, we only considered the data for the top 100 genes on each platform (ranked by probability of differential expression). We found both concordant as well as discordant predicted changes in expression between the two platforms. Discordant predictions may be due in part to low expression levels of some genes or the fact that the specific probes employed in the two platforms assay different splice variants or transcripts of a gene (23). While our experiments were designed to analyze both platforms in parallel as a validation tool, the modest transcriptional changes observed with both platforms prompted us to attempt an alternative approach to identify differentially expressed genes in YAC128 mice compared with controls. We subsequently used the two data sets to create a combined rank list. We calculated a combined rank for each gene by averaging the individual ranks generated from the Affymetrix and Illumina platforms (Supplementary Material, Table S1). This approach incorporates a confirmatory identification of differentially expressed genes. Furthermore, this approach helped to elute genes of interest despite small sample sizes and low magnitude of transcriptional changes in our experimental setting. However, by implementing this approach, we down-weighed false-positive changes, i.e. genes that were ranked high on one platform, but not reproduced on the other platform, would tend to be down-weighed.
Transcriptional changes identified in the top 10 genes from Affymetrix and Illumina genome-wide expression profiling
We next decided to validate the separate findings from the Illumina and Affymetrix results using qPCR. We assessed the top 10 differentially expressed genes based on the Illumina and Affymetrix results, respectively, on mRNAs from a new cohort of 24-month-old YAC128 and control mice (Supplementary Material, Fig. S1). This analysis resulted in confirmation of three genes out of 10 from the Illumina results to be differentially expressed, i.e. Wilm's tumor 1 homolog (Wt1), DAZ interacting protein 1-like (Dzip1l) and Retrotransposon gag domain containing 4 (Rgag4), where Wt1 was up-regulated, and both Dzip1l and Rgag4 were down-regulated in YAC128 compared with controls (Supplementary Material, Fig. S2; Table 1). Netrin G1 (Ntng1) showed a trend for up-regulation in the YAC128 mice compared with controls that was not statistically significant. The mRNAs for the RAB21 member of the RAS oncogene family (Rab21), Docking protein 3 (Dok3), Dopa decarboxylase (Ddc) and up-regulated during skeletal muscle growth 5 (Usmg5) were not found to be differentially expressed between YAC128 and controls in the qPCR analyses (Supplementary Material, Fig. S2). Three different primer sets were tested for olfactory receptor 1502 (Olfr1502) and fibronectin type III domain containing 8 (Fndc8), but we were unable to amplify and quantify these genes from striatal RNA.
Table 1.
Summary of validated genes in YAC128 and control mice using qPCR
| 24 months | 24 months | 24 months | 12 months |
|---|---|---|---|
| Illumina | Affymetrix | Combined rank | Combined rank |
| Wt1 | Wt1 | Wt1 | Mllt10 |
| Ntng1 | Ddit4l | Ntng1 | Surf4 |
| Rab21 | DMPK | Tob2 | Actn2 |
| Dzip1l | Gsg1l | Pcdh20 | Foxj3 |
| Dok3 | Dleu7 | Sfmbt2 | Tnpo1 |
| Olfr1502 | Spata5 | Pogk | Ddah1 |
| Fndc8 | Entpd7 | Acy3 | N4bp1 |
| Rgag4 | Grasp | Hrbl | Wfdc1 |
| Ddc | Slc45a3 | Polr2a | Zfp371 |
| Usmg5 | Rtel1 | Ddit4l | Ppp1r9a |
| 3/10 | 4/10 | 6/10 | 3/10 |
The top 10 genes from the Affymetrix, Illumina and the combined rank lists for 12 and 24 months were validated by qPCR in independent cohorts of YAC128 and control mice. Genes in bold were differentially expressed between YAC128 and controls. We created combined rank lists for the 12- and 24-month data by averaging the individual ranks generated from the Affymetrix and Illumina results for each target.
Similar results obtained with four genes out of 10 detected as differentially expressed based on the Affymetrix results confirmed that differentially expressed RNAs comprised Wt1, DNA damage inducible transcript 4-like (Ddit4l), germ cell-specific gene 1-like protein (Gsg1l) and solute carrier family 45, member 3 (Slc45a3; Supplementary Material, Fig. S3; Table 1). In addition to Wt1, which was top ranked in both the Illumina and Affymetrix rank results, Slc45a3 was also up-regulated, whereas Ddit4l and Gsg1l were down-regulated in YAC128 compared with control mice. DMPK also showed a non-statistically significant trend towards down-regulation in YAC128 compared with controls. Deleted in lymphocytic leukemia, 7 (Dleu7), spermatogenesis associated 5 (Spata5), ectonucleoside triphosphate diphosphohydrolase 7 (Entpd7), GRP1-associated scaffold protein (Grasp) and regulator of telomere elongation helicase 1 (Rtel1) were not differentially expressed in YAC128 compared with controls (Supplementary Material, Fig. S3).
Identification of additional genes from the combined rank list validated as differentially expressed in YAC128
We next assessed the differential expression of the genes from the created combined rank list for 24-month-old YAC128 and control striatal mRNA samples. The qPCR analysis of the top 10 genes from the combined rank list showed differential expression in YAC128 compared with controls for six out of the 10 genes analyzed (Table 1). Wt1, Ntng1 and Ddit4l were included in the top 10 for both Affymetrix and/or Illumina and in the created combined rank list. Additional genes from the combined rank list that were shown to be differentially expressed were: Protocadherin 20 (Pcdh20), Scm-like with four MBT domains protein 2 (Sfmbt2), Aspartoacylase 3 (Acy3) and Polymerase (RNA) II (DNA directed) polypeptide A (Polr2a) (Supplementary Material, Fig. S4). All of these genes were significantly up-regulated in YAC128 compared with controls. There was no significant difference in gene expression for Transducer of ERBB2 (Tob2), Pogo transposable element with KRAB domain (Pogk) or HIV-1 Rev binding protein-like (Hrbl) between YAC128 and controls at 24 months of age (Table 1; Supplementary Material, Fig. S4).
Transcriptional alterations revealed using the combined rank strategy on 12-month-old YAC128 and controls
We next studied the YAC128 mice compared with controls at the earlier 12-month time point. Both neuropathological and behavioral abnormalities are established in the transgenic mice at this time point. YAC128 mice display approximately 10–15% striatal atrophy and 9% neuronal loss at 12 months of age (24). We used the same approach as for the 24-month-old mice to calculate a combined rank by averaging the ranks obtained from the individual platforms (Supplementary Material, Table S2). We again performed qPCR analyses of the top 10 genes from the established combined rank list using a new cohort of 12-month-old YAC128 and control mice. Three out of 10 genes were validated to be differentially expressed: α-actinin 2 (Actn2), dimethylarginine dimethylaminohydrolase 1 (Ddah1) and protein phosphatase 1, regulatory (inhibitor) subunit 9A (Ppp1r9a) (Fig. S5; Table 1). Interestingly, all three genes were down-regulated in YAC128 at 12 months of age. Myeloid/lymphoid leukemia 10 (Mllt10), Surfeit 4 gene (Surf4), Forkhead box J3 (Foxj3), Transportin 1 (Tnpo1), Nedd4-binding protein 1 (N4bp1) and WAP for-disulfide core domain protein 1 precursor (Wfdc1) were not differentially expressed between YAC128 and controls (Supplementary Material, Fig. S5). Three different primer sets were tested for Zinc finger protein 371 (Zfp371), but we were unable to amplify and quantify the expression of this gene from mouse striatal RNA.
