Abstract
Two common inflammatory skin disorders with impaired barrier, atopic dermatitis (AD) and psoriasis, share distinct genetic linkage to the Epidermal Differentiation Complex (EDC) locus on 1q21. The EDC is comprised of tandemly arrayed gene families encoding proteins involved in skin cell differentiation. Discovery of semi-dominant mutations in filaggrin (FLG) associated with AD and a copy number variation within the LCE genes associated with psoriasis provide compelling evidence for the role of EDC genes in the pathogenesis of these diseases. To date, little is known about the potentially complex regulatory landscape within the EDC. Here, we report a computational approach to identify conserved non-coding elements (CNEs) in the EDC queried for regulatory function. Coordinate expression of EDC genes during mouse embryonic skin development and a striking degree of synteny and linearity in the EDC locus across a wide range of mammalian (placental and marsupial) genomes suggests an evolutionary conserved regulatory milieu in the EDC. CNEs identified by comparative genomics exhibit dynamic regulatory activity (enhancer or repressor) in differentiating or proliferating conditions. We further demonstrate epidermal-specific, developmental in vivo enhancer activities (DNaseI and transgenic mouse assays) in CNEs, including one within the psoriasis-associated deletion, LCE3C_LCE3B-del. Together, our multidisciplinary study features a network of regulatory elements coordinating developmental EDC gene expression as an unexplored resource for genetic variants in skin diseases.
INTRODUCTION
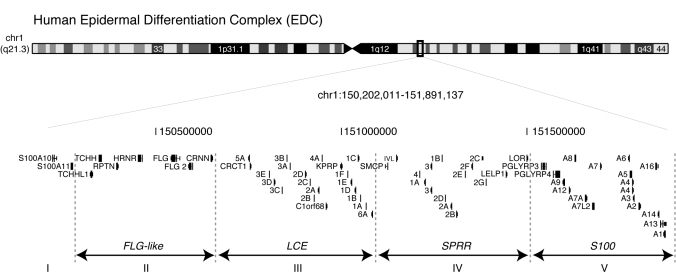
The Epidermal Differentiation Complex (EDC) spanning 1.6 Mb on human 1q21 (mouse 3q) harbors four clusters of tandem gene families: Filaggrin (FLG)-like, Late Cornified Envelope (LCE), Small Proline Rich Region (SPRR) and the S100 genes (Fig. 1) (1–5). FLG-like, LCE and SPRR genes encode structural proteins which are cross-linked to form the essential epidermal barrier at the surface of the skin, although S100 genes encode chemoattractant proteins expressed when the barrier is impaired (6). Recently, the EDC has been implicated in two common inflammatory skin disorders with impaired barrier, atopic dermatitis (AD) and psoriasis, that both share distinct genetic linkage to the EDC suggesting a role for these genes in disease pathogenesis (7–9). Loss-of-function variants in the FLG gene that were first identified in ichthyosis vulgaris, a common dry, scaly skin disorder (10), are also strongly associated with AD with subsequent progression to asthma or allergic rhinitis known as the atopic march (8,11,12). However, only 50% of AD patients possess FLG null alleles (12) and exclusion of the common FLG alleles in familial AD studies continues to demonstrate linkage to the EDC suggesting additional genetic variants in the FLG-like genes and the EDC (9,13). Recent genome-wide association studies identified association of psoriasis to a 30-kb deletion spanning the LCE3C and LCE3B genes (LCE3C_LCE3B-del) (14,15). Although psoriatic skin samples from LCE3C_LCE3B-del genotyped patients demonstrated a decrease in LCE3C and LCE3B expression, it is possible that a regulatory element within the LCE3C_LCE3B-del allele could be a contributing factor as well.
Figure 1.
The EDC is comprised of tandemly arrayed gene families. Human (hg18) EDC on chromosome 1q21 (1.6 Mb).
The spatial and temporal expression of several genes in the EDC during developmental epidermal differentiation and their dense tandem genomic arrangement suggest a genomic mechanism to coordinate their expression. One intriguing model for coordinate expression is a network of cis-regulatory elements in the EDC. Comparative genomics have greatly facilitated the identification of regulatory elements in Conserved Non-coding sequences or Elements (CNEs) (16). We apply this method to identify potential regulatory elements in the EDC locus as an untapped resource for functional genetics studies. Using this bioinformatics approach, we observe remarkable evolutionary conservation of the EDC locus across phylogenetically distinct mammalian genomes. Furthermore, we identified 48 CNEs in the EDC that exhibited dynamic regulatory activity during differentiating and proliferating conditions. We demonstrate epidermal-specific developmental in vivo enhancer activity in two CNEs, especially one in the psoriasis-associated LCE3C_LCE3B-del allele. These results highlight a genomic mechanism to coordinate developmental expression of the EDC genes via cis-regulatory elements that could likely play a role in human skin disease.
