Abstract
Hymenopsin A (1), hymenopsin B (2), and a new macrophorin analogue, 2′,3′-epoxy-13-hydroxy-4′-oxomacrophorin A (3), have been isolated from a fungicolous isolate of Hymenopsis sp. (MYC-1703; NRRL 37638). The structures and relative configurations of these compounds were assigned on the basis of 2D NMR and MS data, and the identity of 1 was confirmed by X-ray crystallographic analysis. The absolute configuration of 2 was proposed on the basis of CD analysis using both empirical and computational methods. Compounds 2 and 3 showed antibacterial activity against Staphylococcus aureus and Bacillus subtilis. Compound 3 was also active against Aspergillus flavus and Fusarium verticillioides.
Fungi continue to serve as excellent sources of secondary metabolites that display a wide variety of biological activities. However, many ecological groups of fungi have not been extensively explored as sources of novel chemistry. One such group consists of the fungicolous and mycoparasitic fungi—species that colonize and/or parasitize others. Fermentation extracts of fungicolous and mycoparasitic fungi often display antifungal activity, and our prior studies of such organisms have afforded a variety of new bioactive natural products.1-3 As part of this ongoing project, a fungicolous isolate of Hymenopsis sp. (MYC-1703 = NRRL 37638; incertae sedis) collected in Hawaii was found to produce three new compounds (1–3) that are biogenetically related to the macrophorins.4-7 Details of the isolation, structure elucidation, and antibacterial activity of these metabolites are presented here.
Results and Discussion
The isolate of Hymenopsis sp. was obtained from the surface of a black stroma of an unidentified pyrenomycete on a dead hardwood branch that was collected from a Eucalyptus forest planting near Honoka'a Hamakua District, Hawaii Co., HI. The fungus was cultured by solid-substrate fermentation on rice. The EtOAc extract of the resulting fermentation mixture showed antifungal activity against Aspergillus flavus and Fusarium verticillioides, and was therefore subjected to chemical investigation, leading to the isolation of 1–3.
The molecular formula of hymenopsin A (1) was found to be C22H32O6 (seven unsaturations) on the basis of ESIMS and NMR data. In examining the 1H NMR spectrum, signals for several oxygenated methine and methylene units were observed, as well as two methyl resonances and one olefinic signal. In the 13C NMR spectrum, only two sp2 carbon signals (corresponding to a trisubstituted olefin unit) were observed. The remaining six degrees of unsaturation must be accounted for by the presence of six rings. Of the remaining 20 signals, eight corresponded to oxygenated sp3 carbons. DEPT data accounted for 28 hydrogens bound to carbon atoms, indicating the presence of four OH groups. 13C NMR chemical shift data, together with the availability of only two other oxygen atoms in the molecular formula, indicated that these two oxygen atoms and the other four oxygenated carbon signals comprise two epoxide units. COSY data revealed several spin-systems corresponding to substructures C-1 through C-3, C-5 through C-7, C-9/C-11, and C-4′/C-5′. HMBC correlations observed for singlet methyl signal H3-14 to C-3, C-4, C-5, and C-13, and from H3-15 to C-1, C-5, C-9, and C-10, together with a correlation from H-7 to C-9, permitted connection of three of these spin-systems to form the A- and B-rings of 1. Further correlations of H2-11 to C-8, C-9, C-10, C-1′, C-5′, and C-6′, and from isolated methylene protons H2-12 to C-7, C-8, C-9, C-1′, and C-2′ enabled construction of the C-ring. Correlations were also observed from the isolated epoxide proton H-2′ to C-1′ and C-6′; from H-4′ to C-7′; and from H2-7′ to C-2′ and C-3′, forming the D-ring of 1.
Treatment of 1 with acetic anhydride/pyridine afforded a tetra-O-acetyl derivative, confirming the presence of four OH groups in 1. The H2-13, H-4′, and H2-7′ signals were all shifted significantly downfield in the 1H NMR spectrum of the tetraacetate relative to their positions in the spectrum of 1, indicating that C-13, C-4′, and C-7′ all bear free OH groups in 1. The fourth (tertiary) OH group was more difficult to locate with certainty, and could not be placed on the basis of δC values because the remaining five oxygenated 13C NMR signals (four of which must be epoxide carbons) had very similar chemical shifts. Ultimately, the final OH group was assigned to oxygenated quaternary carbon C-1′ on the basis of NOESY correlations of H-11β, H-12β, and H-5′ with the corresponding OH signal that were observed when the data were recorded in DMSO-d6 (see below). This required the two epoxide units to be located at the C2′-C3′ and C5′-C6′ positions, and ruled out the possibility of a C1′-C2′ epoxide.