Time course analysis of differentially expressed genes show both early and late changes in gene expression
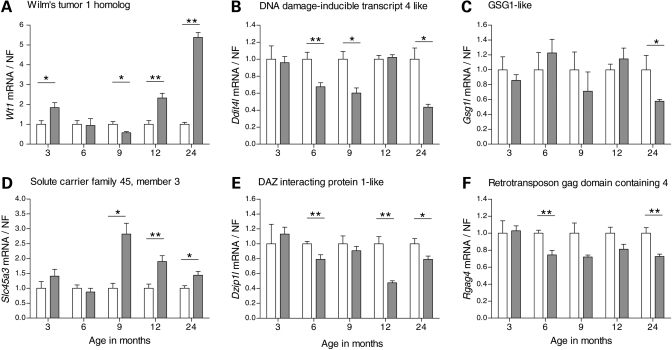
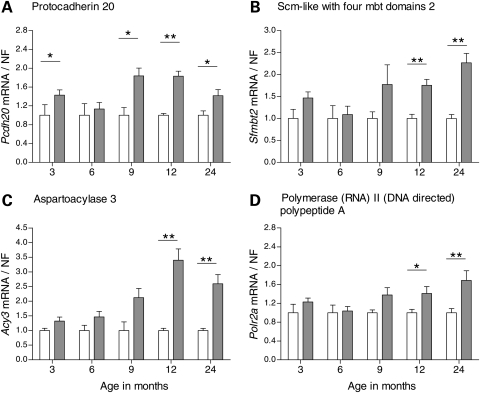
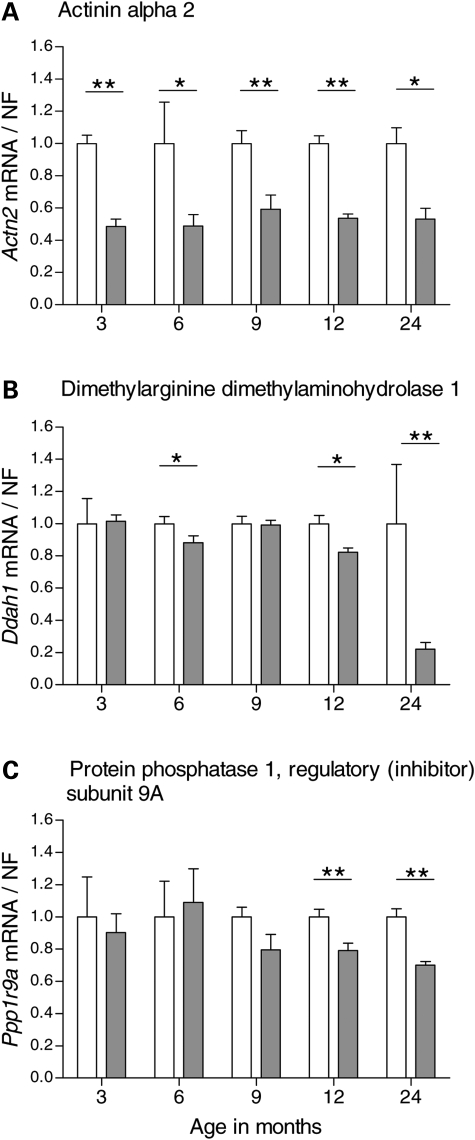
To further characterize the change in expression of the genes that were identified as differentially expressed at 12 or 24 months, we studied additional time points of relevance to disease pathogenesis with regard to both early onset of behavioral abnormalities and neuropathological changes. Taken together, we analyzed the gene expression at 3, 6, 9, 12 and 24 months of age on mRNA from new cohorts of YAC128 and controls using qPCR for each target. Wt1 displayed dynamic transcriptional changes over all five time points analyzed. Initially, YAC128 mice showed an up-regulation of Wt1 at 3 months of age (P = 0.029, Mann–Whitney two-tailed U-test) compared with controls (Fig. 1A). This, however, reverted to similar transcript levels expressed in YAC128 and control mice at 6 months. YAC128 displayed a down-regulation of Wt1 at 9 months of age (P = 0.017), before a distinct 2- and 5-fold up-regulation was observed in YAC128 compared with controls at 12 (P = 0.0025) and 24 months (P = 0.0012), respectively. YAC128 mice showed a continuous down-regulation of Ddit4l (Fig. 1B) from 6 to 24 months of age, except for at 12 months when YAC128 and controls displayed similar transcript levels (P = 0.0079 in 6 months; P = 0.030 in 9 months; P = 0.014 in 24 months YAC128 versus controls). Gsg1l was only down-regulated in YAC128 at 24 months of age (P = 0.018; Fig. 1C). In contrast, Slc45a3 was up-regulated almost 3-fold in the YAC128 mice compared with controls at 9 months of age and continuously up-regulated at later time points (P = 0.029 in 9 months; P = 0.0087 in 12 months; P = 0.015 in 24 months YAC128 versus controls; Fig. 1D). Transcriptional expression profiles with down-regulation in YAC128 compared with controls were observed for Dzip1l (P = 0.0055 in 6 months; P = 0.0012 in 12 months; P = 0.022 in 24 months YAC128 versus controls; Fig. 1E) and Rgag4 (P = 0.0010 in 6 months; P = 0.0082 in 24 months YAC128 versus controls; Fig. 1F). For the genes obtained from the combined ranking lists, YAC128 mice showed a significant up-regulation of Pcdh20 at all time points, except for at 6 months (P = 0.018 at 3 months; P = 0.017 at 9 months; P = 0.0012 at 12 months; P = 0.035 at 24 months YAC128 versus controls; Fig. 2A). Sfmbt2, Acy3 and Polr2a showed up-regulation at the two late-stage time points in YAC128 compared with controls; Sfmbt2 (P = 0.0012 for both 12 and 24 months YAC128 versus controls; Fig. 2B); Acy3 (P = 0.0025 for 12 months; P = 0.0012 for 24 months YAC128 versus controls; Fig. 2C); Polr2a (P = 0.035 for 12 months; P = 0.0047 for 24 months YAC128 versus controls; Fig. 2D). In addition, there were trends toward differential expression for several RNA targets at individual time points even though statistical significance was not reached. Acy3 (P = 0.055; Fig. 2C) and Polr2a (P = 0.051; Fig. 2D) showed borderline significance for up-regulation in 9-month-old YAC128 compared with controls. Several of the analyzed target genes displayed a significant difference in transcriptional expression at the earlier time points. Actn2 was down-regulated in YAC128 as early as 3 months and consistently until 24 months of age (P = 0.0040 in 3 months; P = 0.036 in 6 months; P = 0.0087 in 9 months; P = 0.0012 in 12 months; P = 0.014 in 24-month-old YAC128 versus controls; Fig. 3A). This was the only gene that showed a significant differential expression at every time point analyzed. Ddah1 showed a 5-fold down-regulation in the YAC128 compared with controls at 24 months in addition to changes observed at earlier time points (P = 0.025 for 6 months; P = 0.022 for 12 months; P = 0.0020 for 24-month-old YAC128 versus controls; Fig. 3B). Ppp1r9a showed a down-regulation in YAC128 compared with controls at the two late-stage time points (P = 0.0047 for 12 months; P = 0.0012 for 24 months YAC128 versus controls; Fig. 3C). In general, the Affymetrix platform gave a better estimate than Illumina, of the magnitude of fold change based on our qPCR results (Table 2).
Figure 1.
Time course analysis of differentially expressed genes identified on the Affymetrix and Illumina platforms. We analyzed the top 10 genes from the Affymetrix and Illumina results. YAC128 and wild-type littermate controls were analyzed at 3, 6, 9, 12 and 24 months of age by qPCR. We analyzed Wilm's tumor 1 homolog (Wt1), DNA damage-inducible transcript 4 like (Ddit4l), germ cell-specific gene 1-like protein (Gsg1l) and solute carrier family 45, member 3 (Slc45a3). YAC128 showed an up-regulation of Wt1 transcript levels at 3 months (P = 0.029), 12 months (P = 0.0025) and 24 months (P = 0.0012) and a down-regulation at 9 months (P = 0.0173) compared with controls (A). Ddit4l was down-regulated in YAC128 compared with controls at 6 months (P = 0.0079), 9 months (P = 0.030) and 24 months (P = 0.014) (B). Gsg1l was only down-regulated in YAC128 at 24 months of age compared with controls (P = 0.018) (C). Slc45a3 showed up-regulation at 9 months (P = 0.029), 12 months (P = 0.0087) and 24 months (P = 0.0152) in YAC128 compared with controls (D). In addition to Wt1, DAZ interacting protein 1-like (Dzip1l) and retrotransposon gag domain containing 4 (Rgag4) were identified on the Illumina platform as differentially expressed at 24 months of age. YAC128 showed down-regulation of Dzip1l at 6 months (P = 0.0055), 12 months (P = 0.0012) and 24 months (P = 0.0221) compared with controls (E). Rgag4 was down-regulated in YAC128 compared with controls at 6 months (P = 0.0010) and 24 months (P = 0.0082) (F). YAC128 data were normalized to the calculated average for the wild-type controls for each individual target and time point. The bars show the mean ± SEM for each target. Statistical analysis was performed using Mann–Whitney two-tailed U-test; *P < 0.05; **P < 0.01; ***P < 0.001. White and grey bars indicate wild-type controls and YAC128, respectively.
Figure 2.
Time course analysis of differentially expressed genes identified using the combined rank list for 24-month-old YAC128 and controls. We analyzed the top 10 genes from the established combined rank list. YAC128 and wild-type littermate controls were analyzed at 3, 6, 9, 12 and 24 months of age by qPCR. In addition to Wt1 and Ddit4l, we analyzed Protocadherin 20 (Pcdh20), Scm-like with four mbt domains 2 (Sfmbt2), aspartoacylase 3 (Acy3) and polymerase (RNA) II (DNA directed) polypeptide A (Polr2a) that were included in the top 10 genes of the combined rank list for 24-month-old mice. Pcdh20 was up-regulated in YAC128 compared with controls at 3 months (P = 0.0186), 9 months (P = 0.0173), 12 months (P = 0.0012) and 24 months (P = 0.035) (A). Sfmbt2 was up-regulated in YAC128 compared with controls at 12 months (P = 0.0012) and 24 months (P = 0.0012) (B). Acy3 was up-regulated at 12 months (P = 0.0025) and 24 months (P = 0.0012) (C). There was a tendency for increase of Acy3 transcript levels also at 9 months (P = 0.056). Polr2a transcript levels were increased in YAC128 at 12 months (P = 0.035) and 24 months (P = 0.0047) (D). YAC128 data were normalized to the calculated average for the wild-type controls for each individual target and time point. The bars show the mean ± SEM for each target. Statistical analysis was performed using Mann–Whitney two-tailed U-test; *P < 0.05; **P < 0.01; ***P < 0.001. White and grey bars indicate wild-type controls and YAC128, respectively.
Figure 3.
Time course analysis of differentially expressed genes identified using the combined rank list for 12-month-old YAC128 and controls. We analyzed the top 10 genes from the established combined rank list. YAC128 and wild-type littermate controls were analyzed at 3, 6, 9, 12 and 24 months of age by qPCR. We analyzed actinin alpha 2 (Actn2), dimethylarginine dimethylaminohydrolase 1 (Ddah1) and protein phosphotase 1, regulatory (inhibitor) subunit 9A (Ppp1r9a). Actn2 was down-regulated in YAC128 compared with controls at all five time points; at 3 months (P = 0.0040), 6 months (P = 0.036), 9 months (P = 0.0087), 12 months (P = 0.0012) and 24 months (P = 0.014) (A). Ddah1 transcript levels were lower in YAC128 compared with controls at 6 months (P = 0.021), 12 months (P = 0.022) and 24 months (P = 0.0012) (B). Ppp1r9a was down-regulated in YAC128 compared with controls at 12 months (P = 0.0047) and 24 months (P = 0.0012) (C). YAC128 data were normalized to the calculated average for the wild-type controls for each individual target and time point. The bars show the mean ± SEM for each target. Statistical analysis was performed using Mann–Whitney two-tailed U-test; *P < 0.05; **P < 0.01; ***P < 0.001. White and grey bars indicate wild-type controls and YAC128, respectively.
Table 2.