RESULTS
Developmental coordination of EDC gene expression during epidermal differentiation and skin barrier formation
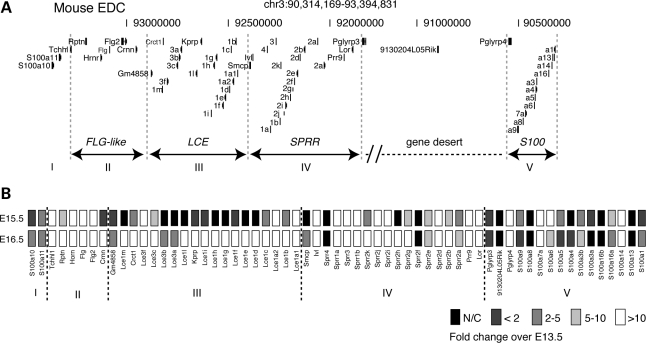
To investigate the degree to which the EDC genes are coordinately regulated during skin epidermal development, we employed a semi-quantitative analysis of pan-EDC gene locus temporal expression in murine embryonic skin (Fig. 2A). In mice, epidermal differentiation commences at embryonic day (E)15.5 followed by acquisition of skin barrier formation at E16.5 (17). Our results indicate initial expression of a majority of the EDC genes (44/61) genes, at E15.5 compared with control E13.5 (Fig. 2B). By E16.5, 11 additional genes were induced, most notably the LCE genes as well as increased expression of other EDC genes. The LCE genes at the extreme 5′ and 3′ ends of the LCE gene family were induced at E15.5 followed by induction of the more internal LCE genes at E16.5. Although several of the S100 genes demonstrated similar expression levels both at E15.5 and E16.5, not all S100 genes were expressed suggesting a discrete mechanism of regulation. These data confirm and expand the previous SPRR and LCE gene family-centric analyses, which revealed cluster-specific gene expression in various adult human epithelia (18,19). Together, these data support a coordination of EDC temporal expression during epidermal development that can be facilitated by cis-regulatory elements.
Figure 2.
Coordinated expression of EDC genes during epidermal differentiation and barrier formation. (A) Mouse (mm9) EDC (chr3), 3.1 Mb, are comprised of 4 gene families (FLG-like [II], LCE [III], SPRR [IV] and S100 [I and V]). Group I represents a cluster of 2 S100 genes (S100A10, S100A11). (B) Heatmap reflecting a semiquantitative real-time PCR analysis of EDC gene expression from mouse dorsal skin at E15.5 (epidermal differentiation) and E16.5 (barrier formation). Experiments were done in triplicate (n = 2 per embryonic stage) and normalized to β2-microglobulin. Gray scale legend, fold-change over E13.5 expression.
The EDC represents an ultraconserved microsyntenic genomic block in mammals
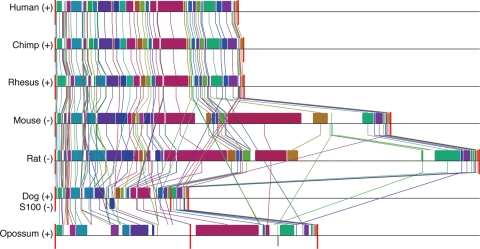
To identify CNEs, we aligned orthologous EDC genes from multiple, phylogenetically distinct mammalian species: human, chimp, rhesus, mouse, rat, dog and opossum (Fig. 3). The metatherian (or marsupial) opossum, representing one of the furthest mammalian phylogenetic branches (diverged 180 million years ago from the human), provides unique insights in ascertaining mammalian biological processes in comparison to these mammals (20–22). For example, opossums exhibit the same patterning of epidermal barrier acquisition observed in other mammals (23,24) yet at an accelerated pace given the shortened gestational age in utero (∼13 days) (Supplementary Material, Fig. 1). Given the observation of conserved patterning of epidermal barrier formation and its unique phylogenetic position in the mammalian lineage, we, therefore, incorporated the opossum genome to empower our comparative genomics studies. We find that the alignment of orthologous EDC genes in multiple mammalian species demonstrated a striking degree of linearity (order or arrangement of the genes) and synteny (genes occurring on the same chromosome) of orthologous EDC genes. Consistent with a shared genome-wide sequence identity to the human (93%) (25), the chimp and rhesus EDC loci are highly conserved (linear and syntenic). This conservation extended to the mammalian order rodentia where mouse and rat EDC loci differed from the human EDC locus in size (3.1 and 3.9 Mb, respectively) owing to a large ∼1.3 Mb ‘gene desert’ insertion (telomeric to HRN and centromeric to the FLG genes). Analysis of orthologous genes in the opossum also revealed conserved linearity with a large 309 Mb insertion separating the FLG-like and SPRR gene families from the LCE and S100 gene families (21). Analysis of the EDC orthologous genes in the dog revealed conserved linearity among the FLG-like, LCE and SPRR gene families with limited EDC synteny as the S100 gene family mapped to another chromosome, suggesting independent regulation of the S100 genes in the dog. Although the evolutionary time between the radiation of mammalian species is too short to produce the null hypothesis of randomly distributed genes in the genomes, the existence of orthologous EDC genes as linear and syntenic loci across mammalian genomes suggests a genetic mechanism to maintain the EDC as a regulatory block.
