Abstract
Nearly every DNA polymerase characterized to date exclusively catalyzes the incorporation of mononucleotides into a growing primer using a DNA or RNA template as a guide to direct each incorporation event. There is, however, one unique DNA polymerase designated terminal deoxynucleotidyl transferase that performs DNA synthesis using only single-stranded DNA as the nucleic acid substrate. In this chapter, we review the biological role of this enigmatic DNA polymerase and the biochemical mechanism for its ability to perform DNA synthesis in the absence of a templating strand. We compare and contrast the molecular events for template-independent DNA synthesis catalyzed by terminal deoxynucleotidyl transferase with other well-characterized DNA polymerases that perform template-dependent synthesis. This includes a quantitative inspection of how terminal deoxynucleotidyl transferase binds DNA and dNTP substrates, the possible involvement of a conformational change that precedes phosphoryl transfer, and kinetic steps that are associated with the release of products. These enzymatic steps are discussed within the context of the available structures of terminal deoxynucleotidyl transferase in the presence of DNA or nucleotide substrate. In addition, we discuss the ability of proteins involved in replication and recombination to regulate the activity of the terminal deoxynucleotidyl transferase. Finally, the biomedical role of this specialized DNA polymerase is discussed focusing on its involvement in cancer development and its use in biomedical applications such as labeling DNA for detecting apoptosis.
Keywords: DNA polymerization, template-independent DNA synthesis, fidelity, recombination, nucleotide analogs
Introduction
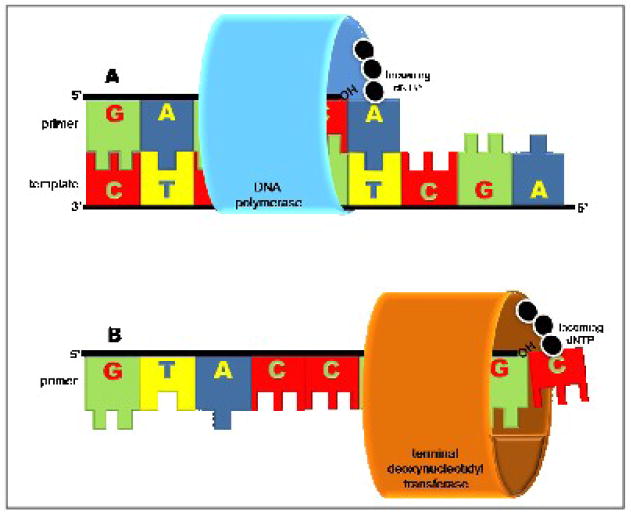
DNA polymerases play essential roles in replication, repair, and recombination of nucleic acid. During each of these biological processes, the polymerase extends a primer using a DNA (or RNA in the case of reverse transcription) template to guide each incorporation event. Even during the bypass of lethal forms of DNA damage, the presence of a templating strand is absolutely essential for polymerase activity. However, the requirement for using a template is not universal as there exists a unique enzyme, denoted as terminal deoxynucleotidyl transferase (TdT), that possesses the unusual ability to incorporate nucleotides in a template-independent manner using only single-stranded DNA as the nucleic acid substrate [1, 2] (Figure 1). The unique ability of TdT to create genomic material de novo makes it one of the most fascinating DNA polymerases found in nature. Although TdT was one of the first DNA polymerase activities identified in mammals [3], it remains one of the most poorly understood enzymes that catalyzes DNA synthesis. Indeed, the specific physiological role for TdT remained elusive for several decades [4–11]. It is now recognized that TdT is responsible for the random addition of nucleotides to single-stranded DNA during V(D)J recombination [12, 13]. By deliberately generating subtle randomization of this genetic material, TdT plays a crucial role in the evolution and adaptation of the vertebrate immune system [6, 9, 14, 15]. The ability of TdT to randomly incorporate nucleotides increases antigen receptor diversity and aids in generating the ~1014 different immunoglobulins and ~1018 unique T cell antigen receptors that are required for the neutralization of potential antigens [16, 17].
Figure 1.
Simplified models for template-dependent and template-independent DNA polymerase activity. (A) Most DNA polymerases require double-stranded DNA as a substrate, where the 5′→3′ strand is used as a primer and the complementary strand 3′→5′ is used as a template. (B) Terminal deoxynucleotidyl transferase is unique in its ability to catalyze phosphoryl transfer in the absence of a template that can not be accommodated in its active site.
This review explores the cellular and molecular mechanisms accounting for the activity of this specialized DNA polymerase. Our discussion begins by examining the biological role of TdT and how synthesizing DNA without using a templating strand is important for V(D)J recombination. Attention will then focus on understanding the molecular mechanism by which TdT performs template-independent polymerization. In this section, we will compare and contrast the mechanism of TdT with template-dependent polymerases that are involved in normal and translesion DNA synthesis, i.e., replication in the absence of correct templating information. The reported structure of TdT is used to provide biophysical insight into the kinetic properties of the polymerase that include the utilization of various metal ion cofactors, nucleic acids, and nucleotide substrates. Finally, the biomedical importance of TdT will be discussed with emphasis on its potential role in the development of certain forms of leukemia as well as its utilization as a biochemical marker for apoptosis.
The Role of TdT in V(D)J Recombination
Most organisms possess sophisticated defense mechanisms to protect them against the invasion of foreign agents such as viruses, bacteria, and parasites. Simple prokaryotes use a complementary system involving DNA methylation of the host genome and endonuclease degradation of foreign genomic material to differentiate self from non-self [18]. Eukaryotes have developed more sophisticated systems to thwart off the invasion of foreign substances. Indeed, the mammalian immune system is arguably one of the most intricate and ingenious methods for actively seeking out and killing a wide variety of invaders.
The vertebrate immune system is divided into two subcategories, the innate and adaptive immune systems, that differ in their specificity. The innate immune system is generally considered to be less specific due to the promiscuous ability of the immune receptors to recognize a limited number of molecules that are common features to many infectious agents including polysaccharides, peptidoglycans, non-methylated CpG DNA, and double-stranded RNA [19–22]. This promiscuous activity allows the innate immune system to act as the first line of defense against infection by rapidly recognizing and responding to pathogens. If the defensive line of the innate system is breached, then a more specific and highly specialized offense system, the adaptive immune response, is mobilized to its full potential.
Adaptive immunity came into existence in vertebrates roughly 500 million years ago [23]. The cells of the adaptive immune system, namely T- and B-cells, have a diverse repertoire of antigen receptors and antibodies that can recognize any antigen encountered throughout life [24]. After the adaptive immune cell receptors bind an antigen, they mount a rapid and robust protective response by a dramatic expansion in the number of pathogen-specific T cells [25–28]. Over the course of one week, thousands of clones are produced that possess effector functions [29–31]. Approximately 95% of these activated T-cells undergo apoptosis [30, 32]. However, a stable population of long-lived T cells resides in the lymphoid and non-lymphoid tissues [33, 34] and patrol for these previously encountered pathogens. The immunological memory displayed by the adaptive immune system provides the vertebrate host with long-lasting protection against subsequent infection. For example, most individuals remain immune to measles for up to 75 years once exposed to an attenuated form of measles virus [35].
At the molecular level, the cells of the immune system have developed a strategy to increase acquired immunity against subsequent biological assaults [36] (Figure 2). This process, commonly known as V(D)J recombination, plays an essential role in abrogating these antigens. Rearrangement of the variable (V), diversity (D) and joining (J) gene segments creates versatility to a competent immune system by generating a diverse repertoire of antigen receptors with unique antibody specificities [37]. This transaction of breaking, rearranging, and rejoining of the V, D, and J regions of the germline immunoglobulin genes requires the collaborative efforts of the three distinct enzyme activities that include nucleases, polymerases, and ligases. Of the three major types of enzymatic activities, our understanding of how specific DNA polymerases function during V(D)J recombination is not yet firmly established. However, crucial information for understanding the role of specific polymerases in V(D)J recombination has started to emerge. The relative functions of the various members of the pol X family of DNA polymerases (TdT, pol μ, and pol λ) during the processing of DNA in V(D)J recombination are distinct [38] and nonoverlapping [9] in vivo. Ramsden and colleagues have indicated that a “gradient” of weak to strong terminal deoxynucleotidyl transferase activity defines the distinct roles of pol λ, pol μ, and TdT in nonhomologous end joining (NHEJ), respectively [38]. Moreover, Bertocci et al have shown that pol μ participates exclusively in light chain and not in heavy chain gene rearrangement [9, 39]. In contrast, pol λ is reported to be recruited only in the heavy chain junctions during V(D)J recombination and precedes the action of TdT [9], which is primarily involved in the random addition of nucleotides to unpaired primer termini [38]. While pol μ and pol λ play important roles in the immune system, the role of TdT in V(D)J rearrangement is showcased in this review. A more thorough description of pol β, pol μ and pol λ is provided in a chapter by Joann Sweasy and colleagues in this Special Issue.
Figure 2.
Overview of the V(D)J recombination process that generates functional Ig heavy chain from the inactive gene segments in developing B- or T-lymphocytes. Shown is the V(D)J recombination process for the formation of a functional IgH gene where DJ assembly occurs prior to the combination with the V segment. See text for further details.
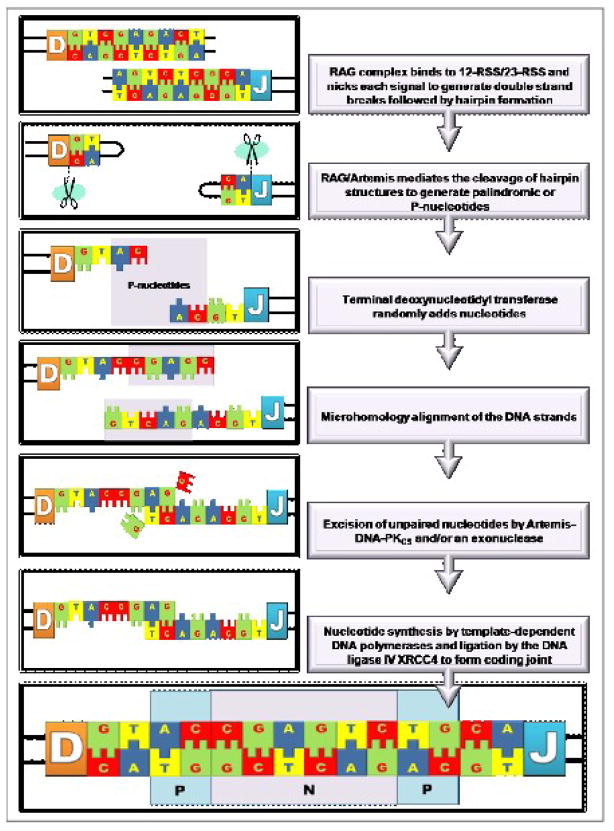
Figure 3A illustrates the crucial steps of V(D)J recombination during the RAG cleavage phase. This process is initiated by introducing a double strand break (DSB) at the edge of the selected gene segment by the RAG-1 and RAG-2 (Recombination-Activating Genes) proteins. These proteins selectively bind to specific and highly conserved recombination signal sequences (RSS) that contain heptamer and nonamer elements separated by a spacer region [40, 41]. There are either 12 bp or 23 bp spacers between the heptamer and nonamer elements of the RSS commonly denoted 12-RSS or 23-RSS [40–43], respectively. Proper recombination demands that one 12-RSS and one 23-RSS be present for efficient cleavage in vivo [40, 41, 44], and this phenomenon is referred to as the “12/23-rule”. After recognition of complementary RSS, the RAG complex introduces a nick between the D and J coding segment and the adjoining recombination signal sequence. The RAG complex also mediates the formation of hairpins at each coding end by using the 3′-OH moiety at each nick as the nucleophile.
Figure 3.
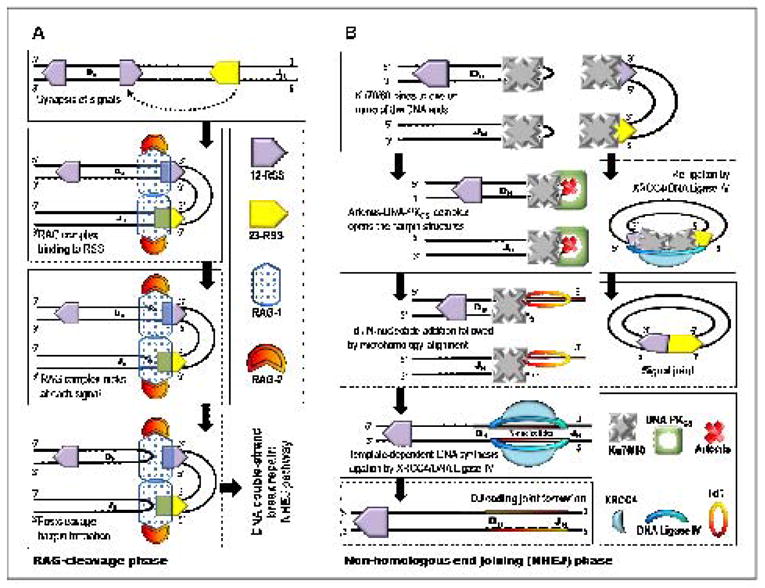
A simplified model for (A) the RAG-cleavage phase generating double-strand breaks and (B) DNA repair through non-homologous end-joining pathway during DJ gene segment assembly of the V(D)J recombination mechanism.