Summary of expressional profiling and transcriptional alterations in YAC128 and HD cases
| Gene symbol | YAC128 Affy P-value | YAC128 Affy FCa | YAC128 Illumina P-value | YAC128 Illumina FCa | YAC128 qPCRb | qPCR FCa,c | Human Affy P-valued | Human Affy FCd | Human qPCRe ↑↓ | YAC128 versus human HD concordance |
|---|---|---|---|---|---|---|---|---|---|---|
| Wt1f | 2.1E−06 | 0.76 | 1.5E−04 | 0.20 | ↑3,12,24 ↓9 | 2.4 | 1.2E−03 | −0.12 | P = 0.08 | – |
| Ddit4lf | 2.9E−06 | −1.2 | 3.3E−02 | −0.39 | ↓6,9,24 | −1.2 | 7.0E−03 | −0.23 | P = 0.66 | – |
| Gsg1lf | 1.5E−05 | −0.53 | 2.8 E−01 | −0.029 | ↓24 | −0.79 | 4.0E−03 | −0.098 | ↓P = 0.052 | Yes |
| Scl45a3f | 1.0E−04 | 1.1 | 9.3E−02 | 0.13 | ↑9,12,24 | 0.53 | 1.1E−02 | 0.17 | ↑P = 0.004 | Yes |
| Dzip1lf | 5.8E−01 | 0.13 | 5.4E−04 | −0.053 | ↓6,12,24 | −0.34 | 3.6E−02 | 0.083 | P = 0.13 | – |
| Rgag4f | 3.7E−01 | −0.30 | 1.1E−03 | −0.16 | ↓6,24 | −0.46 | NA | NA | ND | – |
| Pcdh20f | 3.0E−04 | 1.1 | 8.5E−03 | 0.11 | ↑3,9,12,24 | 0.50 | 2.5E−07 | −0.71 | ↓P = 0.004 | No |
| Sfmbt2f | 3.5E−03 | 0.84 | 1.4E−03 | 0.062 | ↑12,24 | 1.2 | 6.4E−01 | −0.02 | P = 0.08 | – |
| Acy3f | 4.8E−03 | 0.96 | 1.1E−03 | 0.25 | ↑12,24 | 1.4 | 6.2E−01 | −0.02 | P = 0.33 | – |
| Polr2af | 1.8E−03 | 0.53 | 2.5E−02 | 0.18 | ↑12,24 | 0.75 | 5.8E−01 | 0.03 | P = 0.13 | – |
| Actn2g | 1.0E−02 | −0.69 | 2.8E−04 | −0.50 | ↓3,6,9,12,24 | −0.90 | 5.4E−11 | −0.78 | ↓P = 0.017 | Yes |
| Ddah1g | 1.6E−02 | −0.29 | 6.3E−01 | −0.058 | ↓6,12,24 | −0.28 | 2.2E−06 | 0.45 | ↑P = 0.017 | No |
| Ppp1r9ag | 1.8E−02 | −1.4 | 1.0E−02 | −0.26 | ↓12,24 | −0.34 | 8.1E−09 | −0.66 | ↓P = 0.017 | Yes |
ND, not determined; NA, not assigned.
aFold change (FC) is shown as log2 transformed values. Positive values indicate up-regulation in YAC128 compared to controls; Negative values indicate down-regulation in YAC128 compared to controls.
bArrows indicate up or down-regulation of significant transcriptional changes in YAC128 compared to controls at different ages indicated in months.
cFold change indicated shows the magnitude of transcriptional change (log2 transformed) observed with qPCR for 24 month old YAC128 compared to control mice and 12 month old mice for Actn2, Ddah1 and Ppp1r9a.
dHuman HD gene expression data has been previously published in Hodges et al. 2006 (10). Fold change (FC) is shown as log2 transformed values.
eArrows indicate up- or down-regulation of significant transcriptional changes in HD caudate samples compared to controls.
fP-values and fold change data (FC) for Affymetrix and Illumina results are from data set comparing YAC128 and controls at 24 months of age.
gP-values and fold change data (FC) for Affymetrix and Illumina results are from data set comparing YAC128 and controls at 12 months of age.
Transcriptional alterations in HD cases are concordant with changes observed in YAC128
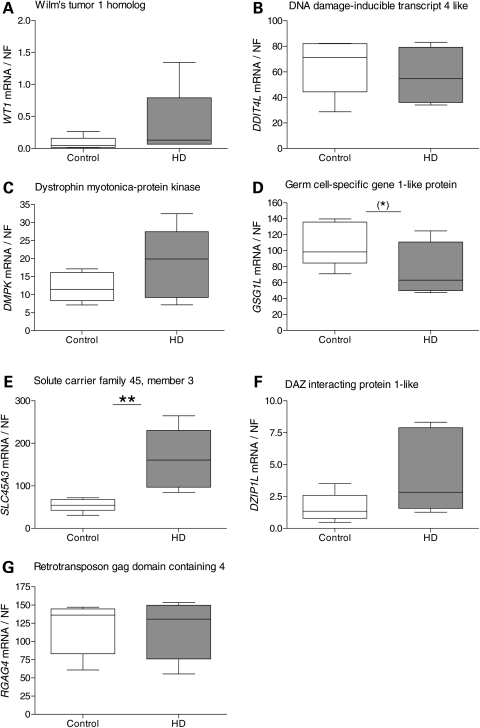
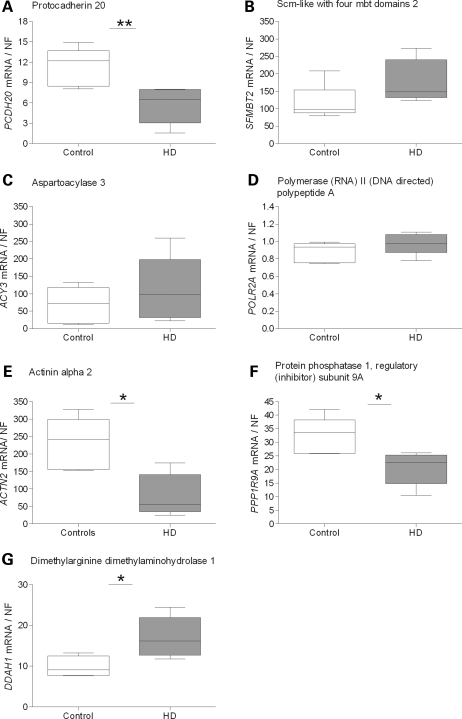
We next studied the genes that were differentially expressed in mouse striatum in caudate samples from human HD and control brains. In total, we analyzed 14 genes that were identified to be differentially expressed on the individual platforms, Affymetrix and Illumina, and the combined rank lists for 12-month and 24-month data, respectively. WT1 showed a tendency for up-regulation in HD caudate, but did not reach statistical significance (P = 0.082, Mann–Whitney two-tailed U-test; Fig. 4A). This tendency to up-regulation was concordant to observations in YAC128 compared with controls at 3, 12 and 24 months of age (Table 2). We decided to analyze DMPK expression in human HD cases, since DMPK showed borderline significance for down-regulation in 24-month-old YAC128 mice compared with controls. However, in our analysis, DMPK showed no transcriptional changes in human HD cases (Fig. 4C). GSG1L showed down-regulation in HD caudate with borderline significance (P = 0.051; Fig. 4D). The gene expression profile was concordant with the YAC128 profile at 24 months of age. SLC45A3 was highly up-regulated in HD caudate compared with controls (P = 0.0043; Fig. 4E), concordant with the expression profile in YAC128 mice compared with controls at 9, 12 and 24 months of age. DDIT4L (Fig. 4B), DZIP1L (Fig. 4F) and RGAG4 (Fig. 4G) did not show any significant changes in HD caudate versus controls. We also studied the additional genes that were identified as differentially expressed from the combined rank lists for the 12- and 24-month YAC128 data sets. PCDH20 was down-regulated in HD caudate compared with controls (P = 0.0043; Fig. 5A). This expression profile was discordant to the changes observed in the YAC128 and controls. SFMTB2, ACY3 and POLR2A (Fig. 5B–D) did not show any significant transcriptional changes between HD cases and controls. All three genes analyzed from the combined rank list created from the 12-month data and validated to be differentially expressed in YAC128 and controls also showed transcriptional changes in human HD caudate samples: ACTN2 (P = 0.017; Fig. 5E), PPP1R9A (P = 0.017; Fig. 5F) and DDAH1 (P = 0.017; Fig. 5G). ACTN2 and PPP1R9A showed down-regulation, whereas DDAH1 showed up-regulation in HD case caudate compared with controls. Gene expression profiles for ACTN2 and PPP1R9A were both concordant with observations in the YAC128 data, in contrast to DDAH1 which was up-regulated in HD caudate and down-regulated in YAC128 at 6, 12 and 24 months compared with controls.
Figure 4.
Transcriptional alterations identified in human HD caudate compared with controls. We analyzed the genes identified as differentially expressed in the YAC128 transgenic model when validating the results from the individual platforms: Affymetrix and Illumina. We quantified WT1, DDIT4L, DMPK, GSG1L, SLC45A3, DZIP1L and RGAG4 in human HD caudate (n = 6) and controls (n = 5) by qPCR. WT1 (A), DDIT4L (B) and DZIP1L (F) did not show differential expression, although showing trends for up-regulation of WT1 and DZIP1L in HD cases compared with controls. GSG1L showed down-regulation in HD caudate with borderline significance (P = 0.051) (D), whereas SLC45A3 was up-regulated in HD cases compared with controls (P = 0.0043) (E). RGAG4 did not show any differential expression between HD cases and controls (G). The middle line of the Box and Whisker plot shows the median, the top and bottom lines show the 75th and 25th percentile, respectively. The top and bottom Whiskers indicate the largest and smallest values. Statistical analysis was performed using Mann–Whitney two-tailed U-test; *P < 0.05; **P < 0.01; ***P < 0.001.
Figure 5.
Transcriptional alterations identified in human HD caudate compared with controls. We analyzed the genes identified as differentially expressed in the YAC128 transgenic mice when validating results from the established 12- and 24-month combined rank lists. In addition to WT1 and DDIT4L, we analyzed PCDH20, SFMBT2, ACY3 and POLR2A mRNA expression in human caudate of HD cases and controls by qPCR. These genes were initially identified to be differentially expressed in the YAC128 compared with controls when validating genes from the combined rank list for 24-month-old mice. PCDH20 was down-regulated in HD cases compared with controls (P = 0.0043) (A). SFMBT2 (B), ACY3 (C) and POLR2A (D) were not differentially expressed in HD cases compared with controls, although SFMBT2 and ACY3 showed tendencies for up-regulation in HD cases compared with controls. We also analyzed ACTN2, DDAH1 and PPP1R9A mRNA expression in human caudate of HD cases and controls. These genes were initially identified to be differentially expressed in the YAC128 compared with controls when validating genes from the combined rank list for 12-month-old mice. All three genes showed transcriptional changes. ACTN2 (E) and PPP1R9A (F) were both down-regulated in HD cases compared with controls (P = 0.017), whereas DDAH1 (G) was up-regulated in HD cases compared with controls (P = 0.017). The middle line of the Box and Whisker plot shows the median, the top and bottom lines show the 75th and 25th percentile, respectively. The top and bottom Whiskers indicate the largest and smallest values. Statistical analysis was performed using Mann–Whitney two-tailed U-test; *P < 0.05; **P < 0.01; ***P < 0.001.