Figure 3.
The EDC represents an ultraconserved microsyntenic block in mammals. Each EDC loci from human, chimp, rhesus, mouse, rat, dog and opossum is depicted as locally collinear blocks (LCBs) (colored boxes) representing homologous regions shared by the aligned genomes and does not contain any rearrangements. Vertical lines trace homologous LCBs between genomes. LCBs below a genome's center black line are in reverse complement orientation relative to the human reference EDC locus. Red vertical lines mark interchromosomal boundaries or chromosomal distances <100 000 kb (opossum). +, forward orientation; −, reverse complement. Note that the S100 genes are in reverse complement in the dog.
Regulatory activities in the CNEs of the EDC
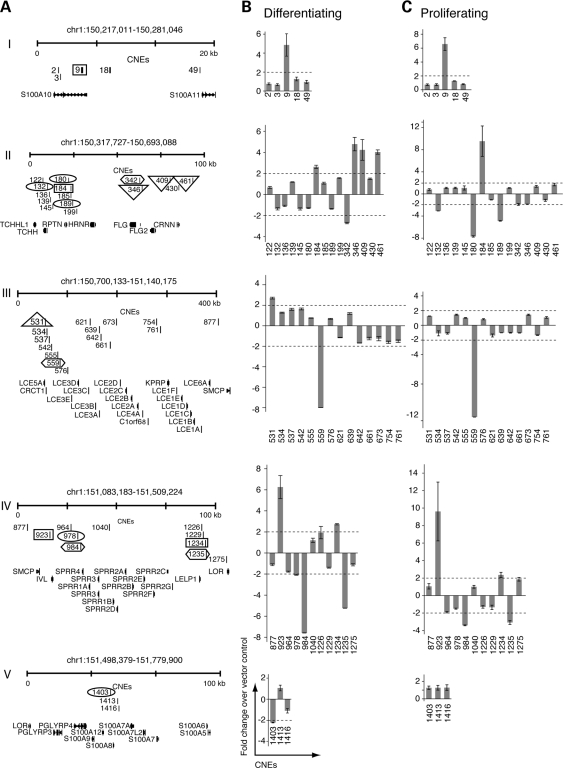
In our search for regulatory elements, we identified 48 CNEs (range of 115–1403 bp; mean = 431 bp) using a conservative alignment of orthologous EDC loci from multiple mammalian species (Fig. 4A, Supplementary Material, Table S1, Methods). Totaling 19 847 bp, CNEs represent 1.2% of the human EDC, a majority of which are located intergenically. To determine the regulatory potential of the CNEs, we performed luciferase reporter assays on cultured epidermal cells (keratinocytes) under terminally differentiating and proliferating conditions (1.2 and 0.05 mm Ca2+, respectively). Fourteen random Non-Conserved Non-coding Elements (NCNE) in the EDC (range of 115–1109 bp; mean = 496 bp) were used as a negative control. Nine CNEs exhibit enhancer activity (mean >2-fold increased luciferase activity) and six CNEs exhibit repressor activity (mean >2-fold decreased luciferase activity) under differentiating conditions (Fig. 4B). Under proliferating conditions, four CNE continued to demonstrate enhancer activity (Fig. 4C) and three CNE continued to demonstrate repressor activity. Three additional CNEs exhibited repressor activity exclusively under proliferating conditions. By comparison, all NCNE failed to exhibit enhancer or repressor activity (data not shown). In summary, these data demonstrate an enrichment of regulatory activity (30%, 15/48) of the EDC CNEs during differentiation compared with 20% (10/48) of the EDC CNEs during proliferation.
Figure 4.
Regulatory activities in the CNEs of the EDC. (A) CNEs (grouped into clusters I–V, as depicted in Fig. 1) are labeled as distance (kb) from the S100A10 transcriptional start site. Identification of CNEs span (hg18) chr1:150 202 011–151 891 137 including −20 kb of the transcriptional start site of S100A10 and +20 kb downstream from S100A1 transcript. EDC CNEs were tested for in vitro enhancer and repressor activity (luciferase reporter assays) in keratinocytes under (B) differentiating and (C) proliferating conditions. CNEs exhibiting >2-fold increased luciferase activity demonstrate enhancer activity and those that exhibit >2-fold decreased luciferase activity demonstrate repressor activity. Columns represent an average of two independent experiments performed in duplicate. Numbered CNEs are highlighted in designated boxes where Rectangle = enhancers (differentiating and proliferating), Hexagon = repressors (differentiating and proliferating), Triangle = enhancers (differentiating only), Oval = repressors (proliferating only), Bars, standard error.
