Abstract
With the discovery that organisms possess multiple DNA polymerases (Pols) displaying different fidelities, processivities, and activities came the realization that mechanisms must exist to manage the actions of these diverse enzymes to prevent gratuitous mutations. Although many of the Pols encoded by most organisms are largely accurate, and participate in DNA replication and DNA repair, a sizeable fraction display a reduced fidelity, and act to catalyze potentially error-prone translesion DNA synthesis (TLS) past lesions that persist in the DNA. Striking the proper balance between use of these different enzymes during DNA replication, DNA repair, and TLS is essential for ensuring accurate duplication of the cell’s genome. This review highlights mechanisms that organisms utilize to manage the actions of their different Pols. A particular emphasis is placed on discussion of current models for how different Pols switch places with each other at the replication fork during high fidelity replication and potentially error-pone TLS.
Keywords: Sliding clamp, DNA polymerase, DNA replication, translesion DNA synthesis, mutagenesis, polymerase switch, toolbelt
INTRODUCTION
Our understanding of the mechanisms by which organisms copy and maintain the fidelity of their genomes has undergone striking changes in the last decade. In contrast to outdated models that envisioned high fidelity leading strand DNA polymerases (Pols) stably associated with the replication fork, and able to persist in a highly processive mode throughout replication, we now realize that the structure and composition of the replisome (the multiprotein complex that catalyzes DNA replication) can be extremely dynamic due in large part to Pol switching. The term Pol switching refers to the process by which one Pol replaces a second Pol at the 3’-OH end of a primed DNA template (Fig. 1). Pol switching is influenced by several factors, including the relative expression levels of the different Pols [1], changes in the sub-cellular localization of the different Pols [2], interactions of Pols with their cognate sliding clamp proteins [2, 3], as well as with other Pols [2, 3], posttranslational modification of Pols and clamp proteins [4], and the relative affinities of the different Pols for the DNA substrate in need of replication [5, 6]. Additional as yet unappreciated factors likely also contribute. In this review, I discuss the factors involved in Pol switching, and have attempted to provide a framework for understanding the biological contexts in which Pol switching can influence genome integrity. A particular emphasis is placed on discussing current models for Pol switching in both prokaryotes and eukaryotes, with a special focus on E. coli. Finally, where appropriate, I refer the reader to other articles in this thematic issue that cover certain topics related to Pol switching in more detail than I do in this review.
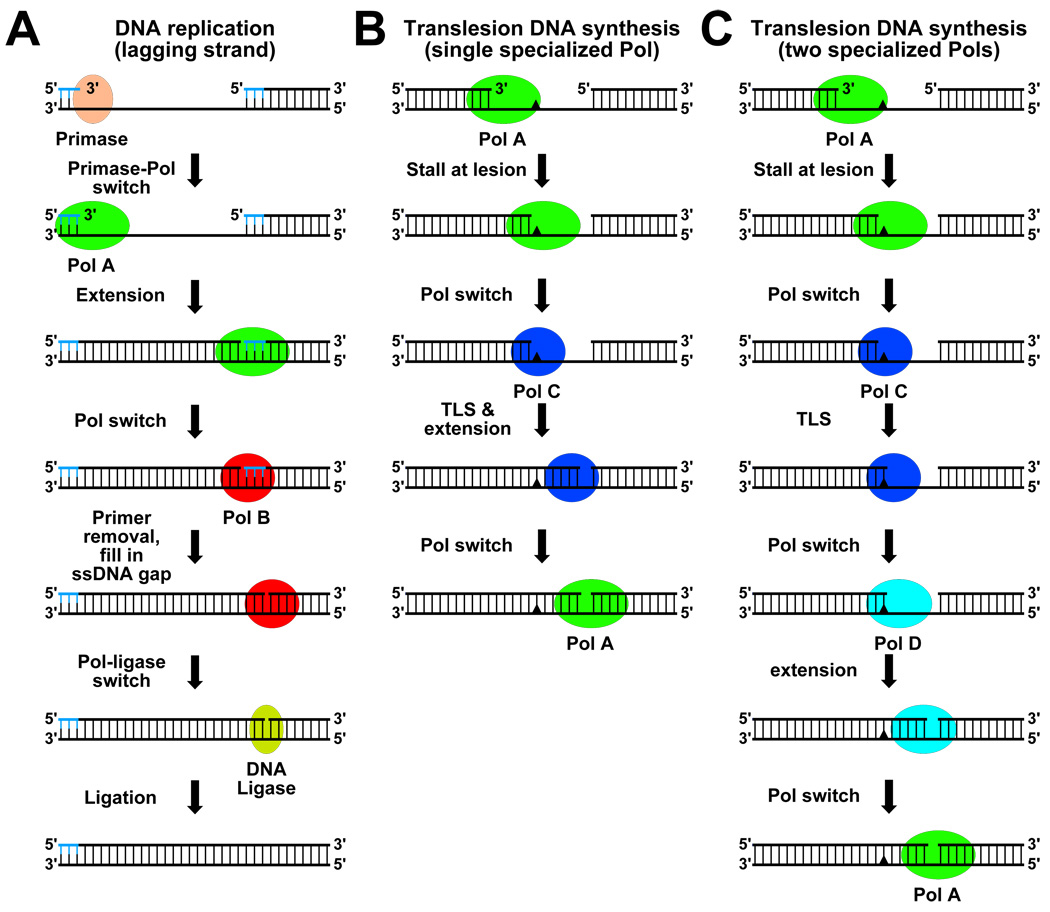
FIGURE 1. Models describing Pol switching during high fidelity DNA replication and potentially error-prone TLS.