Double-strand DNA breaks introduced during the RAG cleavage phase are repaired during the non-homologous end-joining phase (Figure 3B). The nuclease, Artemis:DNA-PKcs, can trim the 5′ and 3′ overhangs [45] and open the hairpins at the coding ends generating palindromic nucleotide sequences (P-nucleotides). At this point, TdT randomly adds nucleotides to available 3′-OH ends increasing the variability of the non-templated nucleotide (N) region of the recombined gene segments [46]. The details of this process are provided in Figure 4. While in vitro studies have shown that TdT can incorporate all four natural nucleotides on to single-stranded DNA (vide infra), there is a definitive bias for the incorporation of dGMP and dCMP versus dAMP or dTMP observed in vivo [8, 47–50]. This preference may offer a plausible explanation for the high G/C content of the Ig and TCR N-regions [48, 49]. In addition, the average N-nucleotide segment length created in vivo is only 2–5 bp per coding joint [47]. This length appears optimal for allowing the DNA strands to undergo microhomology alignment using Watson-Crick base pair recognition patterns. Unpaired nucleotides are trimmed by an exonuclease such as the Artemis:DNA PKcs complex [45]. The gaps are ultimately filled by template-dependent DNA polymerases, and the ligation of the coding ends is carried out by the XRCC4:DNA ligase IV complex [45].
Figure 4.
Simplified overview of the enzymatic steps and the role of terminal deoxynucleotidyl transferase in lymphocyte gene rearrangement. The variability of the recombined gene segments is increased through the random addition of non-templated (N) nucleotides catalyzed by the terminal deoxynucleotidyl transferase prior to complementary pairing and extension by template-dependent DNA polymerases.
The role of TdT in generating immunological diversity is to catalyze the random addition of small numbers of nucleotides at the N regions. This activity has been speculated by virtue of its catalytic properties in addition to its localization to primary lymphoid tissues such as thymus and bone marrow [15, 51, 52]. A wealth of information to support this hypothesis exists and include the correlation between the existence of N regions with TdT expression [13] or mRNA expression [53] and with the inclusion of the TdT gene into a cell line that continuously rearranges immunoglobulin genes but lacks the enzyme [7, 8]. In addition, TdT-knockout mice and TdT-deficient lymphocytes [6, 54] have also been used to validate the role of TdT in N-nucleotide addition during the V(D)J recombination process. As expected, TdT-knockout mice show a ten-fold reduction of T-cell receptor (TCR) diversity compared to the wild-type mice and provides evidence that the addition of N-nucleotides by TdT is of paramount value to establishing a combinatorially diverse antigen receptor repertoire [54].
Biochemical Properties of TdT
Isoforms of TdT
Baltimore and colleagues were amongst the first to show that the occurrence of short nucleotide insertions in the combinatorial junctions of DNA segments during the V(D)J recombination process correlates with the presence of TdT [13, 55]. Over the course of several years, different mRNA splicing variants of the enzyme were identified and studied extensively in mice, bovines, and humans. Thus far, two splice variants of TdT have been observed in mice while bovines and humans each have three.
The two identified splice variants of TdT in mice are TdTS, a short form consisting of 509 amino acids [56], and TdTL, a long form with 529 residues [57]. Both isoforms share all domains that are essential for the binding of nucleotides, DNA, and metal ions [58, 59]. However, the 20 amino acid difference between the two variants reportedly bestows distinct enzymatic behavior that remains controversial [60–62]. For example, Kearney and coworkers have shown that TdTS catalyzes the unbiased and non-templated addition of nucleotides to the combinatorial junctions of antigen receptor genes [60, 62, 63], while TdTL possesses 3′ → 5′ exonuclease activity that can delete nucleotides at the coding ends of the Ig and TCR gene segments [61]. In contrast, Papanicolaou et. al. have argued that TdTL does not possess exonuclease activity and that it exhibits nearly identical enzymatic activity to TdTS in vitro [62, 64]. This discrepancy may be attributed to the lower stability of TdTL compared to TdTS both in vitro [64] as well as in transfected cells [60]. In addition, TdTL can reportedly modulate the amount of nucleotide addition by TdTS [11, 61]. While the mechanism for this regulation remain undefined, these data suggest that the two TdT murine isoforms function to balance the diversification of antigen repertoire assembly to preserve the integrity of the variable region of antigen receptors [61].
Human (h) and bovine (b) TdT isoforms are slightly more complicated as each have three alternative splice variants designated as TdTS (short), TdTL1 (long) and TdTL2 (long) [65–67]. hTdTL1 and hTdTL2 both localize in the nucleus [67]. However, hTdTL2 is expressed more abundantly in normal small lymphocytes compared to hTdTL1 which is readily detected in transformed lymphoid cell lines [67]. Both long isoforms of human TdT possess 3′→5′ exonuclease activity for nucleotide removal whereas the short isoforms perform nucleotide elongation of the coding ends during V(D)J recombination [65, 67]. Overexpression of hTdTS or hTdTL2 independently reduces the efficiency of V(D)J recombination greatly [67]. However, the simultaneous overexpression of hTdTS and hTdTL2 restores recombination frequencies back to normal levels. These observations suggest the possibility of a strong evolutionary selection for co-expressing one transferase (hTdTS) and one exonuclease (hTdTL2) to regulate the length of single-stranded regions generated during NHEJ. In addition, there is evidence for prohibiting the coexpression of two exonucleases (hTdTL1 and hTdTL2) via regulation at the level of mRNA alternative splicing during V(D)J recombination [67]. In fact, the presence of all three human TdT variants during antigen receptor gene rearrangement drastically diminishes the recombination frequency. These results suggest that hTdTL1 may serve to regulate hTdTL2 or hTdTS activity [67].
Sources and Purification of TdT
The calf thymus glands are an abundant source for TdT. In fact, TdT was first purified to apparent homogeneity from calf thymus cell lysate in 1971 [68]. At first, the isolated protein was reported as a proteolysed form containing two polypeptides originally recognized as a heterodimer of α and β subunits [68, 69]. However, subsequent studies established that indigenous TdT is truly a monomeric protein with a molecular weight of 58,000 to 60,000 daltons [70–72]. Unfortunately, obtaining the enzyme in its relatively pure, intact, and active form is extremely laborious and difficult due to proteolysis. Copious amount of the enzyme can be purified from cultured cell lines propagated from patients with acute lymphoblastic leukemia [73]. However, this approach is too expensive and impractical for routine mass production of enzyme. Attempts to overexpress TdT in bacteria systems have generally failed due to three prominent factors: (1) mismatches in the codon frequencies and tRNA pools between E. coli and eukaryotes, (2) low solubility of the protein in these systems, and (3) lower levels of enzyme activity [59, 74, 75]. These complications can be alleviated by overexpressing a rare argU tRNA in the E. coli system and growing cultures at 15°C [64, 76] to boost the production of soluble and active forms of the enzyme. Purification of TdT to apparent homogeneity is accomplished by column chromatography which is facilitated by the presence of a hexa-histidine tag attached to the N-terminus that does not affect its activity [76]. Recombinant human TdT has also been overexpressed in the baculovirus expression system and purified to homogeneity in one step from Trichoplusia ni larvae using a monoclonal antibody affinity column [59, 74]. This last advancement has played an instrumental role in generating significant amounts of protein required for structural characterization of TdT.
Primer Requirement
The unique activity of TdT to incorporate nucleotides in a template-independent manner was reported simultaneously by Karkow and Kamen [77] and Bollum [78] using calf thymus extracts. The template-independent activity of TdT was distinguished from that of other template-dependent DNA polymerases by comparing levels of the incorporation of radiolabeled nucleotides using single-stranded versus double-stranded DNA [1]. Activity measured using single-stranded DNA is attributed only to that of TdT [2]. Subsequent biochemical studies confirmed that TdT requires a single-stranded initiator that is at least three nucleotides long with a free 5′-phosphate end and a free 3′-hydroxyl end for extension [78]. The replication of homopolymers by TdT requires an initiator chain of more than six nucleotides for poly(dA) and more than five nucleotides for poly(dT) [79]. The presence of a ribonucleotide 5′-monophosphate (rNMP) at the 3′-end of the primer does not inhibit the enzyme-primer complex formation [80]. However, further elongation occurs at a slower rate and the addition of more than two ribonucleotides to the single-stranded initiator does inhibit activity [80]. TdT is also unique for its ability to perform de novo synthesis of polynucleotides ranging in size from 2- to 15-mers when provided with dNTPs in absence of a primer [81]. These DNA fragments are hypothesized to act as signals for DNA repair or recombination machinery [81]. There may indeed be credence to this provocative hypothesis since small RNAs, designated as microRNA, influence the activity of many biological pathways [82]. It will prove interesting to firmly test this hypothesis to evaluate if de novo DNA synthesis does indeed occur in vivo and that small DNA fragments can regulate biological processes such as DNA repair and recombination.
Metal Utilization
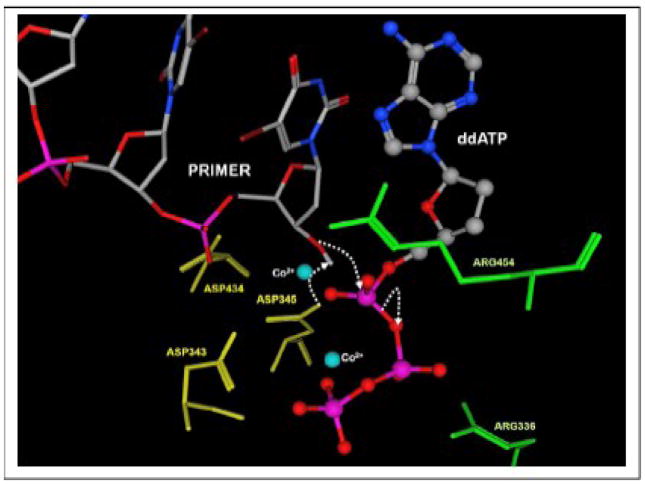
All DNA polymerases require the presence of a divalent metal ion to catalyze the phosphoryl transfer reaction associated with nucleotide incorporation. The general catalytic mechanism for phosphoryl transfer utilized by DNA polymerases is provided in Figure 5. In this mechanism, an aspartate residue near the deoxyribose sugar of the incoming dNTP serves as the general base to abstract the proton from the 3′-OH to generate a more reactive nucleophile. The electron rich 3′-oxygen then attacks the α-phosphate creating a trigonal-bipyramidal penta-coordinated transition state. This step results in the inversion of the α-phosphate stereochemistry [83, 84] and the concerted release of the pyrophosphate leaving group coordinated to another divalent metal ion [85–88].
Figure 5.
The catalytic mechanism model for the nucleotidyl transfer reaction catalyzed by terminal deoxynucleotidyl transferase. See text for details.
TdT, like all DNA polymerases, also requires divalent metal ions for catalysis [73, 89]. However, TdT is unique in its ability to use a variety of divalent cations such as Co2+, Mn2+, Zn2+ and Mg2+. In general, the extension rate of the primer p(dA)n (where n is the chain length from 4 through 50) with dATP in the presence of divalent metal ions is ranked in the following order: Mg2+ > Zn2+ > Co2+ > Mn2+ [73]. In addition, each metal ion has different effects on the kinetics of nucleotide incorporation. For example, Mg2+ facilitates the preferential utilization of dGTP and dATP whereas Co2+ increases the catalytic polymerization efficiency of the pyrimidines, dCTP and dTTP [90]. Zn2+ behaves as a unique positive effector for TdT since reaction rates with Mg2+ are stimulated by the addition of micromolar quantities of Zn2+ [90]. This enhancement may reflect the ability of Zn2+ to induce conformational changes in TdT that yields higher catalytic efficiencies [90]. Polymerization rates are lower in the presence of Mn2+ compared to Mg2+, suggesting that Mn2+ does not support the reaction as efficiently as Mg2+ [73].
Mechanism of Template-Independent Polymerization
When does replication without a template occur?
A DNA polymerase is classically defined as an enzyme that catalyzes template-dependent addition of mononucleotides into a growing primer in the presence of four deoxynucleotides (dNTPs) [91]. Nevertheless, many archaeal, bacterial, viral, and eukaryotic DNA polymerases also catalyze non-templated nucleotide additions to the 3′-termini of blunt-ended DNA [92–96]. In addition, numerous DNA polymerases have been demonstrated to incorporate nucleotides opposite non-instructional DNA lesions such as abasic sites via translesion DNA synthesis [96–103]. In both of these cases, however, these polymerases require the use of duplex DNA as a substrate for efficient catalysis to occur. In fact, the sole polymerase that can add nucleotides using only a single-stranded DNA initiator as a substrate is the template-independent DNA polymerase, TdT [1, 2]. Terminal deoxynucleotidyl transferase activity has also been observed for pol μ [38] and pol λ [104, 105], although DNA synthesis is primarily restricted to the use of a DNA template [38, 104, 105]. In the sections provided below, we compare and contrast the mechanism of nucleotide incorporation catalyzed by TdT with template-dependent DNA polymerases. Particular emphasis is placed on the replication of non-instructional DNA lesions as this represents the most frequent mode of template-independent DNA synthesis catalyzed by these enzymes.