h923 and h621 in LCE3C_LCE3B-del function as epidermal-specific developmental enhancers in vivo
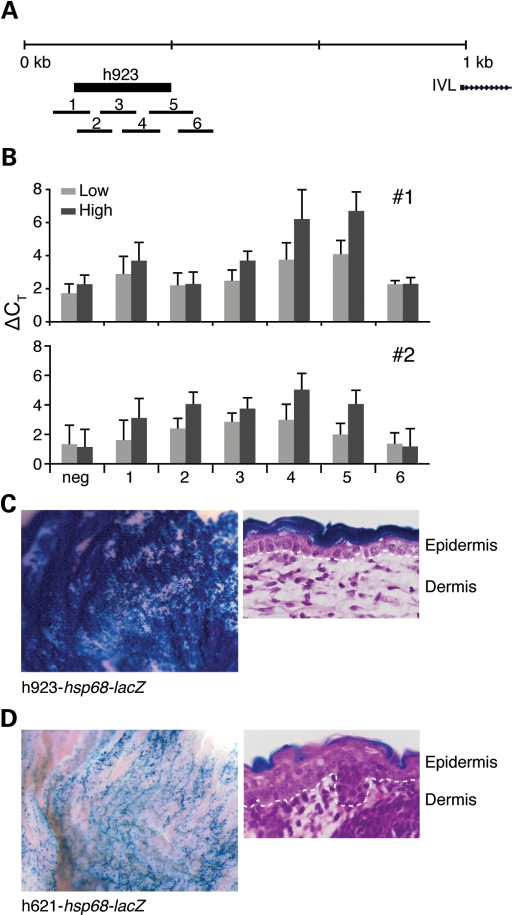
We further characterized the in vivo regulatory activity of two CNEs, selected either by in vitro enhancer activity or by disease relevance, using DNaseI hypersensitivity assays and transgenic mice. h923 is a 657-bp CNE located ∼2.6 kb upstream of the involucrin (IVL) transcriptional start site and demonstrated the highest enhancer activity in vitro. h621 is a 333-bp CNE that maps within the psoriasis-associated LCE3C_LCE3B-del region. Regulatory elements, typically devoid of core nucleosome structure, are hypersensitive to DNaseI treatment (26). We quantified DNaseI sensitivity of human primary epidermal cells by real-time PCR and overlapping amplicons that tile across the targeted CNEs (27). In vivo tissue-specific enhancer activity was assayed using transgenic CNE-hsp68-lacZ reporter mice that only demonstrate β-galactosidase activity when driven by a tissue-specific enhancer (28). We found DNaseI sensitivity in the proximal region of h923 (amplicon 4) in two independent primary keratinocyte samples, mirrored by genome-wide DNaseI hypersensitivity mapping in a proliferating human keratinocyte cell line (Fig. 5A, B and G; Crawford, personal communication). This finding was further corroborated by our observation of h923-hsp68-lacZ transgenic mice (F0) that demonstrated skin-specific β-galactosidase staining localized to the suprabasal layer of the epidermis (Fig. 5C). Consistent with the lack of luciferase activity under differentiating or proliferating conditions, we found no significant DNaseI sensitivity in h621 in two independent primary keratinocyte samples (genotyped as negative for the LCE3C_LCE3B-del) and in genome-wide DNaseI hypersensitivity mapping in a keratinocyte cell line (data not shown). However, h621-hsp68-lacZ transgenic mice (F0) demonstrated epidermal-specific lacZ expression at E16.5 that localized to the upper granular layer of the epidermis (Fig. 5D). Lack of enrichment for cells of the upper epidermal layers in primary tissue potentially contributes to our difficulty obtaining positive DNaseI sensitivity results for h621. Together, these results demonstrate in vivo epidermal-specific enhancer activities in h621 and h923. As well, this data suggests that the loss of enhancer function in the psoriasis-associated LCE3C_LCE3B-del (h621) may function as a contributing disease variant in psoriasis.
Figure 5.
h923 and h621 in LCE3C_LCE3B-del function as enhancers in vivo. Enhancer activity was assayed by DNaseI hypersensitivity and transgenic hsp68-lacZ mice. (A) PCR amplicons of tiling primer sets (1–6) targeting h923 are indicated. (B) DNaseI sensitivity (ΔCT = ΔCT[DNase-treated] − ΔCT[No DNase]) of individual primary keratinocyte samples (#1, #2) of h923. Light grey = low DNaseI, dark grey = hi DNaseI. Results are an average of triplicate readings. Bars, standard error. (C) h923-hsp68-lacZ transgenic mice (F0) demonstrate β-galactosidase-staining in the skin. All β-galactosidase-stained h923-hsp68-lacZ mice (two out of five genotyped-lacZ+ F0 mice) stained skin-specific at E16.5 (shown) and at pnd0. (D) h621-hsp68-lacZ (F0) demonstrate β-galactosidase-staining in the skin at E16.5. Five h621-hsp68-lacZ mice genotyped positive for lacZ. Four were β-galactosidase positive and two were skin-specific (shown). The remaining two β-galactosidase positive mice showed non-specific staining in the paws and brain (data not shown). H&E stains of β-galactosidase-stained cross-sections of the skin, Dotted lines indicate basement membrane demarcating the epidermis from the dermis, 40×.
