Abstract
Proteins and protein networks associated with cochlear pathogenesis in the Ames waltzer (av) mouse, a model for deafness in Usher syndrome 1F (USH1F), were identified. Cochlear protein from wild-type and av mice at postnatal day 30, a time point in which cochlear pathology is well established, was analyzed by quantitative 2D gel electrophoresis followed by mass spectrometry (MS). The analytic gel resolved 2270 spots; 69 spots showed significant changes in intensity in the av cochlea compared with the control. The cochlin protein was identified in 20 peptide spots, most of which were up-regulated, while a few were down-regulated. Analysis of MS sequence data showed that, in the av cochlea, a set of full-length isoforms of cochlin was up-regulated, while isoforms missing the N-terminal FCH/LCCL domain were down-regulated. Protein interaction network analysis of all differentially expressed proteins was performed with Metacore software. That analysis revealed a number of statistically significant candidate protein networks predicted to be altered in the affected cochlea. Quantitative PCR (qPCR) analysis of select candidates from the proteomic and bioinformatic investigations showed up-regulation of Coch mRNA and those of p53, Brn3a and Nrf2, transcription factors linked to stress response and survival. Increased mRNA of Brn3a and Nrf2 has previously been associated with increased expression of cochlin in human glaucomatous trabecular meshwork. Our report strongly suggests that increased level of cochlin is an important etiologic factor leading to the degeneration of cochlear neuroepithelia in the USH1F model.
INTRODUCTION
Usher syndrome is the most common cause of deafness and progressive vision loss in humans. It is clinically subdivided into types 1–3 based on the degree of deafness and the presence of vestibular dysfunction and the age of onset of vision loss due to retinitis pigmentosa. Usher syndrome type 1 (USH1) is the most severe form and is characterized by profound congenital hearing loss and vestibular dysfunction combined with pre-pubertal onset of retinitis pigmentosa (RP). In USH2 hearing loss is milder, the onset of RP is after puberty and vestibular function is unaffected. USH3 patients show progressive hearing loss and variable degrees of vestibular dysfunction. Mutations in different genes cause Usher type 1 phenotype, resulting in different subtype designations such as USH1C, 1D, -1E, -1F and -1G (1–8). For example, a mutation in the protocadherin 15 gene (PCDH15) causes USH1F (7–8). Mice carrying a mutation in Usher gene homologues serve as a good model to study ear disease in each subtype. The Ames waltzer (av) mouse harbors a mutation in Pcdh15 (4), and is a model for inner ear dysfunction in USH1 subtype F (USH1F) (3,5) and nonsyndromic deafness DFNB23 (2). Although the av mouse and other Usher mouse models have been available for many years, none of them have been utilized for inner ear proteomic studies. The analysis of global protein expression will assist in understanding the degenerative processes that take place in USH1F.
In the av model, hair cells develop, but the hair bundles fail to develop properly, rendering the cells non-functional. The hair cells in av mice show signs of degeneration as early as 10 days post-natal (P10) and reach a level of >90% degeneration by P30. Spiral ganglion cell degeneration is first noted around P30, and more than 90% of the cells are lost by P90 (9,10; Fig. 1). In the av model, supporting cells surrounding the hair cells also undergo degeneration, and most of the cells in the organ of Corti in all of the turns of the cochlea disappear by P150. The reason for this is not completely understood. Preventing the degeneration of the organ of Corti will be critical for the success of any therapy aimed at replacing the hair cells in the affected cochlea.
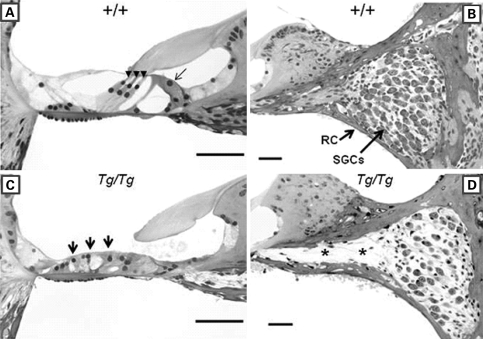
Figure 1.
Light micrographs of cross sections through the basal turn of the cochlear duct from wild-type (+/+) and Pcdh15av-Tg homozygote (Tg/Tg) mouse at P30. (A) and (C) Organ of Corti. (B) and (D) Spiral ganglion cells (SGCs). In the wild-type, the outer hair cells’ (black arrow heads) and inner hair cells’ (black arrow) cochlear neuroepthelium is morphologically normal (A) and the SGCs pack the Rosenthal's Canal (RC) (B). In the mutant, the organ of Corti has collapsed and there are no recognizable hair cells present (arrows in C) and there is a reduction in the number of SGCs. The asterisk marks sites previously occupied by SGCs (D). Sections in (A)–(D) were stained in toluidine blue and the image was captured in black and white. The scale bar in each panel indicates 50 µm. Reproduced with permission from Hearing Research (Alagramam et al. 10).
Proteins are the ultimate effector of phenotype, and have been shown to not always correlate very well with mRNA levels and, thus, should be studied directly (11). By analyzing global protein expression in av mice cochlea at a critical time point (P30) during the pathogenesis, we expect to discover protein networks and biomarkers associated with the degenerative process. Here, we employ proteomic, bioinformatic and targeted gene expression analyses to understand the pathogenic mechanisms in the av mouse cochlea of a mouse model for deafness in Usher syndrome type 1F. Our proteomic study revealed more than 40 protein candidates to be dysregulated in the av cochlea, including variations in the most abundant protein cochlin. Messenger RNA of Coch, as well as transcription factors with binding sites on the Coch gene promoter, was also up-regulated in the av cochlea. In addition to revealing important candidate proteins and networks, our investigation strongly suggests that the over expression of cochlin could be an important factor leading to the observed pathology in the USH1F model.
RESULTS
Differential expression of 69 protein spots detected in the av cochlea
Gels containing proteins from av mutant cochleae and sibling controls were scanned, and the images were uploaded into the DeCyder software. The analytic gel showed an average of 2270 (n = 4 gels) protein spots as shown in image analysis by DeCyder (Fig. 2). The separate images for each sample were compared within gels and across gels to determine the spots that changed significantly for the mutant versus wild type. Spots selected as having statistically significant differential expression were required to have fold changes of at least 1.1 or −1.1 (for P < 0.01), or at least 1.5 or −1.5 (for P < 0.05). Sixty-nine spots were found to pass this statistical threshold and were picked for sequencing analysis. Within these spots, 43 proteins were identified in the spots that were increased in intensity, and 26 proteins were identified in the spots where the intensity was reduced in the cochleae of av mice at P30 compared with controls of the same age (Supplementary Material, Table S1).
Figure 2.
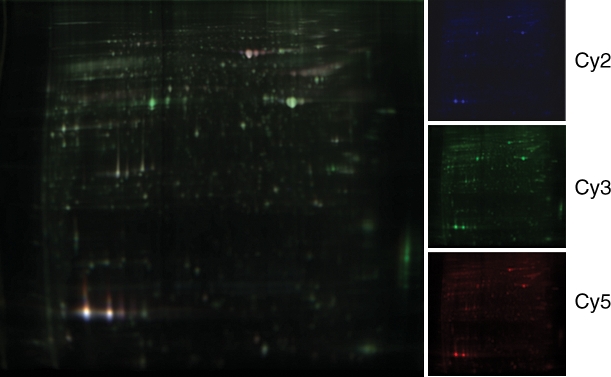
Three channel overlay image of cochlear protein from av mutant and control cochlea. Protein sample from av mouse, wild type and pooled internal standard (pooled from av mouse and wild type cochlea) is shown on the left and individual DIGE images on the right. Proteins were labeled with Cyanine dyes Cy2, Cy3 or Cy5 (spectrally resolvable fluorophores).
Of the spot intensities indicating increased expression in the mutant mouse cochleae, the fold-changes, or average ratio of mutant to wild type protein levels, ranged from 1.12 to 2.55, while those spots indicating decreased intensities had fold-changes that ranged from −1.12 to −1.6. For some spots, more than one protein was identified in the sequencing analysis (Supplementary Material, Table S1). In these situations, the intensity change in the spot could not be unequivocally assigned to a single target. In other cases, the same protein was consistently identified in more than one spot. Some pertinent examples of identified proteins include the protein gelsolin, identified in three peptide spots, and the protein glial fibrillary acidic protein (GFAP), identified in two spots. Gelsolin had abundance changes (fold increases) ranging from 1.31 to 1.46, while GFAP had abundance changes ranging from 1.26 to 1.41 in av mouse cochleae compared with control samples (Supplementary Material, Table S1). Most significant was the data for the protein cochlin, encoded by coagulation factor C homolog (Coch), which was identified in 20 spots. The abundance changes for cochlin ranged from −1.59 to 2.22, depending on the particular protein spot examined (Supplementary Material, Table S1).