(A) Model for Pol switching on lagging strand during maturation of Okazaki fragments. Primase (DnaG in E. coli, Pol α in eukaryotes) synthesizes a short RNA primer (blue), and then undergoes a switch with Pol A (Pol III in E. coli, Pol δ in eukaryotes) for elongation. When Pol A encounters the 5’-end of the previously synthesized Okazaki fragment, it undergoes a switch with Pol B (Pol I in E. coli, FEN1 and Pol δ in eukaryotes) which excises the RNA primer, and fills in the resulting ssDNA gap before switching with DNA ligase. (B & C) Models for Pol switching during TLS. The replicative Pol (Pol A), stalled at a lesion, undergoes a switch with a specialized Pol (Pol C) capable of bypassing the lesion. If Pol C is capable, it will extend the 3’-end of the nascent strand before handing it back to Pol A (B). If Pol C is unable to extend the 3’-end, it must undergo a switch with another specialized Pol (Pol D) for extension before the 3’-end is returned to Pol A (C).
ORGANISMS POSSESS MULTPLE POLS WITH DISTINCT FIDELITIES, PROCESSIVITIES, AND CATALYTIC ABILITIES
Pols are organized into six distinct families based on phylogenetic relationships. Although some organisms, such as Helicobacter pylori, possess only two Pols, most contain far more. For example, the gram-negative bacterium Escherichia coli is currently known to possess five different Pols belonging to four distinct families, the yeast Saccharomyces cerevisiae has at least eight, and Homo sapiens possess at least fourteen (Table 1). Although many of these Pols are accurate, due in part to an intrinsic or associated 3’-to-5’ exonuclease proofreading activity (see Chapter 5), as many as half of the Pols in any given organism possess a more open active site, endowing them with the ability to replicate imperfect DNA templates, and are therefore referred to as ‘specialized’ Pols [7]. Although these specialized Pols participate in a growing number of biological transactions ([8–11]; see Table 1), they are best known for their ability to replicate over lesions that persist in the DNA and act as potent blocks to the normal DNA replication machinery via a process termed translesion DNA synthesis (TLS). As a result of this ability, these Pols typically display a reduced fidelity when replicating undamaged DNA. Likewise, many of these specialized Pols also display modest processivity. For example, the eukaryotic Y-family Pol ι, which lacks an associated proofreading activity, is essentially distributive and catalyzes ~seven misinsertions for every ten bases synthesized in vitro, on average [12, 13]. In contrast, replicative Pols, such as the E. coli C-family Pol III, and the eukaryotic B-family Pol ε and Pol δ, are highly processive, and catalyze less than one error for every 10,000 bases incorporated [14–16]. Structural features of replicative and specialized Pols responsible for their distinct behavior are discussed in Chapters 4 and 7 of this thematic issue.
TABLE 1.
DNA Polymerases identified in E. coli S. cerevisiae, and H. sapiens *.
| Name | Pol family | Proposed biological function(s) |
|---|---|---|
| E. coli | ||
| Pol I | A | DNA replication, Okazaki fragment maturation, DNA repair |
| Pol II | B | TLS |
| Pol III | C | DNA replication, DNA repair |
| Pol IV | Y | TLS, putative DNA damage checkpoint control |
| Pol V | Y | TLS |
| S. cerevisiae | ||
| Pol γ | A | Mitochondrial DNA replication |
| Pol α | B | Priming of nuclear DNA replication |
| Pol δ | B | Nuclear DNA replication |
| Pol ε | B | Nuclear DNA replication |
| Pol ζ | B | TLS |
| Pol IV (λ) | X | Non-homologous end joining, base excision repair |
| Pol η | Y | TLS |
| REV1 | Y | TLS |
| H. sapiens | ||
| Pol γ | A | Mitochondrial DNA replication |
| Pol θ | A | TLS, base excision repair |
| Pol ν | A | DNA repair, TLS |
| Pol α | B | Priming of nuclear DNA replication |
| Pol δ | B | Nuclear DNA replication |
| Pol ε | B | Nuclear DNA replication |
| Pol ζ | B | TLS, somatic hypermutation |
| Pol β | X | Base excision repair |
| Pol λ | X | Non-homologous end joining |
| Pol μ | X | Non-homologous end joining |
| Pol η | Y | TLS, somatic hypermutation, recombination |
| Pol ι | Y | TLS, somatic hypermutation, base excision repair |
| Pol κ | Y | TLS, nucleotide excision repair |
| REV1 | Y | TLS, somatic hypermutation |
Only those Pols possessing catalytic activity in vitro are listed. REV1 is technically a dCMP transferase, but is universally referred to as a Y-family Pol. The Pols listed in this Table have been reviewed by both Burgers et al. [187] and Friedberg et al. [188], and the reader is referred to these sources, as well as references therein, for further details.