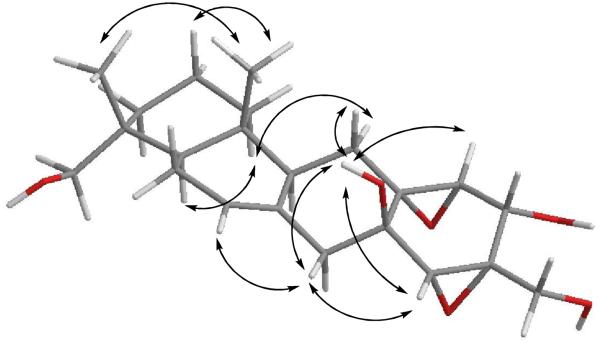
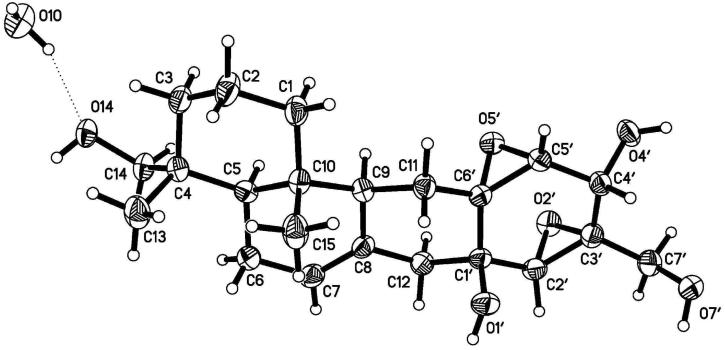
The relative configuration of 1 was assigned by analysis of NOESY data (Figure 1). Stereochemical assignments for the A- and B-rings were relatively straightforward, but establishment of the C- and D-ring configurations was substantially more challenging due to the shortage of proton signals and lack of diagnostic J-values in that region of the molecule. Key correlations of H-9 with H-5 and H-11α indicated the cofacial orientation of these hydrogens. Similarly, correlations of H3-15 with H-11β, H3-14, and H-2β, and of H-12β with H-7 and H-2′ placed these hydrogens on the opposite face. These data enabled assignment of the relative configurations at all of the stereogenic centers except for C-1′, C-5′, and C-6′. However, NMR data collected using DMSO-d6 as the solvent afforded relatively sharp signals for the OH protons, and additional relevant NOESY correlations involving these signals were observed. Correlations of OH-1′ with H-11b, H-12β, and H-5′ permitted assignment of the relative configurations at C-1′, C-5′, and C-6′. Attempts to assign the relative configuration at C-4′ by comparison of the JH4′-H5′ value with those of known macrophorins proved inconclusive.4,6 The 2.7-Hz value observed for 1 was almost identical to those of known macrophorins wherein the 4′-OH group is cis to the 5′,6′ epoxide moiety (2.5 to 3.0 Hz).4,6 Cladospirone bis-epoxide also has a similar bis-epoxide system with an OH in the cis orientation, and has a similar coupling constant for the corresponding protons (3.0 Hz).8 However, there are only a few examples of cases where both possible isomers are available for direct comparison. In one such instance, the sterols (5α,6α;8α,9α-diepoxy-(22E,24R)-ergost-22-ene-3β,7α-diol and its 7β-epimer) were reported. The isomer placing the secondary OH group cis to the epoxide oxygen shows a vicinal coupling of 2.6 Hz, while the corresponding trans epimer displays a 2.9 Hz value.9 Thus, the complete relative configuration of 1 was not assigned with certainty on the basis of NMR data. However, X-ray diffraction analysis (Figure 2) allowed unambiguous assignment of the relative configuration of 1, placing the C-4′ OH cis to the neighboring epoxide unit. These data also confirmed the other stereochemical assignments that had been made on the basis of the NOESY results.
Figure 1.
Selected NOESY correlations for hymenopsin A (1)
Figure 2.
X-ray model of hymenopsin A (1)
Hymenopsin B (2) was assigned the molecular formula C22H30O6 (8 unsaturations) on the basis of MS and NMR data. The NMR data for 2 differed from those of 1 in the absence of the C-4′/H-4′ methine signals, and the appearance of a new ketone carbon signal at δ 203.6 in the 13C NMR spectrum. All other signals were very similar to those of 1, suggesting that 2 differs from 1 only by oxidation of the C-4′ alcohol group to a ketone moiety. Analysis of HMQC, HMBC, and NOESY data enabled confirmation of the structure and relative configuration of hymenopsin B (2) as shown. In the case of 2, however, the absolute configuration was proposed by analysis of CD data. Observation of a negative Cotton effect at 320 nm, and application of the octant rule,10 enabled assignment of the absolute configuration shown (4R, 5R, 9S, 10R, 1′R, 2′R, 3′R, 5′S, 6′R). This conclusion is based on the fact that the majority of substituent atoms closest to the ketone group in the energy-minimized conformation (ChemBio3D 11.0.1) reside in a negative octant. The unusual bond angles and orientations associated with the nearby epoxide groups render application of the method somewhat less straightforward than for a typical cyclohexanone-containing system, however, the absolute configuration proposed is consistent with that originally reported for the biogenetically related compound macrophorin A.4 Related compounds 1 and 3 are presumed to possess absolute configurations analogous to that of 2.