Cochlin is significantly up-regulated in the USH1F model cochlea
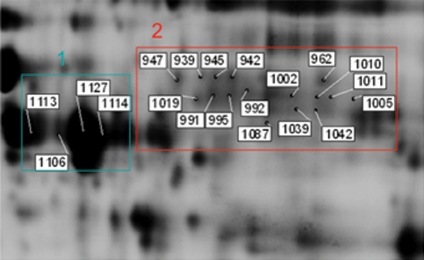
In this study, a specific set of the gel spots identified as cochlin was significantly up-regulated (greater than 2-fold increase) in av mouse cochleae, whereas a second set of spots, at lower overall molecular weight and at a more acidic overall pI, was significantly down-regulated (greater than 1.5-fold decrease). Figure 3 shows 16 protein spots containing cochlin that were up-regulated (box 2) and 4 protein spots containing cochlin that were down-regulated (box 1). This raises the possibility that different isoforms of cochlin may have different roles or respond differently in the pathophysiology of the USH1F model. To assign some structural information to the multiple isoforms observed for cochlin, the tandem MS data for 19 spots were researched using MASCOT software (the data from spot 20 were not of sufficient quality to include in this analysis), and the peptides detected were mapped to the cochlin sequence and its relevant domains (Fig. 4). Figure 5 shows the 36 peptides that were identified in the MASCOT searches of the 19 spots with the sequence of each peptide, the MASCOT score and spot of origin indicated. These peptides cover ∼80% of the total sequence of cochlin. Consensus glycosylation sites are expected at positions 102 and 223. No peptides containing residue 102 are observed, and two spots (962, 1005) show a peptide with residues 223–235. Other posttranslational modifications such as phosphorylation, methylation and acetylation were not detected in any of the protein spots.
Figure 3.
2D gel image showing spots identified as cochlin. This image shows 16 protein spots containing cochlin that were up-regulated (box 2) and 4 protein spots containing cochlin that were down-regulated (box 1).
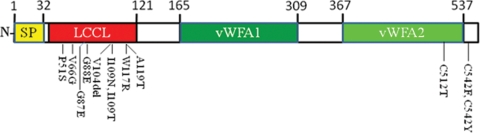
Figure 4.
A schematic showing predicted domains of the cochlin protein and known mutations in patients with DFNA9. The predicted domains are signal peptide (SP), Limulus Factor C, Cochlin and Late gestation lung protein (Lg11) (LCCL), and two von Willerbrand factor A-like domains (vWFA1 and 2). The cochlin protein in mice has 552 amino acid residues and shares ∼94% sequence identity with the human ortholog. The domain structure is illustrated in Figure 4. The protein is secreted and has an SP followed by a region with homology to factor C in Limulus (FCH/LCCL). The remainder of the sequence includes two successive von Willebrand factor A-like domains (vWFA1 and vWFA2) that may function to bind collagen isoforms (52). The nine reported mutations in the N-terminal LCCL/FCH domain and three mutations in the C-terminus of cochlin associated with DFNA9 deafness are shown here (53). In addition, N-linked glycosylation sites were predicted to include residues N-102 and N-223.
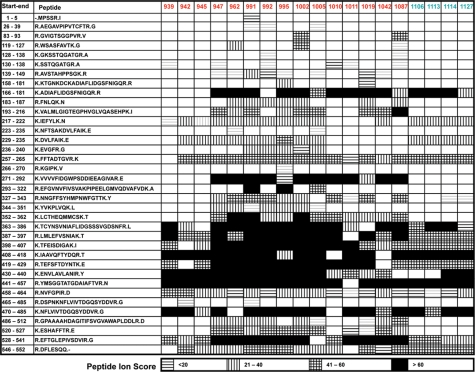
Figure 5.
Sequence of cochlin annotated by peptides identified in the database search. Peptides were detected from tryptic digests of 19 isoforms resolved on 2D gel by LC-MS/MS, followed by MASCOT search. Peptides detected are indicated by filling columns depending on ion score. The peptides in black and grey indicate the best match and non-best match for a corresponding query of MS spectrum, respectively.
Typically, to reject a sufficient number of false-positive identifications when analyzing unknown spots, a high MASCOT score (e.g. over 60) is recommended. However, the presence of the cochlin protein in each of these spots is not in question as every spot has at least three peptides with MASCOT scores above 60. Thus, we can relax the search criteria to permit analysis of peptides with MASCOT scores that would be considered ‘low-scoring’ when using a full-database search. As expected, some peptides, based on their physico-chemical properties that are favorable for mass spectrometry identification, are consistently observed in nearly all the spots. For example, peptide 408–418 is observed in 17 spots, whereas peptide 458–464 is observed in all of the 19 spots. Other peptides are infrequently observed; for example, peptide 465–485 is observed in only two spots. Of course, 20% of the sequence is not observed at all. This is not necessarily due to these sequences being absent or less abundant; they may simply have unfavorable ionization and/or fragmentation characteristics compared with the more frequently observed peptides, or specific regions may be refractory to digestion.
The frequency of observing a MASCOT score of 10 or more for the 684 instances in Figure 5 (36 peptides × 19 spots) is 59%. However, the frequency of observed peptides is not evenly distributed. For example, the von Willebrand domain from residues 367–537 has a number of peptides with very high MASCOT scores. Alternatively, the N-terminal region of cochlin, representing residues 1 to 150–180 in the cochlin sequence (the first eight peptides), has fewer peptides with MASCOT scores above 10; the frequency for all 19 spots is only 18% of the 152 instances (8 peptides × 19 spots). Interestingly, none of the spots from box 1 in Figure 3 (the down-regulated spots) have any MASCOT identified peptides in the N-terminal region whatsoever. We examined whether this lack of MASCOT identified peptides, for these four spots, was significant. We used an unpaired t-test to compare the values of the mean number of peptides (for the first 8 peptides) with significant MASCOT scores for the first 15 spots (mean = 1.80) versus the number of peptides with significant MASCOT scores for the last 4 spots (mean = 0). This comparison gives a two-tailed P-value of 0.0095 (http://www.graphpad.com/quickcalcs/ttest1.cfm). Thus, the absence of MASCOT identified N-terminal peptides for the four spots from box 1 is considered very significant. Each of these four spots has a good MASCOT score for peptide 166–181, confirming that sequence including the first von Willebrand domain is present in these isoforms. The data suggest a hypothesis in which the four spots in box 1 represent cochlin isoforms missing the N-terminal LCCL domain while the spots in box 2 represent full-length cochlin, as they have identified peptides across the full gene sequence. This interpretation suggests that the level of cochlin, especially its full-length isoform, is up-regulated in the av model.
To help determine a cause of the increased cochlin levels detected by proteomic analysis, qPCR assays were performed on 30-day-old av and control mouse cochleae, and levels of Coch mRNA expression were examined. Coch mRNA transcript levels were found to be 2.35-fold higher in av mouse cochleae relative to control samples (Table 1). The ΔΔCt method was employed to calculate fold changes between each normal sample and its paired affected sample (12). A fold change equals 2−ΔΔCt. A higher Ct (cross-over threshold) value represents lower gene expression. The replicate Ct data from the qPCR assays involving Coch are found in Supplementary Material, Table S2. These results demonstrate that the increased levels of full-length cochlin protein detected in the av cochlea by proteomic analysis can be explained by increased transcription of the Coch gene.
Table 1.
QPCR analysis of Coch mRNA expression in the av mouse cochlea at P30
| AV-3J #1 | AV-3J #2 | AV-3J #3 | NC #1 | NC #2 | NC #3 | |
|---|---|---|---|---|---|---|
| Avg Ct: COCH | 19.97 | 19.80 | 19.10 | 20.17 | 19.73 | 18.90 |
| Avg Ct: GAPDH | 24.13 | 21.80 | 22.43 | 21.43 | 22.07 | 21.10 |
| ΔCt: COCH | −4.17 | −2.00 | −3.33 | −1.27 | −2.33 | −2.20 |
| AV-3J - NC | ||||||
| ΔΔCt: COCH | −1.23 | |||||
| Fold change: COCH | 2.35 |
qPCR results measuring Coch mRNA expression in the USH1F model (av-3J mouse cochlea) compared with control mouse cochlea at 30 days. av-3J mouse cochleae were found to have a 2.35-fold increase in Coch mRNA compared with control mouse cochleae. ΔCt represents normalized gene expression level (normalized to its own internal control housekeeping gene GAPDH). A ΔΔCt value (an average value) was obtained by averaging and comparing ΔCt for AV-3J and control samples. A fold change equals 2(−ΔΔCt). A higher Ct value represents lower gene expression.
Protein–protein interaction networks
To utilize all the data obtained through proteomic analysis, every target identified in spots that exhibited statistically significant changes in Supplementary Material, Table S1 was entered into Metacore, a software program that generates protein–protein interaction networks from supplied target lists using curated literature data on protein interactions. The resulting sub-networks include a subset of the target proteins from Supplementary Material, Table S1 connected to each other through other proteins that are related based on the literature mining of the interactions database. These become proteins of interest in the examination of the biological basis of the phenotype. In this case, the analysis yielded seven statistically significant candidate protein networks based on mapping the targets; four of the candidate protein networks with the highest statistically significant P-values are discussed below (Table 2).
Table 2.