With their growing number came the obvious question – how do organisms manage the actions of their different Pols to ensure that the right Pol gets to the right place at the right time? This question takes on added complexity when one considers the fact that bypass of certain classes of DNA lesions requires the actions of multiple specialized Pols. In these cases, organisms must sequentially manage the actions of multiple Pols in order to mediate lesion bypass. Moreover, certain specialized Pols are capable of bypassing specific classes of DNA lesions in a relatively accurate manner, while others bypass the same lesion in a largely error-prone manner [5, 17–19]. Thus, recruitment of the inappropriate specialized Pol during TLS could result in mutations or genome rearrangements. Taken together, these findings highlight the importance of Pol switching to genome integrity.
BIOLOGICAL TRANSACTIONS INVOLVING POL SWITCHING
Several biological processes require the actions of multiple Pols, and thus involve Pol switching. For example, multiple Pols are involved in DNA replication, and the actions of these different Pols must be tightly coordinated to ensure the fidelity of this process (Fig. 1A). Likewise, organisms require a capacity to tolerate lesions that evade repair, or that for whatever reason cannot be repaired. In the absence of such a capacity, these lesions would serve as potent blocks to ongoing replication, resulting in cell death. Since specialized Pols are typically required for replicative bypass of these lesions, Pol switching is also crucial for coordinating high fidelity DNA replication with potentially error-prone TLS (Fig. 1B & C). Finally, certain Pols participate in specific DNA repair functions. In these cases, mechanisms must exist for both recruitment of the appropriate Pol, as well as to coordinate the actions of these Pols with the other proteins involved in the repair function. Although not formally a ‘Pol switch,’ the mechanisms underlying these Pol-partner switches are conceptually similar to Pol switches, and thus, the fundamental rules regarding Pol switching discussed in this review also apply to these types of switches.
CONSEQUENCES OF IMPAIRED POL SWITCHING
Despite our relatively naive understanding of the molecular mechanisms underlying Pol switching, it is evident that recruitment of the inappropriate Pol to a replication fork can result in mutations that contribute to human disease, including cancer. One prominent example of this is the variant form of the human genetic disease Xeroderma pigmentosum (XP-V). Individuals afflicted with XP-V are predisposed to sunlight-induced skin cancers [20, 21]. In 1999, Masutani et al. [22] reported that cells from XP-V patients were deficient in Pol η function. Pol η is a Y-family Pol that displays a remarkably low fidelity on undamaged DNA, but is capable of bypassing UV-induced thymine-thymine cyclobutane dimers in a relatively accurate manner [5, 23, 24]. In the absence of Pol η (e.g., XP-V patients), a different Pol with a reduced fidelity for bypassing thymine-thymine cyclobutane dimers is presumably utilized, leading to an increased level of UV-induced mutations. This elevated frequency of DNA damage-induced mutations is believed to underlie the cancer predisposition of XP-V patients [5, 25].
Proper Pol switching is of utmost importance to genome fidelity and viability in bacteria, as well. For example, several mutations that act to impair function of the replicative E. coli Pol confer conditional lethal phenotypes that are either partially or fully suppressed by genetic inactivation of one or more specialized Pol [26–29]. Taken together, these findings demonstrate that use of the ‘wrong’ Pol can be deleterious in both prokaryotes and eukaryotes.
THE GLOBAL SOS RESPONSE INFLUENCES POL SWITCHING BY REGULATING STEADY STATE LEVELS OF THREE OF THE FIVE E. coli POLS
One obvious mechanism to restrict the timing for a particular Pol switch is to limit expression of one or more of the Pols involved in the switch until such time as their unique activities are required. E. coli and related bacteria regulate expression of their specialized Pols in a DNA damage-responsive manner via the global SOS response. As part of this response, transcription of E. coli Pol II, Pol IV, and Pol V, as well as a variety of other gene products involved in DNA repair and cell cycle regulation, is regulated in a DNA damage-responsive manner by the LexA and RecA proteins [30]. LexA is a transcriptional repressor that binds to a consensus DNA sequence (5’-TACTGT[AT]4ACAGTA-3’) typically located in the operator region of SOS-regulated genes. Upon binding, the LexA homodimer represses transcription of more than 40 unlinked genes [31]. The degree to which LexA represses transcription of the different SOS-regulated genes varies, similar to a rheostat, both in the absence of, and in response to DNA damage. Structurally, the LexA dimer resembles two dumbbells: the C-terminal globules dimerize, while the N-terminal globules bind DNA [32]. Following DNA damage, LexA becomes inactivated, leading to transcriptional derepression of the SOS-regulated genes (i.e., SOS induction). RecA nucleoprotein filaments that assemble on single strand (ss) DNA that is generated by blocked replication forks serve as the signal to the cell that it has suffered DNA damage [33]. Appropriately, inactivation of LexA results from RecA nucleoprotein-facilitated autodigestion of the LexA linker domain that connects the N- and C-terminal globules [30], which targets it for destruction by the Lon and ClpXP proteases [34, 35]. In this way, E. coli ensures that the SOS response is regulated in a DNA damage-responsive manner.