Because the validity of standard empirical interpretation of the CD data might be somewhat questionable in this instance, a more rigorous analysis employing comparison with TDDFT-calculated CD spectra was undertaken. Conformational analysis of structure 2 was performed using a PM3 semiempirical method. The 10 lowest energy conformers obtained were optimized using a 6-31G(d) basis set11 combined with a B3LYP exchange correlation functional,12,13 followed by frequency calculations using the same level of theory to confirm that the conformers correspond to minima on the potential energy curve. CD spectra were calculated for the 10 lowest energy conformers and a weighted Boltzmann-averaged spectrum was also generated. Although peak intensities varied somewhat from one spectrum to another, the calculated spectra for all 10 conformers, as well as the averaged spectrum, displayed a negative Cotton effect in the key region from 320-330 nm, in agreement with the experimental data and the application of the octant rule described above. The same general feature was observed regardless whether the calculations were performed for the gas phase or for simulated methanol solution. Elements of the spectra at lower wavelengths were somewhat more variable, but an accompanying negative Cotton effect centered at 240 nm in the experimental spectrum was also observed in all of the calculated spectra, albeit at slightly higher and varied wavelength. These results provided independent support for the absolute configurational assignment shown.
2′,3′-Epoxy-13-hydroxy-4′-oxomacrophorin A (3) was determined to have the same molecular formula as 2 (C22H30O6) on the basis of HRESIMS data. Analysis of the NMR data revealed that 3 was closely related to 1 and 2, but lacked signals for the trisubstituted olefin and the isolated C-12 methylene unit. Instead, the 1H NMR spectrum contained signals for a terminal olefin characteristic of that observed for the macrophorins,4-7 a series of known compounds with a close biogenetic relationship to 1–2, but lacking the C-ring. In addition, the 13C NMR spectrum for 3 contained a second ketone carbon signal in place of one of the quaternary oxygenated sp3 carbon signals present in the spectrum of 2. Comparison of these data with literature values for macrophorins4-7 indicated that 3 is a new macrophorin analogue that differs from 4′-oxomacrophorin A7 by addition of an epoxide unit involving C-2′ and C-3′ and the presence of an OH group at C-13. The relative configuration was assigned by comparison of chemical shifts and coupling constants with those of hymenopsins A and B (1–2) and with those of known macrophorins. Because of the clear structural relationship between compounds 2 and 3, it was felt that 2 might be formed upon exposure of 3 to acidic conditions. Treatment of 3 with 0.1 N HCl resulted in decomposition to a complex mixture of products that did not include 2, but did lack signals for the terminal olefin and included a minor olefinic signal similar to that observed in the spectrum of 2.
Compounds 1–3 are members of a general biosynthetic class sometimes referred to as meroterpenoids,14 which are formed from a combination of terpenoid and polyketide (or other) units. Compounds 1–3 could be envisioned as arising from a combination of a drimane sesquiterpenoid moiety and a seven-carbon unit that closely resembles the well known metabolite epoxydon, which has a tetraketide origin.6,15 A number of compounds related to 1-3 have been reported previously,4-7, 16,17 including macrophorins, as noted earlier. However, there is only one prior report describing members of this biosynthetic class possessing a carbocyclic C-ring as found in 1 and 2 (from an endophytic isolate of the fungus Phyllosticta spinarum).16 The same report also describes the only other representative of the macrophorin group that displays oxidation at one of the C-4 methyl groups.
Compounds 2 and 3 showed antibacterial activity in standard disk assays against Staphylococcus aureus (ATCC 25923), affording zones of inhibition of 21 and 32 mm, respectively, at 200 μg/disk. A gentamicin sulfate standard showed ca. 25-mm zones of inhibition in both assays at 50 μg/disk. Similar results were observed in assays against Bacillus subtilis (ATCC 6051). Compound 1 showed no activity in either of these assays, suggesting that the C-4′ ketone plays a key role in the activities of 2 and 3. None of the compounds showed activity against Candida albicans (ATCC 14053) when tested at the same level. In other antifungal assays at 200 μg/disk, compounds 1 and 2 showed no activity against A. flavus (NRRL 6541) or F. verticillioides (NRRL 25457), but compound 3 showed 20-mm zones of inhibition in each case. Comparable results in these assays were obtained with a standard of nystatin at 25 μg/disk.