Order of Substrate Binding
Theoretically, the binding of DNA and dNTP substrates to any DNA polymerase can be random, sequential, or strictly ordered. Efficient polymerization for a template-dependent polymerase, however, would be optimal through the strictly ordered binding of DNA substrate prior to dNTP since the converse order of dNTP binding prior to DNA would be correct only once out of four opportunities. Indeed, numerous steady-state and pre-steady state studies have validated that all template-dependent polymerases obey this mechanism (reviewed in [106]).
Not surprisingly, TdT appears to be the lone exception to this rule. The order by which TdT binds DNA and dNTP is indeed random as determined through a series of initial velocity studies performed in the absence and presence of product inhibitors [73]. The double reciprocal plot of 1/rate versus 1/dATP concentration at several fixed concentrations of single-stranded DNA intersected to the left of the y-axis, consistent with either an ordered or random kinetic mechanism. Inhibition by PPi at varying dNTP concentrations gave rise to a competitive inhibition pattern whereas inhibition by PPi at different concentrations of DNA yielded a mixed inhibition pattern. These results are consistent with rapid equilibrium random mechanism in which TdT forms the catalytic competent ternary complex via binding of dNTP prior to DNA or vice versa. It is currently unclear if the ability to randomly bind substrates plays a physiological role in generating random nucleotides during recombination. However, it is possible that the interactions of TdT with PCNA and Ku70/86 [107–109], proteins involved in replication and recombination, may influence its kinetic mechanism.
Mechanism of Nucleotide Selection
An important kinetic step for generating catalytic efficiency and fidelity with template-dependent polymerases occurs during the binding of dNTP to the polymerase:DNA complex. This process is highly influenced by the presence of a templating strand due to hydrogen bonding [110] and steric complementarity [111]. However, since TdT does not rely on a templating strand for polymerization activity, the molecular details regarding nucleotide binding and selection remain elusive.
Several laboratories have demonstrated that TdT displays an unequal bias in the kinetics of nucleotide incorporation. For example, studies from the Coleman laboratory reveal that the Km for dGTP is ~4-fold lower than the Km for dATP (compare Km values of 120 μM versus 540 μM, respectively) [59]. Similar results are reported by the Modak group [112]. More recently, we have demonstrated that recombinant TdT utilizes dGTP, dCTP, and dTTP much more efficiently than dATP [113]. The preferential use of dGTP and dCTP arguably reflects a biophysical bias for more efficient annealing of single-stranded DNA that are intermediates during NHEJ. At the molecular level, the preference in nucleotide utilization could reflect favorable hydrogen-bonding interactions between the incoming dNTP and active site amino acids that guide nucleotide binding. This mechanism would resemble that displayed by the yeast Rev1 protein, an error-prone DNA polymerase that preferentially incorporates dCMP via direct interactions with an active site arginine as opposed to interactions with the templating base [114]. The discrimination against the utilization of dATP is also intriguing especially when compared to the preferential incorporation of dAMP by high fidelity DNA polymerases during the replication of non-coding DNA lesions such as abasic sites. Template-dependent DNA polymerases such as the E. coli Klenow fragment [101] and bacteriophage T4 polymerase [101, 103] incorporate dAMP opposite an abasic site ~100-fold more efficiently than any of the other three natural nucleotides. The preferential incorporation of dAMP by these high-fidelity DNA polymerases is often referred to as the “A-rule” of translesion DNA synthesis [115]. Thus, the preferential usage of dGTP and dCTP by TdT indicates that it does not follow the “A-rule” like the aforementioned well-characterized template-dependent DNA polymerases.
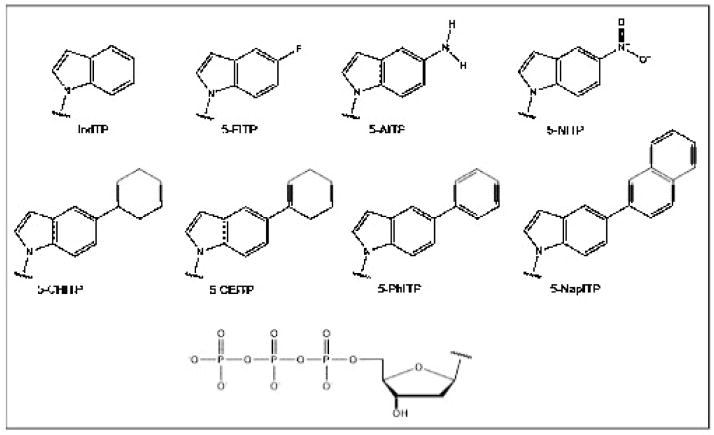
It is also interesting that TdT efficiently incorporates and extends a wide variety of 5-substituted indolyl deoxynucleotides that lack hydrogen-bonding functional groups (Figure 6). TdT uses non-natural nucleotides such as 5-NITP with identical efficiencies as dGTP, the preferred natural nucleotide [113]. This result is intriguing since 5-NITP has been used as a chemical probe to validate the “A-rule” of translesion DNA synthesis with template-dependent DNA polymerases [101]. In fact, several template-dependent polymerases incorporate 5-NIMP opposite an abasic site with remarkably high catalytic efficiencies that approach 106 M−1sec−1 [116]. Thus, TdT presents an interesting paradox by its ability to discriminate against inserting dATP yet effectively incorporating non-natural nucleotides that are used as mechanistic probes for the “A-rule” of translesion DNA synthesis.
Figure 6.
Chemical structures of various 5-substituted indolyl nucleotides used to probe the activity of TdT.
A naïve mechanism to explain the utilization of these non-natural nucleotides is that the primary molecular determinant for nucleotide binding resides simply with interactions of the negatively-charged triphosphate moieties with positively charged amino acids in the polymerase’s active site. This is unlikely to be the sole source for binding as it cannot account for the significant differences observed for the incorporation of various natural and non-natural nucleotide substrates. For example, 5-NIMP is incorporated and extended 10-fold more efficiently than the closely related analog, 5-AIMP, which is poorly incorporated even at concentrations greater than 500 μM [113]. In addition, the replacement of an active site arginine residue (R336) involved in binding the triphosphate moiety of a dNTP with either glutamine (R336Q) or alanine (R336A) does not completely abolish nucleotide binding. Instead, these amino acid substitutions reduce the binding affinities of dGTP and dATP by only 10-fold [59]. These data indicate that ionic interactions between the active site arginine and the triphosphate group are important but not essential for nucleotide binding.
A final point to discuss is with respect to the kinetics of elongation. Our studies with the various 5-substituted indolyl deoxynucleotides provided in Figure 6 reveal an inverse correlation in the kinetics of primer elongation as a function of nucleobase size [113]. For example, large analogs such as 5-PhIMP, 5-CEIMP, and 5-NapIMP are incorporated but not efficiently elongated whereas their smaller counterparts (IndMP, 5-FIMP, and 5-NIMP) are elongated with similar efficiency as natural nucleotides [113]. The inability of TdT to elongate bulky non-natural nucleotides likely results from the steric constraints imposed by the 16-amino acid loop present within TdT (see below) that appears to function as a steric gate to provide selectivity for single-stranded versus duplex DNA.
What Limits Nucleotide Incorporation?
After nucleotide binding, most DNA polymerases undergo an enzymatic conformational change that is proposed to align the incoming dNTP into a precise geometrical shape with the templating nucleobase to allow for phosphoryl transfer. The existence of this conformational change has been demonstrated through kinetic, structural, and spectroscopic studies [93, 98, 117–121] and the reader is recommended to read the article by Kenneth Johnson (this issue) for further details on this subject. Regardless, this conformational change step is believed to impose discrimination against misinserting an incorrect nucleotide into DNA by altering the geometry of the polymerase’s active site to inhibit efficient phosphoryl transfer [122]. Since TdT uses only single-stranded DNA [1, 2], the need for an obligatory conformational change step to ensure fidelity remains in doubt. Put another way, does the lack of “fidelity” displayed by TdT negate the need for a conformational change step? If so, does phosphoryl transfer then become the rate-limiting step for nucleotide incorporation as similarly reported for certain “error-prone” polymerases that have lower constraints in fidelity [123, 124]? Indeed, it is established that with certain DNA polymerases such as pol β, the phosphoryl transfer step as opposed to a conformational change is involved in maintaining fidelity during the nucleotide incorporation step [125].
One way to evaluate the location of the rate-limiting step during polymerization is to measure the effects of thio-substituted nucleotides on the rate of nucleotide incorporation [126]. Since a non-bridging sulfur atom has decreased electronegativity, it is predicted to be less effective than oxygen at stabilizing electron density during the transition-state of phosphoryl transfer. As a result, the magnitude of a “thio effect”, defined as the rate of incorporation using α-O-dNTP versus the rate using an identical concentration of α-S-dNTP, can be used as a diagnostic indication of whether or not chemistry is rate-limiting. If chemistry is the rate-limiting step, then a significant thio-elemental effect (>10) is typically observed while smaller thio-elemental effects of <2 are observed if another kinetic step such as a conformational change preceding phosphoryl transfer is rate-limiting, and thus insensitive to thio substitution. We have preliminary data defining the elemental effect for the incorporation of α-S-dGMP by TdT using different metal cofactors. With Co2+ as the metal cofactor, the rate of α-S-dGTP incorporation is 8-fold slower than that measured using an identical concentration of α-O-dGTP (unpublished results, Berdis, A.J.). The apparent elemental effect of 8 suggests that phosphoryl transfer is indeed the rate-limiting step and is consistent with structural data indicating that TdT exists in a “closed” conformation that alleviates the need for a conformational change for catalysis to occur [127]. In contrast, replacement of Co2+ with Mg2+ gives rise to a smaller elemental effect of ~3 (unpublished results, Berdis, A.J.). The change in the magnitude of the elemental effects suggests that phosphoryl transfer is no longer rate limiting using Mg2+ as the metal cofactor. One implication of these findings is that metal ions directly influence the mechanism of template-independent polymerization by changing the location of the rate-limiting step. However, controversy exists regarding the use of thio-elemental effect to unambiguously define the rate-limiting step for nucleotide incorporation. Much of this controversy involves potential differences in the transition states between the enzyme-catalyzed and non-enzymatic reaction [128]. Therefore, further kinetic and spectroscopic studies on TdT are needed to further investigate this phenomenon
Distributive versus Processive DNA Synthesis
After nucleotide incorporation, template-dependent polymerases show an obligatory release of products in which pyrophosphate is the first product released. At this point, the polymerase can either remain bound to DNA and continue primer elongation or dissociate from the extended primer to re-initiate DNA synthesis on another usable primer. Since TdT functions via a rapid equilibrium random kinetic mechanism [73], it is most likely that TdT catalyzes DNA synthesis in a strictly distributive mode. While this mechanism is reasonable, it has yet to be conclusively established.
Structural Insights into the Mechanism of TdT
The X-Family of DNA Polymerases
TdT is a member of the X-family of DNA polymerases that include DNA polymerase β, (pol β), DNA polymerase λ (pol λ), and DNA polymerase μ (pol μ). Pol β removes the 5′-deoxyribose phosphate moiety and catalyzes gap-filling synthesis during base-excision repair. The mechanism of this polymerase is described in a chapter by Joann Sweasy and colleagues provided in this Special Issue. Pol λ is proposed to have multiple cellular activities. The primary biological role of pol λ is to repair double-stranded breaks (DSBs) and is based upon results showing that recombination does not occur in cell extracts that have been immunodepleted for pol λ [129]. Pol λ has also been demonstrated to perform template-independent DNA synthesis using single-stranded DNA or partially double-stranded DNA with a short 3′ overhang [105]. Finally, pol λ is also believed to be an error prone DNA polymerase since it can bypass certain DNA lesions including abasic sites [130]. However, the physiological role of pol λ in performing translesion DNA synthesis has yet to be validated in vivo. In contrast, pol μ is a bona fide error-prone DNA polymerase that plays an active role in somatic hypermutation [131]. This activity is based on the preferential expression of pol μ in secondary lymphoid tissues as well as its low fidelity during the replication of undamaged DNA [132]. Additionally, the ability of pol μ to bypass several DNA lesions through a deletion mechanism provides circumstantial evidence for its role as a mutase during somatic hypermutation [133]. A perspective on the role of certain X-family DNA polymerases in regulating nucleic acid integrity is provided in a recent review by Ramadan et al [134].
Features of TdT Common with Other X-Family Polymerases
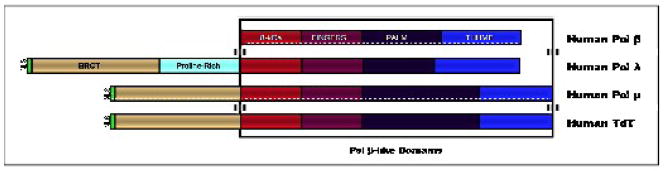
Sequence alignment of the C-termini of the X-family DNA polymerases reveals that all these enzymes possess the fingers, palm, and thumb subdomains that are similar to those described first for pol β (Figure 7). TdT, pol λ, and pol μ contain nuclear localization signal motifs as well as breast cancer susceptibility protein BRCA1 C-terminal (BRCT) domains in their N-termini [127]. BRCT domains function to mediate protein/protein and protein/DNA interactions in DNA repair and cell cycle check-point pathways that are activated by DNA damage [135]. As described later in this review, the BRCT domain of TdT, pol λ, and pol μ may interact with Ku70/80, a heterodimeric protein involved in recognizing and binding the ends of double-strand DNA breaks formed during V(D)J recombination.