Following LexA inactivation, the steady state levels of E. coli Pol II, Pol IV, and Pol V increase dramatically (Table 2). The level of Pol II (encoded by polB) increases from ~50–75 molecules/cell prior to SOS induction to ~350–1,000 molecules/cell following SOS induction ([36, 37]; unpublished results). Likewise, the level of Pol IV (encoded by dinB) increases ~10-fold following SOS induction, going from ~150–250 to more than ~1,200–2,500 molecules/cell ([38]; unpublished results). Pol V (encoded by umuD and umuC) undergoes the most dramatic change. In the absence of SOS induction, Pol V is virtually undetectable, and is estimated to be present at <15 molecules/cell [39]. This is the result of both efficient LexA-dependent transcriptional repression of umuDC [40], as well as Lon-mediated destruction of the modest levels of the umuDC genes expressed in the absence of SOS induction [41]. Following SOS-induction, Pol V levels reach a maximum of ~200 molecules/cell [39]. In addition to transcriptional repression and proteolysis, Pol V is also regulated posttranslationally. UmuD is structurally similar to the C-terminal half of LexA [32, 42–44]. Catalytic activity of the UmuC subunit of Pol V requires that UmuD undergo a RecA-facilitated autodigestion in a manner analogous to LexA [45–47]. Since UmuD bears an extended N-terminal arm rather than a LexA-like N-terminal globule and linker region, UmuD autodigestion serves to remove the N-terminal 24 residues of UmuD, forming UmuD’. Like LexA, both UmuD and UmuD’ exist as dimers. UmuD and UmuD’ can also form a heterodimer, which serves a regulatory role by shuttling UmuD’ into ClpXP-mediated proteolysis to help regulate Pol V levels [41, 48]. However, only the UmuD’ homodimer is capable of activating UmuC for TLS as Pol V [49–51]. Pol V also requires RecA for maximal Pol activity [50–53]. The precise mechanism by which RecA contributes to Pol V-mediated TLS is somewhat controversial, and is a topic of current study in several laboratories [51, 54–58]. One model postulates that RecA-ssDNA nucleoprotein filaments formed adjacent to DNA lesions serve to both recruit and activate Pol V for TLS, while a second model posits that RecA-ssDNA nucleoprotein filaments enable Pol V TLS in trans (i.e., when formed on a ssDNA that is not adjacent to the lesion), but block it in cis (i.e., on the ssDNA adjacent to the lesion). Finally, it is important to note that E. coli Pol V is required for most mutagenesis following exposure to UV or chemical carcinogens [59–61]. As a result, cells that are unable to induce the SOS response are deficient for most DNA damage-induced mutagenesis. Thus, the SOS response plays a critically important role in regulating the actions of Pol V, and, to a lesser extent, Pol II and Pol IV.
TABLE 2.
Affinities and steady state levels of E. coli DNA polymerases for the β sliding clamp.
| Pol | Affinity for β clamp (KD) * |
Approximate steady state concentration# | |
|---|---|---|---|
| SOS-repressed | SOS-induced | ||
| Pol I | ND | 830 Nm | 830 nM |
| Pol II | 32 nM | 80 Nm | 800 nM |
| Pol III | 108 nM | 20 nM | 20 nM |
| Pol IV | 465 nM | 330 nM | 3,300 nM |
| Pol V | NE | <25 nM | 330 nM |
Affinities of the clamp for Pol II [94], the α catalytic subunit of Pol III [100], and Pol IV [94] were measured in solution using surface plasmon resonance (SPR). The affinity of Pol III core for β clamp assembled on DNA is reported to be >5 nM [114]. Although Pol I interacts with the β clamp [74, 76], this interaction was not detected (ND) using SPR (unpublished results). Interactions of the β clamp with Pol V have not yet been examined (NE).
The approximate steady state concentration of each Pol under SOS-repressed and SOS-induced conditions was estimated based on the number of molecules/cell assuming an average cell volume of 10−15 liters. The specific level of each Pol under SOS-repressed and SOS-induced conditions, respectively, used to calculate these estimates are (levels of Pol I and Pol III do not change following SOS induction): Pol I, 500 (unpublished results); Pol II, 50 and 500 ([36, 37]; unpublished results); Pol III, 10 [146]; Pol IV, 200 and 2,000 ([38]; unpublished results); Pol V, <15 and 200 [39].
Several studies have highlighted the contribution of the SOS response to Pol switching [26–29, 62–69]. One recent study by Curti et al. [70] took advantage of the fact that the different E. coli Pols display unique fingerprints with respect to the type(s) of base substitution mutations that they catalyze [71]. Based on nucleotide sequence analysis of spontaneous rifampicin resistant mutations present in isogenic strains bearing deletions of different combinations of the E. coli Pols, the authors concluded that there was significant interplay between all five Pols following SOS induction [70]. For example, most transversion mutations were dependent upon Pol IV and Pol V, most transitions were dependent upon Pol I and Pol III, and that Pol II played a largely antimutagenic role by suppressing mutagenesis at certain hotspots [70]. Thus, following SOS-induction, there appears to be dynamic switching between the different E. coli Pols.