Protein networks of statistical significance generated by Metacore (top four)
| No. | Elements | Processes | Size | Target | P-value ↑ | G-score |
|---|---|---|---|---|---|---|
| 1 | TCP1-beta, TCP1, tubulin alpha, c-myc, VHL, SP1, actin, tubulin gamma 1 etc. | actin filament depolymerization (5.7%; 3.375e − 07); actin filament polymerization (7.5%; 1.068e − 06); microtubule cytoskeleton organization and biogenesis (7.5%; 1.495e − 06); protein folding (13.2%; 1.570e − 06); regulation of progression through cell cycle (17.0%; 3.492e − 06) | 53 | 29 | 1.437e − 61 | 46.6395 |
| 2 | COX Va, cytochrome c oxidase, H2O2, c-myc, AP-1, caspase-3, calpastatin, tear lipocalin etc. | response to oxidative stress (8.2%; 7.512e − 05); induction of apoptosis (10.2%; 8.262e − 05); electron transport (12.2%; 9.994e − 05); vasodilation (4.1%; 2.522e − 04), glycolysis (6.1%; 2.816e − 04) | 51 | 28 | 1.897e − 59 | 46.5686 |
| 3 | HSP47, collagen V, SMAD3, c-myc, insulin, caspase-3, actin, alpha-2/beta-1 integrin etc. | phosphate transport (9.8%; 1.160e − 06); regulation of progression through cell cycle (15.7%; 2.251e − 05); induction of apoptosis by intracellular signals (5.9%; 4.448e − 05); actin filament polymerization (5.9%; 5.820e − 05); positive regulation of cell proliferation(11.8%; 5.839e − 05) | 51 | 28 | 1.897e − 59 | 48.3445 |
| 4 | GDI2, G3P2, RAB2A, huntingtin, OCT-1, SP1, c-myc, RAB-11A etc. | Notch signaling pathway (11.8%; 3.121e − 09); anagen (7.8%; 6.306e − 08); proteolysis during protein maturation (5.9%; 1.203e − 07); cell fate specification (7.8%; 3.371e − 07); chromosome organization and biogenesis (sensu Eukaryota) (9.8%; 5.548e − 07) | 51 | 26 | 6.094e − 54 | 46.5060 |
The table shows the elements (protein) in the network, the important processes involving these proteins, the number of nodes (size), the numbers of the seed target proteins included in the network and the statistical significance as P-value and G-score as defined in Materials and Methods. The networks were ranked based on the hypergeometric distribution representing the probability of a particular mapping of an experiment to a map (or network, or process etc.) to arise by chance, considering the number of proteins in the experiment versus the number of proteins in a map (resp. network, process, etc.) within the ‘full set’ of all proteins in all maps. P-values and G-scores are defined in Materials and Methods.
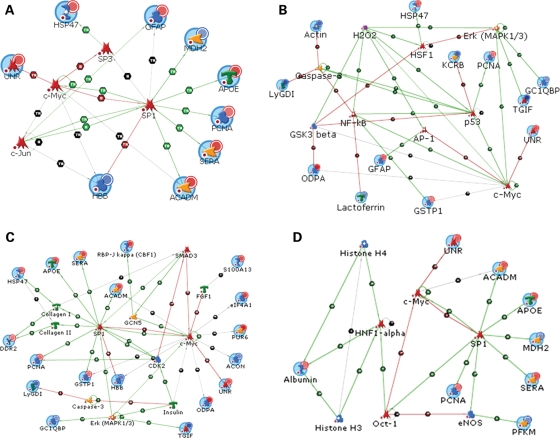
Candidate protein network 1 (P = 1.437e − 61; G-score = 46.6395), shown in Supplementary Material, Figure S1, is implicated in such processes as actin filament polymerization and depolymerization, microtubule cytoskeleton organization and biogenesis, protein folding, regulation of progression through the cell cycle and apoptosis. We used Metacore to extract a sub-network from network 1 based on protein–protein interactions that were well validated in the curated literature, e.g. were not from high-throughput screening data (Fig. 6A). The protein interactomics’ data annotation for this sub-network was done using the Metacore interpretation guidelines. A detailed description for this is given in the Figure 6 legends section. Briefly, identified nodes (based on the 2D DIGE data) and inferred nodes (found by the network analysis algorithm) are constrained to be connected by direct contact. Up- and down-regulation of the inferred nodes is then predicted upon the interpreted biochemical phenomenon. For example, on the periphery of sub-network 1 are seen a number of the targets detected in the 2D gel experiment (as evidenced by the large blue circle above the protein symbol). Connecting the targets in Figure 6A are signaling proteins SP1, SP3 and c-jun, which are all predicted to be increased in the av mouse, whereas c-myc is predicted to be decreased based on the logic of the network defined by the curated interactions database. This prediction comes from the fact that meaningful biological annotation for inferred nodes similar to the one made in Figure 6A is done by considering the cumulative effect of resulting interactions from the neighboring nodes on such inferred nodes. In Figure 6A, although the network object color of the inferred nodes is all red, each of the three nodes SP1, c-jun and SP3 is predicted to be up-regulated considering the net transcriptional control effect with SP1 and c-jun having more pronounced prediction of end-result of up-regulation. c-myc, on the contrary, receives a net down-regulatory prediction outcome due to the same network interpretation logic. This kind of careful annotatory approach has to be resorted to, even in the case of more complex network architecture. SP1, the major hub of this sub-network, is a transcription factor that binds to GC box promoters and activates mRNA synthesis. c-jun, another interesting member of this pathway, is a transcription factor that, when up-regulated, has been implicated in cochlear apoptosis (13,14).
Figure 6.
Sub-networks derived from Metacore analysis. (A)–(D) Detected and predicted proteins and other molecules in predicted interactions. The large blue circle above the protein symbol indicates targets detected in the 2D gel experiment. Proteins shown without this large blue circle are predicted to be present in the network by Metacore analysis. In the upper right quadrant of the proteins with the blue circle is smaller circle which indicates either up-regulation (red) or down-regulation (blue) of the protein in the av mouse compared with control. The symbol at the center of each large blue circle (directly above the protein letter symbol) represents the type of protein (i.e. kinase, receptor ligand, cell membrane protein etc.). The small red circle at the lower left of the identifying symbols refers to a network link termination due to expansion restrictions put when building such a network. The designations TR and B denote relationship labels such as transcription regulation (TR) and binding interaction (B). The line colors indicate the effect of interactions with red indicating down-regulation, green indicating up-regulation and grey indicating no effect. The solidity of the line color indicates the strength of the protein interactions. This explanation applies to all the network data shown here as well as those shown in Supplementary Material. (A) Sub-network derived from network 1. c-jun, SP1 and SP3 levels are predicted to be increased whereas c-myc levels are predicted to be decreased. (B) Sub-network derived from network 2. Protein levels of NF-kB, p53, caspase-3, HSF1 and AP-1 and second messenger molecule H2O2 are predicted to be increased. Protein levels of GSK3 beta, ERK, and c-myc are predicted to be decreased. (C) Sub-network derived from network 3. Protein levels of SP1, SMAD3 and caspase-3 are predicted to be increased. Protein levels of c-myc, ERK and insulin are predicted to be decreased. (D) Sub-network derived from network 4. Protein levels of SP1, OCT-1, HNF1-alpha and histone H4/3 are predicted to be increased. Protein levels of c-myc and eNOS are predicted to be decreased.
Candidate protein network 2 (P = 1.897e − 59; G-score = 46.5686), shown in Supplementary Material, Figure S2, is important in the cells’ response to oxidative stress, induction of apoptosis, electron transport, vasodilation and glycolysis. The sub-network derived from network 2 is shown in Figure 6B. In this pathway, levels of NF-kB, p53, caspase-3, HSF1 and AP-1 are predicted to be increased, while levels of GSK3β, ERK and c-myc are predicted to be decreased in the av model versus wild-type cochleae. Two notable components in these extracted pathways are p53 and caspase-3. p53 is a tumor suppressor protein that plays an important role in the regulation of the cell cycle. Specifically, it functions as a checkpoint for progression from G1 to S. Increased levels of p53 lead to arrest of the cell cycle and cell death in cases of severe stress. Caspase-3 is involved in the activation cascade of caspases that are responsible for the internal pathway of apoptosis. It is responsible for cleaving poly (ADP-ribose) polymerase (PARP), cleaving and activating sterol regulatory element binding proteins (SREBPs) and cleaving caspase-6, -7 and -9.
Candidate protein network 3 (P = 1.897e − 59; G-score = 48.3445), shown in Supplementary Material, Figure S3, is involved in phosphate transport, regulation of progression through the cell cycle, induction of apoptosis, actin filament polymerization and positive regulation of cell proliferation. The sub-network derived from network 3 is shown in Figure 6C. Protein levels of SP1, SMAD3 and caspase-3 in the sub-network are predicted to be increased in the av mouse model, whereas protein levels of c-myc, ERK and insulin are predicted to be decreased. SP1 and caspase-3 have briefly been discussed in networks 1 and 2, respectively. SMAD3 is another protein that serves to modulate transcription. The protein encoded by c-myc is multifunctional and plays a role in cell cycle progression, apoptosis and cellular transformation. It activates growth-related genes, and therefore, it is reasonable that it would be decreased in the process of degeneration. ERK is involved in the initiation and regulation of meiosis, mitosis and post-mitotic functions. It is also logical that this protein would be decreased in pathways related to degeneration.
Candidate protein network 4 (P = 6.094e − 54; G-score = 46.5060), shown in Supplementary Material, Figure S4, is involved in the notch signaling pathway, anagen, proteolysis during protein maturation, cell fate specification, chromosome organization and biogenesis. The sub-network derived from network 4 is shown in Figure 6D. Protein levels of SP1, OCT-1, HNF1-α and histone H4/3 are predicted to be increased in this sub-network, while protein levels of c-myc and eNOS are predicted to be decreased. Oct-1 is a transcription factor that activates the promoters of small nuclear RNAs and the gene for H2B. Histones can act globally to affect transcription levels.