The genus Hymenopsis appears to be previously unexplored from a chemical standpoint. A few detailed taxonomic studies18-20 have led to transfer of some other fungal species into Hymenopsis, including a number that were originally characterized as Myrothecium spp., but no secondary metabolites have been reported from fungi having any of these earlier species names. Thus, compounds 1-3 are the first secondary metabolites to be reported from any member of the genus Hymenopsis.
Experimental Section
General Experimental Procedures
Melting points were obtained using a Fisher-Johns micro-melting point apparatus and are uncorrected. Optical rotations were measured with a Rudolph automatic polarimeter, model AP III. UV measurements were performed with a Varian Cary 100 Bio UV-vis spectrophotometer, and CD data were collected using an Olis Cary-17 spectrophotometer (0.1-cm cell). 1H and 13C NMR spectra were measured on Bruker AVANCE-300 and DRX-400 spectrometers. Chemical shift values were referenced to residual solvent signals as follows (δH/δC): acetone-d6 (2.05/29.8), CDCl3 (7.24/77.0), methanol-d4 (3.30/49.0), DMSO-d6 (2.50/39.5). HMQC and HMBC data were recorded on a Bruker AVANCE-600 instrument. HRESIMS data were recorded on a Micromass Autospec instrument. HPLC was carried out using a Beckman System Gold HPLC instrument equipped with a model 166 or a model 168 variable-wavelength UV detector.
Isolation, Cultivation, and Fermentation of Fungal Material
The isolate (MYC-1703) was obtained from the surface of a black stroma of an unidentified pyrenomycete on a dead hardwood branch that was collected from a Eucalyptus forest planting near Honoka'a Hamakua District, Hawaii Co., HI. This isolate was identified as belonging to the genus Hymenopsis Sacc. and a subculture was deposited in the ARS Culture Collection, NCAUR, USDA, Peoria, IL under the accession number NRRL 37638.
We were unable to assign this isolate to any recognized species of Hymenopsis Sacc.18,19 Nag Raj18 has identified the need for a reassessment of the large number of taxa in Hymenopsis and genera of similar appearance, such as Myrothecium,20 using modern taxonomic criteria. Morphologically, our isolate bears resemblance to H. trochlioides, the type species of Hymenopsis. However, no sequence data are presently available for any species of Hymenopsis. While Hymenopsis is not known as a colonist of fungal sporocarps, some species have been recorded from decaying stems and leaves; substrates known to be colonized by fungicolous fungi.
Hymenopsis sp. was grown on 100 g of autoclaved rice for 30 days at 25 °C. The resulting culture material was then extracted with EtOAc, affording 828 mg of crude extract upon evaporation of the solvent.
Extraction and Isolation
The extract was partitioned between MeCN and hexanes (3 mL of each). The MeCN fraction (675 mg) was chromatographed on a silica gel column using progressive gradient elution (hexanes; CH2Cl2; CH2Cl2/MeOH 19:1, 9:1, 4:1, 7:3, 1:1, and MeOH) to give 12 fractions. Fraction 3 (60 mg), eluted with CH2Cl2/MeOH 19:1, was further separated on another silica gel column using a hexanes/EtOAc solvent system (hexanes; hexanes/EtOAc 19:1, 9:1, 1:1, and EtOAc) to yield 11 fractions. Subfraction 4 (eluted with hexanes/EtOAc 9:1) consisting of 2′,3′-epoxy-13-hydroxy-4′-oxomacrophorin A (3; 6 mg). Fraction 8 (49 mg), eluted with CH2Cl2/MeOH 9:1, was separated by reversed-phase HPLC (25% MeCN/H2O isocratic for 9 min, 25–40% over 1 min, and 40–100% over 20 min) on a Rainin Dynamax-60A C18 preparative column (21.4 × 250 mm) at a flow rate of 10 mL/min with UV detection at 215 nm to afford hymenopsin A (1; 27 mg, tR 18.8 min).
A second fermentation was carried out (200 g rice) and the resulting extract (1.69 g) was again partitioned between MeCN and hexanes. The MeCN fraction (1.27 g) was separated on a silica gel column using a hexanes/EtOAc solvent system as above to give 21 fractions. Fraction 10 (43 mg), eluted with hexanes/EtOAc 4:1, was separated by reversed-phase HPLC (20% MeCN/H2O isocratic over 10 min, 20–30% over 1 min, 30–90% over 40 min, and 90–100% over 1 min) on an Alltech HS BDS 8-μm C18 column (4.6 × 250 mm) at a flow rate of 2 mL/min with UV detection at 215 nm to give six subfractions. Subfraction 3 (17 mg) was then purified by reversed-phase HPLC (55% MeOH/H2O isocratic for 45 min and 55–100% over 5 min) on the same column with UV detection at 215 nm to afford hymenopsin B (2; 9 mg, tR 27.3 min).
