Abstract
Protease-activated receptors (PARs) are G-protein-coupled receptors with an active role in host defense. The two most highly expressed members of the PAR family in gingival epithelial cells (GECs) are PAR1 and PAR2. The major virulence factors of periodontal pathogen Porphyromonas gingivalis are its proteases which can activate PAR2. However, little is known about the function of PARs in GECs when they are activated by their endogenous agonist enzymes. The purpose of this study was to characterize how the expression of innate immune markers is modulated when PAR1 and PAR2 are activated by their agonist enzymes, thrombin and trypsin, respectively. Here, we report that activation of PAR1 and PAR2 induces cell proliferation at low concentration. Activation of PAR via proteolytic activity of thrombin and trypsin induces expression of CXCL5/ENA-78 and CCL20/MIP3α in a concentration-dependent manner. Induction of CXCL5 via PAR1 was inhibited in the presence of PAR1 cleavage blocking antibodies and by PAR1 siRNA. The induction of CXCL5 and CCL20 via PAR2 was inhibited by PAR2 siRNA. These findings indicate an active role in innate immune responses by PAR1 and PAR2 in GECs. Modulation of innate immunity by PARs may contribute to co-ordinated and balanced immunosurveillance in GECs.
Keywords: Protease-activated receptors, innate immunity, oral keratinocytes
Introduction
Oral epithelium is constantly exposed to different pathogenic and non-pathogenic bacteria, but it maintains homeostasis with a sophisticated immune system which relies on cellular receptors and on the expression of innate immune markers. These markers of innate immunity include antimicrobial peptides, a range of chemokines and cytokines, as well as adhesion molecules that initiate cell–cell interaction. Receptors that contribute to innate immunity in oral epithelial cells are pattern recognition receptors (PRRs) including Toll-like receptors (TLRs), NOD-like receptors (NLRs),1 and protease-activated receptors (PARs).
Protease-activated receptors are a family of G-protein-coupled receptors with a unique mechanism of activation. Cleavage of the receptor within the N-terminal exodomain unmasks a cryptic sequence which serves as a tethered ligand and interacts with the receptor to initiate coupling to G proteins.2 Molecular cloning has identified four PARs with distinct N-terminal cleavage sites. Thrombin, the classical activator of PARs, is a key regulator of PAR1 and PAR4 with a co-factor effect provided by PAR3,3,4 but it cannot activate PAR2.5–7 Trypsin-like enzymes such as trypsin, mast cell tryptase and neutrophil proteinase-3 are the activators of PAR2. Among the known members of this family of receptors, PAR1 and PAR2 are prominently expressed in the gastrointestinal tract8 and gingiva,9 while no expression of PAR4 has been detected in gingival epithelial cells (GECs).9 In gingival epithelium, arginine-specific cysteine proteases (gingipains Rgp) from Porphyromonas gingivalis, a major pathogen associated with adult periodontal disease, activate PARs and induce expression of IL-6, the antimicrobial peptide β-defensin-2, and CCL20.9–11 Activation of PAR2 in human GECs and fibroblasts by neutrophil proteinase-3 induces expression of inflammatory markers IL-8 and MCP-1.12,13 Also, it has been shown that topical application of a PAR2 agonist peptide causes periodontitis and exacerbates existing periodontitis in rat models.14 In addition to their pro-inflammatory function, the anti-inflammatory effects of PAR2 activation in murine model of colitis has been reported.15 Despite the elucidation of many aspects of PARs in different cell types, little is known about their function when they are activated by their agonist enzymes, thrombin and trypsin or trypsin-like enzymes in gingival epithelium.
Thrombin, a member of coagulation cascade, is a serine protease. The active form of thrombin is present at the site of gingival tissue injury and inflammation.16 In addition, gingipains (HRgpA and RgpB) release thrombin from pro-thrombin,17 which indicates the likelihood of increased levels of thrombin during periodontal disease. Also, the presence of trypsin-like enzymes in gingival crevicular fluid in chronic periodontitis18 implies the importance of trypsin-like proteases as signaling molecules through activation of PAR2. Because PARs are activated by both physiological and pathogenic products, understanding the consequences of their activation in oral epithelium is of special interest in periodontal health and disease.
In this study, we aimed to investigate the function of PARs by using thrombin and trypsin as the endogenous proteases which are the classical activators of PAR1 and PAR2, respectively. We used a microarray analysis as a tool to explore the major functions of PAR1 and PAR2 and further examined the role of PAR1 and PAR2 activation in cell proliferation and innate immune responses in GECs.
Materials and Methods
Reagents
Human α-thrombin (Haematologic Technologies Inc., Essex Junction, VT, USA) and recombinant human trypsin (Polymun Scientific Immunobiologische Forschung GmbH, Vienna, Austria) were used to stimulate GECs in order to activate PAR1 and PAR2, respectively. The inhibitor for thrombin, D-Phe-Pro-Arg-chloromethyl ketone dihydrochloride (PPACK-HCL) was purchased from Calbiochem (La Jolla, CA, USA). Assessment of the efficiency of inhibitory effect of the PPACK on thrombin was assayed with the chromogenic substrate for thrombin, S-2238 (Chromogenix, Lexington, MA, USA). Trypsin activity was inhibited by tosyl-L-lysine chloromethyl ketone (TLCK; Sigma, St Louis, MO, USA). Both thrombin and trypsin were screened for endotoxin contamination using Limulus amebocyte lysate Pyrotell (Cape Cod Inc., Falmouth, MA, USA). Screening thrombin (1 U/ml) and trypsin (1 nM) verified the endotoxin contamination level was lower than 0.03 EU/ml. The anti-PAR1 monoclonal antibodies WEDE15 and ATAP2 were purchased from Coulter Company (Fullerton, CA, USA) and Santa Cruz Biotechnology (Santa Cruz, CA, USA), respectively.