Figure 7.
Schematic representations of the different domains found in the four X-family DNA polymerases. Each domain is labeled and colored for clarity. NLS represents the nuclear localization signal motif, and BRCT indicates the BRCA1 carboxy terminus domain.
Considering this information, it is quite surprising that members of this family show little primary amino acid sequence identity. For instance, TdT and pol μ share only 42% amino acid identity yet are considered the most closely related members of the X-family of polymerases [136]. Despite this low identity, it is proposed that TdT and pol μ arose from a common ancestor that was a template-dependent polymerase [137]. The strict template-independent activity of TdT appears to be a recent evolutionary event that coincides with the development of V(D)J recombination in mammals.
Tertiary Structure of TdT
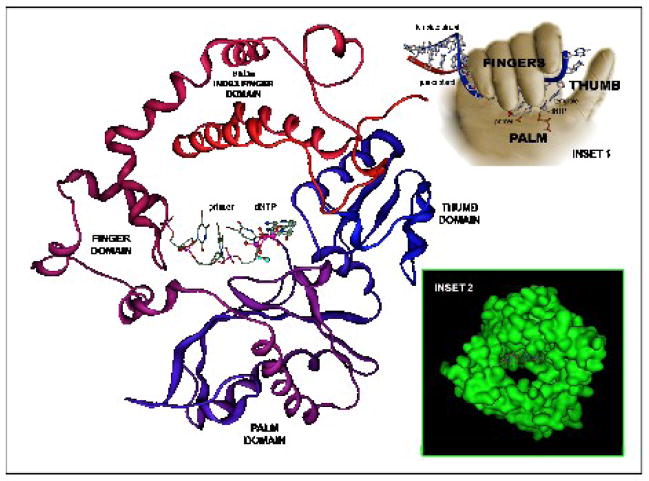
All template-dependent DNA polymerases characterized to date by structural methods show an overall molecular architecture that is analogous to a right hand containing thumb, fingers, and palm subdomains (Figure 8, inset 1) (reviewed in [86]). Although these subdomains work synergistically during the polymerization process, it is easier to describe their functions independently. The palm subdomain is considered to be the heart of the polymerase as this is where the phosphoryl transfer reaction takes place. This subdomain contains at least two carboxylate residues that are highly conserved amongst all DNA polymerases [138] which function to coordinate metal ions that act as Lewis acids. These metal ions lower the activation energy barrier for the phosphoryl transfer reaction by stabilizing the build-up of the negative charge that accumulates on the α-phosphate and the oxyanion of the β- and γ-phosphate leaving group of the incoming dNTP substrate [88]. The function of the fingers subdomain is to coordinate interactions between the templating base and the incoming dNTP. These interactions are designed to align the nucleobases properly prior to catalysis. With template-dependent DNA polymerases such as pol β [87, 139, 140], the thumb subdomain serves a dual role by positioning duplex DNA for accepting the incoming dNTP as well as for translocation of the polymerase to the next templating base position after catalysis.
Figure 8.
Crystal structure of terminal deoxynucleotidyl transferase showing the finger, thumb, palm and index finger (8kDa) subdomains that work synergistically to catalyze nucleotide incorporation. Inset 1 shows the hand-like morphology of most DNA polymerases. Inset 2 shows a surface model for the ring-like structure of TdT in which the primer strand is located perpendicular to the hole where dNTPs presumably diffuse to enter the active site. The ternary complex structure was prepared using the available binary crystal structures of murine TdT (PDB ID codes 1KEJ (TdT•ddATP) and 1KDH (TdT•ssDNA) [116]). MOE (www.chemcomp.com) was used for all structural modeling.
The structure of TdT shown in Figure 8 shows similarities to template-dependent DNA polymerases by the presence of thumb, fingers, and palm subdomains. The catalytic site of TdT also shows the presence of two Co2+ ions in the palm domain [127]. This feature is consistent with the proposed “two divalent metal ion” mechanism [88] associated with all polymerases currently identified. Finally, these metal ions are coordinated by the oxygen atoms of the triphosphate moiety of the incoming dNTP as well as by three aspartate residues [127].
Despite these similarities, there are some notable structural variations in TdT that distinguish it from template-dependent polymerases. In particular, TdT possesses a “lariat-like” loop that prevents the enzyme from interacting with a template strand [127]. In addition, TdT contains an 8kDa domain, also known as the “index finger” domain, which contacts the thumb subdomain to form a hole that may allow dNTPs to diffuse into the enzyme’s active site [127]. This feature defines the general morphology of TdT as a ring-like structure in which the primer lies perpendicularly to the axis of the hole on the palm domain (Figure 8, inset 2).
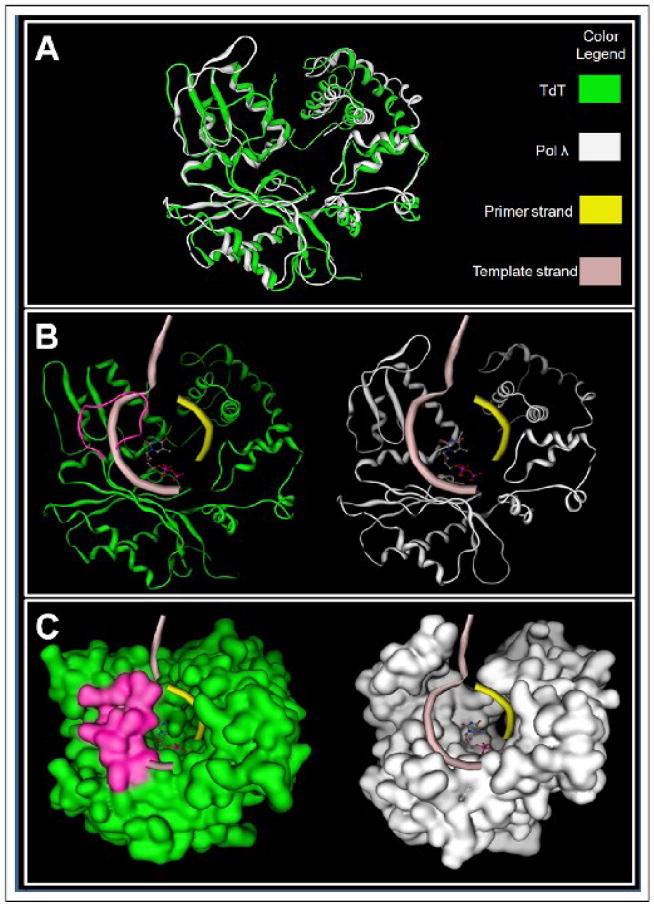
Superimposing the structure of TdT (PDB ID code 1KEJ) [127] with the closed form of pol λ (PDB ID code 1XSN) [141] shows that the “lariat-like” loop of 16 amino acids in TdT crystal prevents the polymerase from interacting with duplex DNA (Figure 9). This provides a physical explanation for the unique ability of TdT to catalyze the nucleotidyl transfer reaction in a template-independent fashion. Delarue and colleagues originally pointed out that TdT appears to be sealed in a closed conformation that is similar to pol β [127], suggesting that TdT can add nucleotides without undergoing an obligatory conformational change [127].
Figure 9.
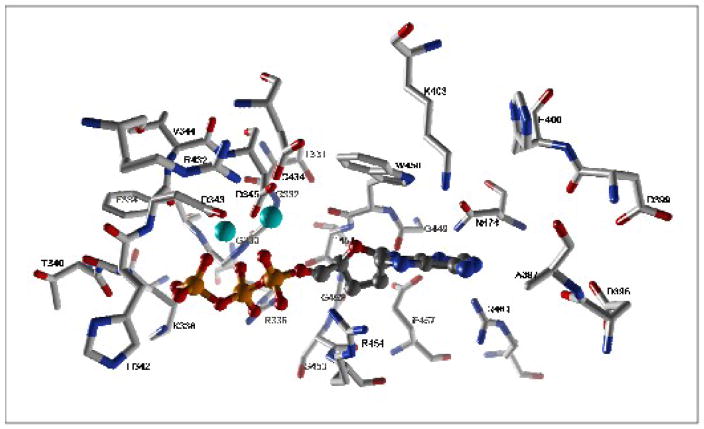
The active site of TdT (PDB ID code: 1KEJ) as defined by amino acids that exist within 6 Å of the bound nucleotide substrate, ddATP. The incoming nucleotide is shown in ball-stick representation in CPK color scheme. The two cobalt ions are colored as cyan. This figure was prepared using the UCSF Chimera package (http://www.cgl.ucsf.edu/chimera).
Insight into the mechanism of nucleotide selection comes from an examination of the available binary structures of the TdT•ddATP (PDB ID code 1KEJ) and TdT•ssDNA (PDB ID code 1KDH) complexes [127]. An inspection of amino acids that reside within 6Å of the bound ddATP (Figure 10) reveals the presence of three positively charged residues (Lys338, Arg336, and Arg454) that point toward the triphosphate moiety to neutralize the negatively charged phosphate molecules. In addition, three conserved aspartates (Asp343, Asp345, and Asp434) reside in the catalytic palm subdomain and function to position two Co2+ ions for catalysis. The aromatic ring of Trp450 appears to provide favorable pi-pi stacking interaction as it lies parallel to the adenine ring of the incoming nucleotide and resides only 3.6 Å away. The positively charged e amino group of Lys403 side chain is only 4 Å away and can provide favorable pi-cation interactions with the adenine nucleobase. Although Arg454 interacts with the triphosphate moiety, it may also participate in pi-cation interactions with the aromatic adenine to provide additional stability. No other side chains are in close proximity with either the 2′ or the 3′ position of the sugar of ddATP. The absence of amino acids that could function as a “steric gate” could account for the rather promiscuous nature of TdT in its ability to incorporate both ribo- and deoxyribonucleotides [142].
Figure 10.
Comparing the crystal structures of template-independent TdT enzyme (PDB code: 1KEJ) and template-dependent DNA polymerase, pol λ (PDB code: 1XSN). (A) Superimposed structures of TdT and pol λ. The superimposed structures show an incredibly high degree of similarity between the two polymerases of the family X despite the fact that they catalyze different modes of DNA polymerization (template-independent versus template-dependent). (B) The ribbon structure of TdT (left) with the putative model of primer-template duplex derived from the human pol λ ternary complex (right) to show the “lariat-like” loop (in magenta) that prevents the ability of TdT to accommodate a templating strand. (C) Molecular surface model of TdT (left) with the putative model of primer-template duplex (shown in ribbon) derived from the human pol λ ternary complex (right). The dNTP is shown in ball and stick model in CPK color scheme. The models were prepared using the MOE package (www.chemcomp.com).
Regulating the Activity of TdT
Since V(D)J recombination is required for a competent immune system, it is imperative that the activities of the enzymes involved in this process be tightly regulated. TdT is no exception as it is regulated by transcriptional control, post-translational modifications, and protein-protein interactions [143].
Expression of TdT is confined to primary lymphoid tissues including thymus and bone marrow [15, 51, 52]. Transcriptional control is regulated by proteins such as AP-1 [144] as well as through the expression of the RAG genes [145]. The activity of TdT may also be regulated at the post-translational level by phosphorylation. It has been demonstrated that TdT is phosphorylated in lymphoblastoid cells when treated with [32P]-phosphate [146]. In addition, recombinant human TdT is phosphorylated in vitro by protein kinase C [147] while calf thymus TdT can be phosphorylated by beef heart cAMP-dependent protein kinase [148]. In the latter case, calf thymus TdT is phosphorylated at multiple sites that correspond to amino acids Ser7 and Thr19 in human TdT [74, 146]. While TdT can be phosphorylated at multiple sites, it is unclear as to how phosphorylation regulates its activity in vivo.
Protein-protein interactions between TdT and other DNA binding proteins have been shown to produce both positive and negative effects on the catalytic activity of TdT [108, 149]. One group of proteins reported to regulate the activity of TdT are referred to as TdT interacting factors (TdiFs). TdiF1 is a protein that binds to the C-terminus of TdT and increases its polymerase activity by ~4-fold [108]. TdiF2 is another protein that binds TdT through its C-terminus, possibly through interactions with the proline-rich and pol β-like domains. However, the interaction of TdiF2 with TdT decreases the polymerase activity of TdT by ~2-fold [108].
PCNA is another protein that physically interacts with TdT [146]. By encircling DNA, PCNA acts as a general processivity factor for various template-dependent DNA polymerases including pol δ and ε [150]. In fact, PCNA orchestrates nearly every aspect of DNA synthesis ranging from chromosomal DNA replication to DNA repair and recombination [151]. PCNA decreases the polymerase activity of TdT by ~2-fold. This effect on TdT activity is similar to that observed by TdiF2 [146]. One possibility is that PCNA and TdiF2 compete with TdiF1 for binding to the C-terminus of TdT which provides a mechanism to reversibly regulate TdT activity.