Validation of proteomic and bioinformatic analysis
To provide some validation of the proteomic analysis that was performed and the predictions that were given by Metacore, select candidates from this analysis were further scrutinized by focused testing. Namely, we performed qPCR analysis to measure the levels of p53 transcript in av mouse cochleae versus controls at day 30. Transcript levels of p53 were found to be consistently elevated by 38% (1.38-fold increase) in av mouse cochleae compared with controls at day 30 (Table 3). These results confirm the up-regulation of p53 predicted by Metacore analysis. qPCR experiments were also carried out to measure the transcript levels of two transcription factors, Nrf2 (Nfe2l2 gene) and Brn3a (Pou4f1 gene), that have roles in cell stress response and survival (15–18). Results of these qPCR experiments are shown in Table 3. Nrf2 and Brn3a transcript levels were shown to be increased by 11% (1.11-fold) and 71% (1.71-fold), respectively, relative to controls at P30. The replicate Ct data from the qPCR assays involving p53, Nrf2 and Brn3a are given in Supplementary Material, Table S3. It is important to note that the co-efficient of variation 0.00–0.02 (Tables 2 and 3) indicates negligible variation between replicates, indicating that the results are reliable and, therefore, biologically significant.
Table 3.
qPCR analysis of candidate genes
| Average Cts | AV-3J #1 | AV-3J #2 | AV-3J #3 | NC #1 | NC #2 | NC #3 |
|---|---|---|---|---|---|---|
| TRP53 | 26.87 | 26.67 | 26.63 | 26.57 | 26.33 | 25.73 |
| NFE2L2 | 28.73 | 26.90 | 27.07 | 27.00 | 26.90 | 26.30 |
| POU4F1 | 33.83 | 32.90 | 32.90 | 33.87 | 33.17 | 32.00 |
| GAPDH | 26.03 | 23.53 | 24.40 | 23.87 | 24.20 | 22.97 |
| ΔCts | ||||||
| TRP53 | 0.83 | 3.13 | 2.23 | 2.70 | 2.13 | 2.77 |
| NFE2L2 | 2.70 | 3.37 | 2.67 | 3.13 | 2.70 | 3.33 |
| POU4F1 | 7.80 | 9.37 | 8.50 | 10.00 | 8.97 | 9.03 |
| ΔΔCts | AV-3J - NC | |||||
| TRP53 | −0.47 | |||||
| NFE2L2 | −0.14 | |||||
| POU4F1 | −0.78 | |||||
| Fold change | AV-3J - NC | |||||
| TRP53 | 1.38 | |||||
| NFE2L2 | 1.11 | |||||
| POU4F1 | 1.71 | |||||
qPCR results measuring Pou4f1 (encoding Brn3a), Nfe2l2 (encoding Nrf2) and Trp53 (encoding p53) mRNA expression in the USH1F model (AV-3J mouse cochlea) compared with control mouse cochlea at 30 days. AV-3J mouse cochleae were found to have a 1.38-fold increase in Trp53 mRNA, a 1.11-fold increase in Nfe2l2 mRNA and a 1.71-fold increase in Pou4f1 mRNA compared with control mouse cochleae. ΔCt represents normalized gene expression level (normalized to its own internal control housekeeping gene GAPDH). A ΔΔCt value (an average value) was obtained by averaging and comparing ΔCt for AV-3J and control samples. A fold change equals 2(−ΔΔCt). A higher Ct value represents lower gene expression.
DISCUSSION
The role of cochlin
Supplementary Material, Table S1, listing proteins identified from differentially expressed spots in the USH1F model, provides an initial step in the development of a mouse auditory protein database. Of the proteins identified in multiple spots on the gel, cochlin was consistently identified in 20 spots, warranting further discussion. Cochlin, the encoded protein product of COCH, is a secreted protein whose function is not completely understood, but it is believed to have a role in the extracellular matrix (ECM) and the structure of the basilar membrane (19). Cochlin appears to have numerous isoforms with heterogeneity caused partly by N-glycosylation and proteolytic cleavage [as shown by the presence of the smaller isoforms after secretion into media of transfected cells but not in the cell pellets (20)], and potentially by alternative splicing as well (19,21). Robertson et al. (20) identified two major isoforms, a presumptive, full-length cochlin at 60 kDa and an isoform with the N-terminal FCH domain truncated with an apparent molecular weight (MW) of ∼40 kDa. Previous 2D gel work also identified a full-length isoform, as well as smaller isoforms that exhibited successive truncations at the N-terminus with MW values of the predominant isoforms seen at 44 and 40 kDa (22). Recent data have also indicated that the abundance of the full-length isoform decreases relative to the abundance of the N-terminally truncated form as a function of age in the rat with the full-length isoform predominant at day 24 but decreased relative to the truncated form at day 70 (23).
Cochlin was found to be the most abundant protein in the bovine inner ear (22), and more recently in the mouse and human cochleae (24), by proteomic analysis. Cochlin immunostaining is restricted to structures of mesodermal origin and is not found in neuroectodermally derived structures. The organ of Corti, neuronal cell bodies and central and peripheral axons lack cochlin (24). Mutations in COCH have been shown to cause DFNA9, an autosomal dominant disease characterized by nonsyndromic deafness with vestibular dysfunction (25). In DFNA9, cochlin was localized to insoluble, homogeneous eosinophilic deposits in the ECM (24). It is unknown whether the cochlin deposits are a primary cause or result of the pathologic state. Additionally, a 64 kDa isoform of cochlin interacts with Choline transporter-like protein 2 (CTL2), which is a multi-transmembrane protein expressed in inner ear supporting cells and is a target of antibody-induced hearing loss (26). Cochlin is also a prominent target antigen in CD4+ T-cell mediated autoimmune hearing loss (27,28). Additionally, increased levels of cochlin have been found in the trabecular meshwork of glaucomatous mice and are presumed to have a causal role in the development of increased intraocular pressure and glaucoma (29). Cochlin is clearly involved in various pathologies of the inner ear and eye.
Overall, the available evidence indicates that the up-regulated cochlin spots appear to correspond to full-length protein isoforms, while the down-regulated spots appear to lack N-terminal sequences, consistent with truncation or splice variation that removes the FCH/LCCL domain. This is supported by the overall lower pI of the down-regulated spots, as the N-terminus is basic; the tighter distribution of the spots, as truncation of the N-terminus would delete one of the two putative glycosylation sites; and the overall size differences between the two spot clusters. The qPCR assays shows concomitant increase in Coch mRNA expression supporting proteomic analysis of the av cochlea. It should be noted that the qPCR primers were designed toward the 5′ end of Coch mRNA and the corresponding amino acid sequence is represented at the N-terminus of the full-length cochlin protein isoform. Therefore, the qPCR results reflect the expression of Coch mRNA that encodes full-length cochlin. The potential deleterious effects that are conveyed by this molecular phenotype are currently unknown, but the observed change is counter to what has been reported for normal development. For example, normal development in a rat is accompanied by increases in an N-terminally truncated form with age (23). This increase in Coch expression and cochlin protein levels may or may not be unique to the USH1F model. Increased Coch transcription may in fact be a generalized response to stress that is present in all of the Usher syndromes as well as other hearing disorders.
The transcription factors p53, Nrf2 and Brn3a, all associated with cell stress response and survival, were analyzed in this study, and transcript levels for p53 and Brn3a are significantly increased in the av model. Furthermore, increased expression of Nrf2 and Brn3a has been shown to be associated with increased cochlin expression in human glaucomatous trabecular meshwork, while mutations in the Brn3a and Nrf2 transcription factor binding sequences of the COCH gene have been associated with lower cochlin expression (30). Nrf2 is known to play a role in coordinating the up-regulation of cytoprotective genes involved in combating oxidative stress (17) and enhancing cell survival (18). Brn3a plays an important role in the survival of neurons in the inner ear (31,32), in part by regulating apoptosis (15,16). The fact that a relative abundance of transcription factors Nrf2 and Brn3a has been shown to be associated with increased cochlin expression is very interesting and provides support for the results obtained in our study. Our results show increased cochlin expression, along with certain transcription factors, is associated with stress response signaling and cell survival in the USH1F model at 30 days. It follows that cochlin expression may be associated with stress response signaling and cell survival and, therefore, affected by the levels of transcription factors similarly associated with these processes. The up-regulation of cochlin, combined with the up-regulation of transcription factors—linked to Coch mRNA expression and cell stress response and survival—suggests a unifying hypothesis to explain the observed degenerative pathology in the av mouse cochlea. Hypothesis: loss of hair cells in the av cochlea triggers activation of transcription factors linked to stress response and survival, including p53, Nrf2 and Brn3a, which in turn leads to increased expression of cochlin protein. We propose that the increased level of cochlin may lead to a secondary autoimmune response, as occurs in some autoimmune hearing disorders. A secondary autoimmune response could account for the loss of supporting cells in the av cochlea (USH1F model). Altered levels of cochlin isoforms may also lead to deposits of cochlin, as occurs in DFNA9. An interesting note about individuals with DFNA9 is that hearing loss and vestibular deterioration occur over decades, presumably due to gradual changes in the extracellular matrix containing cochlin deposits. Cochlin is not detected in the eye, but its expression is detected in the eye with glaucoma (25). Individuals with USH1F are deaf and have vestibular dysfunction at birth or in early infancy, and then gradually become blind over the first few decades. Due to cochlin's pathologic role in the inner ear in individuals with DFNA9 and in the eye in individuals with glaucoma, it may be possible that cochlin has a role in the gradual blindness that occurs in USH1F. It needs to be pointed out that the function of the down-regulated, truncated Cochlin isoform is unknown and deciphering the role of this Cochlin form may improve our understanding in relation to this hypothesis.