Gingival epithelial cell culture
Primary human GECs were isolated from healthy human gingival tissue samples from patients undergoing third molar extraction at the Department of Oral Surgery, School of Dentistry, University of Washington, in accordance with the Institutional Review Board-approved study. Tissue preparation and cell culture method have been previously described in detail by our group.19 Briefly, cells were grown in serum-free supplemented Keratinocyte Basal Medium (KBM; Cambrex, Walkersville, MD, USA) plus 0.03 mM Ca2+. After 24 h, the medium was replaced with serum-free supplemented KBM plus 0.15 mM Ca2+ and cells were grown to 75–80% confluence. Fourth passage cells were used for all experiments. Due to the possible variation between individual donors, we looked for consistent results in GECs from at least three donors with technical duplicate for each set of experiments, unless otherwise stated.
DNA microarray analysis
Cells were stimulated with thrombin and trypsin or their inactivated forms for 6 h and then harvested for RNA extraction. Quality of RNA was checked with the Agilent 2100 Bioanalyzer. Samples were processed for microarrays at the Center of Array Technology at University of Washington, using Affymetrix GeneChip® Human Genome U133A 2.0 arrays (Affymetrix, Inc., Santa Clara, CA, USA) following the manufacturer’s protocols for labeling and hybridization onto GeneChips (HG-U133A).
Array normalization and data analysis for the microarray experiments were carried out with R statistical software package that is specific for microarray analysis and with the microarray software analysis program Bioconductor,20 and the Bioconductor package gcrma.21 Subsequently, the genes were filtered based upon a 1.5-fold change cut-off and genes with similar change in expression in the presence of inhibitors (PPACK or TLCK) were excluded. Functional enrichment analysis for comparing lists of genes altered by thrombin or trypsin was performed by FATIGO (Fast Assignment and Transference of Information by GO).22
Cell proliferation assay
Cell proliferation was determined using WST-1 reagent (Roche Diagnostics, Indianapolis, IN, USA). Assays were performed in 96-well cell culture plates with initial seeding density of 5 × 103 cells/well in 100 μl of culture medium. The cells were allowed to attach to the plate; after 24 h, the medium was changed and cells were challenged with stimulants. The WST-1 assay was carried out in cells from five different donors and each experiment was performed in triplicate for each experimental condition.
Cell transfection with small-interfering RNA (siRNA)
For gene silencing, we used guaranteed small interfering RNA (siRNA) tagged with Alexa Fluor 488 (Qiagen, Valencia, CA, USA) specific for PAR1 and PAR2.11 Cells were plated in 24-well plates; after 24 h, medium was changed and cells were transfected with 25 nM siRNA against PAR1, PAR2 or non-silencing siRNA using HiPerFect (Qiagen) for 48 h, and subsequently stimulated with thrombin (10 U/ml) or trypsin (10 nM) for 6 h. Non-transfected cells as well as cells treated with non-silencing siRNA or lipid carrier HiPerFect alone served as negative controls. Specific and efficient knock-down of the target genes was assessed by Quantitative RT-PCR (QRT-PCR).
RNA isolation, reverse transcription and quantitative RT-PCR
Total RNA was extracted from cells using RNeasy mini kit (Qiagen). The reverse transcription reaction was performed using 300–1000 ng of total RNA with Oligo(dT)15 primer and the reverse transcription reagent kit (Promega, Madison, WI, USA) in a total volume of 20 μl. Quantitative RT-PCR was performed using 1 μl cDNA, 250 nM primers and 12.5 μl iQ SYBR Green Supermix (Bio-Rad, Hercules, CA, USA) in a total volume of 25 μl. All reactions were carried out in duplicate. Samples without cDNA served as a negative control. Melting curve was performed at the end of each QRT-PCR to ensure the gene product was specific. Glyceraldeyde-3-phosphate dehydrogenase (GAPDH) was used as a house-keeping gene for normalization. The primers for PAR1, PAR2 and CCL20 have been described previously.11 The primer sequences for GAPDH and CXCL5 are as follow: GAPDH-5′, CAAAGTTGTCATGGATGACC; GAPDH-3′, CCATGGAGAAGGCGGGG; CXCL5-5′, TCTGCAAGTGTTCGCCATAG; and CXCL5-3′, TGTCTTCCCTGGGTTCAGAG. Data were analyzed using the Pfaffl method23 to calculate fold change in gene expression.
Enzyme-linked immunosorbent assay (ELISA)
The presence of CCL20 and CXCL5 in GEC culture supernatants was determined by ELISA method following the manufacturer’s protocol (R&D, Minneapolis, MN, USA).
Statistical analysis
To analyze data from QRT-PCR and ELISA, statistical comparisons among groups were performed using one-way analysis of variance followed by Student’s t-test. Data are shown as mean with the SD. A P-value of less than 0.05 was considered as significant.
Results
Genes are differentially expressed when GECs are activated with thrombin versus trypsin
In order to investigate differential gene expression regulated by PAR1 or PAR2 in GECs, global gene expression was analyzed using Affymetrix HG-U133A microarrays. GECs were stimulated with thrombin (10 U/ml which equals approximately 60 nM), thrombin inactivated by PPACK (180 nM), PPACK (180 nM), trypsin (10 nM), trypsin inactivated by TLCK (10 μM), and TLCK (10 μM) or left unstimulated to serve as controls. To minimize possible variation between individual donors, equal amounts of RNA from five different donors, duplicate for each condition, were combined. Data analysis revealed that thrombin altered expression of 1211 genes with more than 1.5-fold changes (406 genes up-regulated and 805 genes down-regulated). The 10 probes with the greatest up-regulation by thrombin were RPS11, RPL38, RPL27, RPS19, RPL38, H3F3A, RPLP2, B4GALT1, RPS20 and TXN. Among them, seven were ribosomal protein (RP) encoding genes, suggesting changes in ribosomal function after PAR1 activation.24,25 Other highly up-regulated genes include cortactin (CTTN), which has been reported as a relevant marker of head and neck squamous cell carcinoma,26 and syndecan4 (SDC4), which is an intrinsic regulator of inflammatory reactions (data not shown).27
Activation by trypsin altered expression of 716 genes with more than 1.5-fold change (320 genes up-regulated and 396 genes down-regulated). IL8 and CCL20 were among the group of the 10 most highly up-regulated genes. Others included H3F3A, COX5B, EEF1D and FTL as well as three ribosomal protein encoding genes (RPS11, RSP19 and RPL14; data not shown).