Other proteins such as the Ku proteins and template-dependent DNA polymerases may also regulate the activity of TdT. For example, the “N” regions generated by TdT are unusually longer when Ku80 is knocked-out compared to when it is normally expressed [152]. Since Ku80 aids in the recruitment of TdT to the site of V(D)J recombination, these knock-down experiments suggests that Ku80 also regulates the catalytic activity of TdT once it is bound to DNA [61, 153]. TdT activity can also be regulated indirectly by other members of the X-family of polymerases. For example, TdT and Pol μ can efficiently compete for the same DNA substrate [152]. It has subsequently been argued that both proteins, if present during V(D)J recombination, could compete for DNA ends such that Pol μ could affect the activity of TdT during “N” region synthesis [132].
Biomedical Applications of TdT
TdT as an Anti-Cancer Target
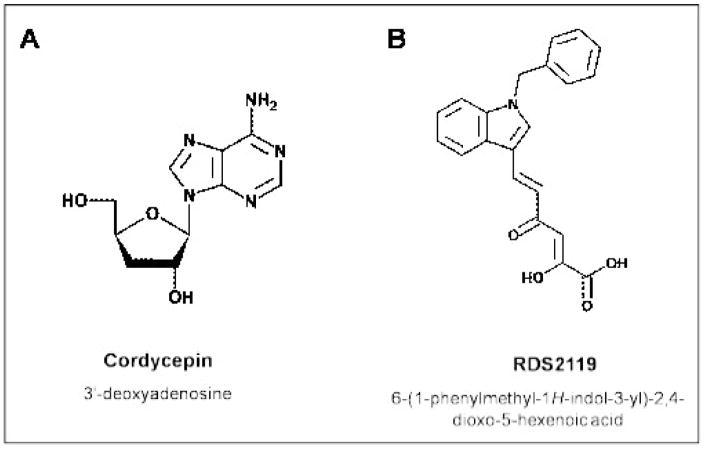
There is mounting clinical evidence that alterations in TdT activity and/or its level of expression play significant roles in cancer initiation, progression, and response to chemotherapy. TdT is overexpressed in B- and T-cell acute lymphocytic leukemias (ALL) and in acute myelocytic leukemias (AML) [154–156]. About 90% of patients with ALL show variable levels of TdT expression as well as multiple isoforms of TdT in their blast cells [154–156]. While the frequency of TdT overexpression in AML is less (~20%), it is still significantly higher than levels found in lymphoid malignancies such as chronic lymphocytic leukemia (CLL) [156]. In addition, higher levels of TdT activity correlate with a poor prognosis due to sub-optimal responses to chemotherapy that culminates in reduced survival times. Prognosis and survival studies show that remission rates are two-fold lower in leukemia patients that are TdT-positive compared to TdT-negative patients[157]. These findings have driven attempts to develop selective inhibitors against TdT that could be employed as chemotherapeutic agents against these forms of leukemia. One example is the nucleoside analog cordycepin (3′-deoxy adenosine) (Figure 11A). This nucleoside analog is cytotoxic against TdT-positive leukemias, especially when used in combination with the adenosine deaminase inhibitor deoxycoformycin [158, 159]. The cytotoxic activity of cordycepin correlates with the ability of the triphosphate form to inhibit single-stranded DNA synthesis by TdT in vitro [160]. Since cordycepin lacks a 3′-OH moiety, incorporation of this analog terminates primer extension and generates abortive intermediates along the recombination pathway that may induce apoptosis.
Figure 11.
Chemical structures for different inhibitors of TdT. (A) Cordycepin. (B) RDS 2119.
Unfortunately, cordycepin is not widely used in chemotherapy as it causes side effects that are associated with the inhibition of various enzymes involved in nucleos(t)ide metabolism [161]. As a consequence, there is significant effort in developing selectively inhibitors of TdT that do not resemble nucleos(t)ide analogs. Indeed, encouraging progress has been made by the groups of DiSanto and Maga as evident in their recent demonstration that certain aryldiketo hexenoic acids inhibits the catalytic activity of pol λ and TdT without acting as chain-terminators [162]. One analog, designated RDS 2119 (Figure 11B), displays higher cytotoxicity against a TdT-positive leukemia cell line (Molt4) compared to a cell line derived from cervical cancer (HeLa) [162].
TdT as a Biochemical Tool
TdT utilizes a wide variety of nucleotide analogs such as 2′,3′-dideoxynucleotides [163], p-nitrophenylethyl triphosphate [164], p-nitrophenyl triphosphate [164], 2′-deoxy-L-ribonucleoside 5′-triphosphates [165], and dinucleoside 5′,5′-tetraphosphates [166]. The ability of TdT to toleratebulky modifications to the nucleobase moeity has led to an effective method for in vivo and in vitro labeling of double-strand DNA breaks. One technique called TUNEL (TdT-mediated dUTP-biotin nick end-labeling) is based on the ability of TdT to efficiently incorporate biotinylated dUMP on to 3′-ends of single-stranded DNA that occur at the sites of DNA breaks. The incorporated biotin dUMP is easily visualized by fluorescently labeled avidin or streptavidin and allows for direct conformation and quantitative measurements of the number and location of DNA breaks. This technology is widely used for the detection of apoptosis, a form of programmed cell death in eukaryotic cells [167]. In addition, TdT can be used as a biocatalyst to label the 3′-termini of synthetic oligonucleotides with a radioactive nucleotide or a variety of fluorescent probes [168]. These labeled primers can then be annealed to a complementary strand and used as radioactive substrates to monitor the activity of enzymes involved in nucleic acid metabolism that include restriction endonucleases, DNA glycosylases, and template-dependent DNA polymerases.
Conclusions
Amongst all DNA polymerases identified to date, TdT remains the most enigmatic. This remarkable polymerase can truly be called the “black sheep” of the polymerase family since it breaks (or at least bends) nearly every rule established for template-dependent polymerases. Most notably, TdT is defiant in its inability to perform polymerization with duplex DNA while incorporating nucleotides using single-stranded DNA as the substrate. The template-independent activity of TdT predicts that it should incorporate natural dNMPs with equal efficiencies. Again, TdT bends the rules by displaying an unexplained bias towards incorporating dGMP and dCMP. In fact, TdT even discriminates against the incorporation of dAMP, the preferred nucleotide for template-dependent polymerase during the replication of certain forms of damaged DNA. Finally, TdT is unique in its ability to randomly bind DNA and dNTP to form the catalytically competent ternary complex. It could be argued that TdT must be rebellious in order to perform its unique biological function for generating random coding information during V(D)J recombination.
Although TdT is unique, it does share many features that are common amongst most template-dependent DNA polymerases. From a structural perspective, TdT contains the fingers, thumb, and palm subdomains that are present in all DNA polymerases characterized to date. The most notable difference is the inclusion of the “lariat loop” that blocks the ability of TdT to utilize duplex DNA. Furthermore, TdT can interact with other replicative proteins such as PCNA that function to coordinate polymerase activity during replication, repair, and recombination. Although the functional outcome of these interactions remain poorly understood, future biochemical and cell-based studies will undoubtedly shed more insight into the biological role and molecular mechanism of this fascinating and enigmatic DNA polymerase.
Abbreviations and Textual Footnotes
- TdT
terminal deoxynucleotidyl transferase
- CpG
cytidine guanine base pair
- RAG-1
recombination-activating gene 1
- RAG-2
recombination-activating gene 2
- RSS
recombination signal sequences
- Ig
immunoglobulin
- NHEJ
non-homologous end joining
- TCR
T-cell receptor
- PK
protein kinase
- PPi
inorganic pyrophosphate
- 5-NIMP
5-nitro-indolyl-2′-deoxyriboside-5′-monophosphate
- 5-AIMP
5-amino-indolyl-2′-deoxyriboside-5′-monophosphate
- 5-PhIMP
5-phenyl-indolyl-2′-deoxyriboside-5′-monophosphate
- 5-CEIMP
5-cyclohexenyl-indolyl-2′-deoxyriboside-5′-monophosphate
- TdiFs
TdTase interacting factors
- PCNA
Proliferating cell nuclear antigen
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bollum FJ. Thermal conversion of nonpriming deoxyribonucleic acid to primer. J Biol Chem. 1959;234:2733–2734. [PubMed] [Google Scholar]
- 2.Bollum FJ. Chemically Defined Templates and Initiators for Deoxypolynucleotide Synthesis. Science. 1964;144:560. doi: 10.1126/science.144.3618.560-b. [DOI] [PubMed] [Google Scholar]
- 3.Bollum FJ. Calf thymus polymerase. J Biol Chem. 1960;235:2399–2403. [PubMed] [Google Scholar]
- 4.Kallenbach S, Goodhardt M, Rougeon F. A rapid test for V(D)J recombinase activity. Nucleic Acids Res. 1990;18:6730. doi: 10.1093/nar/18.22.6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilfillan S, Dierich A, Lemeur M, Benoist C, Mathis D. Mice lacking TdT: mature animals with an immature lymphocyte repertoire. Science. 1993;261:1175–1178. doi: 10.1126/science.8356452. [DOI] [PubMed] [Google Scholar]
- 6.Komori T, Okada A, Stewart V, Alt FW. Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science. 1993;261:1171–1175. doi: 10.1126/science.8356451. [DOI] [PubMed] [Google Scholar]
- 7.Landau NR, Schatz DG, Rosa M, Baltimore D. Increased frequency of N-region insertion in a murine pre-B-cell line infected with a terminal deoxynucleotidyl transferase retroviral expression vector. Mol Cell Biol. 1987;7:3237–3243. doi: 10.1128/mcb.7.9.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lieber MR, Hesse JE, Mizuuchi K, Gellert M. Lymphoid V(D)J recombination: nucleotide insertion at signal joints as well as coding joints. Proc Natl Acad Sci U S A. 1988;85:8588–8592. doi: 10.1073/pnas.85.22.8588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertocci B, De Smet A, Weill JC, Reynaud CA. Nonoverlapping functions of DNA polymerases mu, lambda, and terminal deoxynucleotidyltransferase during immunoglobulin V(D)J recombination in vivo. Immunity. 2006;25:31–41. doi: 10.1016/j.immuni.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 10.Haeryfar SM, Hickman HD, Irvine KR, Tscharke DC, Bennink JR, Yewdell JW. Terminal deoxynucleotidyl transferase establishes and broadens antiviral CD8+ T cell immunodominance hierarchies. J Immunol. 2008;181:649–659. doi: 10.4049/jimmunol.181.1.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benedict CL, Gilfillan S, Thai TH, Kearney JF. Terminal deoxynucleotidyl transferase and repertoire development. Immunol Rev. 2000;175:150–157. [PubMed] [Google Scholar]
- 12.Baltimore D. Is terminal deoxynucleotidyl transferase a somatic mutagen in lymphocytes? Nature. 1974;248:409–411. doi: 10.1038/248409a0. [DOI] [PubMed] [Google Scholar]
- 13.Desiderio SV, Yancopoulos GD, Paskind M, Thomas E, Boss MA, Landau N, Alt FW, Baltimore D. Insertion of N regions into heavy-chain genes is correlated with expression of terminal deoxytransferase in B cells. Nature. 1984;311:752–755. doi: 10.1038/311752a0. [DOI] [PubMed] [Google Scholar]
- 14.Kepler TB, Borrero M, Rugerio B, McCray SK, Clarke SH. Interdependence of N nucleotide addition and recombination site choice in V(D)J rearrangement. J Immunol. 1996;157:4451–4457. [PubMed] [Google Scholar]
- 15.Kunkel TA, Gopinathan KP, Dube DK, Snow ET, Loeb LA. Rearrangements of DNA mediated by terminal transferase. Proc Natl Acad Sci U S A. 1986;83:1867–1871. doi: 10.1073/pnas.83.6.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sadofsky MJ. The RAG proteins in V(D)J recombination: more than just a nuclease. Nucleic Acids Res. 2001;29:1399–1409. doi: 10.1093/nar/29.7.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janeway CA. Immunobiology: the immune system in health and disease. 4. Current Biology Publications; London: 1999. [Google Scholar]