DISCUSSION
We performed genome-wide expression profiling on striatal tissue from YAC128 and controls at 12 and 24 months of age. We utilized two powerful platforms in parallel, Illumina and Affymetrix arrays, with the ultimate goal to study transcriptional changes in striatum and to identify genes of importance for disease pathogenesis. We identified 13 genes that were differentially expressed in YAC128 compared with controls, as confirmed in independent groups of YAC128 samples. We also used qPCR to further assess time-dependent alterations in the expression of these genes with respect to the onset and progression of disease phenotype. Most importantly, we studied these genes in HD cases and we showed transcriptional alterations consistent with the findings in YAC128. Transcriptional changes identified may derive from a variety of different mechanisms involved in disease and may therefore lead to the identification of crucial pathways involved in HD pathogenesis.
It was first shown that long polyglutamine stretches activate transcription in vitro (25,26) and many transcription factors contain glutamine-rich activation domains such as CREB-binding protein (CBP), TATA-box binding protein (TBP) and specificity protein 1 (Sp1). Accordingly, mutant huntingtin protein has been suggested to act directly as a transcription factor by mimicking or interfering with the actions of transcription factors that contain glutamine-rich activation domains (27). More recent studies showed that huntingtin protein, both wild-type and mutant, has the ability to bind DNA without additional transcription factors (28). Mutant huntingtin, however, increased the transcriptional binding factor activity overall. In addition, mutant huntingtin displayed increased occupancy at gene promoters in vivo compared with wild-type huntingtin. Mutant huntingtin is thus suggested to alter the transcriptional profile through modulation of the DNA confirmation and altered binding of transcriptional factors (28).
Several studies indicate that mutant huntingtin interacts with sequesters and/or compromises the normal function of transcription factors such as Sp1 (29,30), the nuclear receptor co-repressor (N-CoR) (31), CBP (32,33), p53 (34,35), TBP (36) and TAFII130 (37) or disrupts the core transcriptional machinery by interacting with the pre-initiation complex (38). Furthermore, studies have provided evidence that huntingtin may modulate chromatin structure by interfering with the acetylated and deacetylated states of histones, which led to the testing of HDAC inhibitors for therapeutic intervention in HD. It has also been suggested that intranuclear inclusions may non-specifically alter gene expression by reducing the association of transcription factors to DNA binding sites (39), although this was shown not to be the case in the R6/2 mouse model of HD (40).
Previously, Kuhn et al. (12) performed a meta-analysis on seven different mouse models of HD and human postmortem caudate. In this study, it was concluded that short N-terminal mouse models exhibit rapid effects and transcriptional changes similar to what is observed in human brain. Nevertheless, knock-in (CHL2Q150/Q150, HdhQ92/Q92) and the YAC128 transgenic full-length huntingtin mouse models also displayed significant HD-like transcript profiles at an older age. No distinct transcriptional changes could be assigned to the differences in expression of full-length huntingtin and N-terminal huntingtin fragments (12). Polyglutamine diseases share many features including a polyQ-dependent neurodegeneration and inclusion body pathology (41,42). Transcriptional alterations observed could thus be caused by the polyglutamine expansion alone and be independent of the huntingtin protein context (15). Moreover, the transcriptional changes observed may be due to secondary bystander effects from affected tissue as well as surrounding tissue, and/or changes not related to disease. Regardless, transcriptional dysregulation may be an important mechanism in HD pathogenesis. Several genome-wide expression studies have been performed and it has been difficult to extrapolate what changes are relevant to disease pathogenesis. We validated that Wt1, Ddit4l, Gsg1l, Slc45a3, Dzip1l and Rgag4, which were initially identified on either the Affymetrix or Illumina platform, were differentially expressed. Remarkably, by combining the results from the Affymetrix and Illumina arrays we were able to pinpoint additional genes that were differentially expressed. These genes were Pcdh20, Sfmbt2, Acy3, Polr2a, Ddah1, Actn2 and Pppr1r9a. This is partly due to the down-weighting of false-positive results (reduction of Type I error) when combining the two data sets, but more notably, this approach helped in eluting genes of interest despite small sample sizes and low magnitude of transcriptional changes in our experimental setting.
In our study, we showed transcriptional changes for selected genes in our YAC128 mouse model of HD already at 3 months of age. This indicates that the changes observed in Wt1, Pcdh20 and Actn2 might play an important role in disease pathogenesis, since neuropathological changes are not manifested at this early time point. Interestingly, Wt1 displayed transcriptional changes with some distinct fluctuations observed throughout the time points measured, whereas Actn2 was consistently down-regulated in YAC128 compared with controls. In contrast, the alterations in Ppp1r9a, which show late transcriptional changes from 12 months on, may reflect a secondary response to neuronal loss and additional bystander effects from both affected as well as adjacent brain regions.
We studied the gene expression of several genes in HD cases and controls and showed transcriptional changes in SLC45A3, PCDH20, ACTN2, DDAH1 and PPP1R9A. The genes showing a differential transcriptional expression profile in YAC128 and controls, but not reaching statistical significance in human samples, might still be involved in HD pathogenesis. In our study, discrepancies in transcriptional expression profiles between the YAC128 mouse and human HD case data might be due to biological inter-variability in the human samples. The HD grades analyzed ranged from 1 to 3 in the human samples, which might reduce the power to detect differences due to sample variability. In addition, differences in the postmortem interval might have an effect on the expression of some genes together with general biological variability in the samples analyzed (43,44).
Several of the genes we have identified to be differentially expressed are suggested to be involved in RNA regulation, i.e. Wt1, Dzip1l, Sfmbt2 and Polr2a. The gene encoding fused in sarcoma (FUS) was recently linked to amyotrophic lateral sclerosis (ALS) and shown to be involved in RNA regulation (45,46). This discovery is an example of neurodegenerative disease caused by a mutation in a gene that is predominantly involved in DNA repair, and regulation of RNA transcription, splicing and transport.
Wilm's tumor 1 homolog (Wt1) plays an essential role in the normal development of the urogenital system, and the human gene is mutated in a small subset of patients with Wilm's tumors (47). Wt1 belongs to a family of zinc-finger transcription factors for which α-actinin 1 (Actn1) has been identified as an mRNA target (48).The WT1(+KTS) isoform is suggested to bind close to or at the start codon of Actn1 mRNA (48). More recently, it was shown that WT1 associated with transcripts encoding actin-binding proteins and other cytoskeletal proteins including α-actinins and Ppp1r9a (49). We performed a sequence comparison of the region around the transcriptional start site for murine Actn1 and Actn2 (data not shown). There was ∼70% homology between these regions, suggesting Wt1 could potentially act as a transcription factor involved in Actn2 regulation. Additional experiments are required to study putative interactions and to confirm whether Wt1 directly regulates Actn2 and Ppp1r9a. Wt1 mRNA was in low abundance compared with Actn2 in the striatum of YAC128 and control mice. We observed a significant reduction of Actn2 mRNA levels at all time points measured compared with levels at 3 months in both YAC128 and controls (data not shown). Apart from this observation, there is no obvious correlation in the expression profiles of Wt1 and Actn2 in the YAC128 or control mice. Transcript levels in human HD caudate showed a tendency for up-regulation in HD cases compared with controls, which correlates to increased levels observed in the YAC128 mice at 3, 12 and 24 months of age. This concordance strengthens the hypothesis that WT1 may play an important role in HD pathogenesis.
Actn2 was found to be down-regulated in YAC128 mice compared with controls as early as 3 months of age. This gene displays a consistent down-regulation at all time points analyzed. More importantly, this down-regulation was confirmed in human HD caudate. Our findings in YAC128 mice and HD patients are in agreement with observations made in R6/2 mice both at 6 and 12 weeks of age, in R6/1 mice and in previous analysis of human HD samples (9,10,14). Actn2 is involved in cell adhesion and cytoskeletal arrangement and has been suggested to play a crucial role for the anchoring of NMDA receptors in central neurons (50–52). It has previously been shown that Actn2 co-immunoprecipitates with the protein RGS9-2 in rat striatum, suggesting a functional relationship between RGS9-2 and α-actinin 2 in calcium-mediated inactivation of NMDA receptors (53).
Slc45a3 belongs to a family of solute carriers with over 25 members. Slc14a1, which belongs to this same family, has previously been shown to be differentially expressed both in human HD caudate (10) as well as in the R6/2 model (54). We observed up-regulation of Slc45a3 in YAC128 at 9, 12 and 24 months of age as well as in HD caudate compared with controls. The protein is predicted to be a plasma membrane protein and has been shown to constitute the majority of erythroblast transformation-specific family member gene fusions observed in prostate cancers. Slc45a3 gene expression is not unique to prostate tissue and may play an important role in solute regulation in striatal neurons (55).
Protocadherin 20 (Pcdh20) belongs to the cadherin superfamily, of which most members are expressed predominantly in the central nervous system. Protocadherins are involved in Ca2+-mediated cell–cell adhesion and are suggested to be involved in the formation and maintenance of synaptic connections. In situ hybridization studies performed on rat brain during early postnatal stage (P3), a critical period for establishment of specific synaptic connections, provide evidence for region-dependent expression patterns in cerebral cortex of several protocadherins, including Pcdh20 (56). Data suggest that there is a correlation between regional expression patterns of different protocadherins and development of specific synaptic connections between cerebral cortex and other communicating brains regions (56). YAC128 exhibited an up-regulation of Pcdh20 as early as 3 months of age and also at 9, 12 and 24 months, while we observed a down-regulation of PCDH20 in the caudate of HD cases compared with controls. This data suggests that alterations in Pcdh20 expression may lead to impairment of synaptic connections in striatum.