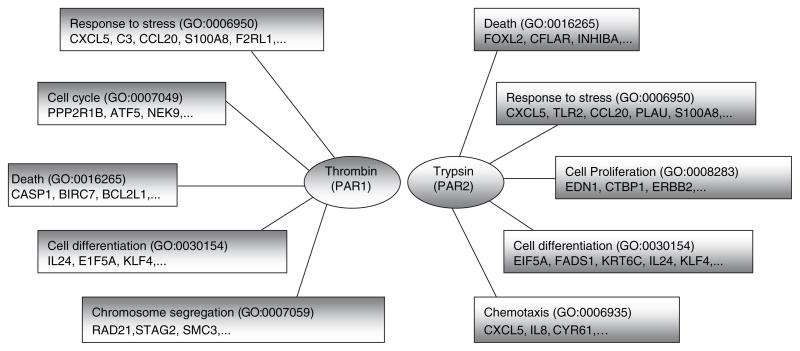
Comparison of genes regulated by thrombin and trypsin using functional analysis suggested both thrombin and trypsin induce changes in regulation of genes related to immune responses, apoptosis and cell differentiation. Thrombin activation had greater effects on cell cycle and chromosome segregation, while trypsin activation evoked changes in genes involved in cell proliferation and chemotaxis (Fig. 1). Results showed that trypsin-stimulated cells have a higher percentage of genes in the category of inflammatory and defense responses compared to cells stimulated with thrombin (data not shown). Within the category of inflammatory and defense responses, both thrombin and trypsin induced expression of numerous chemokines and cytokines (Table 1).
Fig. 1.
Functional gene ontology (GO) analysis using FatiGo. Genes regulated by proteolytic activity of thrombin and trypsin which had more than 1.5-fold change in expression compared to unstimulated cells were used for functional analysis.
Table 1.
Thrombin- and trypsin-induced change in expression of genes associated with inflammatory or defense responses
| Gene Symbol | Thrombin (Fold Change) | Trypsin (Fold Change) | Gene Name |
|---|---|---|---|
| IL8 | 2.3 | 9.1 | interleukin 8 |
| CCL20 | 1.8 | 5.4 | chemokine (C-C motif) ligand 20 |
| CSF2 | 3.0 | 4.4 | colony stimulating factor 2 |
| IL1RN | 1.6 | 3.9 | interleukin 1 receptor antagonist |
| CXCL2 | 3.3 | 3.8 | chemokine (C-X-C motif) ligand 2 |
| CXCL1 | 2.7 | 3.2 | chemokine (C-X-C motif) ligand 1 |
| CXCL3 | 2.4 | 2.9 | chemokine (C-X-C motif) ligand 3 |
| CXCL5 | 1.7 | 2.3 | chemokine (C-X-C motif) ligand 5 |
| IL1B | 1.5 | 2.3 | interleukin 1β |
| TNIP1 | - | 2.2 | TNFAIP3 interacting protein 1 |
| INHBA | 1.7 | 2.2 | inhibin, beta A |
| S100A8 | 1.5 | 2.1 | S100 calcium binding protein A8 |
| SPN | 2.2 | 1.7 | sialophorin (leukosialin, CD43) |
| CD59 | 2.1 | - | complement regulatory protein |
| CSF3 | 2.0 | 1.9 | colony stimulating factor 3 |
Based on these results, we further studied the mitogenic effect of thrombin and trypsin on GECs by using cell proliferation assay. Also, to verify the inflammatory responses triggered in response to these proteases, we focused on CXCL5 and CCL20, as they were up-regulated by PAR1 and PAR2 activation consistently in both preliminary and in the present microarray studies.
Continuous stimulation with thrombin and trypsin increases GEC proliferation
The microarray results suggested a role for PARs in GEC proliferation. To test the mitogenic effect of thrombin and trypsin on GECs, cells were stimulated with increasing doses of thrombin or trypsin, either transiently (1 h) or continuously (24 h). After 24 h, the metabolic activity of the GECs was assessed by WST-1 assay as a measure of proliferation.
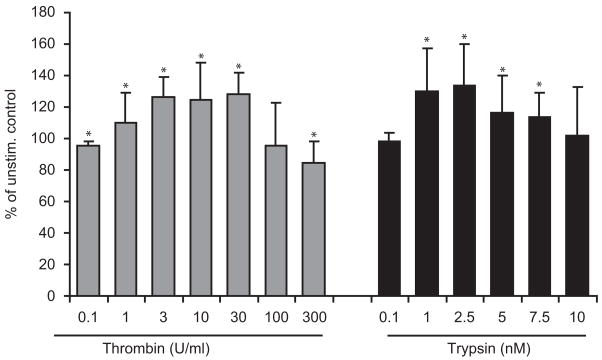
Continuous versus transient stimulation with thrombin and trypsin resulted in different effects. When cells were continuously stimulated, both thrombin and trypsin showed biphasic effects with stimulation at low doses but had no effect or growth inhibition at high doses. Thrombin at 1 U/ml induced a significant increase in cell proliferation (110 ± 4%) which increased up to 30 U/ml (128 ± 3%). Thrombin at 300 U/ml had a significant negative effect on cell proliferation (85 ± 4%). In trypsin-stimulated cells, the maximum mitogenic effect was observed at 2.5 nM (134 ± 7%); higher concentration of trypsin (up to 7.5 nM) induced proliferation at lower rate (113 ± 4%; Fig. 2). With transient stimulation (1 h), neither thrombin nor trypsin had a significant mitogenic effect on GECs (data not shown).
Fig. 2.
The proliferative effect of continued (24 h) stimulation with thrombin (0.1–300 U/ml) and trypsin (0.1–10 nM) after 24 h incubation was assessed by WST-1 assay. Continued application of thrombin for 24 h at 1–30 U/ml significantly increased cell proliferation and at 300 U/ml had negative effect on cell proliferation. Similarly, trypsin at concentration 1–7.5 nM increased cell proliferation significantly with more prominent effect at lower concentration (1, 2.5 nM). Data represent mean ± SD from at least four different donors. *P<0.05 versus unstimulated control.