- 18.Jeltsch A. Maintenance of species identity and controlling speciation of bacteria: a new function for restriction/modification systems? Gene. 2003;317:13–16. doi: 10.1016/s0378-1119(03)00652-8. [DOI] [PubMed] [Google Scholar]
- 19.Beutler B. Not “molecular patterns” but molecules. Immunity. 2003;19:155–156. doi: 10.1016/s1074-7613(03)00212-7. [DOI] [PubMed] [Google Scholar]
- 20.Beutler B. Inferences, questions and possibilities in Toll-like receptor signalling. Nature. 2004;430:257–263. doi: 10.1038/nature02761. [DOI] [PubMed] [Google Scholar]
- 21.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 22.Raulet DH. Interplay of natural killer cells and their receptors with the adaptive immune response. Nat Immunol. 2004;5:996–1002. doi: 10.1038/ni1114. [DOI] [PubMed] [Google Scholar]
- 23.Pancer Z, Cooper MD. The evolution of adaptive immunity. Annu Rev Immunol. 2006;24:497–518. doi: 10.1146/annurev.immunol.24.021605.090542. [DOI] [PubMed] [Google Scholar]
- 24.Schatz DG, Oettinger MA, Schlissel MS. V(D)J recombination: molecular biology and regulation. Annu Rev Immunol. 1992;10:359–383. doi: 10.1146/annurev.iy.10.040192.002043. [DOI] [PubMed] [Google Scholar]
- 25.Harty JT, Badovinac VP. Shaping and reshaping CD8+ T-cell memory. Nat Rev Immunol. 2008;8:107–119. doi: 10.1038/nri2251. [DOI] [PubMed] [Google Scholar]
- 26.Kaech SM, Wherry EJ, Ahmed R. Effector and memory T-cell differentiation: implications for vaccine development. Nat Rev Immunol. 2002;2:251–262. doi: 10.1038/nri778. [DOI] [PubMed] [Google Scholar]
- 27.Sprent J, Surh CD. T cell memory. Annu Rev Immunol. 2002;20:551–579. doi: 10.1146/annurev.immunol.20.100101.151926. [DOI] [PubMed] [Google Scholar]
- 28.Williams MA, Bevan MJ. Effector and memory CTL differentiation. Annu Rev Immunol. 2007;25:171–192. doi: 10.1146/annurev.immunol.25.022106.141548. [DOI] [PubMed] [Google Scholar]
- 29.Murali-Krishna K, Altman JD, Suresh M, Sourdive DJ, Zajac AJ, Miller JD, Slansky J, Ahmed R. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity. 1998;8:177–187. doi: 10.1016/s1074-7613(00)80470-7. [DOI] [PubMed] [Google Scholar]
- 30.Goldrath AW, Bevan MJ. Selecting and maintaining a diverse T-cell repertoire. Nature. 1999;402:255–262. doi: 10.1038/46218. [DOI] [PubMed] [Google Scholar]
- 31.Butz EA, Bevan MJ. Massive expansion of antigen-specific CD8+ T cells during an acute virus infection. Immunity. 1998;8:167–175. doi: 10.1016/s1074-7613(00)80469-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Badovinac VP, Porter BB, Harty JT. Programmed contraction of CD8(+) T cells after infection. Nat Immunol. 2002;3:619–626. doi: 10.1038/ni804. [DOI] [PubMed] [Google Scholar]
- 33.Masopust D, Vezys V, Marzo AL, Lefrancois L. Preferential localization of effector memory cells in nonlymphoid tissue. Science. 2001;291:2413–2417. doi: 10.1126/science.1058867. [DOI] [PubMed] [Google Scholar]
- 34.Lefrancois L, Masopust D. T cell immunity in lymphoid and non-lymphoid tissues. Curr Opin Immunol. 2002;14:503–508. doi: 10.1016/s0952-7915(02)00360-6. [DOI] [PubMed] [Google Scholar]
- 35.Gourley TS, Wherry EJ, Masopust D, Ahmed R. Generation and maintenance of immunological memory. Semin Immunol. 2004;16:323–333. doi: 10.1016/j.smim.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 36.Papermaster BW, Condie RM, Finstad J, Good RA. Evolution of the Immune Response. I. the Phylogenetic Development of Adaptive Immunologic Responsiveness in Vertebrates. J Exp Med. 1964;119:105–130. doi: 10.1084/jem.119.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schatz DG. V(D)J recombination. Immunol Rev. 2004;200:5–11. doi: 10.1111/j.0105-2896.2004.00173.x. [DOI] [PubMed] [Google Scholar]
- 38.Nick McElhinny SA, Havener JM, Garcia-Diaz M, Juarez R, Bebenek K, Kee BL, Blanco L, Kunkel TA, Ramsden DA. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 39.Bertocci B, De Smet A, Berek C, Weill JC, Reynaud CA. Immunoglobulin kappa light chain gene rearrangement is impaired in mice deficient for DNA polymerase mu. Immunity. 2003;19:203–211. doi: 10.1016/s1074-7613(03)00203-6. [DOI] [PubMed] [Google Scholar]
- 40.Early P, Huang H, Davis M, Calame K, Hood L. An immunoglobulin heavy chain variable region gene is generated from three segments of DNA: VH, D and JH. Cell. 1980;19:981–992. doi: 10.1016/0092-8674(80)90089-6. [DOI] [PubMed] [Google Scholar]
- 41.Sakano H, Maki R, Kurosawa Y, Roeder W, Tonegawa S. Two types of somatic recombination are necessary for the generation of complete immunoglobulin heavy-chain genes. Nature. 1980;286:676–683. doi: 10.1038/286676a0. [DOI] [PubMed] [Google Scholar]
- 42.Sakano H, Kurosawa Y, Weigert M, Tonegawa S. Identification and nucleotide sequence of a diversity DNA segment (D) of immunoglobulin heavy-chain genes. Nature. 1981;290:562–565. doi: 10.1038/290562a0. [DOI] [PubMed] [Google Scholar]
- 43.Kurosawa Y, von Boehmer H, Haas W, Sakano H, Trauneker A, Tonegawa S. Identification of D segments of immunoglobulin heavy-chain genes and their rearrangement in T lymphocytes. Nature. 1981;290:565–570. doi: 10.1038/290565a0. [DOI] [PubMed] [Google Scholar]
- 44.Steen SB, Gomelsky L, Roth DB. The 12/23 rule is enforced at the cleavage step of V(D)J recombination in vivo. Genes Cells. 1996;1:543–553. doi: 10.1046/j.1365-2443.1996.d01-259.x. [DOI] [PubMed] [Google Scholar]
- 45.Mayer RT, McCollum TG, Niedz RP, Hearn CJ, McDonald RE, Berdis E, Doostdar H. Characterization of seven basic endochitinases isolated from cell cultures of Citrus sinensis (L.) Planta. 1996;200:289–295. doi: 10.1007/BF00200295. [DOI] [PubMed] [Google Scholar]
- 46.Janeway CA, Travers P, Walport M, Schlomchik M. Immunobiology: the immune system in health and disease. 6. Garland Publishing; New York: 2005. [Google Scholar]
- 47.Mickelsen S, Snyder C, Trujillo K, Bogue M, Roth DB, Meek K. Modulation of terminal deoxynucleotidyltransferase activity by the DNA-dependent protein kinase. J Immunol. 1999;163:834–843. [PubMed] [Google Scholar]
- 48.Feeney AJ. Lack of N regions in fetal and neonatal mouse immunoglobulin V-D-J junctional sequences. J Exp Med. 1990;172:1377–1390. doi: 10.1084/jem.172.5.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bangs LA, Sanz IE, Teale JM. Comparison of D, JH, and junctional diversity in the fetal, adult, and aged B cell repertoires. J Immunol. 1991;146:1996–2004. [PubMed] [Google Scholar]
- 50.Basu M, Hegde MV, Modak MJ. Synthesis of compositionally unique DNA by terminal deoxynucleotidyl transferase. Biochem Biophys Res Commun. 1983;111:1105–1112. doi: 10.1016/0006-291x(83)91413-4. [DOI] [PubMed] [Google Scholar]
- 51.Coleman MS, Hutton JJ, De Simone P, Bollum FJ. Terminal deoxyribonucleotidyl transferase in human leukemia. Proc Natl Acad Sci U S A. 1974;71:4404–4408. doi: 10.1073/pnas.71.11.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bollum FJ. Terminal deoxynucleotidyl transferase as a hematopoietic cell marker. Blood. 1979;54:1203–1215. [PubMed] [Google Scholar]
- 53.Yancopoulos GD, Blackwell TK, Suh H, Hood L, Alt FW. Introduced T cell receptor variable region gene segments recombine in pre-B cells: evidence that B and T cells use a common recombinase. Cell. 1986;44:251–259. doi: 10.1016/0092-8674(86)90759-2. [DOI] [PubMed] [Google Scholar]
- 54.Cabaniols JP, Fazilleau N, Casrouge A, Kourilsky P, Kanellopoulos JM. Most alpha/beta T cell receptor diversity is due to terminal deoxynucleotidyl transferase. J Exp Med. 2001;194:1385–1390. doi: 10.1084/jem.194.9.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alt FW, Baltimore D. Joining of immunoglobulin heavy chain gene segments: implications from a chromosome with evidence of three D-JH fusions. Proc Natl Acad Sci U S A. 1982;79:4118–4122. doi: 10.1073/pnas.79.13.4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Doyen N, d’Andon MF, Bentolila LA, Nguyen QT, Rougeon F. Differential splicing in mouse thymus generates two forms of terminal deoxynucleotidyl transferase. Nucleic Acids Res. 1993;21:1187–1191. doi: 10.1093/nar/21.5.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koiwai O, Yokota T, Kageyama T, Hirose T, Yoshida S, Arai K. Isolation and characterization of bovine and mouse terminal deoxynucleotidyltransferase cDNAs expressible in mammalian cells. Nucleic Acids Res. 1986;14:5777–5792. doi: 10.1093/nar/14.14.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farrar YJ, Evans RK, Beach CM, Coleman MS. Interactions of photoactive DNAs with terminal deoxynucleotidyl transferase: identification of peptides in the DNA binding domain. Biochemistry. 1991;30:3075–3082. doi: 10.1021/bi00226a014. [DOI] [PubMed] [Google Scholar]
- 59.Yang B, Gathy KN, Coleman MS. Mutational analysis of residues in the nucleotide binding domain of human terminal deoxynucleotidyl transferase. J Biol Chem. 1994;269:11859–11868. [PubMed] [Google Scholar]
- 60.Bentolila LA, Fanton d’Andon M, Nguyen QT, Martinez O, Rougeon F, Doyen N. The two isoforms of mouse terminal deoxynucleotidyl transferase differ in both the ability to add N regions and subcellular localization. Embo J. 1995;14:4221–4229. doi: 10.1002/j.1460-2075.1995.tb00096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thai TH, Purugganan MM, Roth DB, Kearney JF. Distinct and opposite diversifying activities of terminal transferase splice variants. Nat Immunol. 2002;3:457–462. doi: 10.1038/ni788. [DOI] [PubMed] [Google Scholar]
- 62.Doyen N, Boule JB, Rougeon F, Papanicolaou C. Evidence that the long murine terminal deoxynucleotidyltransferase isoform plays no role in the control of V(D)J junctional diversity. J Immunol. 2004;172:6764–6767. doi: 10.4049/jimmunol.172.11.6764. [DOI] [PubMed] [Google Scholar]
- 63.Benedict CL, Kearney JF. Increased junctional diversity in fetal B cells results in a loss of protective anti-phosphorylcholine antibodies in adult mice. Immunity. 1999;10:607–617. doi: 10.1016/s1074-7613(00)80060-6. [DOI] [PubMed] [Google Scholar]
- 64.Boule JB, Rougeon F, Papanicolaou C. Comparison of the two murine terminal [corrected] deoxynucleotidyltransferase terminal isoforms. A 20-amino acid insertion in the highly conserved carboxyl-terminal region modifies the thermosensitivity but not the catalytic activity. J Biol Chem. 2000;275:28984–28988. doi: 10.1074/jbc.M005544200. [DOI] [PubMed] [Google Scholar]
- 65.Thai TH, Kearney JF. Isoforms of terminal deoxynucleotidyltransferase: developmental aspects and function. Adv Immunol. 2005;86:113–136. doi: 10.1016/S0065-2776(04)86003-6. [DOI] [PubMed] [Google Scholar]
- 66.Takahara K, Hayashi N, Fujita-Sagawa K, Morishita T, Hashimoto Y, Noda A. Alternative splicing of bovine terminal deoxynucleotidyl transferase cDNA. Biosci Biotechnol Biochem. 1994;58:786–787. doi: 10.1271/bbb.58.786. [DOI] [PubMed] [Google Scholar]
- 67.Thai TH, Kearney JF. Distinct and opposite activities of human terminal deoxynucleotidyltransferase splice variants. J Immunol. 2004;173:4009–4019. doi: 10.4049/jimmunol.173.6.4009. [DOI] [PubMed] [Google Scholar]
- 68.Chang LM, Bollum FJ. Deoxynucleotide-polymerizing enzymes of calf thymus gland. V. Homogeneous terminal deoxynucleotidyl transferase. J Biol Chem. 1971;246:909–916. [PubMed] [Google Scholar]
- 69.Deibel MR, Jr, Coleman MS. Limited proteolysis of calf thymus terminal deoxynucleotidyl transferase. Arch Biochem Biophys. 1980;202:414–419. doi: 10.1016/0003-9861(80)90445-2. [DOI] [PubMed] [Google Scholar]