Eukaryotes also appear to regulate the levels of their different Pol levels as part of a mechanism to regulate Pol switching. DNA damage-induced mutagenesis in Schizosaccharomyces pombe is regulated as part of a DNA damage checkpoint response [72]. The purpose of these checkpoint responses is to arrest the cell cycle, activate DNA repair and DNA damage tolerance pathways, or, if the cell has suffered severe DNA damage, induce apoptosis. Thus, these responses are functionally equivalent to the E. coli SOS response. One function of the checkpoint response in S. pombe is to increase transcription of the gene encoding Pol κ, which together with Pol ζ acts in DNA damage-induced mutagenesis [72].
ACCESS OF POLS TO THE REPLICATION FORK IS REGULATED IN PART THROUGH THEIR INTERACTIONS WITH SLIDING CLAMP PROTIENS
Sliding clamp proteins play critically important roles in targeting Pols to replication forks, as well as enabling Pol switching. The E. coli β sliding clamp was initially identified as a subunit of the replicative Pol (Pol III) based on its ability to dramatically increase processivity of the holoenzyme [73]. However, over the last decade, it has become evident that the clamp interacts with all five E. coli Pols, as well as additional proteins involved in DNA replication and cell cycle progression [3, 74–81]. The archael and eukaryotic clamp proteins, known as proliferating cell nuclear antigen (PCNA), similarly interact with a variety of proteins involved in DNA replication, DNA repair, TLS, DNA damage checkpoint controls, and cell cycle progression (recently reviewed in [82]). Thus, sliding clamps are now widely recognized as playing key roles in coordinating multiple critically important dynamic events at replication forks. One current challenge is to define the mechanisms controlling the spatial and temporal regulation of these diverse interactions.
In general, bacterial clamps (β clamps) exist as homodimers, whereas eukaryotic and archael clamps (PCNA) are homotrimers. One notable exception to this rule is the Sulfolobus solfataricus PCNA clamp, which is a heterotrimer [83]. Despite the lack of amino acid sequence similarity between the different clamps, crystallographic studies have revealed that their three-dimensional structures are remarkably similar [84–87]. The head-to-tail arrangement of the protomers forms a ring with two distinct surfaces. The so-called N-side bears the N-terminus, while the C-side contains a hydrophobic cleft that serves as a common docking site for most of its different partner proteins [88–91]. The structure of the β clamp on DNA was recently reported [92]. In the crystal, the clamp makes specific contacts with DNA, consistent with computational modeling studies [93]. These contacts are proposed to play important roles in proper clamp function and loading [92, 94], and are absolutely required for viability [94]. They also contribute to Pol switching, as discussed in detail later on in this review.
Clamps are loaded around DNA by a conserved multi-subunit AAA+ ATPase (expanded group of ATPases Associated with various cellular Activities) referred to as the clamp loader (reviewed in [95]). The mechanism of clamp loading is well conserved between organisms (Fig. 2A). The eukaryotic clamp loader complex is called RFC, while bacterial clamp loaders are referred to as the DnaX (or γ) complex. Like clamp loaders in other organisms, E. coli DnaX adopts an arc-shaped structure that is proposed to interact with and stabilize the open form of the β clamp [96]. This form of the clamp, in which a single interface is broken, adopts a lock washer-like structure [97]. The structure of the clamp loader is reported to match the contour of both the open clamp and the DNA [96]. A stable DnaX-β complex requires that DnaX first bind ATP. This complex then associates with nicked or primed DNA, reportedly ratcheting itself over the 3’-end of the primer, much like a screw cap on a bottle. Once properly positioned, the clamp loader complex is proposed to hydrolyze bound ATP, resulting in its immediate dissociation, leaving the clamp behind to close over the duplex DNA. Despite the ability of the clamp to interact with DNA, an activity that contributes to its loading onto DNA, it nevertheless slides along double strand (ds) DNA [92]. Once loaded onto a circular DNA, the clamp persists with a half-life of >100 min in vitro [98]. Due to this remarkable stability, clamps accumulate on lagging strand during DNA replication. As there are only ~300–600 clamps/cell [98, 99], and a new clamp is required for replication of each of the 4–5,000 Okazaki fragments, it is postulated that the clamp must be actively unloaded from DNA during replication [98]. The DnaX complex, as well as the δ subunit of DnaX, which is present in excess over the other DnaX subunits in vivo, can unload β from DNA [98, 100]. Likewise, both the eukaryotic RFC clamp loader, as well as the RFC2 subunit of RFC catalyze unloading of PCNA [101]. Although not yet formally tested, it stands to reason that clamp recycling plays an important role in Pol switching, as well as in regulating access of specialized Pols to the DNA.