Thrombin and trypsin induce mRNA expression of CXCL5/ENA-78 and CCL20/MIP3α in a concentration-dependent manner
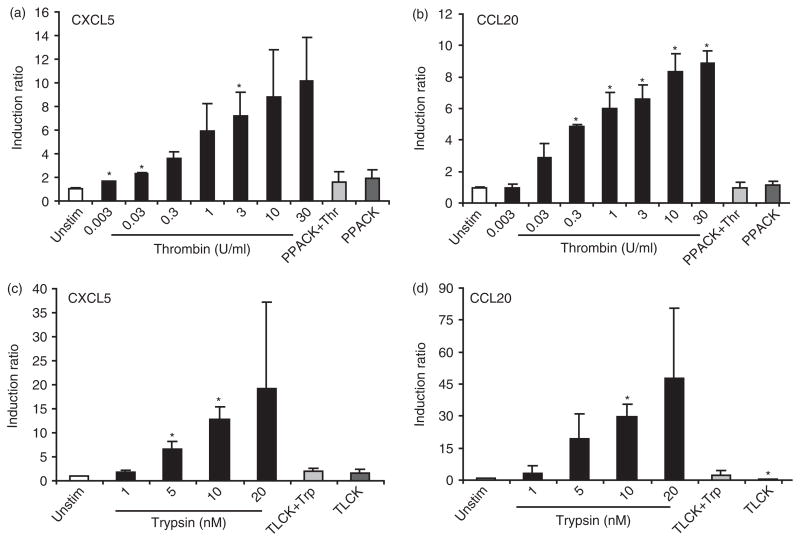
The microarray data showed CXCL5 and CCL20 were both up-regulated in GECs stimulated with thrombin and trypsin. CXCL5 is a potent neutrophil chemo-attractant and CCL20 has antimicrobial function and also attracts immature dendritic cells. Thus, we further verified the concentration-dependent up-regulation of CXCL5 and CCL20 mRNA by QRT-PCR analysis (Fig. 3). A concentration-dependent expression of CXCL5 and CCL20 was induced by thrombin and trypsin. A similar response was seen in the OKF6 cell line, an immortalized oral keratinocyte cell line (hTERT; kindly provided by Dr James Rheinwald; data not shown). Pre-incubation of thrombin and trypsin with their corresponding inhibitors, PPACK and TLCK, respectively, resulted in a blocking effect, confirming that induction of CCL20 and CXCL5 is a result of proteolytic activity of thrombin and trypsin (Fig. 3). The concentration of thrombin and trypsin used here did not show effects on cell morphology. However, trypsin at 20 nM was the maximum concentration the cells could tolerate without detaching from the dish. For subsequent experiments, we used thrombin at 10 U/ml and trypsin at 10 nM.
Fig. 3.
Thrombin and trypsin induce mRNA expression of CXCL5 (a,c) and CCL20 (b,d) in a concentration-dependent manner in GECs. Cells were stimulated with increasing concentration of thrombin (0.003–30 U/ml) or trypsin (1–20 nM) or their inactivated forms with appropriate inhibitors for 6 h. GAPDH was used as an internal control for normalization. The results are expressed as mean ± SD of cells from three different donors with each in duplicate. *P<0.05 versus unstimulated control.
Thrombin and trypsin induce secretion of CXCL5 and CCL20
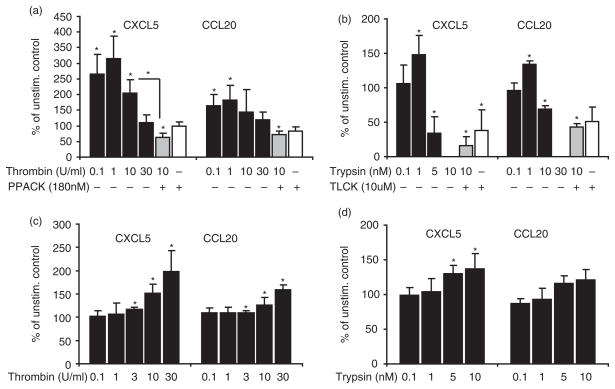
Next, we verified whether the effect of thrombin and trypsin on gene expression was reflected in protein secretion. To investigate this effect, we stimulated GECs at 75–80% confluence with thrombin (0.1–30 U/ml) or trypsin (0.1–10 nM) for 24 h and assessed CXCL5 and CCL20 in the culture supernatant by ELISA. Results showed that thrombin induced the release of CXCL5 and CCL20 from GECs with a peak at 0.1–1 U/ml but, at higher concentration, this effect was reversed; pretreatment of thrombin with the specific inhibitor blocked the release. When cells were stimulated with trypsin, the same pattern was observed; lower doses of trypsin (1 nM) increased secretion of CXCL5 and CCL20 but higher concentrations resulted in decreased CXCL5 and CCL20 in the culture supernatant (Fig. 4A,B). Thus, these results suggest these chemokines may have been degraded in the supernatant especially at higher protease concentration. Therefore, we treated cells with thrombin or trypsin for 1 h, then removed the culture medium and washed the cells thoroughly with fresh medium, prior to incubation for 24 h in fresh medium. The results confirmed concentration-dependent secretion of CXCL5 and CCL20 in culture supernatant in response to both thrombin and trypsin (Fig. 4C,D). Since in similar conditions thrombin and trypsin showed no mitogenic effect in GECs, our data suggest that chemokine induction is part of the immune response and not simply a result of increased cell number.
Fig. 4.
Thrombin and trypsin secrete CXCL5 and CCL20 in the culture supernatant. Cells were stimulated with increased doses of thrombin or trypsin for 24 h. Supernatant was collected and concentration of CXCL5 and CCL20 was determined by ELISA. Higher concentration of stimulants was associated with lower amount of CXCL5 and CCL20 in the supernatant. Pretreatment of thrombin (10 U/ml) and trypsin (10 nM) with appropriate inhibitors abolished the effect of enzymes (a,b). A concentration-dependent pattern was observed 24 h after the onset of short term (1 h) stimulation (c,d). Data from three separate donors each in duplicate are presented. Data are given as percent of mean values of normalized samples to unstimulated control ± SD. *P<0.05 versus unstimulated control.