- 70.Chang LM, Plevani P, Bollum FJ. Proteolytic degradation of calf thymus terminal deoxynucleotidyl transferase. J Biol Chem. 1982;257:5700–5706. [PubMed] [Google Scholar]
- 71.Bollum FJ, Chang LM. Immunological detection of a conserved structure for terminal deoxynucleotidyltransferase. J Biol Chem. 1981;256:8767–8770. [PubMed] [Google Scholar]
- 72.Nakamura H, Tanabe K, Yoshida S, Morita T. Terminal deoxynucleotidyltransferase of 60,000 daltons from mouse, rat, and calf thymus. Purification by immunoadsorbent chromatography and comparison of peptide structures. J Biol Chem. 1981;256:8745–8751. [PubMed] [Google Scholar]
- 73.Deibel MR, Jr, Coleman MS. Biochemical properties of purified human terminal deoxynucleotidyltransferase. J Biol Chem. 1980;255:4206–4212. [PubMed] [Google Scholar]
- 74.Chang LM, Rafter E, Rusquet-Valerius R, Peterson RC, White ST, Bollum FJ. Expression and processing of recombinant human terminal transferase in the baculovirus system. J Biol Chem. 1988;263:12509–12513. [PubMed] [Google Scholar]
- 75.Peterson RC, Cheung LC, Mattaliano RJ, White ST, Chang LM, Bollum FJ. Expression of human terminal deoxynucleotidyl transferase in Escherichia coli. J Biol Chem. 1985;260:10495–10502. [PubMed] [Google Scholar]
- 76.Boule JB, Johnson E, Rougeon F, Papanicolaou C. High-level expression of murine terminal deoxynucleotidyl transferase in Escherichia coli grown at low temperature and overexpressing argU tRNA. Mol Biotechnol. 1998;10:199–208. doi: 10.1007/BF02740839. [DOI] [PubMed] [Google Scholar]
- 77.Karkow JS, Kamen LHO. DNA synthesis in thymus gland. 1966;2:307. [Google Scholar]
- 78.Kato KI, Goncalves JM, Houts GE, Bollum FJ. Deoxynucleotide-polymerizing enzymes of calf thymus gland. II. Properties of the terminal deoxynucleotidyltransferase. J Biol Chem. 1967;242:2780–2789. [PubMed] [Google Scholar]
- 79.Chang LM, Cassani GR, Bollum FJ. Deoxynucleotide-polymerizing enzymes of calf thymus gland. VII. Replication of homopolymers. J Biol Chem. 1972;247:7718–7723. [PubMed] [Google Scholar]
- 80.Roychoudhury R. Enzymic synthesis of polynucleotides. Oligodeoxynucleotides with one 3′-terminal ribonucleotide as primers for polydeoxynucleotide synthesis. J Biol Chem. 1972;247:3910–3917. [PubMed] [Google Scholar]
- 81.Ramadan K, Shevelev IV, Maga G, Hubscher U. De novo DNA synthesis by human DNA polymerase lambda, DNA polymerase mu and terminal deoxyribonucleotidyl transferase. J Mol Biol. 2004;339:395–404. doi: 10.1016/j.jmb.2004.03.056. [DOI] [PubMed] [Google Scholar]
- 82.Vineyard D, Patterson-Ward J, Berdis AJ, Lee I. Monitoring the timing of ATP hydrolysis with activation of peptide cleavage in Escherichia coli Lon by transient kinetics. Biochemistry. 2005;44:1671–1682. doi: 10.1021/bi048618z. [DOI] [PubMed] [Google Scholar]
- 83.Gupta AP, Benkovic SJ. Stereochemical course of the 3′----5′-exonuclease activity of DNA polymerase I. Biochemistry. 1984;23:5874–5881. doi: 10.1021/bi00319a029. [DOI] [PubMed] [Google Scholar]
- 84.Burgers PM, Eckstein F. A study of the mechanism of DNA polymerase I from Escherichia coli with diastereomeric phosphorothioate analogs of deoxyadenosine triphosphate. J Biol Chem. 1979;254:6889–6893. [PubMed] [Google Scholar]
- 85.Lin P, Pedersen LC, Batra VK, Beard WA, Wilson SH, Pedersen LG. Energy analysis of chemistry for correct insertion by DNA polymerase beta. Proc Natl Acad Sci U S A. 2006;103:13294–13299. doi: 10.1073/pnas.0606006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brautigam CA, Steitz TA. Structural and functional insights provided by crystal structures of DNA polymerases and their substrate complexes. Curr Opin Struct Biol. 1998;8:54–63. doi: 10.1016/s0959-440x(98)80010-9. [DOI] [PubMed] [Google Scholar]
- 87.Sawaya MR, Pelletier H, Kumar A, Wilson SH, Kraut J. Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism. Science. 1994;264:1930–1935. doi: 10.1126/science.7516581. [DOI] [PubMed] [Google Scholar]
- 88.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci U S A. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chang LM, Bollum FJ. Doxynucleotide-polymerizing enzymes of calf thymus gland. IV. Inhibition of terminal deoxynucleotidyl transferase by metal ligands. Proc Natl Acad Sci U S A. 1970;65:1041–1048. doi: 10.1073/pnas.65.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chang LM, Bollum FJ. Multiple roles of divalent cation in the terminal deoxynucleotidyltransferase reaction. J Biol Chem. 1990;265:17436–17440. [PubMed] [Google Scholar]
- 91.Kornberg A, Baker TA. DNA replication. 2. W. H. Freeman and Company; New York: 1992. [Google Scholar]
- 92.Clark JM. Novel non-templated nucleotide addition reactions catalyzed by procaryotic and eucaryotic DNA polymerases. Nucleic Acids Res. 1988;16:9677–9686. doi: 10.1093/nar/16.20.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Clark JM, Joyce CM, Beardsley GP. Novel blunt-end addition reactions catalyzed by DNA polymerase I of Escherichia coli. J Mol Biol. 1987;198:123–127. doi: 10.1016/0022-2836(87)90462-1. [DOI] [PubMed] [Google Scholar]
- 94.Hwang H, Taylor JS. Role of base stacking and sequence context in the inhibition of yeast DNA polymerase eta by pyrene nucleotide. Biochemistry. 2004;43:14612–14623. doi: 10.1021/bi0489558. [DOI] [PubMed] [Google Scholar]
- 95.Peliska JA, Benkovic SJ. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science. 1992;258:1112–1118. doi: 10.1126/science.1279806. [DOI] [PubMed] [Google Scholar]
- 96.Vineyard D, Zhang X, Donnelly A, Lee I, Berdis AJ. Optimization of non-natural nucleotides for selective incorporation opposite damaged DNA. Org Biomol Chem. 2007;5:3623–3630. doi: 10.1039/b712480e. [DOI] [PubMed] [Google Scholar]
- 97.Devadoss B, Lee I, Berdis AJ. Enhancing the “A-rule” of translesion DNA synthesis: promutagenic DNA synthesis using modified nucleoside triphosphates. Biochemistry. 2007;46:13752–13761. doi: 10.1021/bi701328h. [DOI] [PubMed] [Google Scholar]
- 98.Goodman MF, Creighton S, Bloom LB, Petruska J. Biochemical basis of DNA replication fidelity. Crit Rev Biochem Mol Biol. 1993;28:83–126. doi: 10.3109/10409239309086792. [DOI] [PubMed] [Google Scholar]
- 99.Hatahet Z, Zhou M, Reha-Krantz LJ, Ide H, Morrical SW, Wallace SS. In vitro selection of sequence contexts which enhance bypass of abasic sites and tetrahydrofuran by T4 DNA polymerase holoenzyme. J Mol Biol. 1999;286:1045–1057. doi: 10.1006/jmbi.1998.2520. [DOI] [PubMed] [Google Scholar]
- 100.Loeb LA, Preston BD. Mutagenesis by apurinic/apyrimidinic sites. Annu Rev Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- 101.Sheriff A, Motea E, Lee I, Berdis AJ. Mechanism and dynamics of translesion DNA synthesis catalyzed by the Escherichia coli Klenow fragment. Biochemistry. 2008;47:8527–8537. doi: 10.1021/bi800324r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shibutani S, Takeshita M, Grollman AP. Translesional synthesis on DNA templates containing a single abasic site. A mechanistic study of the “A rule”. J Biol Chem. 1997;272:13916–13922. doi: 10.1074/jbc.272.21.13916. [DOI] [PubMed] [Google Scholar]
- 103.Berdis AJ. Dynamics of translesion DNA synthesis catalyzed by the bacteriophage T4 exonuclease-deficient DNA polymerase. Biochemistry. 2001;40:7180–7191. doi: 10.1021/bi0101594. [DOI] [PubMed] [Google Scholar]
- 104.Maga G, Ramadan K, Locatelli GA, Shevelev I, Spadari S, Hubscher U. DNA elongation by the human DNA polymerase lambda polymerase and terminal transferase activities are differentially coordinated by proliferating cell nuclear antigen and replication protein A. J Biol Chem. 2005;280:1971–1981. doi: 10.1074/jbc.M411650200. [DOI] [PubMed] [Google Scholar]
- 105.Crespan E, Zanoli S, Khandazhinskaya A, Shevelev I, Jasko M, Alexandrova L, Kukhanova M, Blanca G, Villani G, Hubscher U, Spadari S, Maga G. Incorporation of non-nucleoside triphosphate analogues opposite to an abasic site by human DNA polymerases beta and lambda. Nucleic Acids Res. 2005;33:4117–4127. doi: 10.1093/nar/gki723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Benkovic SJ, Cameron CE. Kinetic analysis of nucleotide incorporation and misincorporation by Klenow fragment of Escherichia coli DNA polymerase I. Methods Enzymol. 1995;262:257–269. doi: 10.1016/0076-6879(95)62022-2. [DOI] [PubMed] [Google Scholar]
- 107.Ibe S, Fujita K, Toyomoto T, Shimazaki N, Kaneko R, Tanabe A, Takebe I, Kuroda S, Kobayashi T, Toji S, Tamai K, Yamamoto H, Koiwai O. Terminal deoxynucleotidyltransferase is negatively regulated by direct interaction with proliferating cell nuclear antigen. Genes Cells. 2001;6:815–824. doi: 10.1046/j.1365-2443.2001.00460.x. [DOI] [PubMed] [Google Scholar]
- 108.Fujita K, Shimazaki N, Ohta Y, Kubota T, Ibe S, Toji S, Tamai K, Fujisaki S, Hayano T, Koiwai O. Terminal deoxynucleotidyltransferase forms a ternary complex with a novel chromatin remodeling protein with 82 kDa and core histone. Genes Cells. 2003;8:559–571. doi: 10.1046/j.1365-2443.2003.00656.x. [DOI] [PubMed] [Google Scholar]
- 109.Fujisaki S, Sato A, Toyomoto T, Hayano T, Sugai M, Kubota T, Koiwai O. Direct binding of TReP-132 with TdT results in reduction of TdT activity. Genes Cells. 2006;11:47–57. doi: 10.1111/j.1365-2443.2005.00916.x. [DOI] [PubMed] [Google Scholar]
- 110.Beckman RA, Loeb LA. Multi-stage proofreading in DNA replication. Q Rev Biophys. 1993;26:225–331. doi: 10.1017/s0033583500002869. [DOI] [PubMed] [Google Scholar]
- 111.Kool ET. Active site tightness and substrate fit in DNA replication. Annu Rev Biochem. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]
- 112.Modak MJ. Biochemistry of terminal deoxynucleotidyltransferase: mechanism of inhibition by adenosine 5′-triphosphate. Biochemistry. 1978;17:3116–3120. doi: 10.1021/bi00608a027. [DOI] [PubMed] [Google Scholar]
- 113.Berdis AJ, McCutcheon D. The use of non-natural nucleotides to probe template-independent DNA synthesis. Chembiochem. 2007;8:1399–1408. doi: 10.1002/cbic.200700096. [DOI] [PubMed] [Google Scholar]
- 114.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005;309:2219–2222. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- 115.Strauss BS. The ‘A rule’ of mutagen specificity: a consequence of DNA polymerase bypass of non-instructional lesions? Bioessays. 1991;13:79–84. doi: 10.1002/bies.950130206. [DOI] [PubMed] [Google Scholar]
- 116.Reineks EZ, Berdis AJ. Evaluating the contribution of base stacking during translesion DNA replication. Biochemistry. 2004;43:393–404. doi: 10.1021/bi034948s. [DOI] [PubMed] [Google Scholar]
- 117.Echols H, Goodman MF. Fidelity mechanisms in DNA replication. Annu Rev Biochem. 1991;60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- 118.Kiefer JR, Mao C, Braman JC, Beese LS. Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal. Nature. 1998;391:304–307. doi: 10.1038/34693. [DOI] [PubMed] [Google Scholar]
- 119.Hogg M, Wallace SS, Doublie S. Crystallographic snapshots of a replicative DNA polymerase encountering an abasic site. Embo J. 2004;23:1483–1493. doi: 10.1038/sj.emboj.7600150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bloom LB, Otto MR, Beechem JM, Goodman MF. Influence of 5′-nearest neighbors on the insertion kinetics of the fluorescent nucleotide analog 2-aminopurine by Klenow fragment. Biochemistry. 1993;32:11247–11258. doi: 10.1021/bi00092a039. [DOI] [PubMed] [Google Scholar]