DISCUSSION
Our multidisciplinary study demonstrates a network of interspersed cis-regulatory elements in the EDC to coordinate gene expression during mammalian epidermal differentiation. Furthermore, we report a striking degree of synteny and linearity in EDC across eutherian and metatherian mammalian phylogenetic species. Discovery of extant metatherian evolutionary origins in the development of a unique mammalian tissue empowered our comparative genomics studies ascertaining CNEs as regulatory elements. We demonstrate the dynamic nature of regulatory activity of EDC CNEs with enhancer and repressor activity in differentiating versus proliferating conditions. Moreover, we show that two CNEs (h621 and h923) demonstrate in vivo epidermal-specific, developmental enhancer activity using DNaseI hypersensitivity assays and transgenic mice.
h923 enhancer is sufficient to direct epidermal tissue specificity
h923 demonstrated consistent high enhancer activity in our cell reporter assays suggesting a dual regulatory role in proliferating and differentiating conditions. Although h923 partially overlaps with a previously described distal regulatory region in the human IVL promoter required for tissue-specificity and expression when tested with its endogenous promoter in transgenic mice (29), our results demonstrate that h923 is sufficient to direct epidermal tissue specificity without its native IVL promoter.
h621 enhancer maps to the psoriasis-associated LCE3C_LCE3B-del
Despite our observation of negative cell-based reporter results in h621, we were able to demonstrate in vivo epidermal-specific enhancer activity in our transgenic mice. Our findings are consistent with previous reports of regulatory polymorphisms that fail to recapitulate their in vivo effects in cell-based assays (30). Identification of in vivo developmental enhancer activity in h621 mapping within the psoriasis-associated LCE3C_LCE3B-del allele suggests an alternative disease mechanism in LCE3C_LCE3B-del-psoriasis, a loss of regulatory activity affecting global transcription of the EDC.
Cluster of regulatory elements (Group II): association with AD?
Interestingly, the first genome-wide association study for AD recently identified a tagging SNP in linkage disequilibrium (LD) with the FLG-like gene family within the EDC even when excluding individuals with the two common mutations in the FLG locus (R501X and 2282del4) (9). Another study for AD also demonstrated linkage to the EDC even after excluding the FLG common mutations (31). Since these studies only genotyped subjects for the two common FLG mutations, it is possible that either other FLG variants or alternatively, other genetic variants within the LD block account for the residual association in these studies (12). To that end, we observe a cluster of CNEs with some of the highest regulatory activity under differentiating conditions in this LD block (Group II, Fig. 5A), suggesting possible FLG-like gene regulatory regions to analyze in AD etiology.
Developmental enhancers in disease
This study augments the evidence that developmental enhancers may be implicated and causative in disease (32). In addition to their role in development, they may also play an extensive role in regulating adult tissue repair and in the case of skin diseases, response to barrier impairment or environmental stress. As we learn more about the role of lincRNAs and miRNAs that are also evolutionarily conserved (33,34), it would be intriguing to investigate whether our CNEs encode these small RNA molecules as well. In summary, our study not only provides a much-anticipated source of potential genetic variants in AD and psoriasis but also underscores the importance of extensive genomic and complementary functional studies in conjunction with genetic studies.
MATERIALS AND METHODS
Semiquantitative real-time PCR
RNA was extracted from the dorsal skin of E13.5, E15.5 and E16.5 mice with TRIzol/chloroform (Invitrogen) and RNeasy purification (Qiagen). cDNA was generated from the RNA (5 µg) using Superscript VILO cDNA kit (Invitrogen). Semiquantitative real-time PCR on cDNA was performed in triplicate (ABI 7300) using primers specific for target sequence (35) (Supplementary Material, Table S2) and SYBR Green (Invitrogen) to measure increased fluorescence of targeted amplicon. Primers used in this study amplify across intron boundaries where available and were confirmed for specificity using UCSC's ‘in silico PCR’ feature. Real-time PCR analysis (using the ΔΔCT method per manufacturer's instructions) included experimental data with single-peak dissociation curves and normalization to β2-microglobulin expression.
Comparative genomic sequence analysis
The EDC loci from human (hg18), chimp (panTro2), rhesus (rheMac2), mouse (mm8), rat (rn4), dog (canFam2) and opossum (monDom4) sequences were obtained from UCSC using the ‘convert’ option from the human reference sequence and aligned using Multispecies Percent Identity Plot Maker, MultiPipMaker (36) to obtain CNEs. ‘Single coverage’ and ‘repeat masked sequence’ options were selected in MultiPipMaker to exclude false positive regions that could be generated from paralogous gene sequences represented in the EDC. Sequences identified by MultiPipMaker as alignable with at least 100 bp and >50% nucleotide identity across all seven mammalian genomes in non-coding regions were designated as CNEs. CNEs immediately adjacent to exon and introns were excluded. For synteny block analysis, the EDC loci were aligned using Mauve (version 2.3.0) (37) with the default settings and seed families enabled.