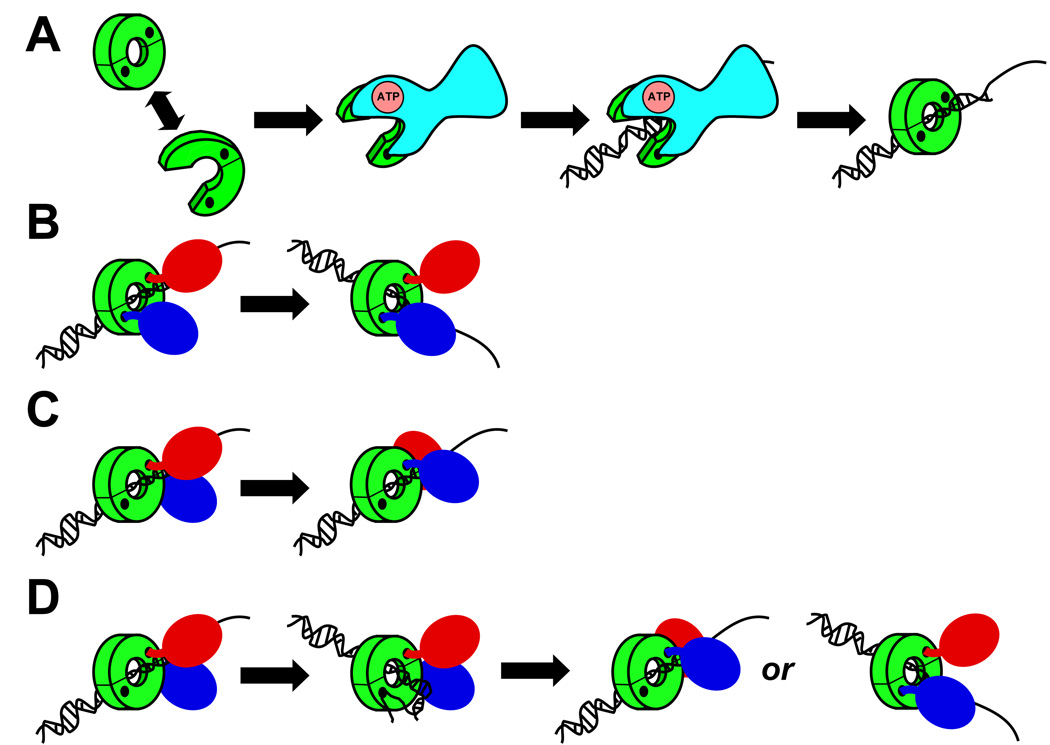
FIGURE 2. Models depicting possible roles of the homodimeric E. coli β clamp in mediating Pol switching.
(A) Cartoon depiction of the mechanism by which the E. coli DnaX (or γ) complex catalyzes loading of the β clamp onto the 3’-OH end of a primed DNA. The β clamp is depicted in green, and DnaX is in blue. The black circle depicts the hydrophobic cleft on each β clamp protomer, and the line represents the clamp dimer interface. (B) Cartoon depiction of the ‘toolbelt’ model, as originally proposed by Pages and Fuchs [184]. The replicative Pol is in red, and the specialized Pol is in blue. (C) Cartoon depiction of the role of non-cleft surfaces on the clamp in Pol switching. In this particular example, the replicative (red) and specialized (blue) Pols are switching via a single cleft on the clamp. (D) Possible mechanisms by which DNA and specialized Pols might compete with each other for interaction with the β clamp, and subsequent access to the replication fork. See text for further details regarding the models presented in this Figure.
Interactions of the different Pols with the clamp involve multiple distinct surfaces. This important point is discussed in greater detail later on in this review. One of the more heavily studied contacts involves association of a consensus Clamp Binding Motif (CBM; QL[S/D]LF) present in bacterial Pols, or a PCNA Interacting Protein box (PIP box; Qxx[M/L/I]xxF[F/Y/W]) present in eukaryotic Pols, with a hydrophobic cleft located near the C-terminus of each clamp protomer [88, 102]. Mutations disrupting the CBM in E. coli Pol II, Pol III, Pol IV, or Pol V [77, 81, 103], or the PIP box in eukaryotic Pol δ, Pol η, Pol ι, Pol κ, or REV1 [104–111], serve to severely impair the biological activity of these Pols, thus underscoring the importance of these clamp-Pol interactions. Importantly, clamp does not only serve to tether Pols to the DNA template, but can also act to stimulate catalytic activity of the Pol. For example, interactions of eukaryotic Pol κ with PCNA, as well as E. coli Pol IV with β clamp markedly reduces their apparent Km for dNTPs [110, 112].
Early models proposed that the ability of a Pol to compete with another for access to the replication fork correlated with its affinity for the clamp. However, PCNA displays a similar affinity (~10–30 nM) for several of its partners, including RFC, Pol δ, Pol ε, and Pol λ (reviewed in [113]). In contrast, the affinity of the E. coli β clamp for its different Pols can vary more than 10-fold (Table 2). Likewise, the steady state concentrations of the different E. coli Pols can vary as much as ~40-fold prior to SOS-induction, and as much as ~160-fold following SOS-induction (Table 2). Interestingly, with the exception of Pol III (which to binds a clamp/primed DNA complex ~20-fold more tightly than clamp alone in solution [114]), each Pol is present at a steady state level greater than its apparent KD. However, there does not appear to be an obvious relationship between their relative affinities for the clamp, and their steady state levels. For example, Pol II binds the clamp very tightly in solution (KD=32 nM), but is present at an ~4-fold higher level than Pol III in the absence of SOS-induction. Likewise, Pol IV binds the clamp ~4-fold less tightly than does Pol III, yet is present at an ~16-fold higher level than Pol III. Thus, although clamp-Pol interactions are crucial for the biological roles of the different Pols, the precise relationship between affinity of the different Pols for the β clamp and their ability to gain access to the replication fork are presently unclear.