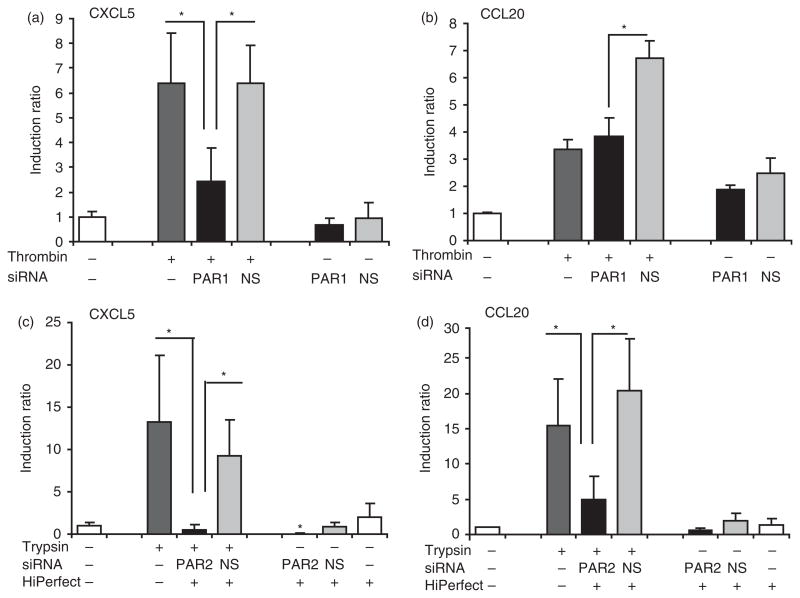
PAR1 knock-down inhibits induction of CXCL5 by thrombin: PAR2 knock-down inhibits trypsin-induced CXCL5 and CCL20 expression
In order to verify the involvement of PARs in the induction of CXCL5 and CCL20 by thrombin and trypsin, we utilized gene silencing using siRNA specific for PAR1 and PAR2. Screening the cells under the microscope 24 h after transfection suggested that more than 80% of cells had taken up the Alexa Fluor 488-labeled siRNA into the nucleus. Utilizing QRT-PCR confirmed less than 10% of mRNA expression of PAR1 and PAR2 remained after transfection with respective siRNAs, compared to transfection with non-silencing siRNA or non-transfected cells and neither PAR1 nor PAR2 siRNA had an off-target effect on the expression of the other one (data not shown). When cells were transfected with PAR1 siRNA and subsequently stimulated with thrombin, CXCL5 gene expression was significantly lower compared to either untransfected cells (P = 0.02) or cells transfected with non-silencing-siRNA (P = 0.02; Fig. 5A), suggesting that thrombin regulates CXCL5 via PAR1. In addition, CXCL5 is also regulated by trypsin via PAR2, as knock-down of PAR2 by siRNA blocked induction of CXCL5 by trypsin compared to non-transfected cells (P = 0.007) and non-silencing siRNA (P = 5.1E-05; Fig. 5C). On the other hand, siRNA for PAR1 and PAR2 had different effects on CCL20 expression. Knocking-down PAR2 resulted in statistically significant reduction of CCL20 expression compared to either non-silencing-siRNA (P = 0.003) or untransfected cells stimulated with trypsin (P = 0.001; Fig. 5D). Although PAR1 siRNA compared to non-silencing siRNA significantly reduced the induction of CCL20 (P = 0.015) in response to thrombin, compared to untransfected cells there was no significant difference (P = 0.45; Fig. 5B). These results confirm activation of PAR1 and PAR2 play a role in the induction of the selected markers of innate immunity in GECs, but suggest there may be differences between PAR1 and PAR2.
Fig. 5.
Silencing PAR1 by PAR1 siRNA inhibits thrombin-induced CXCL5 with no effect on CCL20; silencing PAR2 inhibits induction of CXCL5 and CCL20 by trypsin. Forty-eight hours after transfection with PAR1, PAR2 or non-silencing (NS) siRNA, cells were stimulated with thrombin 10 U/ml or trypsin 10 nM for 6 h. The mRNA expression of CXCL5 (a,c) and CCL20 (b,d) was assessed by QRT-PCR. Data represent mean ± SD from three different donors. *P<0.05.
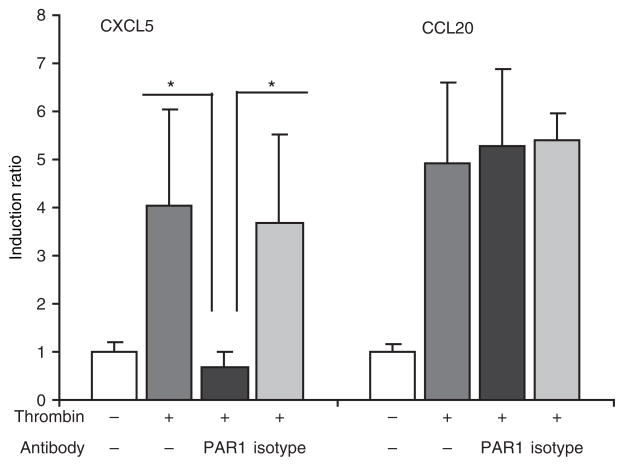
Antibodies against PAR1 inhibit induction of CXCL5 by thrombin
In order to study further the role of PAR1 in the induction of inflammatory markers, we used antibodies against PAR1 which inhibit its cleavage and activation. Pre-incubation of GECs with the mixtures of WEDE15 and ATAP2 antibodies28 completely inhibited thrombin responses for CXCL5 up-regulation, suggesting PAR1 cleavage is required for induction of CXCL5 by thrombin. No change in induction of CCL20 was seen when PAR1 cleavage was inhibited (Fig. 6). Neither PAR1 antibodies nor isotype control had effect on induction of the selected markers on their own (data not shown). This result was in agreement with the lack of significant effect on CCL20 induction with PAR1 siRNA and the possibility that thrombin is working via a different mechanism to induce CCL20.