Luciferase assay
A library of CNE and NCNE from the EDC was generated by PCR amplification from human BAC clones using Fast Start Fidelity Taq (Roche) and subsequent cloning into Gateway (Invitrogen) compatible pGL3 (Promega) plasmids upstream of a mouse Sprr1a promoter driving firefly luciferase expression. NCNE were selected using a sliding window view of the human EDC locus to identify non-conserved (no shared alignment between mouse, rat, dog and opossum) DNA sequences across the EDC locus in intergenic regions that represented a range from 115 to 1109 bp and a median size of 496 bp similar to the CNEs. All CNEs and NCNEs were sequence verified. Dual luciferase assays (Promega) were performed in duplicate in two independent experiments on mouse SP-1 keratinocytes plated on 6-well plates under proliferating (0.05 mm Ca2+) or terminal differentiating (1.3 mm Ca2+) conditions in S-MEM/10% chelex-treated fetal bovine serum (Lifeblood Medical) and measured 24 and 72 h post-transfection (Fluorskan Ascent FL fluorimeter, Thermo Scientific), respectively (35). For each well, firefly luciferase activity was normalized to the co-transfected Renilla luciferase activity and empty vector control.
DNaseI hypersensitivity assay
Human skin samples were obtained with appropriate informed consent and reviewed by the NIH Office of Human Subject Review. Only human newborn foreskin samples that screened negative for the LCE3C_LCE3B-del (positive for LCE3CF/LCE3CR PCR allele and negative for the EDCDELL/EDCDELR PCR allele) (14) were analyzed for the h621 DNaseI assay. Foreskin samples were cut into 1 cm pieces and incubated on dispase:HBSS (1:1) (BD Biosciences) with dermis down at 4°C overnight. Epidermal sheets were peeled off the next day and incubated in 0.25% trypsin/EDTA at 37°C for 10′ with frequent shaking and pipetting to obtain single cell isolation. Single cells were lysed with 0.03% NP-40/10 mm Tris–HCl (pH 7.5)/10 mm NaCl/3 mm MgCl2 buffer and digested with increasing amounts of DNaseI (Roche) at 37°C to obtain DNaseI-treated nuclei. After removal of proteins with overnight proteinase K digestion, DNaseI-digested DNA was subsequently isolated via phenol/chloroform extraction. Double-stranded DNA was quantitated using PicoGreen (Invitrogen) according to the manufacturer's instructions. DNaseI sensitivity was assayed by real-time PCR (ABI 7300) using 9.5 ng of DNaseI-digested DNA, SYBR Green (Qiagen) and tiling primers (200 bp amplicons with 50 bp overlap) to amplify targeted sequence (Supplementary Material, Table S3) (27). A non-conserved region (∼8.7 kb downstream of SPRR3) was designed as a negative DNaseI control.
In vivo mouse enhancer assay
All animals were maintained in an AAALAC accredited, murine pathogen-free facility at the National Institutes of Health (Bethesda, MD, USA) in accordance with institutional protocols and the Guide for the Care and Use of Laboratory Animals. All animal procedures were approved by the NHGRI Animal Care and Use Committee. CNEs were cloned into the Gateway-compatible hsp68-lacZ reporter vector (gift from Marcelo Nobrega, University of Chicago), sequence-verified, purified by cesium chloride gradient (Loftstrand) and linearized with SalI (NEB) prior to injection into fertilized eggs that were implanted into pseudopregnant females. Analysis was done on transgenic founder embryos (F0) at the corresponding embryonic stage post-transplantation (where transplantation day is designated as embryonic day [E] 0.5) and were genotyped using lacZ primers. Whole-mount embryos were stained overnight for β-galactosidase staining activity using X-gal (Fermentas) after cold fixation as previously described (38). β-galactosidase stained tissue sections were obtained from paraffin blocks (Histoserv).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Human Genome Research Institute Intramural Research Program and the National Institute of Health K99AR055948 to C.G.S.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Cherry Yang, Yicong Liu and Roli Mandana for technical expertise, Carole Yee for primary keratinocyte samples, Peter Chines for primer tile design, Elsa Escobar and Laurentine Sop for mouse husbandry and Julia Fekecs for assistance in preparing the figures. We thank Greg Crawford and David Sankoff for helpful discussions with the genome-wide DNase hypersensitivity data and genomic synteny, respectively. We also thank Anne Bowcock, Elizabeth Grice and Stacie Loftus for manuscript critiques.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Volz A., Korge B.P., Compton J.G., Ziegler A., Steinert P.M., Mischke D. Physical mapping of a functional cluster of epidermal differentiation genes on chromosome 1q21. Genomics. 1993;18:92–99. doi: 10.1006/geno.1993.1430. [DOI] [PubMed] [Google Scholar]