Dimethylarginine dimethylaminohydrolase 1 (Ddah1) is one of two described enzymes (Ddah1 and Ddah2) that hydrolyzes methylated arginine analogues and asymmetric dimethylarginine (ADMA) produced during cellular turnover of methylated proteins. ADMA is an endogenous inhibitor of nitric oxide synthase (NOS), which catalyzes NO production (57,58). Thus, Ddah1 is a target for novel therapeutic agents designed to modulate NO generation. Neuronal nitric-oxide synthase-positive interneurons constitute ∼10% of the cells in the striatum. These interneurons persist in HD, in contrast to the GABAergic medium spiny neurons (MSNs) which are susceptible to neurodegeneration in the striatum. In one study, it was hypothesized that NO nitrosylates the NMDA receptors in the interneurons and render them less sensitive to activation, whereas NO diffusing into the MSNs lead to toxicity when it reacts with superoxide anion (O2−) (59,60). Administration of NOS inhibitors in R6/2 transgenic mice accelerated the onset of disease symptoms (61). R6/1 transgenic mice with only one copy of nNOS showed a delayed onset of disease, whereas mice lacking both copies displayed an acceleration of the disease (61). We showed decreased levels of Ddah1 in YAC128 compared with controls which might cause decreased hydrolysis of ADMA, and reduced inhibition of nitric oxide synthase (NOS), resulting in increased levels of NO in the YAC128. We observed increased levels of DDAH1 in HD cases which might suggest an up-regulation of the enzyme to compensate for reduced levels of NO production due to increased levels of ADMA in the brain. Alternatively, increased DDAH1 levels might be a primary or secondary contributor to the pathological state of HD. We need to further investigate the relationship between DDAH1, ADMA, NOS and NO and the potential role these molecular targets might play in HD pathogenesis.
PPP1R9A [protein phosphatase 1, regulatory (inhibitor) subunit 9A] encodes Neurabin I (neural tissue-specific F-actin-binding protein I) whose name stems from the fact that Neurabin I has been shown to bind and inhibit the function of protein phosphatase I (PPI) (62,63). In human, PPP1R9A is located in a cluster of imprinted genes and the protein has been shown to be involved in actin cytoskeleton dynamics and in synaptic formation and function (63,64). It has been shown that Ppp1r9a is imprinted mainly in skeletal muscle (maternally expressed), but not in brain (64). Furthermore, it has also been shown that Neurabin I is highly concentrated in the synapses of developed neurons and is involved in neurite formation (65). We showed a down-regulation of Ppp1r9a in striatum in YAC128 mice as well as HD cases compared with controls. Down-regulation of both Ppp1r9a and Actn2 strengthens the evidence that deficiencies in cytoskeletal dynamics play a role in HD pathogenesis.
We have identified protocadherin 20 (Pcdh20), neurabin I (Ppp1r9a) and actinin alpha 2 (Actn2) to be transcriptionally altered in the striatum of the YAC128 mice and in human HD caudate. These genes are known to play a role in synaptic formation, synaptic plasticity and cytoskeletal arrangement. Both Actn2 and Pcdh20 were altered already at 3 months in the YAC128 mice. Our results suggest that transcriptional changes of these genes reflect neuronal dysfunction of pathways involved in axonal transport, synaptic plasticity and dendrite integrity. The hallmark of HD is selective degeneration of vulnerable medium spiny neurons, whereas interneurons are spared in the striatum. Prior to cell death, however, there are neuropathological changes occurring that indicate early synaptic pathology. Morphological changes, including dystrophic neurites, have been described in spiny striatal and cortical pyramidal neurons in HD (66–68). HD mice have been employed to study the correlation between dysmorphic changes in neurons and behavioural abnormalities. Dysmorphic dendrites in the striatal spiny and the cortical pyramidal neurons were shown to be predictive of onset and severity of behavioural deficits in HD mice (69). Dystrophic neurites have been suggested to correlate with alterations in synaptic plasticity in HD. For example, the R6/2 mice exhibit altered synaptic plasticity, which has been suggested to contribute to the pre-symptomatic changes in cognition (70). Furthermore, both YAC46 and YAC72 HD mice exhibit early electrophysiological abnormalities indicative of altered synaptic function, including excitatory NMDA receptor activity (16,71). It has been suggested that cell–cell interactions between cortical and striatal neurons are critical for HD pathogenesis through the study of BAC transgenic mice (72). In addition, mutant huntingtin has been shown to inhibit both fast axonal transport and elongation of neuritic processes (73). We present novel transcriptional changes in several genes that are involved in synaptic integrity and function. The finding that these genes are transcriptionally dysregulated in the YAC128 mice and human HD caudate emphasizes the importance of these pathways in HD pathogenesis. Moreover, modulation of synaptic influences may have therapeutic potential in HD.
In our study, we have identified several genes that are differentially expressed in both the YAC128 model as well as in human HD caudate. Transcriptional changes identified in this study might reflect: (1) genes that lie within the pathogenic pathways leading to neurodegeneration; (2) genes that reflect the cellular attempts to block the disease process; and (3) genes that reflect non-specific responses to neurodegeneration. The first group of genes will provide novel insights into the specific downstream mechanisms of neurodegeneration. Knowledge of these pathways will provide new experimental starting points from which to discover novel approaches directed specifically to modify these disease-causing pathways in HD. The second group of genes represents components of pathways that are up- or down-regulated in an attempt to reduce neuronal death. These might be exploited in the future as a way to protect neurons. The profiling of YAC128 mice has illuminated several molecular changes in HD brain that have received little or no attention in previous studies.
Transcriptional changes are likely to be important in HD pathogenesis. Full-length models have less obvious transcriptional changes than HD fragment models as described previously (74). Nevertheless, in this paper, we present an experimental approach that has successfully identified novel transcriptional changes in YAC128 striatum and human HD caudate that have been validated by qPCR. Additional studies can be envisaged to better understand the contribution of these genes and their potential role in HD pathogenesis. Ultimately, further characterization of the role of these genes may lead to the identification of new pathways involved in HD.
MATERIALS AND METHODS
Mice
Transgenic HD mice expressing human HD huntingtin with 120 CAG repeats (YAC128) and wild-type littermates were used for the described experiments (18). The mice were group housed in polystyrene cages under a normal light–dark cycle (6 am to 8 pm) in a clean facility and with free access to water and standard rodent chow. All experiments were performed in accordance with the University of British Columbia animal care committee.
RNA isolation for genome-wide expression analysis using the Affymetrix and Illumina platforms
Striatal tissue was collected from 12- and 24-month-old YAC128 mice and wild-type littermates. Animals were anaesthetized using Avertine, and tissue was collected in RNAlater (Ambion) and stored at −80°C prior to RNA isolation. Homogenization of tissue was performed using a Fastprep Homogenizer (ThermoScientific). Total RNA from mouse striatum was extracted using the Qiagen RNeasy mini kit and eluted in 30 µl RNase free water according to the manufacturer's instructions. For 12-month-old mice, total RNA from four YAC128 and four wild-type littermates was extracted. For 24-month-old mice, total RNA from six YAC128 and four wild-type littermates was extracted. Total RNA was quantified using the Nanodrop spectrophotometer (ThermoScientific) and the integrity of the total RNA was determined electrophoretically on the RNA Nano Assay Chip run on the Bioanalyzer 2100 (Agilent Tech).
Illumina Bead arrays
Biotinylated antisense RNA was generated using the Illumina Totalprep RNA amplification kit (Illumina®) and hybridized to an Illumina Sentrix® Mouse-6 sample BeadChip (Illumina®). The BeadChip used contained six arrays and interrogated 47 769 transcripts derived from the National Center for Biotechnology Information (NCBI) Reference Sequence (RefSeq) database, the RIKEN® FANTOM™2 database and the Mouse Exonic Evidence Based Oligonucleotide (MEEBO) set. The BeadChips were scanned on the Bead express platform (Illumina®) and gene expression analysis was undertaken using the Illumina BeadStudio software (Illumina®).
Affymetrix
RNA was prepared according to the manufacturer's two-cycle target labeling procedure (Affymetrix, Inc.) with a starting amount of 25 ng of total RNA per sample. We used the Mouse Genome 430 2.0 Arrays (Affymetrix, Inc.) for all samples. This mouse array is a single GeneChip comprising over 45 000 probe sets representing more than 34 000 well-substantiated mouse genes.
Statistical analysis of Illumina and Affymetrix genome-wide expression data
Quality assessment was performed using the affy R package (75). Pre-processing of the Illumina and Affymetrix microarrays was done using Bead array (76) and Affymetrix GeneChip® Operating Software (GCOS), respectively. In the analysis, the data was split into two based on age, 12 and 24 months, respectively. Chips were normalized using Robust Multiarray Average (RMA). Within each subset, we performed a t-test between the wild-type and transgenic YAC128 data. The genes were ranked based on P-values and assigned to probes using Gemma (22) and Illumina and Affymetrix provided annotations. These ranks were subsequently used to create the combined rank list described. We calculated the combined rank for each target by averaging the individual ranks on the Affymetrix and the Illumina platforms, creating a new gene list based on the calculated combined rank value obtained. Corrections for multiple testing were performed using the false discovery rate according to Benjamini and Hochberg (21,77).
Human material
RNA was extracted from fresh-frozen samples (stored in −80°C) of human caudate collected with minimal postmortem interval to autopsy from six HD-gene-positive cases and five age- and sex-matched controls as described previously (10). All samples were carefully selected on the basis of RNA quality, and the HD cases were additionally graded according to the Vonsattel grade of disease pathology (scale 0–4) with two samples each representing HD grade 1, 2 and 3, respectively (78). RNA was extracted from human caudate using TRIzol (Invitrogen) followed by RNeasy column cleanup (Qiagen) using the manufacturer's protocols.