Protein network analysis
Of the networks generated by Metacore, networks 1 and 2 deserve particular mention. Network 1 has the highest P-value and G-score of the generated candidate networks. One particular protein in this network, the transcription factor c-jun, has been implicated in cochlear apoptosis in the setting of noise exposure (13) and hair cell apoptosis in the setting of neomycin toxicity (14). This is very interesting, especially considering that caspase-3 and p53 (two other proteins involved in apoptosis), are key proteins in network 2. Of even greater significance is that c-jun inhibitors, such as D-JNK-1, have been shown to prevent hair cell death caused by gentamicin (33), and hearing loss induced by neomycin and electrode insertion trauma (34). Additionally, all-trans retinoic acid, a known inhibitor of the JNK signaling pathway, has been shown to inhibit apoptosis (35) and recover hearing loss due to noise exposure (36) in mice. The fact that c-jun has been shown to be involved in the apoptotic pathway leading to hair cell death in noise exposure, ototoxicity and trauma makes it an interesting candidate biomarker and therapeutic target for USH1F.
Network 2 has the second highest P-value and G-score of the candidate networks. It has two proteins, caspase-3 and p53, in its extracted sub-network that are known to be involved in the cells’ response to stress and the initiation of apoptosis. Caspase-3 has been shown to be involved in the apoptotic pathways that result from noise-induced hearing loss (37) and from cisplatin ototoxicity (38). In fact, the caspase-3 inhibitor, z-DEVD-fmk, has been shown to prevent hearing loss in guinea pigs treated with ototoxic concentrations of cisplatin (38). p53 has been implicated in initiating apoptosis in cochlear and vestibular hair cells exposed to cisplatin and other stresses. Pifithrin-α, a synthetic inhibitor of p53, has been shown to protect hair cells from the toxic effects of cisplatin (39). This inhibitor may hold promise in the setting of USH1F early in development to prevent the death of hair cells and subsequent degeneration of spiral ganglion neurons.
Another interesting candidate biomarker for USH1F is the protein gelsolin, which was shown to be up-regulated in av cochleae when compared with controls. Gelsolin is a known substrate of caspase-3 and may serve as a marker of apoptosis and the subsequent morphologic changes in mutant cochleae (40). Alternatively, it has also been shown that gelsolin may help prevent apoptosis, and thus, may represent a compensatory response to neurosensory degeneration in the inner ear (41). Gelsolin concentration in cerebrospinal fluid from patients with certain neurologic disorders has been shown to be altered and may serve as an important diagnostic biomarker (42). Additionally, GFAP, which is normally expressed in supporting cells (43), was shown to be up-regulated in av cochleae and may serve as a marker of a compensatory response before the supporting cells degenerate in the USH1F model. Serum-GFAP has been shown to be up-regulated in cases of cerebral hemorrhage (44) and traumatic brain injury (45) and may indeed serve as a marker of clinical severity in USH1F.
The four candidate networks described in this study provide the initial framework for elucidating the pathways involved in hair cell and spiral ganglion cell degeneration. If either of these cell types can be spared, then it may be possible to prevent complete deafness or restore hearing (e.g. using cochlear prostheses) to patients with USH1F and other hearing disorders.
A thorough analysis of the four candidate networks revealed several proteins or molecules whose levels are either predicted to be elevated or depressed in av mouse cochleae, an important model for deafness for USH1F. Proteins and other molecules with predicted increased expression include NF-kB, p53, collagens 1 and 2, SP1, Ca2+, c-jun, SP3, caspase-3, HSF1, AP-1, SMAD3, GCN5, OCT-1, HNF-1 and 2, histone and H2O2. Proteins with predicted decreased expression include PXR, c-fos, ESR1, c-myc, GSK3 beta, ERK, insulin, CDK2, eNOS and STAT3. In addition to cochlin, which was detected by MS, a number of these molecules may serve as useful biomarkers for the diagnosis or progression of USH1F, or hearing diseases in general. Future studies will be needed to confirm the value of these candidate biomarkers.
Role of p53 in the degenerative process
p53, as described above, is a tumor suppressor protein that has the ability to activate DNA repair enzymes, initiate apoptosis and serve as a checkpoint for G1/S transition in the cell cycle. The transcript levels of p53, as determined by qPCR, were shown to be consistently increased in av mouse cochleae on day 30, when 90% of the hair cells have degenerated and the spiral ganglion cells are starting to degenerate. It thus appears that p53 has a central role in hair cell degeneration and in initiating apoptosis in spiral ganglion cells. p53 inhibitors such as pifithrin-α may have therapeutic value in USH1F and should be examined in future studies. The fact that transcripts of Brn3a, another transcription factor associated with cell stress response and survival, were elevated in the USH1F model at 30 days provides additional support for the role of the stress response in the USH1F model at this time point. As discussed above, the elevation in the levels of these transcripts may play a role in the cochlin up-regulation that was detected by proteomic analysis (30).
In summary, this study provides both relevant and important information about global protein expression in the cochlea of the USH1F mouse model for deafness, and it serves as a guide for future studies of USH1F. Since other mouse models for deafness in Usher syndrome type 1 (e.g. USH1B, 1C and 1D) show a similar degenerative profile as the USH1F model, namely the loss of hair cells followed by the loss of spiral ganglion cells, the proteomic analysis reported here may also be useful for other subtypes of Usher syndrome type 1.
MATERIALS AND METHODS
Sample preparation
Mice
The Ames waltzer (av) mouse, a model for deafness in USH1F and DFNB23, harbors a mutation in the Pcdh15 gene (Pcdh15). In our previous work, we characterized different mutations in Pcdh15 (or alleles of Pcdh15) including Pcdh15av-Tg and Pcdh15av-3J (4,9,10). The protein coded by Pcdh15 (PCDH15) is predicted to be a transmembrane protein. Pcdh15av-Tg and Pcdh15av-3J alleles are presumed to be functional null alleles because mutation in these alleles is predicted to code for Pcdh15 protein lacking the entire transmembrane domain and all of the cytoplasmic domains (4). Pcdh15av-Tg allele arose in and is maintained in the FVB/N background, whereas Pcdh15av-3J allele arose in and is maintained in the C57BL/6J (B6) background. B6 background carries a mutation in cadherin 23, which is associated with age-related hearing (Ahl) loss; to avoid this confounding factor, we used Pcdh15av-Tg in the FVB/N background for this study. The use of mice was approved by the Institutional Animal Care and Use Committee at Case Western Reserve University.
Mouse tissue harvest
After euthanasia, mouse heads were severed, wrapped in aluminum foil and frozen in −80°C for 1 min. This allowed for the rapid cooling of tissue in the head, including the inner ear. Working quickly through dissection, the brain was removed. The temporal bone was placed on a pre-chilled petri dish, and the brain cavity was bathed with ice-cold wash buffer (5 mm magnesium acetate, 10 mm Tris, pH 8.0). The petri dish was placed under the dissection microscope, and the cochlea was dissected from both ears (under 5 min) and transferred to a microfuge tube containing 1 ml of ice-cold wash buffer. The tube was inverted a few times to wash off blood or tissue debris sticking to the cochlea. The tissue was then transferred to a dry tube (pre-chilled on ice) and stored at −20°C for later use (protein extraction). We found these steps to be important to minimize protein degradation and obtain consistent yields from small tissue such as cochlea.
Protein sample preparation
Normal and affected cochleae of 30-day-old mice were used in this study. The cochleae from three normal or av mice (6 cochleae) were pooled to have a sufficient amount of protein to run a gel. In total, cochleae from 24 mice (12 wild type and 12 mutants) were used to run four gels. The cochleae were harvested from the animal as rapidly as possible and placed on a petri dish containing ice-cold lysis buffer and excess tissue surrounding the cochlea was removed under a dissection micoroscope. Then, homogenization was carried out in an Eppendorf tube with liquid nitrogen using a pestle. Proteins were extracted in 120–150 µl of lysis buffer, consisting of 4% CHAPS, 30 mm tris, 2 m thiourea, and 7 m urea. A three-cycle ice/water bath sonication was used to aid in cell disruption. Protein degradation and dephosphorylation were inhibited with a protease inhibitor cocktail (Sigma Aldrich, St. Louis, MO, USA) and a wide-spectrum phosphatase inhibitor (Roche, Nutley, NJ, USA), respectively. The protease inhibitor cocktail and phosphatase inhibitor were added to the lysis buffer per the manufacturer's recommended concentration. The lysate was then centrifuged for 10 min at 12 000 rpm, after which the protein fraction was withdrawn by pipette. Colorimetric assay was performed using the 2D-Quant kit (GE Healthcare, Waukesha, WI, USA) to determine the protein concentration of the lysate. Protein fractions were stored at −80°C.