Hymenopsin A (1)
white crystals (from CHCl3/MeOH/hexanes 2:1:1); mp153–155 °C; [α]25D +31 (c 0.7, MeOH); 1H and 13C NMR data, see Tables 1 and 2; key HMBC data (CDCl3): H2-1 → C-2, 3, 5, 10, 15; H2-2 → C-1, 3, 4, 10; H2-3 → C-1, 2, 4, 5, 13, 14; H-5 → C-4, 6, 7, 9, 10, 13, 14, 15; H2-6 → C-4, 5, 7, 8; H-7 → C-5, 6, 9, 12; H-9 → C-7, 8, 10, 15; H2-11 → C-8, 9, 10, 1′, 5′, 6′; H2-12 → C-7, 8, 9, 1′, 2′; H2-13 → C-3, 4, 5, 14; H3-14 → C-3, 4, 5, 13; H3-15 → C-1, 5, 9, 10; H-2′ → C-11, 12, 1′, 6′; H-4′ → C-3′, 7′; H-5′ → C-11, 3′, 4′; H2-7′ → C-2′, 3′; NOESY data (DMSO-d6): H-2β ↔ H3-14; H-2β ↔ H3-15; H-5 ↔ H2-13; H-5 ↔ H-9; H-5 ↔ H2-6;H-7 ↔ H-12β; H-9 ↔ H-11α; H-11β ↔ H3-15; H-11β ↔ OH-1′; H-12β ↔ OH-1′; H-12β ↔ H-2′; OH-1′↔ H-5′; H-2′↔ OH-7′; OH-4′↔ H-5′;
Table 1.
1H NMR Data [δH (mult, JH)] for Compounds 1–3
| position | 1a | 2b | 3c |
|---|---|---|---|
| 1α | 1.49 (ddd, 13, 13, 2.5) | 1.39 (ddd 13, 13, 4.9) | 1.05 (m) |
| 1β | 1.26 (br d, 13) | 1.33 (m) | 1.63 (m) |
| 2 | 1.44 (m), 1.53 (m) | 1.50 (br m) | 1.59 (br m) |
| 3α | 0.95 (ddd, 13, 13, 5.7) | 1.02 (ddd, 13, 13, 5.7) | 1.28 (m) |
| 3β | 1.69 (br d, 13) | 1.66 (br d, 13) | 1.42 (dd, 12, 2.6) |
| 5 | 1.60 (dd, 11, 5.5) | 1.50 (m) | 1.05 (dd, 12, 5.5) |
| 6 | 1.95 (br m) | 1.97 (br m) | 1.29 (m), 1.70 (m) |
| 7 (or 7α) | 5.44 (br s) | 5.72 (br s) | 1.98 (ddd, 13, 13, 4.9) |
| 7β | -- | -- | 2.35 (ddd, 13, 4.0, 2.3) |
| 9 | 2.08 (br d, 13) | 2.18 (br d, 13) | 1.69 (m) |
| 11α | 0.96 (dd, 13, 5.7) | 1.10 (dd, 13, 5.7) | 2.14 (dd, 15, 11) |
| 11β | 2.35 (t, 13) | 2.35 (t, 13) | 2.25 (dd, 15, 2.3) |
| 12α | 2.54 (br d, 14) | 2.94 (br d, 15) | 4.81 (s) |
| 12β | 2.31 (d, 14) | 2.48 (d, 15) | 4.44 (s) |
| 13a | 3.02 (dd, 11, 5.6) | 3.14 (dd, 11, 6.0) | 3.08 (d, 11) |
| 13b | 3.33 (dd, 11, 5.6) | 3.36 (dd, 11, 6.0) | 3.39 (d, 11) |
| 13-OH | 3.60 (t, 5.6) | * | * |
| 14 | 0.85 (s) | 0.88 (s) | 0.71 (s) |
| 15 | 0.91 (s) | 0.90 (s) | 0.72 (s) |
| 1′-OH | 3.78 (br s) | 2.06 (s) | |
| 2′ | 3.01 (s) | 3.45 (s) | 3.63 (s) |
| 4′ | 4.38 (br d, 7.1) | -- | -- |
| 4′-OH | 3.88 (d, 7.1) | -- | -- |
| 5′ | 3.15 (d, 2.7) | 3.33 (s) | 3.82 (s) |
| 7′α | 3.45 (d, 12) | 3.88 (dd, 13, 5.7) | 3.91 (d, 14) |
| 7′β | 3.84 (d, 12) | 3.96 (dd, 13, 8.0) | 4.03 (d, 14) |
| 7′-OH | 4.31 (br s) | 1.81 (dd, 8.0, 5.7) | * |
Acetone-d6, 600 MHz.
CDCl3, 600 MHz.
CDCl3, 300 MHz.