POSTTRANSLATIONAL MODIFICATION OF THE EUKARYOTIC PCNA CLAMP CONTRIBUTES TO POL SWITCHING
As discussed in Chapter 16 of this thematic issue, the different biological activities of the eukaryotic PCNA clamp are regulated in part via posttranslational mechanisms. For example, ubiquitination of PCNA is critically important for enabling TLS. S. cerevisiae Rad6, Rad18, Mms2, Ubc13, and Rad5 proteins participate in replication of damaged DNA. Although Rad6 and Rad18 are required for both accurate and error-prone replication-associated pathways, Mms2, Ubc13, and Rad5 are specific for an accurate tolerance pathway [115]. Rad18 and Rad5 are E3 ubiquitin ligases, while Rad6, Mms2, and Ubc13 are E2 ubiquitin conjugating enzymes capable of forming ubiquitin chains that are linked via lysine-63 as opposed to the classical lysine-48 linkage that targets proteins for proteolysis. In 2002, Hoege et al. [4] demonstrated that, following exposure to methyl methanesulfonate, yeast PCNA first became mono-ubiquitinated at lysine-164 by Rad6 and Rad18, followed by subsequent poly-ubiquitinatination by Mms2, Ubc13, and Rad5. Mono-ubiquitination of PCNA enabled TLS, while poly-ubiquitination promoted an as yet poorly understood accurate DNA damage avoidance pathway [4]. Although important differences in the ubiquitination process exist, a similar series of events appears to take place in other eukaryotic organisms [109, 116–122]. In contrast, less is known regarding the regulation of PCNA ubiquitination. In mammals, Ubiquitin Specific Protease 1 (USP1) removes ubiquitin from mono-ubiquitinated PCNA [123]. In addition, both p53 and p21 appear to regulate ubiquitination of PCNA in response to UV irradiation [119, 124]. p53 is transcription factor that serves a well-established role as a tumor suppressor, while p21 is a cyclin-dependent kinase inhibitor that acts to regulate cell cycle progression in G1. p21 also interacts with PCNA, and may influence Pol switching under certain conditions by competing with Pols (as well as possibly other PCNA partners) for binding to the clamp (reviewed in [82]).
Results discussed above, taken together with the finding that the eukaryotic Y-family Pols possess a ubiquitin binding motif that enhances their respective affinities for mono-ubiquitinated PCNA, and contributes to their biological functions [125], suggests the following model for Pol switching when the replicative Pol is blocked at a lesion. Upon stalling, ssDNA is generated, due to continued progression of the helicase. This accumulated ssDNA contributes to Rad18 activation, which together with Rad6 acts to mono-ubiquitinate PCNA [126]. The specialized Pols gain access to the mono-ubiquitinated PCNA, due in part to their enhanced affinity for this form of the clamp, which may enable them to out compete Pol ε or Pol δ. Following bypass of the lesion, the specialized Pol may release the DNA template, or simply dissociate from PCNA, enabling access of Pol ε and/or δ.
POL-POL INTERACTIONS CONTRIBUTE TO SWITCHING
In 1999, at about the same time that the umuDC gene products were realized to encode the fifth E. coli Pol (Pol V), both UmuD and UmuD’ were determined to interact, albeit differently, with the α catalytic, ε proofreading, and β clamp subunits of Pol III [3]. These interactions were suggested to contribute to both a primitive DNA damage checkpoint control involving UmuD and UmuC, as well as a switch between a stalled Pol III and Pol V (UmuD’2-UmuC), enabling TLS [3, 127–129]. However, this model has not yet been formally tested. More recently, overexpressed levels of Pol IV were determined to inhibit replication in E. coli cells [130]. This activity of Pol IV was likened to a checkpoint that acts to slow replication in response to DNA damage, thereby allowing time for lesions to be repaired in the absence of ongoing replication. Inhibition of replication correlated with the ability of elevated levels of Pol IV to displace Pol III from a β clamp encircling a primed DNA template in vitro [130, 131]. This ability of Pol IV relied largely on its N-terminal 230 residues, and hence did not appear to absolutely require Pol IV-β clamp interactions. Nonetheless, full length Pol IV was most effective at inhibiting replication in vivo [130]. Thus, this putative checkpoint appears to involve interaction of Pol IV with both the clamp and Pol III. However, a direct interaction between Pol IV and Pol III has not yet been described. The relationship of this process to Pol switching is also unclear. Nonetheless, the finding that only overexpressed levels of Pol IV inhibited DNA synthesis in vivo suggests that this putative checkpoint role of Pol IV is restricted to SOS-induced conditions [130]. The mechanism(s) acting to prevent Pol IV from displacing Pol III during replication of undamaged DNA have yet to be defined. In contrast to this study, Indiani et al. [132] demonstrated that Pol III and Pol IV bind simultaneously to a single β clamp. These authors also described conditions under which modest levels of Pol IV (e.g., ~6-fold molar excess over Pol III) switched with a stalled Pol III in vitro [132]. For these experiments, Pol III was stalled via omission of two of the four dNTPs. We have recently reproduced these findings using a similar assay, and determined that a 2-fold molar excess of Pol IV was sufficient to promote efficient switching with a stalled Pol III* [133].