Fig. 6.
Blocking antibodies for PAR1 inhibit expression of CXCL5 induced by thrombin, but do not have any effect on expression of CCL20. Cleavage of PAR1 by thrombin was inhibited by 20 min incubation of the cells with the mixture of two antibodies, ATAP2 and WEDE15 (each 25 μg/ml) and then cells were stimulated with thrombin 10 U/ml for 6 h. Mouse monoclonal IgG1 (50 μg/ml) was used as the negative control. Expression of CXCL5 and CCL20 was measured by QRT-PCR. Data represent mean ± SD from three different donors. *P<0.05.
Discussion
The protease-activated receptor (PAR) family is structurally unrelated to the pattern recognition receptor family (PRR) but, like PRRs, they signal potential danger in the environment by activating innate immune markers and inflammatory responses. These receptors are clearly important for GEC responses to the environment that may include pathogenic or physiological products. Here, we documented the overall function of PAR1 and PAR2 activation in GECs, and demonstrated the active role of PARs in cell proliferation and induction of innate immune responses in GECs.
Chronic inflammation in periodontitis is associated with epithelial proliferation and migration; moreover, cell adhesion and proliferation during the wound-healing process is an important step toward tissue repair. The effect of thrombin and trypsin-like enzymes on cell proliferation via PAR1 and PAR2 activation in various cell types has been reported.29–33 Here, results showed that continuous stimulation of GECs at low concentration of agonist enzymes for PAR1 and PAR2 enhanced cell growth. The up-regulation of ribosomal-associated genes by thrombin, which was observed in data from microarray analysis, may be related to this proliferation and wound-healing response, which was also clearly indicated by the gene ontology analysis. Other specific markers for proliferative response are the thrombin up-regulated EGR1 (early growth response 1), a transcription factor associated with proliferation,34 and the trypsin up-regulated EDN1 (endothelin 1), that has a role in proliferation and protection from apoptosis.35 It has been reported that the signaling cascade initiated by thrombin includes the activation of EGF receptor as well.34 Whether the effects on growth are induced directly by PARs or via transactivation of EGF receptors await further investigation. In contrast to our findings, an inhibitory effect on keratinocyte growth was reported using an exogenous agonist peptide for PAR2.29 Different experimental conditions and different means of activation (agonist peptide versus enzyme) may account for this difference.
Our microarray data show that many similar functional groups of genes are regulated by thrombin and trypsin, although trypsin under the conditions used here has more potent effects, especially on the cytokine activity, receptor binding and immune response. Nevertheless, both thrombin and trypsin resulted in the induction of the subfamily of CXC chemokines including CXCL1, CXCL2, CXCL3, CXCL5 and CXCL8 (IL8), which are chemo-attractants for neutrophils and have important role in wound healing. In these studies, we investigated induction of CXCL5 by PAR1 and PAR2 as a representative of the CXC chemokines, and one of the CXCR2 ligands that is involved in neutrophil attraction and activation, inflammatory responses, tumor growth and angiogenesis.36–38 We also investigated CCL20 which is a multifunctional cytokine with broad-spectrum antimicrobial properties similar to defensins.39,40 CXCL5 and CCL20 are distinct types of cytokines that are induced in oral epithelial cells stimulated with oral bacteria.11,41 Our follow-up studies confirmed that CXCL5 and CCL20, induced by trypsin are regulated via PAR2. Similarly, PAR1-modulated expression of CXCL5 was induced by proteolytic activity of thrombin. These effects were observed both at mRNA and protein levels, although the magnitude of the secreted protein response was not correlated with magnitude of the mRNA response. The absence of correlation between mRNA and protein responses may be related to the experimental conditions for determining mRNA and protein expression, possible degradation of secreted protein, and to mRNA stability and translational regulation of cytokine and chemokine expression.42 Our finding that CCL20 up-regulation is via PAR2 is consistent with previous findings that GECs up-regulate CCL20 in response to P. gingivalis and gingipains via PAR2.11
In the present study, the proteolytic activity of thrombin induced expression of CCL20, but this did not occur via PAR1 as shown by results of both blocking antibody and siRNA studies. This suggests that PAR1 is not the only receptor responsive to thrombin in GECs. These cells also express PAR3, but little or no PAR4 has been demonstrated.9 A low concentration of thrombin can cleave PAR1 and PAR3 which have the hirudin-like sequence in the amino-terminal exodomain.3 Considering that CCL20 induction by thrombin is inhibited in the presence of PPACK and that thrombin proteolytic effects are most likely PAR mediated, our results suggest that CCL20 induction by thrombin may be via other thrombin receptors than PAR1, possibly a direct contribution of PAR3. There is some controversy about the functional role of PAR3. Some investigators have described PAR3 as a non-signaling, thrombin-activated receptor that acts along with PAR1 and PAR4,3,43 while other reports have claimed that PAR3 can signal autonomously and independently of PAR1 activation.44 Thus, although PAR1 is more abundant than PAR3 in GECs, and we anticipated that major responses to thrombin are via PAR1, the possible function of PAR3 and PAR4 in GECs needs to be investigated.
The studies described here confirm a role for both PAR1 and PAR2 in gingival epithelial responses that may be related to periodontal health. Studies of periodontitis in a rat model suggested that activation of PAR2 induces bone loss and increases myeloperoxidase activity, an index of granulocyte infiltration into gingival tissues.14 Our data suggest that activation of PAR1 and PAR2 leads to induction of chemokines that attract granulocytes, consistent with data from the rat model. Nevertheless, although studies of the rat model suggest that chronic activation of PAR2 led to periodontitis via its effect on bone resorption, we cannot rule out the possible protective role of PARs activation in gingival epithelial cells. Activation of PARs by endogenous proteases at physiological level may be beneficial to maintain GECs in an active surveillance mode, primed for innate immune responses to pathogenic stimulants. In the presence of pathogenic stimulants, PAR activation in GECs serves to initiate innate immunity. On the other hand, accumulation of proteases with the ability to over-activate PARs will be destructive for periodontal tissues. Similarly, the proliferative effect of PAR1 and PAR2 at low levels of proteases may be beneficial for wound healing and gingival tissue turnover, but uncontrolled activity could be harmful for maintenance of gingival health. These considerations lead us to suggest that PARs, like other components of innate immunity, act as a double-edged sword with both protective and destructive effects. Understanding the mechanisms that keep their functions in balance can shed new light on the role of PARs in progression and treatment of inflammatory disease in the gingival epithelium as well as other epithelial tissues.