- 2.Zhao X.P., Elder J.T. Positional cloning of novel skin-specific genes from the human epidermal differentiation complex. Genomics. 1997;45:250–258. doi: 10.1006/geno.1997.4952. [DOI] [PubMed] [Google Scholar]
- 3.Mischke D., Korge B.P., Marenholz I., Volz A., Ziegler A. Genes encoding structural proteins of epidermal cornification and S100 calcium-binding proteins form a gene complex (‘epidermal differentiation complex’) on human chromosome 1q21. J. Invest. Dermatol. 1996;106:989–992. doi: 10.1111/1523-1747.ep12338501. [DOI] [PubMed] [Google Scholar]
- 4.Marshall D., Hardman M.J., Nield K.M., Byrne C. Differentially expressed late constituents of the epidermal cornified envelope. Proc. Natl. Acad. Sci. USA. 2001;98:13031–13036. doi: 10.1073/pnas.231489198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eckert R.L., Broome A.M., Ruse M., Robinson N., Ryan D., Lee K. S100 proteins in the epidermis. J. Invest. Dermatol. 2004;123:23–33. doi: 10.1111/j.0022-202X.2004.22719.x. [DOI] [PubMed] [Google Scholar]
- 6.Segre J.A. Epidermal barrier formation and recovery in skin disorders. J. Clin. Invest. 2006;116:1150–1158. doi: 10.1172/JCI28521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cookson W.O., Ubhi B., Lawrence R., Abecasis G.R., Walley A.J., Cox H.E., Coleman R., Leaves N.I., Trembath R.C., Moffatt M.F., et al. Genetic linkage of childhood atopic dermatitis to psoriasis susceptibility loci. Nat. Genet. 2001;27:372–373. doi: 10.1038/86867. [DOI] [PubMed] [Google Scholar]
- 8.Palmer C.N., Irvine A.D., Terron-Kwiatkowski A., Zhao Y., Liao H., Lee S.P., Goudie D.R., Sandilands A., Campbell L.E., Smith F.J., et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat. Genet. 2006;38:441–446. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- 9.Esparza-Gordillo J., Weidinger S., Folster-Holst R., Bauerfeind A., Ruschendorf F., Patone G., Rohde K., Marenholz I., Schulz F., Kerscher T., et al. A common variant on chromosome 11q13 is associated with atopic dermatitis. Nat. Genet. 2009;41:596–601. doi: 10.1038/ng.347. [DOI] [PubMed] [Google Scholar]
- 10.Smith F.J., Irvine A.D., Terron-Kwiatkowski A., Sandilands A., Campbell L.E., Zhao Y., Liao H., Evans A.T., Goudie D.R., Lewis-Jones S., et al. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat. Genet. 2006;38:337–342. doi: 10.1038/ng1743. [DOI] [PubMed] [Google Scholar]
- 11.Baurecht H., Irvine A.D., Novak N., Illig T., Buhler B., Ring J., Wagenpfeil S., Weidinger S. Toward a major risk factor for atopic eczema: meta-analysis of filaggrin polymorphism data. J. Allergy Clin. Immunol. 2007;120:1406–1412. doi: 10.1016/j.jaci.2007.08.067. [DOI] [PubMed] [Google Scholar]
- 12.Sandilands A., Terron-Kwiatkowski A., Hull P.R., O'Regan G.M., Clayton T.H., Watson R.M., Carrick T., Evans A.T., Liao H., Zhao Y., et al. Comprehensive analysis of the gene encoding filaggrin uncovers prevalent and rare mutations in ichthyosis vulgaris and atopic eczema. Nat. Genet. 2007;39:650–654. doi: 10.1038/ng2020. [DOI] [PubMed] [Google Scholar]
- 13.Morar N., Cookson W.O., Harper J.I., Moffatt M.F. Filaggrin mutations in children with severe atopic dermatitis. J. Invest. Dermatol. 2007;127:1667–1672. doi: 10.1038/sj.jid.5700739. [DOI] [PubMed] [Google Scholar]
- 14.de Cid R., Riveira-Munoz E., Zeeuwen P.L., Robarge J., Liao W., Dannhauser E.N., Giardina E., Stuart P.E., Nair R., Helms C., et al. Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat. Genet. 2009;41:211–215. doi: 10.1038/ng.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X.J., Huang W., Yang S., Sun L.D., Zhang F.Y., Zhu Q.X., Zhang F.R., Zhang C., Du W.H., Pu X.M., et al. Psoriasis genome-wide association study identifies susceptibility variants within LCE gene cluster at 1q21. Nat. Genet. 2009;41:205–210. doi: 10.1038/ng.310. [DOI] [PubMed] [Google Scholar]
- 16.Visel A., Bristow J., Pennacchio L.A. Enhancer identification through comparative genomics. Semin. Cell Dev. Biol. 2007;18:140–152. doi: 10.1016/j.semcdb.2006.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blanpain C., Fuchs E. Epidermal homeostasis: a balancing act of stem cells in the skin. Nat. Rev. Mol. Cell. Biol. 2009;10:207–217. doi: 10.1038/nrm2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson B., Tilli C.M., Hardman M.J., Avilion A.A., MacLeod M.C., Ashcroft G.S., Byrne C. Late cornified envelope family in differentiating epithelia–response to calcium and ultraviolet irradiation. J. Invest. Dermatol. 2005;124:1062–1070. doi: 10.1111/j.0022-202X.2005.23699.x. [DOI] [PubMed] [Google Scholar]