Relative quantification of mRNA by qPCR
Tissue samples from striatum were sampled from naïve YAC128 and wild-type littermates at 3, 6, 9, 12 and 24 months of age, respectively. Striatal tissue from 3-, 6- and 9-month-old samples were frozen immediately at −80°C. Striatal samples from 12- and 24-month-old mice were stored in RNAlater at −80°C. Homogenization of each individual tissue was performed using a Fastprep Homogenizer (ThermoScientific). We isolated total RNA from homogenized tissues using the Qiagen RNeasy minikit. We carried out reverse transcription cDNA synthesis using the Quantitect Reverse Transcription kit (Qiagen) according to the manufacturer's instructions. In total, striatal RNA was extracted from eight wild-type and five YAC128 mice for 3-month-old mice; nine wild-type and eight YAC128 mice for 6-month-old mice; five wild-type and six YAC128 mice for 9-month-old mice; six wild-type and seven YAC128 mice for 12-month-old mice; and seven wild-type and six YAC128 mice for 24-month-old mice. Quantitative analyses of mRNA expression were performed using FastSYBR®green mix according to the manufacturer's instructions (Applied Biosystems). Amplification of cDNA was performed using the 7500 Fast Real-Time PCR System (Applied Biosystems). Primers were constructed over exon/exon boundaries to avoid amplification of contaminating genomic DNA. All primers were designed using the Primer Express software version 3.0 (Applied Biosystems). Primer sequences are available as Supplementary Material (Table S3 for mouse and Table S4 for human qPCR analysis). Relative quantification of mRNA levels was calculated using the standard curve method, with amplification of target mRNA and control genes in separate wells. Standard curves were created using 10-fold serial dilutions of either mouse liver, cortex or kidney cDNA. Each sample was run in duplicate. The relative amount of mRNA in each well was calculated as the ratio between the target mRNA and a normalization factor calculated from the endogenous levels of the reference genes analyzed. In Figures 1–3, each sample ratio was divided by the mean of the wild-type littermate control samples for each individual time point. The following targets were analyzed for all five time points according to above: Wilm's tumor homolog 1 (Wt1), DNA-damage-inducible transcript 4-like (Ddit4), germ cell-specific gene 1-like protein (Gsg1l), solute carrier family 45 member 3 (Slc45a3), DAZ interacting protein 1-like (Dzip1l), retrotransposon gag domain containing 4 (Rgag4), Protocadherin-20 (Pcdh20), Scm-like with four MBT domains protein 2 (Sfmbt2), aspartoacylase-2 (Aminoacylase3) (Acy3), polymerase (RNA) II (DNA directed) polypeptide A (Polr2a), actinin alpha-2 (Actn2), dimethylarginine dimethylaminohydrolase 1 (Ddah1) and protein phosphatase 1, regulatory (inhibitor) subunit 9A (Ppp1r9a). Genes that were not annotated or only described as cDNA clones at the time of analysis were not included in the qRT–PCR analysis of the top 10 genes. These genes include Dio3os (Affymetrix rank list) and D830041|17Rik (Illumina rank list) (Supplementary Material, Fig. S1).
Calculation of normalization factor for qPCR data
We analyzed multiple reference genes for normalization of the qPCR data; 18S ribosomal RNA (Rn18s), beta-2 microglobulin (B2m), glyceraldehyde-3-phosphate dehydrogenase (Gapdh), actin-beta (Actb), ribosomal protein, large, P0 (Rplp0) and hypoxanthine guanine phosphoribosyl transferase 1 (Hprt1). We applied the GeNorm software analysis for calculation of the most accurate normalization factor for our data measurements at 3, 6, 9, 12 and 24 months, respectively (79). The normalization factor (NF) was based on geometric averaging of multiple internal control genes. Calculations were based on the average expression stability and pairwise variation analysis using the GeNorm-software (http://medgen.ugent.be/~jvdesomp/genorm/). Accurate NFs were calculated based on Gapdh, Rn18s and Rplp0 for 3-month data; Gapdh, Actb and Hprt1 for 6-, 9-and 24-month data; Rn18s, Actb and Hprt1 for 12-month data; ACTB, GAPDH and PGK1 (phosphoglycerate kinase 1) for human HD and control caudate samples.
Statistical analysis
Mann–Whitney two-tailed U-test was performed using GraphPad Prism version 4 for statistical analysis of the mRNA expression.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Swedish Research Council (K.B.), Canadian Institutes of Health Research (B.R.L., M.R.H., M.A.P.), CHDI Foundation (B.R.L., M.R.H.), the EPFL (R.L.-C., A.K.) and Michael Smith Foundation for Health Research (B.R.L., M.A.P.). This work was supported by the National Institutes of Health (GMO76990 to P.P.).
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Ge Lu, Catherine Carter, Nagat Bissada, Laila Al-Alwan and the Lausanne DNA array facility for technical assistance.
Conflict of Interest statement. B.R.L. is a Canadian Institutes of Health Research Clinician-Scientist and Michael Smith Foundation for Health Research Scholar. P.P. is a Canadian Institutes of Health Research New Investigator and Michael Smith Foundation for Health Research Scholar. M.R.H. is a Canada Research Chair in Human Genetics.
REFERENCES
- 1.Gusella J.F. Group H.D.C. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. [see comment] Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.Furtado S., Suchowersky O., Rewcastle B., Graham L., Klimek M.L., Garber A. Relationship between trinucleotide repeats and neuropathological changes in Huntington's disease. Ann. Neurol. 1996;39:132–136. doi: 10.1002/ana.410390120. [DOI] [PubMed] [Google Scholar]
- 3.Sotrel A., Paskevich P.A., Kiely D.K., Bird E.D., Williams R.S., Myers R.H. Morphometric analysis of the prefrontal cortex in Huntington's disease. Neurology. 1991;41:1117–1123. doi: 10.1212/wnl.41.7.1117. [DOI] [PubMed] [Google Scholar]
- 4.Gil J.M., Rego A.C. Mechanisms of neurodegeneration in Huntington's disease. Eur. J. Neurosci. 2008;27:2803–2820. doi: 10.1111/j.1460-9568.2008.06310.x. [DOI] [PubMed] [Google Scholar]
- 5.Imarisio S., Carmichael J., Korolchuk V., Chen C.W., Saiki S., Rose C., Krishna G., Davies J.E., Ttofi E., Underwood B.R., et al. Huntington's disease: from pathology and genetics to potential therapies. Biochem. J. 2008;412:191–209. doi: 10.1042/BJ20071619. [DOI] [PubMed] [Google Scholar]
- 6.Richfield E.K., Maguire-Zeiss K.A., Cox C., Gilmore J., Voorn P. Reduced expression of preproenkephalin in striatal neurons from Huntington's disease patients. Ann. Neurol. 1995;37:335–343. doi: 10.1002/ana.410370309. [DOI] [PubMed] [Google Scholar]
- 7.Augood S.J., Faull R.L., Emson P.C. Dopamine D1 and D2 receptor gene expression in the striatum in Huntington's disease. Ann. Neurol. 1997;42:215–221. doi: 10.1002/ana.410420213. [DOI] [PubMed] [Google Scholar]
- 8.Augood S.J., Faull R.L., Love D.R., Emson P.C. Reduction in enkephalin and substance P messenger RNA in the striatum of early grade Huntington's disease: a detailed cellular in situ hybridization study. Neuroscience. 1996;72:1023–1036. doi: 10.1016/0306-4522(95)00595-1. [DOI] [PubMed] [Google Scholar]
- 9.Desplats P.A., Kass K.E., Gilmartin T., Stanwood G.D., Woodward E.L., Head S.R., Sutcliffe J.G., Thomas E.A. Selective deficits in the expression of striatal-enriched mRNAs in Huntington's disease. J. Neurochem. 2006;96:743–757. doi: 10.1111/j.1471-4159.2005.03588.x. [DOI] [PubMed] [Google Scholar]
- 10.Hodges A., Strand A.D., Aragaki A.K., Kuhn A., Sengstag T., Hughes G., Elliston L.A., Hartog C., Goldstein D.R., Thu D., et al. Regional and cellular gene expression changes in human Huntington's disease brain. Hum. Mol. Genet. 2006;15:965–977. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- 11.Thomas E.A. Striatal specificity of gene expression dysregulation in Huntington's disease. J. Neurosci. Res. 2006;84:1151–1164. doi: 10.1002/jnr.21046. [DOI] [PubMed] [Google Scholar]
- 12.Kuhn A., Goldstein D.R., Hodges A., Strand A.D., Sengstag T., Kooperberg C., Becanovic K., Pouladi M.A., Sathasivam K., Cha J.H., et al. Mutant Huntingtin’s effects on striatal gene expression in mice recapitulate changes observed in human Huntington's disease brain and do not differ with mutant Huntingtin length or wild-type Huntingtin dosage. Hum. Mol. Genet. 2007;16:1845–1861. doi: 10.1093/hmg/ddm133. [DOI] [PubMed] [Google Scholar]
- 13.Luthi-Carter R., Hanson S.A., Strand A.D., Bergstrom D.A., Chun W., Peters N.L., Woods A.M., Chan E.Y., Kooperberg C., Krainc D., et al. Dysregulation of gene expression in the R6/2 model of polyglutamine disease: parallel changes in muscle and brain. Hum. Mol. Genet. 2002;11:1911–1926. doi: 10.1093/hmg/11.17.1911. [DOI] [PubMed] [Google Scholar]
- 14.Luthi-Carter R., Strand A., Peters N.L., Solano S.M., Hollingsworth Z.R., Menon A.S., Frey A.S., Spektor B.S., Penney E.B., Schilling G., et al. Decreased expression of striatal signaling genes in a mouse model of Huntington's disease. Hum. Mol. Genet. 2000;9:1259–1271. doi: 10.1093/hmg/9.9.1259. [DOI] [PubMed] [Google Scholar]