2D gel electrophoresis
The protein samples were labeled with Cydyes (spectrally resolvable fluorophores) and multiplexed on gels using the 2D DIGE system, which has previously been described by our group and others (46–48). This system allows the simultaneous analysis of up to three distinct samples and contains an internal standard when one of the CyDyes is used to label a pooled sample, containing a proportional amount of each sample in the experiment. This helps to account for variations in protein loading during the image analysis step. Image analysis was performed using 50 µg protein samples labeled by Cydyes. If a gel was to be used for spot excision, it was loaded with an additional 350 µg of unlabeled, pooled sample adequate for trypsin digestion and peptide detection by LC-MS (2). It should be noted that the 2D DIGE method, although it provides a very precise measure of relative abundance, has a few limitations. Generally, only proteins in the highest 25% of abundance are typically detected by the method. Often, biological regulatory proteins of interest may be of low abundance and can be missed in the analysis. Also, membrane proteins are generally poorly represented in 2D gels, as are highly basic proteins (49,50).
First dimension
Each Cydye (Cy2, Cy3 and Cy5) was reconstituted in N,N-dimethylforamide (DMF), and a 400 pmol quantity was used to label 50 µg of protein at a pH of 8–9. The pooled internal standard (PIS) described above was labeled with Cy2, whereas normal and affected cochlea samples were labeled with either Cy3 or Cy5. Cy3 and Cy5 were alternately swapped for normal and affected cochlea on subsequent samples to eliminate dye-specific effects that may bias image analysis. Labeling of the protein samples was performed in the dark for 30 minutes, followed by quenching of the reaction by 10 mm lysine. Labeled protein samples were mixed with 2× sample buffer [8 mm urea, 2% dithiothreitol (DTT), 4% CHAPS, 2% Pharmalyte] and placed on ice for 10 min. The samples were then loaded onto non-linear 3–10 Immobiline DryStrips (GE Healthcare), placed in a strip holder, and focused with an IPGphor system following a step gradient protocol ranging from 30 to 8000 volts for 27 h. The strips were stored at −80°C until second dimension analysis was performed. Also, for the protein identification experiment, two pooled, unlabeled 350 µg samples were focused and stored individually until they could be separated in the second dimension on different gels for the purpose of spot excision. The unlabeled sample was stained with Deep Purple gel stain (GE Healthcare).
Second dimension
The Ettan DALT Twelve apparatus was used for separation by molecular weight. The stored DryStrips were rehydrated in 10 ml of re-equilibration buffer [8 m urea, 100 mm Tris–HCl (pH 6.8), 30% glycerol, 1% SDS] along with 20 mm DTT and 45 mg/ml iodoacetamide, respectively, for 10 min each in two separate steps. Afterward, the DryStrips were placed on a 12.5% polyacrylamide gel between two glass plates submerged in running buffer (40% acylamide-Bis, 150 mm Tris, 10% SDS, 10% APS, 10% TEMED, along with a few crystals of bromophenol blue), and then covered with a 0.5% agarose solution. Separation occurred at 15°C at 0.5 watts/gel, and then 1.0 watts/gel for 15 h, and was stopped when the bromophenol dye reached the bottom of the gel.
Gel fixing
Gels used for spot excision were previously fixed to one of the glass plates covering the gel with silane (GE Healthcare). After the experiment had stopped, the gels were fixed with 50% methanol and 7.5% acetic acid.
Image analysis
The Typhoon 9400 variable mode imager (GE Healthcare) was used to scan the gels. Laser lights specific for each CyDye fluorophore excitation wavelength were used so that each fluorophore would be excited separately (Fig. 2). An initial tuning was performed at a resolution of 1000 µm, after which a final scan at a 100 µm resolution was performed. Gels to be used for spot excision were stained with Deep Purple stain as described above, imaged using 532/560 wavelength light, and then stored in the dark at 4°C. After imaging each gel for the three fluorophores, the images were imported into the imaging analysis software DeCyder (GE Healthcare) for spot detection, spot matching (intra-gel) and determination of statistically significant biological variation (inter-gel) based on the measurement of relative abundance change after background subtraction and normalization to the internal standard. A statistically significant spot has a mean fold change greater than or equal to ±50%, for a paired t-test P-value of less than or equal to 0.05, or greater than or equal to ±10%, for a paired t-test P-value of less than or equal to 0.01. A manual check was performed on each statistically significant spot to ensure that it was likely a protein spot and not a gel artifact. After passing statistical significance and the manual check, a protein spot pick list was created and exported to the software controlling the Ettan robotic spot picker (GE Healthcare). The spots were excised with a 3 mm core and placed in a 96-well plate for digestion.
In-gel digestion
The excised gel spots were washed in 50 µl of both 25 mm ammonium bicarbonate (ABC) and 50% acetonitrile (ACN) for 10 min in quadruplicate. A volume of 10 mm dithiothreitol (DTT) prepared fresh in 30 µl of 25 mm ABC was added to each gel spot well. The samples were incubated at 56°C for 45 min and then cooled at room temperature for 20 min. The DTT was removed, and 30 µl of 55 mm iodoacetamide (IAM) was added to each gel spot well. The samples were then incubated at room temperature in the dark for 45 min. Following the incubation, the IAM was removed and the samples were washed in 50 µl of both 25 mm ABC and 50% ACN. Excised gel spots were covered with 10 µl of a 100 ng trypsin solution, incubated at room temperature for 10 min, immersed with 15 µl of 25 mm ABC and placed in a 37°C water bath overnight for protein digestion. The reaction was quenched the following day by adding 7 µl of 1% (final concentration) formic acid. The digested peptides were extracted by adding 30 µl of 50% ACN/5% formic acid, and then submitting the samples to vortexing for 30 min, centrifugation and then sonication for 5 min.
Mass spectrometry and database software
An LTQ mass spectrometer (Thermo Electron Corp., Bremen, Germany) equipped with an Ettan MDLC (GE Healthcare) was used to analyze most samples by tandem MS. A Finnigan LTQ FT hybrid mass spectrometer (Thermo Electron Corp.) operating in a positive ion mode was used to run six samples. A volume of 2.5 µl of tryptic peptides were desalted on a C-18 pre-column (PepMap 100, 300 × 5 µm particle size, 100 Å, Dionex), then separated on a reverse phase column (C-18, 75 µm × 150 nm, 3 µm, Dionex) using mobile phase A (0.1% formic acid) and B (84% acetonitrile, 0.1% formic acid) with a linear gradient of 2% per min, beginning with 100% A. Peptides were subsequently infused at a flow rate of 100 nl/min via a Pico Tip emitter (New Objective, Inc., Woburn, MA, USA) at a voltage of 1.8 kV. Mass spectra were recorded in the ion trap, and MS2 spectra were acquired for the five most intense ions in the LTQ employing collision energy of 35 eV and an isolation width of 2.5 Da.
A search against an indexed mouse peptide database was performed using Bioworks version 3.2 (Thermo Electron Corp.) employing the SEQUEST software. A peptide mass tolerance of 2.5 Da and a fragment tolerance of 1.0 Da were used in the search. Search parameters included complete carbamidomethylation of cysteine, partial methionine oxidation and two missed cleavage sites. Statistically significant peptides had a P-value of <0.001, and cross correlation (Xcorr) values of 1.9, 2.5 and 3.0 for 1+, 2+ and 3+ ions, respectively. The protein probability cutoff was P < 0.001, and each ‘hit’ necessarily required at least three peptides for consideration, with rare exceptions for low-molecular-weight proteins. The theoretical pI and molecular weight were compared with the observed values on the gel image for each peptide that had not been filtered out by the search parameters. Post-translational modifications and cleavage products for the 19 cochlin-containing spots were re-searched against the rodent NCBInr database using MASCOT v.2.1.03 (51). Searches were performed with carbamidomethylation of cysteine, with partial oxidation of methionine, with up to three missed cleavages allowed, and with mass tolerance of 1.5 and 0.8 Da for MS and MS/MS, respectively. The data are illustrated in Figure 5 with relative shading; MASCOT scores from 10 to 20 provide the least dark shading while those with a score of 60 or more are darkest.
Network analysis
We used Metacore from GeneGo Inc. (version 4.6 build 12332) to search for protein–protein interaction networks. We imported all targets from Supplementary Material, Table S1 and the corresponding mean fold change values represented by the significantly changing proteins identified in our experiments. We used the ‘Analyze Network’ feature in MetaCore to build our networks; 70 targets were mapped in the software and subsequently analyzed. This option is selective for networks most saturated with the targets in our import list, and it returns an array of unbiased small networks that are likely to be statistically significant and biologically relevant, the two possibilities we wanted to assess (52,53). Pre-filtering options were species (mouse) and tissue (cochlea). Networks generated were sorted by P-value and G-score. The P-value is based on the level of saturation of each sub-network with proteins from the original user selected set. The G-score is used to downgrade networks that result from proteins with data values being assembled around nodes with high degree (many interactions to a gene) without protein data values. Therefore, in general, sub-networks with high positive G-scores would be highly saturated with the uploaded list while excluding high degree nodes without data values. Only manually curated evidence for interactions were considered, and networks were limited to 50 nodes or less. We chose the top four networks from the search results as ranked by both G-score and P-value. MetaCore is proprietary software. More information about this software can be obtained from www.genego.com.
qPCR of cochlin, p53, Nrf2 and Brn3a
Normal and affected mice were euthanized, and cochlea were dissected and stored at −80°C immediately. Total RNA was isolated by Trizol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. Extracted RNA was quantitated with a NanoDrop spectrophotometer, and RNA quality analyzed using Agilent Bioanalyzer with RNA Nano Chip 6000. Reverse transcription was performed with SABiosciences (Frederick, MD, USA) RT2 First Strand Kit (C-03), which contains an effective genomic DNA elimination step and a built-in external RNA control. RNA input was 1000 ng per RT reaction. qRT-PCR was performed on the cDNA templates generated in an iCycler Bio-Rad in 96 well format in a volume of 24 µl per reaction. SYBR green dye was used for the qPCR assay. Primer catalog numbers used from SABiosciences are PPM26157A for Cochlin, PPM34723A for Brn3a, PPM24614A for Nrf2, PPM02931B for TRP53 (p53) and PPM02946A for GAPDH (used as an internal control). The amount of cDNA template in each reaction was equivalent to 10 ng of input RNA. Assays were performed in triplicate. Data analysis was performed via Excel. The ΔΔCt method was employed to calculate fold changes between each normal sample and its paired affected sample (12). ΔCt represents normalized gene expression level (normalized to its own internal control housekeeping gene GAPDH). ΔCt for test and control samples were averaged and compared to obtain the ΔΔCt value (an average value). CV, or coefficient of variation, is calculated using the formula STDEV/average.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by a grant from Rainbow Board of Trustees, Rainbow Babies and Children's Hospitals, University Hospitals Case Medical Center, R21-DC07866 from the NIDCD to K.A. and by the Translational Technologies Core of the CTSA, UL1-RR024989, from the National Institutes of Health to M.C.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Nahid Robertson and Cynthia Morton from Harvard Medical School for critical reading and helpful comments during the preparation of the revised manuscript.