- 121.Hariharan C, Reha-Krantz LJ. Using 2-aminopurine fluorescence to detect bacteriophage T4 DNA polymerase-DNA complexes that are important for primer extension and proofreading reactions. Biochemistry. 2005;44:15674–15684. doi: 10.1021/bi051462y. [DOI] [PubMed] [Google Scholar]
- 122.Johnson KA. Conformational coupling in DNA polymerase fidelity. Annu Rev Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- 123.Dunlap CA, Tsai MD. Use of 2-aminopurine and tryptophan fluorescence as probes in kinetic analyses of DNA polymerase beta. Biochemistry. 2002;41:11226–11235. doi: 10.1021/bi025837g. [DOI] [PubMed] [Google Scholar]
- 124.Arnold JJ, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2+ Biochemistry. 2004;43:5126–5137. doi: 10.1021/bi035212y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sucato CA, Upton TG, Kashemirov BA, Batra VK, Martinek V, Xiang Y, Beard WA, Pedersen LC, Wilson SH, McKenna CE, Florian J, Warshel A, Goodman MF. Modifying the beta, gamma leaving-group bridging oxygen alters nucleotide incorporation efficiency, fidelity, and the catalytic mechanism of DNA polymerase beta. Biochemistry. 2007;46:461–471. doi: 10.1021/bi061517b. [DOI] [PubMed] [Google Scholar]
- 126.Schray KJ, Benkovic SJ. Mechanisms of hydrolysis of phosphate ester derivatives of phosphoenolpyruvic acid. J Am Chem Soc. 1971;93:2522–2529. doi: 10.1021/ja00739a027. [DOI] [PubMed] [Google Scholar]
- 127.Delarue M, Boule JB, Lescar J, Expert-Bezancon N, Jourdan N, Sukumar N, Rougeon F, Papanicolaou C. Crystal structures of a template-independent DNA polymerase: murine terminal deoxynucleotidyltransferase. Embo J. 2002;21:427–439. doi: 10.1093/emboj/21.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Herschlag D, Piccirilli JA, Cech TR. Ribozyme-catalyzed and nonenzymatic reactions of phosphate diesters: rate effects upon substitution of sulfur for a nonbridging phosphoryl oxygen atom. Biochemistry. 1991;30:4844–4854. doi: 10.1021/bi00234a003. [DOI] [PubMed] [Google Scholar]
- 129.Zhang X, Lee I, Berdis AJ. Evaluating the contributions of desolvation and base-stacking during translesion DNA synthesis. Org Biomol Chem. 2004;2:1703–1711. doi: 10.1039/b401732c. [DOI] [PubMed] [Google Scholar]
- 130.Blanca G, Villani G, Shevelev I, Ramadan K, Spadari S, Hubscher U, Maga G. Human DNA polymerases lambda and beta show different efficiencies of translesion DNA synthesis past abasic sites and alternative mechanisms for frameshift generation. Biochemistry. 2004;43:11605–11615. doi: 10.1021/bi049050x. [DOI] [PubMed] [Google Scholar]
- 131.Ruiz JF, Dominguez O, Lain de Lera T, Garcia-Diaz M, Bernad A, Blanco L. DNA polymerase mu, a candidate hypermutase? Philos Trans R Soc Lond B Biol Sci. 2001;356:99–109. doi: 10.1098/rstb.2000.0754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mahajan KN, Nick McElhinny SA, Mitchell BS, Ramsden DA. Association of DNA polymerase mu (pol mu) with Ku and ligase IV: role for pol mu in end-joining double-strand break repair. Mol Cell Biol. 2002;22:5194–5202. doi: 10.1128/MCB.22.14.5194-5202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Reynaud CA, Frey S, Aoufouchi S, Faili A, Bertocci B, Dahan A, Flatter E, Delbos F, Storck S, Zober C, Weill JC. Transcription, beta-like DNA polymerases and hypermutation. Philos Trans R Soc Lond B Biol Sci. 2001;356:91–97. doi: 10.1098/rstb.2000.0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ramadan K, Shevelev I, Hubscher U. The DNA-polymerase-X family: controllers of DNA quality? Nature Reviews Molecular Cell Biology. 2004;5:1038–1043. doi: 10.1038/nrm1530. [DOI] [PubMed] [Google Scholar]
- 135.Rodriguez MC, Songyang Z. BRCT domains: phosphopeptide binding and signaling modules. Front Biosci. 2008;13:5905–5915. doi: 10.2741/3125. [DOI] [PubMed] [Google Scholar]
- 136.Braithwaite DK, Ito J. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 1993;21:787–802. doi: 10.1093/nar/21.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Uchiyama Y, Takeuchi R, Kodera H, Sakaguchi K. Distribution and roles of X-family DNA polymerases in eukaryotes. Biochimie. 2009;91:165–170. doi: 10.1016/j.biochi.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 138.Steitz TA. DNA polymerases: structural diversity and common mechanisms. J Biol Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 139.Sawaya MR, Prasad R, Wilson SH, Kraut J, Pelletier H. Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry. 1997;36:11205–11215. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- 140.Pelletier H, Sawaya MR, Wolfle W, Wilson SH, Kraut J. Crystal structures of human DNA polymerase beta complexed with DNA: implications for catalytic mechanism, processivity, and fidelity. Biochemistry. 1996;35:12742–12761. doi: 10.1021/bi952955d. [DOI] [PubMed] [Google Scholar]
- 141.Garcia-Diaz M, Bebenek K, Krahn JM, Kunkel TA, Pedersen LC. A closed conformation for the Pol lambda catalytic cycle. Nat Struct Mol Biol. 2005;12:97–98. doi: 10.1038/nsmb876. [DOI] [PubMed] [Google Scholar]
- 142.Pandey VN, Modak MJ. Biochemistry of terminal deoxynucleotidyltransferase. Identification and unity of ribo- and deoxyribonucleoside triphosphate binding site in terminal deoxynucleotidyltransferase. J Biol Chem. 1989;264:867–871. [PubMed] [Google Scholar]
- 143.Fowler JD, Suo Z. Biochemical, Structural, and Physiological Characterization of Terminal Deoxynucleotidyl Transferase. Chemical Reviews. 2006;106:2092–2110. doi: 10.1021/cr040445w. [DOI] [PubMed] [Google Scholar]
- 144.Peralta-Zaragoza O, Recillas-Targa F, Madrid-Marina V. Terminal deoxynucleotidyl transferase is down-regulated by AP-1-like regulatory elements in human lymphoid cells. Immunology. 2004;111:195–203. doi: 10.1111/j.0019-2805.2003.01791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.McBlane JF, van Gent DC, Ramsden DA, Romeo C, Cuomo CA, Gellert M, Oettinger MA. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 146.Elias L, Longmire J, Wood A, Ratliff R. Phosphorylation of terminal deoxynucleotidyl transferase in leukemic cells. Biochem Biophys Res Commun. 1982;106:458–465. doi: 10.1016/0006-291x(82)91132-9. [DOI] [PubMed] [Google Scholar]
- 147.Trubiani O, Bollum FJ, Di Primio R. Terminal deoxynucleotidil transferase is a nuclear PKC substrate. FEBS Lett. 1995;374:367–370. doi: 10.1016/0014-5793(95)01148-8. [DOI] [PubMed] [Google Scholar]
- 148.Chang LM, Bollum FJ. Cyclic AMP-dependent phosphorylation of terminal deoxynucleotidyl transferase. J Biol Chem. 1982;257:9588–9592. [PubMed] [Google Scholar]
- 149.Yamashita N, Shimazaki N, Ibe S, Kaneko R, Tanabe A, Toyomoto T, Fujita K, Hasegawa T, Toji S, Tamai K, Yamamoto H, Koiwai O. Terminal deoxynucleotidyltransferase directly interacts with a novel nuclear protein that is homologous to p65. Genes Cells. 2001;6:641–652. doi: 10.1046/j.1365-2443.2001.00449.x. [DOI] [PubMed] [Google Scholar]
- 150.Vivona JB, Kelman Z. The diverse spectrum of sliding clamp interacting proteins. FEBS Lett. 2003;546:167–172. doi: 10.1016/s0014-5793(03)00622-7. [DOI] [PubMed] [Google Scholar]
- 151.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 152.Sandor Z, Calicchio ML, Sargent RG, Roth DB, Wilson JH. Distinct requirements for Ku in N nucleotide addition at V(D)J- and non-V(D)J-generated double-strand breaks. Nucleic Acids Res. 2004;32:1866–1873. doi: 10.1093/nar/gkh502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hodgson DR, Sanderson JM. The synthesis of peptides and proteins containing non-natural amino acids. Chem Soc Rev. 2004;33:422–430. doi: 10.1039/b312953p. [DOI] [PubMed] [Google Scholar]
- 154.Greaves M, Paxton A, Janossy G, Pain C, Johnson S, Lister TA. Acute lymphoblastic leukaemia associated antigen. III ALterations in expression during treatment and in relapse. Leuk Res. 1980;4:1–14. doi: 10.1016/0145-2126(80)90043-0. [DOI] [PubMed] [Google Scholar]
- 155.Hoffbrand AV, Ganeshaguru K, Janossy G, Greaves MF, Catovsky D, Woodruff RK. Terminal deoxynucleotidyl-transferase levels and membrane phenotypes in diagnosis of acute leukaemia. Lancet. 1977;2:520–523. doi: 10.1016/s0140-6736(77)90662-6. [DOI] [PubMed] [Google Scholar]
- 156.Kung PC, Long JC, McCaffrey RP, Ratliff RL, Harrison TA, Baltimore D. Terminal deoxynucleotidyl transferase in the diagnosis of leukemia and malignant lymphoma. Am J Med. 1978;64:788–794. doi: 10.1016/0002-9343(78)90518-1. [DOI] [PubMed] [Google Scholar]
- 157.Venditti A, Del Poeta G, Buccisano F, Tamburini A, Aronica G, Bruno A, Cox-Froncillo MC, Maffei L, Simone MD, Papa G, Amadori S. Biological pattern of AML-M0 versus AML-M1: response. Blood. 1997;89:345–346. [PubMed] [Google Scholar]
- 158.McCaffrey R, Bell R, Lillquist A, Wright G, Baril E, Minowada J. Selective killing of leukemia cells by inhibition of TdT. Haematol Blood Transfus. 1983;28:24–27. doi: 10.1007/978-3-642-68761-7_4. [DOI] [PubMed] [Google Scholar]
- 159.Spigelman Z, Duff R, Beardsley GP, Broder S, Cooney D, Landau NR, Mitsuya H, Ullman B, McCaffrey R. 2′,3′-Dideoxyadenosine is selectively toxic for TdT-positive cells. Blood. 1988;71:1601–1608. [PubMed] [Google Scholar]
- 160.Kodama EN, McCaffrey RP, Yusa K, Mitsuya H. Antileukemic activity and mechanism of action of cordycepin against terminal deoxynucleotidyl transferase-positive (TdT+) leukemic cells. Biochem Pharmacol. 2000;59:273–281. doi: 10.1016/s0006-2952(99)00325-1. [DOI] [PubMed] [Google Scholar]
- 161.Plunkett W, Gandhi V. Purine and pyrimidine nucleoside analogs. Cancer Chemother Biol Response Modif. 2001;19:21–45. [PubMed] [Google Scholar]
- 162.Locatelli GA, Di Santo R, Crespan E, Costi R, Roux A, Hubscher U, Shevelev I, Blanca G, Villani G, Spadari S, Maga G. Diketo hexenoic acid derivatives are novel selective non-nucleoside inhibitors of mammalian terminal deoxynucleotidyl transferases, with potent cytotoxic effect against leukemic cells. Mol Pharmacol. 2005;68:538–550. doi: 10.1124/mol.105.013326. [DOI] [PubMed] [Google Scholar]
- 163.Ono K. Inhibitory effects of various 2′,3′-dideoxynucleoside 5′-triphosphates on the utilization of 2′-deoxynucleoside 5′-triphosphates by terminal deoxynucleotidyltransferase from calf thymus. Biochim Biophys Acta. 1990;1049:15–20. doi: 10.1016/0167-4781(90)90078-g. [DOI] [PubMed] [Google Scholar]
- 164.Arzumanov AA, Victorova LS, Jasko MV, Yesipov DS, Krayevsky AA. Terminal deoxynucleotidyl transferase catalyzes the reaction of DNA phosphorylation. Nucleic Acids Res. 2000;28:1276–1281. doi: 10.1093/nar/28.5.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sosunov VV, Santamaria F, Victorova LS, Gosselin G, Rayner B, Krayevsky AA. Stereochemical control of DNA biosynthesis. Nucleic Acids Res. 2000;28:1170–1175. doi: 10.1093/nar/28.5.1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Krayevsky AA, Victorova LS, Arzumanov AA, Jasko MV. Terminal deoxynucleotidyl transferase. catalysis of DNA (oligodeoxynucleotide) phosphorylation. Pharmacol Ther. 2000;85:165–173. doi: 10.1016/s0163-7258(99)00070-4. [DOI] [PubMed] [Google Scholar]
- 167.Steensma DP, Timm M, Witzig TE. Flow cytometric methods for detection and quantification of apoptosis. Methods Mol Med. 2003;85:323–332. doi: 10.1385/1-59259-380-1:323. [DOI] [PubMed] [Google Scholar]
- 168.Gorczyca W, Gong J, Darzynkiewicz Z. Detection of DNA strand breaks in individual apoptotic cells by the in situ terminal deoxynucleotidyl transferase and nick translation assays. Cancer Res. 1993;53:1945–1951. [PubMed] [Google Scholar]