- 15.Luthi-Carter R., Strand A.D., Hanson S.A., Kooperberg C., Schilling G., La Spada A.R., Merry D.E., Young A.B., Ross C.A., Borchelt D.R., et al. Polyglutamine and transcription: gene expression changes shared by DRPLA and Huntington's disease mouse models reveal context-independent effects. Hum. Mol. Genet. 2002;11:1927–1937. doi: 10.1093/hmg/11.17.1927. [DOI] [PubMed] [Google Scholar]
- 16.Hodgson J.G., Agopyan N., Gutekunst C.A., Leavitt B.R., LePiane F., Singaraja R., Smith D.J., Bissada N., McCutcheon K., Nasir J., et al. A YAC mouse model for Huntington's disease with full-length mutant huntingtin, cytoplasmic toxicity, and selective striatal neurodegeneration. Neuron. 1999;23:181–192. doi: 10.1016/s0896-6273(00)80764-3. [DOI] [PubMed] [Google Scholar]
- 17.Hodgson J.G., Smith D.J., McCutcheon K., Koide H.B., Nishiyama K., Dinulos M.B., Stevens M.E., Bissada N., Nasir J., Kanazawa I., et al. Human Huntingtin derived from YAC transgenes compensates for loss of murine Huntingtin by rescue of the embryonic lethal phenotype. Hum. Mol. Genet. 1996;5:1875–1885. doi: 10.1093/hmg/5.12.1875. [DOI] [PubMed] [Google Scholar]
- 18.Slow E.J., van Raamsdonk J., Rogers D., Coleman S.H., Graham R.K., Deng Y., Oh R., Bissada N., Hossain S.M., Yang Y.Z., et al. Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum. Mol. Genet. 2003;12:1555–1567. doi: 10.1093/hmg/ddg169. [DOI] [PubMed] [Google Scholar]
- 19.Lerch J.P., Carroll J.B., Dorr A., Spring S., Evans A.C., Hayden M.R., Sled J.G., Henkelman R.M. Cortical thickness measured from MRI in the YAC128 mouse model of Huntington's disease. Neuroimage. 2008;41:243–251. doi: 10.1016/j.neuroimage.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 20.Graham R.K., Deng Y., Slow E.J., Haigh B., Bissada N., Lu G., Pearson J., Shehadeh J., Bertram L., Murphy Z., et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 21.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B. 1995;57:289–300. [Google Scholar]
- 22.Barnes M., Freudenberg J., Thompson S., Aronow B., Pavlidis P. Experimental comparison and cross-validation of the Affymetrix and Illumina gene expression analysis platforms. Nucleic Acids Res. 2005;33:5914–5923. doi: 10.1093/nar/gki890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stalteri M.A., Harrison A.P. Interpretation of multiple probe sets mapping to the same gene in Affymetrix GeneChips. BMC Bioinformatics. 2007;8:13. doi: 10.1186/1471-2105-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Raamsdonk J.M., Pearson J., Slow E.J., Hossain S.M., Leavitt B.R., Hayden M.R. Cognitive dysfunction precedes neuropathology and motor abnormalities in the YAC128 mouse model of Huntington's disease. J. Neurosci. 2005;25:4169–4180. doi: 10.1523/JNEUROSCI.0590-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seipel K., Georgiev O., Gerber H.P., Schaffner W. Basal components of the transcription apparatus (RNA polymerase II, TATA-binding protein) contain activation domains: is the repetitive C-terminal domain (CTD) of RNA polymerase II a ‘portable enhancer domain’? Mol. Reprod. Dev. 1994;39:215–225. doi: 10.1002/mrd.1080390215. [DOI] [PubMed] [Google Scholar]
- 26.Gerber H.P., Seipel K., Georgiev O., Hofferer M., Hug M., Rusconi S., Schaffner W. Transcriptional activation modulated by homopolymeric glutamine and proline stretches. Science. 1994;263:808–811. doi: 10.1126/science.8303297. [DOI] [PubMed] [Google Scholar]
- 27.Sugars K.L., Rubinsztein D.C. Transcriptional abnormalities in Huntington disease. Trends Genet. 2003;19:233–238. doi: 10.1016/S0168-9525(03)00074-X. [DOI] [PubMed] [Google Scholar]
- 28.Benn C.L., Sun T., Sadri-Vakili G., McFarland K.N., DiRocco D.P., Yohrling G.J., Clark T.W., Bouzou B., Cha J.H. Huntingtin modulates transcription, occupies gene promoters in vivo, and binds directly to DNA in a polyglutamine-dependent manner. J. Neurosci. 2008;28:10720–10733. doi: 10.1523/JNEUROSCI.2126-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunah A.W., Jeong H., Griffin A., Kim Y.M., Standaert D.G., Hersch S.M., Mouradian M.M., Young A.B., Tanese N., Krainc D. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington’s disease. Science. 2002;296:2238–2243. doi: 10.1126/science.1072613. [DOI] [PubMed] [Google Scholar]
- 30.Li S.H., Cheng A.L., Zhou H., Lam S., Rao M., Li H., Li X.J. Interaction of Huntington disease protein with transcriptional activator Sp1. Mol. Cell Biol. 2002;22:1277–1287. doi: 10.1128/mcb.22.5.1277-1287.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boutell J.M., Thomas P., Neal J.W., Weston V.J., Duce J., Harper P.S., Jones A.L. Aberrant interactions of transcriptional repressor proteins with the Huntington's disease gene product, huntingtin. Hum. Mol. Genet. 1999;8:1647–1655. doi: 10.1093/hmg/8.9.1647. [DOI] [PubMed] [Google Scholar]
- 32.McCampbell A., Taylor J.P., Taye A.A., Robitschek J., Li M., Walcott J., Merry D., Chai Y., Paulson H., Sobue G., et al. CREB-binding protein sequestration by expanded polyglutamine. Hum. Mol. Genet. 2000;9:2197–2202. doi: 10.1093/hmg/9.14.2197. [DOI] [PubMed] [Google Scholar]
- 33.Nucifora F.C., Jr, Sasaki M., Peters M.F., Huang H., Cooper J.K., Yamada M., Takahashi H., Tsuji S., Troncoso J., Dawson V.L., et al. Interference by huntingtin and atrophin-1 with CBP-mediated transcription leading to cellular toxicity. Science. 2001;291:2423–2428. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- 34.Steffan J.S., Kazantsev A., Spasic-Boskovic O., Greenwald M., Zhu Y.Z., Gohler H., Wanker E.E., Bates G.P., Housman D.E., Thompson L.M. The Huntington’s disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc. Natl Acad. Sci. USA. 2000;97:6763–6768. doi: 10.1073/pnas.100110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bae B.I., Xu H., Igarashi S., Fujimuro M., Agrawal N., Taya Y., Hayward S.D., Moran T.H., Montell C., Ross C.A., et al. p53 mediates cellular dysfunction and behavioral abnormalities in Huntington’s disease. Neuron. 2005;47:29–41. doi: 10.1016/j.neuron.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 36.van Roon-Mom W.M., Reid S.J., Jones A.L., MacDonald M.E., Faull R.L., Snell R.G. Insoluble TATA-binding protein accumulation in Huntington's disease cortex. Brain Res. Mol. Brain Res. 2002;109:1–10. doi: 10.1016/s0169-328x(02)00450-3. [DOI] [PubMed] [Google Scholar]
- 37.Shimohata T., Nakajima T., Yamada M., Uchida C., Onodera O., Naruse S., Kimura T., Koide R., Nozaki K., Sano Y., et al. Expanded polyglutamine stretches interact with TAFII130, interfering with CREB-dependent transcription. Nat. Genet. 2000;26:29–36. doi: 10.1038/79139. [DOI] [PubMed] [Google Scholar]
- 38.Zhai W., Jeong H., Cui L., Krainc D., Tjian R. In vitro analysis of huntingtin-mediated transcriptional repression reveals multiple transcription factor targets. Cell. 2005;123:1241–1253. doi: 10.1016/j.cell.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 39.Chen-Plotkin A.S., Sadri-Vakili G., Yohrling G.J., Braveman M.W., Benn C.L., Glajch K.E., DiRocco D.P., Farrell L.A., Krainc D., Gines S., et al. Decreased association of the transcription factor Sp1 with genes downregulated in Huntington's disease. Neurobiol. Dis. 2006;22:233–241. doi: 10.1016/j.nbd.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 40.Sadri-Vakili G., Menon A.S., Farrell L.A., Keller-McGandy C.E., Cantuti-Castelvetri I., Standaert D.G., Augood S.J., Yohrling G.J., Cha J.H. Huntingtin inclusions do not down-regulate specific genes in the R6/2 Huntington's disease mouse. Eur. J. Neurosci. 2006;23:3171–3175. doi: 10.1111/j.1460-9568.2006.04871.x. [DOI] [PubMed] [Google Scholar]
- 41.Zoghbi H.Y., Orr H.T. Glutamine repeats and neurodegeneration. Annu. Rev. Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- 42.Ross C.A., Wood J.D., Schilling G., Peters M.F., Nucifora F.C., Jr, Cooper J.K., Sharp A.H., Margolis R.L., Borchelt D.R. Polyglutamine pathogenesis. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 1999;354:1005–1011. doi: 10.1098/rstb.1999.0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chevyreva I., Faull R.L., Green C.R., Nicholson L.F. Assessing RNA quality in postmortem human brain tissue. Exp. Mol. Pathol. 2008;84:71–77. doi: 10.1016/j.yexmp.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 44.Ervin J.F., Heinzen E.L., Cronin K.D., Goldstein D., Szymanski M.H., Burke J.R., Welsh-Bohmer K.A., Hulette C.M. Postmortem delay has minimal effect on brain RNA integrity. J. Neuropathol. Exp. Neurol. 2007;66:1093–1099. doi: 10.1097/nen.0b013e31815c196a. [DOI] [PubMed] [Google Scholar]
- 45.Vance C., Rogelj B., Hortobagyi T., De Vos K.J., Nishimura A.L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P., et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kwiatkowski T.J., Jr, Bosco D.A., Leclerc A.L., Tamrazian E., Vanderburg C.R., Russ C., Davis A., Gilchrist J., Kasarskis E.J., Munsat T., et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 47.Fukuzawa R., Anaka M.R., Weeks R.J., Morison I.M., Reeve A.E. Canonical WNT signalling determines lineage specificity in Wilms tumour. Oncogene. 2009;28:1063–1075. doi: 10.1038/onc.2008.455. [DOI] [PubMed] [Google Scholar]