Exchangeable signals—variable or not clearly observed.
Table 2.
13C NMR Data (δC) for Compounds 1–3
| position | 1a | 2b | 3b |
|---|---|---|---|
| 1 | 36.6 | 38.1 | 38.5 |
| 2 | 19.1 | 20.6 | 18.5 |
| 3 | 40.5 | 41.9 | 37.4 |
| 4 | 36.0 | 37.9 | 35.3 |
| 5 | 44.3 | 46.1 | 48.4 |
| 6 | 24.4 | 26.1 | 24.0 |
| 7 | 124.8 | 130.1 | 38.1 |
| 8 | 133.7 | 133.3 | 148.1 |
| 9 | 50.4 | 51.3 | 51.0 |
| 10 | 38.4 | 40.0 | 39.3 |
| 11 | 29.0 | 31.1 | 20.9 |
| 12 | 42.5 | 45.8 | 107.0 |
| 13 | 72.0 | 74.7 | 71.9 |
| 14 | 18.5 | 20.8 | 17.6 |
| 15 | 16.0 | 18.3 | 15.0 |
| 1′ | 67.6 | 70.1 | 196.2 |
| 2′ | 64.0 | 69.8 | 67.0 |
| 3′ | 62.9 | 64.0 | 65.2 |
| 4′ | 65.0 | 203.6 | 197.9 |
| 5′ | 63.5 | 65.0 | 58.8 |
| 6′ | 66.1 | 74.1 | 62.6 |
| 7′ | 62.2 | 62.5 | 60.9 |
Methanol-d4, 125 MHz.
CDCl3, 100 MHz
HRESIMS obsd m/z 415.2095 (M + Na)+, calcd for C22H32O6Na, 415.2096.
Hymenopsin B (2)
white glass; [α]25D +14 (c 1.1, MeOH); UV (MeOH) λmax (log ε) 206 (4.3); 230 (3.8); 315 (2.8); CD (MeOH) Δε 240 (−3.4), 320 (−11); 1H and 13C NMR data, see Tables 1 and 2; key HMBC data (CDCl3): H2-1 → C-2, 3, 5, 10; H2-2 → C-1, 3, 4, 10; H2-3 → C-1, 2, 4, 5, 14; H-5 → C-4, 6, 7, 9, 1; H2-6 → C-4, 5, 7, 8; H-7 → C-12; H-9 → C-7, 8, 11, 1′; H2-11 → C-8, 9, 1′, 5′, 6′; H2-12 → C-7, 8, 9, 1′, 2′, 6′; H2-13 → C-3, 4, 5, 14; H3-14 → C-3, 4, 5, 13; H3-15 → C-1, 5, 9, 10; H-2′ → C-1′, 3′, 6′, 7′; H-5′ → C-3′, 4′, 6′; H2-7′ → C-2′, 3′, 4′; selected NOESY data (CDCl3): H-2β ↔ H3-14; H-2β ↔ H3-15; H-5 ↔ H2-13; H-5 ↔ H-9; H-9 ↔ H-12β; H-9 ↔ H-11α; H-11β ↔ H3-15; H-12β ↔ H-2′; HRESIMS obsd m/z 413.1940 (M + Na)+, calcd for C22H30O6Na, 413.1940.
2′,3′-Epoxy-13-hydroxy-4′-oxomacrophorin A (3)
colorless oil; [α]25D +12 (c 0.4, MeOH); 1H and 13C NMR data, see Tables 1 and 2; HRESIMS obsd m/z 391.2133 (M + H)+, calcd for C22H31O6, 391.2121.
Preparation of Tetra-O-acetyl hymenopsin A
A sample of 1 (1.0 mg) was reacted with acetic anhydride (0.2 mL) and pyridine (0.2 mL). The reaction mixture was stirred for 24 h, and then evaporated, to afford 1.1 mg of the tetra-O-acetyl derivative. ESIMS m/z 561. (M + H)+; 1H NMR (CD3OD) δ 0.94 (s, H3-14), 0.98 (s, H3-15), 1.00 (m, H-11α), 1.14 (ddd, J = 13, 13, 6.1 Hz, H-3α), 1.39 (m, H-1α), 1.41 (m, H-1β), 1.41 (m, H-2α), 1.52 (m, H-2β and H-5), 1.69 (br d, J = 13 Hz, H-3β), 1.86 (dd, J = 13, 13 Hz, H-11β), 1.99 (m, H2-6 and H-9), 2.03 (s, CH3CO), 2.04 (s, CH3CO), 2.19 (s, CH3CO), 2.20 (s, CH3CO), 2.57 (d, J = 15 Hz, H-12b), 2.69 (br d, J = 15 Hz, H-12α), 3.23 (s, H-2′), 3.42 (d, J = 2.7 Hz, H-5′), 3.67 (d, J = 11 Hz, H-13a), 3.85 (d, J = 11 Hz, H-13b), 3.98 (d, J = 12 Hz, H-7′a), 4.14 (d, J = 12 Hz, H-7′b), 5.57 (br s, H-7), 5.81 (d, J = 2.7 Hz, H-4′).