Acknowledgments
This work was supported by NIDCR grant R01DE16961. We kindly thank Dr Theo Bammler (Department of Environmental and Occupational Health Sciences, University of Washington) for help in microarray analysis and Dr James Rheinwald for providing the TERT cells.
References
- 1.Sugawara Y, Uehara A, Fujimoto Y, et al. Toll-like receptors, NOD1, and NOD2 in oral epithelial cells. J Dent Res. 2006;85:524–529. doi: 10.1177/154405910608500609. [DOI] [PubMed] [Google Scholar]
- 2.Vu TK, Hung DT, Wheaton VI, Coughlin SR. Molecular cloning of a functional thrombin receptor reveals a novel proteolytic mechanism of receptor activation. Cell. 1991;64:1057–1068. doi: 10.1016/0092-8674(91)90261-v. [DOI] [PubMed] [Google Scholar]
- 3.Nakanishi-Matsui M, Zheng YW, Sulciner DJ, Weiss EJ, Ludeman MJ, Coughlin SR. PAR3 is a cofactor for PAR4 activation by thrombin. Nature. 2000;404:609–613. doi: 10.1038/35007085. [DOI] [PubMed] [Google Scholar]
- 4.Coughlin SR. Protease-activated receptors in hemostasis, thrombosis and vascular biology. J Thromb Haemost. 2005;3:1800–1814. doi: 10.1111/j.1538-7836.2005.01377.x. [DOI] [PubMed] [Google Scholar]
- 5.Macfarlane SR, Seatter MJ, Kanke T, Hunter GD, Plevin R. Proteinase-activated receptors. Pharmacol Rev. 2001;53:245–282. [PubMed] [Google Scholar]
- 6.Steinhoff M, Buddenkotte J, Shpacovitch V, et al. Proteinase-activated receptors: transducers of proteinase-mediated signaling in inflammation and immune response. Endocr Rev. 2005;26:1–43. doi: 10.1210/er.2003-0025. [DOI] [PubMed] [Google Scholar]
- 7.Coughlin SR, Camerer E. PARticipation in inflammation. J Clin Invest. 2003;111:25–27. doi: 10.1172/JCI17564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacNaughton WK. Epithelial effects of proteinase-activated receptors in the gastrointestinal tract. Mem Inst Oswaldo Cruz. 2005;100:211–215. doi: 10.1590/s0074-02762005000900036. [DOI] [PubMed] [Google Scholar]
- 9.Lourbakos A, Potempa J, Travis J, et al. Arginine-specific protease from Porphyromonas gingivalis activates protease-activated receptors on human oral epithelial cells and induces interleukin-6 secretion. Infect Immun. 2001;69:5121–5130. doi: 10.1128/IAI.69.8.5121-5130.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung WO, Hansen SR, Rao D, Dale BA. Protease-activated receptor signaling increases epithelial antimicrobial peptide expression. J Immunol. 2004;173:5165–5170. doi: 10.4049/jimmunol.173.8.5165. [DOI] [PubMed] [Google Scholar]
- 11.Dommisch H, Chung WO, Rohani MG, et al. Protease-activated receptor 2 mediates human beta-defensin 2 and CC chemokine ligand 20 mRNA expression in response to proteases secreted by Porphyromonas gingivalis. Infect Immun. 2007;75:4326–4333. doi: 10.1128/IAI.00455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uehara A, Sugawara S, Muramoto K, Takada H. Activation of human oral epithelial cells by neutrophil proteinase 3 through protease-activated receptor-2. J Immunol. 2002;169:4594–4603. doi: 10.4049/jimmunol.169.8.4594. [DOI] [PubMed] [Google Scholar]
- 13.Uehara A, Muramoto K, Takada H, Sugawara S. Neutrophil serine proteinases activate human nonepithelial cells to produce inflammatory cytokines through protease-activated receptor 2. J Immunol. 2003;170:5690–5696. doi: 10.4049/jimmunol.170.11.5690. [DOI] [PubMed] [Google Scholar]
- 14.Holzhausen M, Spolidorio LC, Vergnolle N. Proteinase-activated receptor-2 (PAR2) agonist causes periodontitis in rats. J Dent Res. 2005;84:154–159. doi: 10.1177/154405910508400209. [DOI] [PubMed] [Google Scholar]
- 15.Fiorucci S, Mencarelli A, Palazzetti B, et al. Proteinase-activated receptor 2 is an anti-inflammatory signal for colonic lamina propria lymphocytes in a mouse model of colitis. Proc Natl Acad Sci USA. 2001;98:13936–13941. doi: 10.1073/pnas.241377298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tatakis DN. Blood coagulation factors in periodontal pathophysiology: a review with emphasis on the role of thrombin. Semin Thromb Hemost. 1992;18:28–33. doi: 10.1055/s-2007-1002407. [DOI] [PubMed] [Google Scholar]
- 17.Imamura T, Banbula A, Pereira PJ, Travis J, Potempa J. Activation of human prothrombin by arginine-specific cysteine proteinases (Gingipains R) from Porphyromonas gingivalis. J Biol Chem. 2001;276:18984–18991. doi: 10.1074/jbc.M006760200. [DOI] [PubMed] [Google Scholar]
- 18.Eley BM, Cox SW. Cathepsin B/L-, elastase-, tryptase-, trypsin- and dipeptidyl peptidase IV-like activities in gingival crevicular fluid: a comparison of levels before and after periodontal surgery in chronic periodontitis patients. J Periodontol. 1992;63:412–417. doi: 10.1902/jop.1992.63.5.412. [DOI] [PubMed] [Google Scholar]
- 19.Chung WO, Dale BA. Innate immune response of oral and foreskin keratinocytes: utilization of different signaling pathways by various bacterial species. Infect Immun. 2004;72:352–358. doi: 10.1128/IAI.72.1.352-358.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Z, Irizarry R, Gentleman RC, Murillo FM, Spencer F. A model based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc. 2004;99:909–917. [Google Scholar]