- 48.Morrison A.A., Venables J.P., Dellaire G., Ladomery M.R. The Wilms tumour suppressor protein WT1 (+KTS isoform) binds alpha-actinin 1 mRNA via its zinc-finger domain. Biochem. Cell Biol. 2006;84:789–798. doi: 10.1139/o06-065. [DOI] [PubMed] [Google Scholar]
- 49.Nabet B., Tsai A., Tobias J.W., Carstens R.P. Identification of a putative network of actin-associated cytoskeletal proteins in glomerular podocytes defined by co-purified mRNAs. PLoS One. 2009;4:e6491. doi: 10.1371/journal.pone.0006491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luthi-Carter R., Apostol B.L., Dunah A.W., DeJohn M.M., Farrell L.A., Bates G.P., Young A.B., Standaert D.G., Thompson L.M., Cha J.H. Complex alteration of NMDA receptors in transgenic Huntington's disease mouse brain: analysis of mRNA and protein expression, plasma membrane association, interacting proteins, and phosphorylation. Neurobiol. Dis. 2003;14:624–636. doi: 10.1016/j.nbd.2003.08.024. [DOI] [PubMed] [Google Scholar]
- 51.Dunah A.W., Wyszynski M., Martin D.M., Sheng M., Standaert D.G. Alpha-actinin-2 in rat striatum: localization and interaction with NMDA glutamate receptor subunits. Brain Res. Mol. Brain Res. 2000;79:77–87. doi: 10.1016/s0169-328x(00)00102-9. [DOI] [PubMed] [Google Scholar]
- 52.Wyszynski M., Lin J., Rao A., Nigh E., Beggs A.H., Craig A.M., Sheng M. Competitive binding of alpha-actinin and calmodulin to the NMDA receptor. Nature. 1997;385:439–442. doi: 10.1038/385439a0. [DOI] [PubMed] [Google Scholar]
- 53.Bouhamdan M., Yan H.D., Yan X.H., Bannon M.J., Andrade R. Brain-specific regulator of G-protein signaling 9-2 selectively interacts with alpha-actinin-2 to regulate calcium-dependent inactivation of NMDA receptors. J. Neurosci. 2006;26:2522–2530. doi: 10.1523/JNEUROSCI.4083-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crocker S.F., Costain W.J., Robertson H.A. DNA microarray analysis of striatal gene expression in symptomatic transgenic Huntington's mice (R6/2) reveals neuroinflammation and insulin associations. Brain Res. 2006;1088:176–186. doi: 10.1016/j.brainres.2006.02.102. [DOI] [PubMed] [Google Scholar]
- 55.Rickman D.S., Pflueger D., Moss B., VanDoren V.E., Chen C.X., de la Taille A., Kuefer R., Tewari A.K., Setlur S.R., Demichelis F., et al. SLC45A3-ELK4 is a novel and frequent erythroblast transformation-specific fusion transcript in prostate cancer. Cancer Res. 2009;69:2734–2738. doi: 10.1158/0008-5472.CAN-08-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim S.Y., Chung H.S., Sun W., Kim H. Spatiotemporal expression pattern of non-clustered protocadherin family members in the developing rat brain. Neuroscience. 2007;147:996–1021. doi: 10.1016/j.neuroscience.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 57.Cardounel A.J., Cui H., Samouilov A., Johnson W., Kearns P., Tsai A.L., Berka V., Zweier J.L. Evidence for the pathophysiological role of endogenous methylarginines in regulation of endothelial NO production and vascular function. J. Biol. Chem. 2007;282:879–887. doi: 10.1074/jbc.M603606200. [DOI] [PubMed] [Google Scholar]
- 58.Murray-Rust J., Leiper J., McAlister M., Phelan J., Tilley S., Santa Maria J., Vallance P., McDonald N. Structural insights into the hydrolysis of cellular nitric oxide synthase inhibitors by dimethylarginine dimethylaminohydrolase. Nat. Struct. Biol. 2001;8:679–683. doi: 10.1038/90387. [DOI] [PubMed] [Google Scholar]
- 59.Zucker B., Luthi-Carter R., Kama J.A., Dunah A.W., Stern E.A., Fox J.H., Standaert D.G., Young A.B., Augood S.J. Transcriptional dysregulation in striatal projection- and interneurons in a mouse model of Huntington's disease: neuronal selectivity and potential neuroprotective role of HAP1. Hum. Mol. Genet. 2005;14:179–189. doi: 10.1093/hmg/ddi014. [DOI] [PubMed] [Google Scholar]
- 60.Ahern G.P., Klyachko V.A., Jackson M.B. cGMP and S-nitrosylation: two routes for modulation of neuronal excitability by NO. Trends Neurosci. 2002;25:510–517. doi: 10.1016/s0166-2236(02)02254-3. [DOI] [PubMed] [Google Scholar]
- 61.Deckel A.W., Tang V., Nuttal D., Gary K., Elder R. Altered neuronal nitric oxide synthase expression contributes to disease progression in Huntington's disease transgenic mice. Brain Res. 2002;939:76–86. doi: 10.1016/s0006-8993(02)02550-7. [DOI] [PubMed] [Google Scholar]
- 62.McAvoy T., Allen P.B., Obaishi H., Nakanishi H., Takai Y., Greengard P., Nairn A.C., Hemmings H.C., Jr Regulation of neurabin I interaction with protein phosphatase 1 by phosphorylation. Biochemistry. 1999;38:12943–12949. doi: 10.1021/bi991227d. [DOI] [PubMed] [Google Scholar]
- 63.Oliver C.J., Terry-Lorenzo R.T., Elliott E., Bloomer W.A., Li S., Brautigan D.L., Colbran R.J., Shenolikar S. Targeting protein phosphatase 1 (PP1) to the actin cytoskeleton: the neurabin I/PP1 complex regulates cell morphology. Mol. Cell. Biol. 2002;22:4690–4701. doi: 10.1128/MCB.22.13.4690-4701.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakabayashi K., Makino S., Minagawa S., Smith A.C., Bamforth J.S., Stanier P., Preece M., Parker-Katiraee L., Paton T., Oshimura M., et al. Genomic imprinting of PPP1R9A encoding neurabin I in skeletal muscle and extra-embryonic tissues. J. Med. Genet. 2004;41:601–608. doi: 10.1136/jmg.2003.014142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakanishi H., Obaishi H., Satoh A., Wada M., Mandai K., Satoh K., Nishioka H., Matsuura Y., Mizoguchi A., Takai Y. Neurabin: a novel neural tissue-specific actin filament-binding protein involved in neurite formation. J. Cell Biol. 1997;139:951–961. doi: 10.1083/jcb.139.4.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ferrante R.J., Kowall N.W., Richardson E.P., Jr Proliferative and degenerative changes in striatal spiny neurons in Huntington's disease: a combined study using the section-Golgi method and calbindin D28k immunocytochemistry. J. Neurosci. 1991;11:3877–3887. doi: 10.1523/JNEUROSCI.11-12-03877.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jackson M., Gentleman S., Lennox G., Ward L., Gray T., Randall K., Morrell K., Lowe J. The cortical neuritic pathology of Huntington's disease. Neuropathol. Appl. Neurobiol. 1995;21:18–26. doi: 10.1111/j.1365-2990.1995.tb01024.x. [DOI] [PubMed] [Google Scholar]
- 68.DiFiglia M., Sapp E., Chase K.O., Davies S.W., Bates G.P., Vonsattel J.P., Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 69.Laforet G.A., Sapp E., Chase K., McIntyre C., Boyce F.M., Campbell M., Cadigan B.A., Warzecki L., Tagle D.A., Reddy P.H., et al. Changes in cortical and striatal neurons predict behavioral and electrophysiological abnormalities in a transgenic murine model of Huntington's disease. J. Neurosci. 2001;21:9112–9123. doi: 10.1523/JNEUROSCI.21-23-09112.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Murphy K.P., Carter R.J., Lione L.A., Mangiarini L., Mahal A., Bates G.P., Dunnett S.B., Morton A.J. Abnormal synaptic plasticity and impaired spatial cognition in mice transgenic for exon 1 of the human Huntington's disease mutation. J. Neurosci. 2000;20 doi: 10.1523/JNEUROSCI.20-13-05115.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Di Filippo M., Tozzi A., Picconi B., Ghiglieri V., Calabresi P. Plastic abnormalities in experimental Huntington's disease. Curr. Opin. Pharmacol. 2007;7:106–111. doi: 10.1016/j.coph.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 72.Gu X., Li C., Wei W., Lo V., Gong S., Li S.H., Iwasato T., Itohara S., Li X.J., Mody I., et al. Pathological cell–cell interactions elicited by a neuropathogenic form of mutant Huntingtin contribute to cortical pathogenesis in HD mice. Neuron. 2005;46:433–444. doi: 10.1016/j.neuron.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 73.Szebenyi G., Morfini G.A., Babcock A., Gould M., Selkoe K., Stenoien D.L., Young M., Faber P.W., MacDonald M.E., McPhaul M.J., et al. Neuropathogenic forms of huntingtin and androgen receptor inhibit fast axonal transport. Neuron. 2003;40:41–52. doi: 10.1016/s0896-6273(03)00569-5. [DOI] [PubMed] [Google Scholar]
- 74.Chan E.Y., Luthi-Carter R., Strand A., Solano S.M., Hanson S.A., DeJohn M.M., Kooperberg C., Chase K.O., DiFiglia M., Young A.B., et al. Increased huntingtin protein length reduces the number of polyglutamine-induced gene expression changes in mouse models of Huntington's disease. Hum. Mol. Genet. 2002;11:1939–1951. doi: 10.1093/hmg/11.17.1939. [DOI] [PubMed] [Google Scholar]
- 75.Gautier L., Cope L., Bolstad B.M., Irizarry R.A. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 76.Dunning M.J., Smith M.L., Ritchie M.E., Tavare S. Bead array: R classes and methods for Illumina bead-based data. Bioinformatics. 2007;23:2183–2184. doi: 10.1093/bioinformatics/btm311. [DOI] [PubMed] [Google Scholar]
- 77.Hochberg Y., Benjamini Y. More powerful procedures for multiple significance testing. Stat. Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 78.Vonsattel J.P., Myers R.H., Stevens T.J., Ferrante R.J., Bird E.D., Richardson E.P., Jr Neuropathological classification of Huntington's disease. J. Neuropathol. Exp. Neurol. 1985;44:559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- 79.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.