- 19.Cabral A., Voskamp P., Cleton-Jansen A.M., South A., Nizetic D., Backendorf C. Structural organization and regulation of the small proline-rich family of cornified envelope precursors suggest a role in adaptive barrier function. J. Biol. Chem. 2001;276:19231–19237. doi: 10.1074/jbc.M100336200. [DOI] [PubMed] [Google Scholar]
- 20.Margulies E.H., Maduro V.V., Thomas P.J., Tomkins J.P., Amemiya C.T., Luo M., Green E.D. Comparative sequencing provides insights about the structure and conservation of marsupial and monotreme genomes. Proc. Natl. Acad. Sci. USA. 2005;102:3354–3359. doi: 10.1073/pnas.0408539102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mikkelsen T.S., Wakefield M.J., Aken B., Amemiya C.T., Chang J.L., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- 22.Samollow P.B. The opossum genome: insights and opportunities from an alternative mammal. Genome Res. 2008;18:1199–1215. doi: 10.1101/gr.065326.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hardman M.J., Sisi P., Banbury D.N., Byrne C. Patterned acquisition of skin barrier function during development. Development. 1998;125:1541–1552. doi: 10.1242/dev.125.8.1541. [DOI] [PubMed] [Google Scholar]
- 24.Hardman M.J., Moore L., Ferguson M.W., Byrne C. Barrier formation in the human fetus is patterned. J. Invest. Dermatol. 1999;113:1106–1113. doi: 10.1046/j.1523-1747.1999.00800.x. [DOI] [PubMed] [Google Scholar]
- 25.Gibbs R.A., Rogers J., Katze M.G., Bumgarner R., Weinstock G.M., Mardis E.R., Remington K.A., Strausberg R.L., Venter J.C., Wilson R.K., et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 26.Felsenfeld G., Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- 27.Crawford G.E., Holt I.E., Mullikin J.C., Tai D., Blakesley R., Bouffard G., Young A., Masiello C., Green E.D., Wolfsberg T.G., et al. Identifying gene regulatory elements by genome-wide recovery of DNase hypersensitive sites. Proc. Natl. Acad. Sci. USA. 2004;101:992–997. doi: 10.1073/pnas.0307540100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kothary R., Clapoff S., Darling S., Perry M.D., Moran L.A., Rossant J. Inducible expression of an hsp68-lacZ hybrid gene in transgenic mice. Development. 1989;105:707–714. doi: 10.1242/dev.105.4.707. [DOI] [PubMed] [Google Scholar]
- 29.Crish J.F., Gopalakrishnan R., Bone F., Gilliam A.C., Eckert R.L. The distal and proximal regulatory regions of the involucrin gene promoter have distinct functions and are required for in vivo involucrin expression. J. Invest. Dermatol. 2006;126:305–314. doi: 10.1038/sj.jid.5700019. [DOI] [PubMed] [Google Scholar]
- 30.Cirulli E.T., Goldstein D.B. In vitro assays fail to predict in vivo effects of regulatory polymorphisms. Hum. Mol. Genet. 2007;16:1931–1939. doi: 10.1093/hmg/ddm140. [DOI] [PubMed] [Google Scholar]
- 31.Morar N., Edster P., Street T.L., Weidinger S., Di W., Dixon A.L., Taylor M., Holt R., Broxholme J., Kloop N., et al. Positional cloning of susceptibility genes for atopic dermatitis in the epidermal differentiation complex. J. Invest. Dermatol. 2007;127:531. [Google Scholar]
- 32.Epstein D.J. Cis-regulatory mutations in human disease. Brief Funct. Genomic Proteomic. 2009;8:310–316. doi: 10.1093/bfgp/elp021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guttman M., Amit I., Garber M., French C., Lin M.F., Feldser D., Huarte M., Zuk O., Carey B.W., Cassady J.P., et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 35.Martin N., Patel S., Segre J.A. Long-range comparison of human and mouse Sprr loci to identify conserved noncoding sequences involved in coordinate regulation. Genome Res. 2004;14:2430–2438. doi: 10.1101/gr.2709404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwartz S., Zhang Z., Frazer K.A., Smit A., Riemer C., Bouck J., Gibbs R., Hardison R., Miller W. PipMaker—a web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Darling A.C., Mau B., Blattner F.R., Perna N.T. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hou L., Panthier J.J., Arnheiter H. Signaling and transcriptional regulation in the neural crest-derived melanocyte lineage: interactions between KIT and MITF. Development. 2000;127:5379–5389. doi: 10.1242/dev.127.24.5379. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.