- 22.Al-Shahrour F, Minguez P, Tarraga J, et al. BABELOMICS: a systems biology perspective in the functional annotation of genome-scale experiments. Nucleic Acids Res. 2006;34:W472–W476. doi: 10.1093/nar/gkl172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brodersen DE, Nissen P. The social life of ribosomal proteins. FEBS J. 2005;272:2098–2108. doi: 10.1111/j.1742-4658.2005.04651.x. [DOI] [PubMed] [Google Scholar]
- 25.Wool IG. Extraribosomal functions of ribosomal proteins. Trends Biochem Sci. 1996;21:164–165. [PubMed] [Google Scholar]
- 26.Hofman P, Butori C, Havet K, et al. Prognostic significance of cortactin levels in head and neck squamous cell carcinoma: comparison with epidermal growth factor receptor status. Br J Cancer. 2008;98:956–964. doi: 10.1038/sj.bjc.6604245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kon S, Ikesue M, Kimura C, et al. Syndecan-4 protects against osteopontin-mediated acute hepatic injury by masking functional domains of osteopontin. J Exp Med. 2008;205:25–33. doi: 10.1084/jem.20071324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Brien PJ, Prevost N, Molino M, et al. Thrombin responses in human endothelial cells. Contributions from receptors other than PAR1 include the transactivation of PAR2 by thrombin-cleaved PAR1. J Biol Chem. 2000;275:13502–13509. doi: 10.1074/jbc.275.18.13502. [DOI] [PubMed] [Google Scholar]
- 29.Derian CK, Eckardt AJ, Andrade-Gordon P. Differential regulation of human keratinocyte growth and differentiation by a novel family of protease-activated receptors. Cell Growth Differ. 1997;8:743–749. [PubMed] [Google Scholar]
- 30.Algermissen B, Sitzmann J, Nurnberg W, Laubscher JC, Henz BM, Bauer F. Distribution and potential biologic function of the thrombin receptor PAR-1 on human keratinocytes. Arch Dermatol Res. 2000;292:488–495. doi: 10.1007/s004030000168. [DOI] [PubMed] [Google Scholar]
- 31.Fager G. Thrombin and proliferation of vascular smooth muscle cells. Circ Res. 1995;77:645–650. doi: 10.1161/01.res.77.4.645. [DOI] [PubMed] [Google Scholar]
- 32.Moller T, Hanisch UK, Ransom BR. Thrombin-induced activation of cultured rodent microglia. J Neurochem. 2000;75:1539–1547. doi: 10.1046/j.1471-4159.2000.0751539.x. [DOI] [PubMed] [Google Scholar]
- 33.Ahmad R, Knafo L, Xu J, Sindhu ST, Menezes J, Ahmad A. Thrombin induces apoptosis in human tumor cells. Int J Cancer. 2000;87:707–715. [PubMed] [Google Scholar]
- 34.Kaufmann K, Thiel G. Epidermal growth factor and thrombin induced proliferation of immortalized human keratinocytes is coupled to the synthesis of Egr-1, a zinc finger transcriptional regulator. J Cell Biochem. 2002;85:381–391. doi: 10.1002/jcb.10145. [DOI] [PubMed] [Google Scholar]
- 35.Kim TH, Xiong H, Zhang Z, Ren B. beta-Catenin activates the growth factor endothelin-1 in colon cancer cells. Oncogene. 2005;24:597–604. doi: 10.1038/sj.onc.1208237. [DOI] [PubMed] [Google Scholar]
- 36.Walz A, Burgener R, Car B, Baggiolini M, Kunkel SL, Strieter RM. Structure and neutrophil-activating properties of a novel inflammatory peptide (ENA-78) with homology to interleukin 8. J Exp Med. 1991;174:1355–1362. doi: 10.1084/jem.174.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rubie C, Frick VO, Wagner M, et al. ELR+ CXC chemokine expression in benign and malignant colorectal conditions. BMC Cancer. 2008;8:178. doi: 10.1186/1471-2407-8-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park JY, Park KH, Bang S, et al. CXCL5 overexpression is associated with late stage gastric cancer. J Cancer Res Clin Oncol. 2007;133:835–840. doi: 10.1007/s00432-007-0225-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Starner TD, Barker CK, Jia HP, Kang Y, McCray PB., Jr CCL20 is an inducible product of human airway epithelia with innate immune properties. Am J Respir Cell Mol Biol. 2003;29:627–633. doi: 10.1165/rcmb.2002-0272OC. [DOI] [PubMed] [Google Scholar]
- 40.Yang D, Chen Q, Hoover DM, et al. Many chemokines including CCL20/MIP-3alpha display antimicrobial activity. J Leukoc Biol. 2003;74:448–455. doi: 10.1189/jlb.0103024. [DOI] [PubMed] [Google Scholar]
- 41.Yin L, Dale BA. Activation of protective responses in oral epithelial cells by Fusobacterium nucleatum and human beta-defensin-2. J Med Microbiol. 2007;56:976–987. doi: 10.1099/jmm.0.47198-0. [DOI] [PubMed] [Google Scholar]
- 42.Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003;4:117. doi: 10.1186/gb-2003-4-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McLaughlin JN, Patterson MM, Malik AB. Protease-activated receptor-3 (PAR3) regulates PAR1 signaling by receptor dimerization. Proc Natl Acad Sci USA. 2007;104:5662–5667. doi: 10.1073/pnas.0700763104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ostrowska E, Reiser G. The protease-activated receptor-3 (PAR-3) can signal autonomously to induce interleukin-8 release. Cell Mol Life Sci. 2008;65:970–981. doi: 10.1007/s00018-008-7555-y. [DOI] [PMC free article] [PubMed] [Google Scholar]