Conflict of Interest statement: None declared.
REFERENCES
- 1.Adato A., Michel V., Kikkawa Y., Reiners J., Alagramam K.N., Weil D., Yonekawa H., Wolfrum U., El-Amraoui A., Petit C. Interactions in the network of Usher syndrome type 1 proteins. Hum. Mol. Genet. 2005;14:347–356. doi: 10.1093/hmg/ddi031. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed Z.M., Riazuddin S., Ahmad J., Bernstein S.L., Guo Y., Sabar M.F., Sieving P., Riazuddin S., Griffith A.J., Friedman T.B., et al. PCDH15 is expressed in the neurosensory epithelium of the eye and ear and mutant alleles are responsible for both USH1F and DFNB23. Hum. Mol. Genet. 2003;12:3215–3223. doi: 10.1093/hmg/ddg358. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed Z.M., Riazuddin S., Bernstein S.L., Ahmed Z., Khan S., Griffith A.J., Morell R.J., Friedman T.B., Riazuddin S., Wilcox E.R. Mutations of the protocadherin gene PCDH15 cause Usher syndrome type 1F. Am. J. Hum. Genet. 2001;69:25–34. doi: 10.1086/321277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alagramam K.N., Murcia C.L., Kwon H.Y., Pawlowski K.S., Wright C.G., Woychik R.P. The mouse Ames waltzer hearing-loss mutant is caused by mutation of Pcdh15, a novel protocadherin gene. Nat. Genet. 2001;27:99–102. doi: 10.1038/83837. [DOI] [PubMed] [Google Scholar]
- 5.Alagramam K.N., Yuan H., Kuehn M.H., Murcia C.L., Wayne S., Srisailpathy C.R., Lowry R.B., Knaus R., Van Laer L., Bernier F.P., et al. Mutations in the novel protocadherin PCDH15 cause Usher syndrome type 1F. Hum. Mol. Genet. 2001;10:1709–1718. doi: 10.1093/hmg/10.16.1709. [DOI] [PubMed] [Google Scholar]
- 6.Becirovic E., Ebermann I., Nagy D., Zrenner E., Seeliger M.W., Bolz H.J. Usher syndrome type 1 due to missense mutations on both CDH23 alleles: investigation of mRNA splicing. Hum. Mutat. 2008;29:452. doi: 10.1002/humu.9526. [DOI] [PubMed] [Google Scholar]
- 7.Overlack N., Maerker T., Latz M., Nagel-Wolfrum K., Wolfrum U. SANS (USH1G) expression in developing and mature mammalian retina. Vision Res. 2008;48:400–412. doi: 10.1016/j.visres.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 8.Reiners J., van Wijk E., Marker T., Zimmermann U., Jurgens K., te Brinke H., Overlack N., Roepman R., Knipper M., Kremer H., et al. Scaffold protein harmonin (USH1C) provides molecular links between Usher syndrome type 1 and type 2. Hum. Mol. Genet. 2005;14:3933–3943. doi: 10.1093/hmg/ddi417. [DOI] [PubMed] [Google Scholar]
- 9.Alagramam K.N., Kwon H.Y., Cacheiro N.L., Stubbs L., Wright C.G., Erway L.C., Woychik R.P. A new mouse insertional mutation that causes sensorineural deafness and vestibular defects. Genetics. 1999;152:1691–1699. doi: 10.1093/genetics/152.4.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alagramam K.N., Zahorsky-Reeves J., Wright C.G., Pawlowski K.S., Erway L.C., Stubbs L., Woychik R.P. Neuroepithelial defects of the inner ear in a new allele of the mouse mutation Ames waltzer. Hear. Res. 2000;148:181–191. doi: 10.1016/s0378-5955(00)00152-0. [DOI] [PubMed] [Google Scholar]
- 11.Zheng Q.Y., Rozanas C.R., Thalmann I., Chance M.R., Alagramam K.N. Inner ear proteomics of mouse models for deafness, a discovery strategy. Brain Res. 2006;1091:113–121. doi: 10.1016/j.brainres.2006.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Murai N., Kirkegaard M., Jarlebark L., Risling M., Suneson A., Ulfendahl M. Activation of JNK in the inner ear following impulse noise exposure. J. Neurotrauma. 2008;25:72–77. doi: 10.1089/neu.2007.0346. [DOI] [PubMed] [Google Scholar]
- 14.Sugahara K., Rubel E.W., Cunningham L.L. JNK signaling in neomycin-induced vestibular hair cell death. Hear. Res. 2006;221:128–135. doi: 10.1016/j.heares.2006.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiggins A.K., Wei G., Doxakis E., Wong C., Tang A.A., Zang K., Luo E.J., Neve R.L., Reichardt L.F., Huang E.J. Interaction of Brn3a and HIPK2 mediates transcriptional repression of sensory neuron survival. J. Cell. Biol. 2004;167:257. doi: 10.1083/jcb.200406131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eng S.R., Kozlov S., Turner E.E. Unaltered expression of Bcl-2 and TAG-1/axonin-1 precedes sensory apoptosis in Brn3a knockout mice. Neuroreport. 2003;14:173. doi: 10.1097/00001756-200302100-00002. [DOI] [PubMed] [Google Scholar]
- 17.Calkins M.J., Jakel R.J., Johnson D.A., Chan K., Kan Y.W., Johnson J.A. Protection from mitochondrial complex II inhibition in vitro and in vivo by Nrf2-mediated transcription. Proc. Natl Acad. Sci. USA. 2005;102:244. doi: 10.1073/pnas.0408487101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kensler T.W., Wakabayashi N., Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 2007;47:89. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 19.Kommareddi P.K., Nair T.S., Raphael Y., Telian S.A., Kim A.H., Arts H.A., El-Kashlan H.K., Carey T.E. Cochlin isoforms and their interaction with CTL2 (SLC44A2) in the inner ear. J. Assoc. Res. Otolaryngol. 2007;8:435–446. doi: 10.1007/s10162-007-0099-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robertson N.G., Hamaker S.A., Patriub V., Aster J.C., Morton C.C. Subcellular localisation, secretion, and post-translational processing of normal cochlin, and of mutants causing the sensorineural deafness and vestibular disorder, DFNA9. J. Med. Genet. 2003;40:479–486. doi: 10.1136/jmg.40.7.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sekine K., Ikezono T., Matsumura T., Shindo S., Watanabe A., Li L., Pawankar R., Nishino T., Yagi T. Expression of cochlin mRNA splice variants in the inner ear. Audiol. Neurootol. 2009;15:88–96. doi: 10.1159/000231634. [DOI] [PubMed] [Google Scholar]
- 22.Ikezono T., Omori A., Ichinose S., Pawankar R., Watanabe A., Yagi T. Identification of the protein product of the Coch gene (hereditary deafness gene) as the major component of bovine inner ear protein. Biochem. Biophys. Res. Commun. 2001;1535:258–265. doi: 10.1016/s0925-4439(00)00101-0. [DOI] [PubMed] [Google Scholar]
- 23.Shindo S., Ikezono T., Ishizaki M., Sekiguchi S., Mizuta K., Li L., Takumida M., Pawankar R., Yagi T. Spatiotemporal expression of cochlin in the inner ear of rats during postnatal development. Neurosci. Lett. 2008;444:148–152. doi: 10.1016/j.neulet.2008.07.091. [DOI] [PubMed] [Google Scholar]
- 24.Robertson N.G., Cremers C.W.R.J., Huygen P.L.M., Ikezono T., Krastins B., Kremer H., Kuo S.F., Liberman M.C., Merchant S.N., Miller C.E., et al. Cochlin immunostaining of inner ear pathologic deposits and proteomic analysis in DFNA9 deafness and vestibular dysfunction. Hum. Mol. Genet. 2006;15:1071–1085. doi: 10.1093/hmg/ddl022. [DOI] [PubMed] [Google Scholar]
- 25.Robertson N.G., Lu L., Heller S., Merchant S.N., Eavey R.D., McKenna M., Nadol J.B., Miyamoto R.T., Linthicum F.H., Lubianca Neto J.F., et al. Mutations in a novel cochlear gene cause DFNA9, a human nonsyndromic deafness with vestibular dysfunction. Nat. Genet. 1998;20:299–303. doi: 10.1038/3118. [DOI] [PubMed] [Google Scholar]
- 26.Nair T.S., Kozma K.E., Hoefling N.L., Kommareddi P.K., Ueda Y., Gong T.-W., Lomax M.I., Lansford C.D., Telian S.A., Satar B., et al. Identification and characterization of choline transporter-like protein 2, an inner ear glycoprotein of 68 and 72kDa that is the target of antibody-induced hearing loss. J. Neurosci. 2004;24:1772–1779. doi: 10.1523/JNEUROSCI.5063-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baek M.-J., Park H.-M., Johnson J.M., Altuntas C.Z., Jane-Wit D., Jaini R., Solares C.A., Thomas D.M., Ball E.J., Robertson N.G., et al. Increased frequencies of cochlin-specific T cells in patients with autoimmune sensorineural hearing loss. J. Immunol. 2006;177:4203–4210. doi: 10.4049/jimmunol.177.6.4203. [DOI] [PubMed] [Google Scholar]