X-ray Crystallographic Analysis of Hymenopsin A (1):21
A colorless plate (0.28 × 0.17 × 0.02 mm) was isolated from the sample and mounted with grease on the tip of a glass capillary epoxied to a brass pin and placed on the diffractometer with the long crystal dimension (unit cell b-axis) approximately parallel to the diffractometer phi axis. Data were collected with a Nonius Kappa CCD diffractometer (Mo K-alpha radiation, graphite monochromator) at 200(2) K (cold N2 gas stream) using standard CCD techniques yielding 15399 data. Lorentz and polarization corrections were applied. A correction for absorption using the multi-scan technique was also applied (Tmax = 0.999, Tmin = 0.975). Equivalent data were averaged yielding 1936 unique data (R-int = 0.044, 1689 F > 4*Sig(F), Friedel pairs averaged). Based on a preliminary examination of the crystal, the space group P21 was assigned (no exceptions to the systematic absence: 0k0, k=odd was noted). The computer programs from the HKL package were used for data reduction. The preliminary model of the structure was obtained using XS, a direct methods program. Least-squares refining of the model vs. the data was performed with the XL computer program. Illustrations were made with the XP program and tables were made with the XCIF program. All are in the SHELXTL v6.1 package. Thermal ellipsoids shown in the illustration are at the 50% level. All non-hydrogen atoms were refined with anisotropic thermal parameters. All H atoms were included with the riding model using the XL program default values. No further restraints or constraints were imposed on the refinement model. A water molecule of crystallization is included in the structure. The water O-H bonds were restrained to be 0.84 Å and the H-H distance to be 1.35 Å. The water H atoms were assigned isotropic thermal parameters, U(iso)(H) = 1.5*U(iso,eq)(O). The final refinement gave R1 = 0.0372, wR2 = 0.0866.
Calculation Methods
Conformers of hymenopsin B (2) were initially optimized using the MM2 molecular force field built into Chem 3D Pro 11.0 (CambridgeSoft, Cambridge, MA). Conformational analysis was performed using a PM3 semiempirical method as implemented in Spartan 06 version 129int9e (Wavefunction, Inc., Irvine, CA). The ten lowest energy conformers obtained were optimized using a 6-31G(d) basis set11 combined with B3LYP exchange correlation functional,12,13 followed by frequency calculations using the same level of theory. The absence of imaginary frequencies confirmed that all conformers were minima on the potential energy surface. No symmetry constraints were used during the optimization.
Time-dependent density functional calculations (TDDFT) calculations were carried out using Gaussian 03 Rev. D.01 (Gaussian, Inc., Wallingford, CT) employing the B3LYP functional and the 6-31G(d) basis set. CD spectra were calculated for the 10 lowest energy conformers at geometries obtained from the B3LYP/6-31G(d) calculations. The lowest possible symmetry was used, and a total of 20 excited states were calculated for each conformer. Only singlet excited states were considered. Both gas phase and solution phase calculations were performed, with the latter using methanol with a PCM solvation model.22 Single-point calculations were done at B3LYP/6-31G(d) geometries using the RI-SCS-MP2 method23,24 with TZVP25 basis set in both gas phase and methanol solvent using the COSMO26 solvation model, as implemented in ORCA version 2.7.0.27 The resulting RI-SCS-MP2/TZVP//B3LYP/6-31G(d)-calculated single point energies were employed in conformational averaging via Boltzmann weighting using SpecDis version 1.44,28 which was also used to visualize the CD spectra.
Supplementary Material
Acknowledgment
We thank Dr. J. M. Verkley of the Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands, for examining the culture of Hymenopsis. Research grants from the National Science Foundation (CHE-0315591, CHE-0718315, and AGS-0927944) and the National Institutes of Health (GM 60600) are gratefully acknowledged. The authors also thank Dr. Torsten Bruhn, Universität Würzburg, for helpful discussions regarding CD calculations.
Footnotes
Dedicated to the late Dr. John W. Daly of NIDDK, NIH, Bethesda, Maryland and to the late Dr. Richard E. Moore of the University of Hawaii at Manoa in honor of their pioneering work on bioactive natural products.
Supporting Information Available: 1H and 13C NMR spectra for 1–3, an illustration of the calculated minimum energy conformation of 2, and calculated and experimental CD spectra for 2. This material is available free of charge on the Internet at http://pubs.acs.org.