- 28.Solares C.A., Edling A.E., Johnson J.M., Baek M.-J., Hirose K., Hughes G.B., Tuohy V.K. Murine autoimmune hearing loss mediated by CD4+ T cells specific for inner ear peptides. J. Clin. Invest. 2004;113:1210–1217. doi: 10.1172/JCI18195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhattacharya S.K., Rockwood E.J., Smith S.D., Bonilha V.L., Crabb J.S., Kuchtey R.W., Robertson N.G., Peachey N.S., Morton C.C., Crabb J.W. Proteomics reveal Cochlin deposits associated with glaucomatous trabecular meshwork. J. Biol. Chem. 2005;280:6080–6084. doi: 10.1074/jbc.M411233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Picciani R.G., Diaz A., Lee R.K., Bhattacharya S.K. Potential for transcriptional up-regulation of cochlin in glaucomatous trabecular meshwork: a combinatorial bioinformatic and biochemical analytical approach. Invest. Ophthalmol. Vis. Sci. 2008;50:3106–3111. doi: 10.1167/iovs.08-3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang S.W., Mu X., Bowers W.J., Kim D.-S., Plas D.J., Crair M.C., Federoff H.J., Gan L., Klein W.H. Brn3b/Brn3c double knockout mice reveal an unsuspected role for Brn3c in retinal ganglion cell axon outgrowth. Development. 2002;129:467–477. doi: 10.1242/dev.129.2.467. [DOI] [PubMed] [Google Scholar]
- 32.Ma L., Lei L., Eng S.R., Turner E., Parada L.F. Brn3a regulation of TrkA/NGF receptor expression in developing sensory neurons. Development. 2003;130:3525–3524. doi: 10.1242/dev.00582. [DOI] [PubMed] [Google Scholar]
- 33.Ylikoski J., Xing-Qun L., Virkkala J., Pirvola U. Blockade of c-Jun N-terminal kinase pathway attenuates gentamicin-induced cochlear and vestibular hair cell death. Hear. Res. 2002;163:71–81. doi: 10.1016/s0378-5955(01)00380-x. [DOI] [PubMed] [Google Scholar]
- 34.Eshraghi A.A., Wang J., Adil E., He J., Zine A., Bublik M., Bonny C., Puel J.-L., Balkany T.J., Van De Water T.R. Blocking c-Jun-N-terminal kinase signaling can prevent hearing loss induced by both electrode insertion trauma and neomycin ototoxicity. Hear. Res. 2007;226:168–177. doi: 10.1016/j.heares.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 35.Ahn J.H., Kang H.H., Kim Y.-J., Chung J.W. Anti-apoptotic role of retinoic acid in the inner ear of noise-exposed mice. Biochem. Biophys. Res. Commun. 2005;335:485–490. doi: 10.1016/j.bbrc.2005.07.114. [DOI] [PubMed] [Google Scholar]
- 36.Shim H.J., Kang H.H., Ahn J.H., Chung J.W. Retinoic acid applied after noise exposure can recover the noise-induced hearing loss in mice. Acta. Otolaryngol. 2009;129:233–238. doi: 10.1080/00016480802226155. [DOI] [PubMed] [Google Scholar]
- 37.Nicotera T.M., Hu B.H., Henderson D. The caspase pathway in noise-induced apoptosis of the chinchilla cochlea. J. Assoc. Res. Otolaryngol. 2003;4:466–477. doi: 10.1007/s10162-002-3038-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J., Ladrech S., Pujol R., Brabet P., Van De Water T.R., Puel L. Caspase inhibitors, but not c-Jun NH2-terminal kinase inhibitor treatment, prevent cisplatin-induced hearing loss. Cancer Res. 2004;64:9217–9224. doi: 10.1158/0008-5472.CAN-04-1581. [DOI] [PubMed] [Google Scholar]
- 39.Zhang M., Liu W., Ding D., Salvi R. Pifithrin-alpha suppresses p53 and protects cochlear and vestibular hair cells from cisplatin-induced apoptosis. Neuroscience. 2003;120:191–205. doi: 10.1016/s0306-4522(03)00286-0. [DOI] [PubMed] [Google Scholar]
- 40.Kothakota S., Azuma T., Reinhard C., Klippel A., Tang J., Chu K., McGarry T.J., Kirschner M.W., Koths K., Kwiatkowski D.J., et al. Caspase-3-generated fragment of gelsolin: effector of morphological change in apoptosis. Science. 1997;278:294–298. doi: 10.1126/science.278.5336.294. [DOI] [PubMed] [Google Scholar]
- 41.Harms C., Bosel J., Lautenschlager M., Harms U., Braun J.S., Hortnagl H., Dirnagl U., Kwiatkowski D.J., Fink K., Endres M. Neuronal gelsolin prevents apoptosis by enhancing actin depolymerization. Mol. Cell. Neurosci. 2004;25:69–82. doi: 10.1016/j.mcn.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 42.Kulakowska A., Drozdowski W., Sadzynski A., Bucki R., Janmey P.A. Gelsolin concentration in cerebrospinal fluid from patients with multiple sclerosis and other neurological disorders. Eur. J. Neurol. 2008;15:584–588. doi: 10.1111/j.1468-1331.2008.02133.x. [DOI] [PubMed] [Google Scholar]
- 43.Rio C., Dikkes P., Liberman M.C., Corfas G. Glial fibrillary acidic protein expression and promoter activity in the inner ear of developing and adult mice. J. Comp. Neurol. 2002;442:156–162. doi: 10.1002/cne.10085. [DOI] [PubMed] [Google Scholar]
- 44.Vos P.E., van Gils M., Beems T., Zimmerman C., Verbeek M.M. Increased GFAP and S100beta but not NSE serum levels after subarachnoid haemorrhage are associated with clinical severity. Eur. J. Neurol. 2006;13:632–638. doi: 10.1111/j.1468-1331.2006.01332.x. [DOI] [PubMed] [Google Scholar]
- 45.Nylen K., Ost M., Csajbok L.Z., Nilsson I., Blennow K., Nellgard B., Rosengren L. Increased serum-GFAP in patients with severe traumatic brain injury is related to outcome. J. Neurol. Sci. 2006;240:85–91. doi: 10.1016/j.jns.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 46.Chang J., Chance M.R., Nicholas C., Ahmed N., Guilmeau S., Flandez M., Wang D., Byun D.-S., Nasser S., Albanese J.M., et al. Proteomic changes during intestinal cell maturation in vivo. J. Proteomics. 2008;71:530–546. doi: 10.1016/j.jprot.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marouga R., David S., Hawkins E. The development of the DIGE system: 2D fluorescence difference gel analysis technology. Anal. Bioanal. Chem. 2005;382:669–678. doi: 10.1007/s00216-005-3126-3. [DOI] [PubMed] [Google Scholar]
- 48.Yohannes E., Chang J., Christ G.J., Davies K.P., Chance M.R. Proteomics analysis identifies molecular targets related to diabetes mellitus-associated bladder dysfunction. Mol. Cell. Proteomics. 2008;7:1270–1285. doi: 10.1074/mcp.M700563-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McGuire J.F., Casado B. Proteomics: a primer for otologists. Otol. Neurotol. 2004;25:842–849. doi: 10.1097/00129492-200409000-00032. [DOI] [PubMed] [Google Scholar]
- 50.Wasinger V.C., Corthals G.L. Proteomic tools for biomedicine. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2002;771:33–48. doi: 10.1016/s1570-0232(02)00125-3. [DOI] [PubMed] [Google Scholar]
- 51.Perkins D.N., Pappin D.J., Creasy D.M., Cottrell J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 52.Nagy I., Trexler M., Patthy L. The second von Willebrand type A domain of cochlin has high affinity for type I, type II and type IV collagens. FEBS Lett. 2008;582:4003–4007. doi: 10.1016/j.febslet.2008.10.050. [DOI] [PubMed] [Google Scholar]
- 53.Robertson N.G., Jones S.M., Sivakumaran T.A., Giersch A.B.S., Jurado S.A., Call L.M., Miller C.E., Maison S.F., Liberman M.C., Morton C.C. A targeted Coch missense mutation: a knock-in mouse model for DFNA9 late-onset hearing loss and vestibular dysfunction. Hum. Mol. Genet. 2008;17:3426–3434. doi: 10.1093/hmg/ddn236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.