References and Notes
- 1.Shim SH, Swenson DC, Gloer JB, Dowd PF, Wicklow DT. Org. Lett. 2006;8:1225–1228. doi: 10.1021/ol060107c. [DOI] [PubMed] [Google Scholar]
- 2.Li C, Gloer JB, Wicklow DT, Dowd PF. J. Nat. Prod. 2005;68:319–322. doi: 10.1021/np0496486. [DOI] [PubMed] [Google Scholar]
- 3.Angawi RF, Swenson DC, Gloer JB, Wicklow DT. J. Nat. Prod. 2005;68:212–216. doi: 10.1021/np049625r. [DOI] [PubMed] [Google Scholar]
- 4.Sassa T, Yoshikoshi H. Agric. Biol. Chem. 1983;47:187–189. [Google Scholar]
- 5.Sassa T, Nukina M. Agric. Biol. Chem. 1984;48:1923–1925. [Google Scholar]
- 6.Sassa T, Ishizaki A, Nukina M, Ikeda M, Sugiyama T. Biosci., Biotechnol., Biochem. 1998;62:2260–2262. doi: 10.1271/bbb.62.2260. [DOI] [PubMed] [Google Scholar]
- 7.Fujimoto H, Nakamura E, Kim Y, Okuyama E, Ishibashi M, Sassa T. J. Nat. Prod. 2001;64:1234–1237. doi: 10.1021/np010152n. [DOI] [PubMed] [Google Scholar]
- 8.Thiergardt R, Hug P, Rihs G, Peter HH. Tetrahedron Lett. 1994;35:1043–1046. [Google Scholar]
- 9.Yaoita Y, Endo M, Tani Y, Machida K, Amemiya K. Chem. Pharm. Bull. 1999;47:847–851. [Google Scholar]
- 10.Kirk DN. Tetrahedron. 1986;42:777–818. [Google Scholar]
- 11.Hariharan PC, Pople JA. Theor. Chim. Acta. 1973;28:213–222. [Google Scholar]
- 12.Becke AD. J. Chem. Phys. 1993;98:5648–5652. [Google Scholar]
- 13.Lee C, Yang W, Parr RG. Phys. Rev. B: Condens. Matter. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 14.Jones RCF. Nat. Prod. Rep. 1985;2:401–26. doi: 10.1039/np9850200401. [DOI] [PubMed] [Google Scholar]
- 15.Nabeta K, Ichihara A, Sakamura S. J. Chem. Soc., Chem. Commun. 1973:814–815. [Google Scholar]
- 16.Wijeratne EMK, Paranagama PA, Marron MT, Gunatilaka MK, Arnold AE, Gunatilaka AAL. J. Nat. Prod. 2008;71:218–222. doi: 10.1021/np070600c. [DOI] [PubMed] [Google Scholar]
- 17.Yamazaki H, Omura S, Tomoda H. J. Antibiot. 2009;62:207–211. doi: 10.1038/ja.2009.19. [DOI] [PubMed] [Google Scholar]
- 18.Nag Raj TR. Coelomycetous Anamorphs with Appendage Bearing Conidia. Mycologue Publ.; Waterloo, Ontario, Canada: 1993. p. 1101. [Google Scholar]
- 19.Sutton BC. The Coelomycetes. Commonwealth Mycological Institute; Kew, Surrey, England: 1980. p. 696. [Google Scholar]
- 20.Tulloch M. Mycological Papers. 1972;130:1–42. [Google Scholar]
- 21.Crystallographic data for compound 1 have been deposited with the Cambridge Crystallographic Data Centre (deposition number CCDC 636283). Copies of the data can be obtained, free of charge, on application to the director, CCDC 12 Union Road, Cambridge CB2 1EZ, UK (fax: +44 1223 336033 or e-mail: deposit@ccdc.cam.ac.uk)
- 22.Dunning TH., Jr. J. Chem. Phys. 1989;90:1007–1023. [Google Scholar]
- 23.Weigend F, Haser M. Theor. Chem. Acc. 1997;97:331–340. [Google Scholar]
- 24.Grimme S. J. Chem. Phys. 2003;118:9095–9102. [Google Scholar]
- 25.Schaefer A, Horn H, Ahlrichs R. J. Chem. Phys. 1992;97:2571–2577. [Google Scholar]
- 26.Sinnecker S, Rajendran A, Klamt A, Diedenhofen M, Neese F. J. Phys. Chem. A. 2006;110:2235–2245. doi: 10.1021/jp056016z. [DOI] [PubMed] [Google Scholar]
- 27.Neese F. ORCA, Version 2.7.0. Universität Bonn; Bonn, Germany: 2009. [Google Scholar]
- 28.Bruhn T, Hemberger Y, Schaumlöffel A, Maksimenka K, Bringmann G. SpecDis, version 1.44, 2009. Universität Würzburg; Würzburg, Germany: 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.