Several eukaryotic Pols also interact physically with each other. For example, Pol η interacts with Pol ι [134], while REV1 interacts with Pol η, Pol ι, Pol κ, as well as Pol ζ [135, 136]. Importantly, these interactions appear to be crucial for proper function(s) and sub-cellular localization of these Pols. In spite of the finding that DNA damage-induced mutagenesis in eukaryotes is dependent upon REV1, catalytic activity of REV1 is not required [137–139]. Instead, REV1 appears to act as a scaffold to recruit multiple specialized Pols to the clamp to enable their switching. However, the precise mechanisms underlying the switching process remain to be elucidated. One currently popular view is that Pol switching may be largely stochastic, and that by simply maximizing the number of Pols in the vicinity of a lesion, the cell has the ability to sample several different Pols, thereby affording the Pol with the most favorable kinetic properties for that particular lesion the advantage in catalyzing bypass. Results from Becherel & Fuchs [140] support this type of model, at least with respect to those switches involving E. coli Pol II and Pol V during bypass of an N2-acetylaminofluorene adduct within the NarI mutation hot spot. However, in this instance, the switch appears to involve interactions of the different Pols with distinct structural conformations of the NarI-containing DNA template, as well as the N-2-acetylaminofluorene adduct.
DISTINCTIONS BETWEEN SWITCHES INVOLVING ONLY REPLICATIVE POLS AND SWITCHES INVOLVING BOTH REPLICATIVE AND SPECIALIZED POLS
The mechanisms by which Pols switch is likely to be heavily influenced by the biological transaction being catalyzed, as well as the particular Pols involved. Several important mechanistic differences exist between switches involving only high fidelity Pols acting in replication of an undamaged DNA template, and switches involving both high fidelity and low fidelity Pols enabling TLS. For example, switching during replication of an undamaged template is largely unidirectional in that one Pol replaces another via a single switch (Fig. 1A). Examples of these types of switches are discussed in detail in the following two sections. In contrast, switching during TLS is more complex, and involves at least two distinct switches (Fig. 1B). The first switch is initiated by the replicative Pol stalling at a lesion, and involves replacement of this stalled Pol with a specialized Pol. The second switch occurs after the specialized Pol has negotiated the lesion, and involves replacement of the specialized Pol with a replicative Pol for continued high fidelity synthesis. Importantly, bypass of a growing number of DNA lesions require multiple specialized Pols, and thus involve a third switch. These lesions include, for example, (+)-trans-benzo(a)pyrene-N2-dG, which in E. coli requires both Pol IV and Pol V for bypass [62], and in mammalian cells requires Pol κ and Pol ζ [141], apurinic/apyrimidinic sites, which in yeast require both Pol η and Pol ζ [142], and intrastrand dG-dG cisplatin adducts, which in mammalian cells require Pol η and Pol ζ [141]. In these cases, one specialized Pol inserts a base opposite the lesion, but cannot extend the nascent strand further. A second specialized Pol is therefore required to extend the nascent strand, and subsequently returns the 3’-OH end of the DNA template back to the replicative Pol (Fig. 1C).
The second distinction between switches involving only replicative Pols and those involving both replicative and specialized Pols pertains to the length of the patch that must be synthesized by a given Pol. This is particularly critical during TLS. For example, if a specialized Pol synthesizes too long of a patch during lesion bypass (i.e., ≥5 bases), it may contain one or more misinsertions in addition to the TLS event, due to the low fidelity that most specialized Pols display when replicating undamaged DNA. If these mismatches are left unrepaired, they will give rise to what is referred to as ‘untargeted’ mutations [143]. In contrast, if the patch is too short (i.e., ≤4–5 bases) then an abortive process will ensue [144, 145]. For example, if the specialized Pol synthesizes only one or two bases beyond the lesion prior to switching, then the replicative Pol will recognize the discontinuity in the DNA duplex structure caused by the lesion. As a result, the replicative Pol will stall, allowing its associated proofreading activity access to the 3’-OH end of the primer. The 3’-to-5’ exonuclease proofreading activity will degrade the nascent strand, excising the base inserted opposite the lesion by the specialized Pol, necessitating another bypass attempt. In contrast to TLS, replicative Pols possess proofreading activity, and are therefore largely accurate. Thus, the length of the patch that each replicative Pol synthesizes is unlikely to severely effect fidelity.
COORDINATION OF MULTIPLE POLS DURING HIGH FIDELITY REPLICATION IN PROKARYOTES
In light of the fact that multiple Pols cooperate during chromosomal replication, a brief review of what is currently known regarding Pol switching in the context of high fidelity replication in prokaryotes is warranted. Duplication of the 4.6 × 106 bp E. coli chromosome relies on both Pol III and Pol I. Pol III is responsible for the majority of DNA synthesis, while Pol I plays a smaller albeit important role. In contrast to Pol I, which is comprised of a single polypeptide, the holoenzyme form of Pol III (Pol III HE) consists of 17 separate polypeptides, which are organized into three distinct subassemblies, including two core complexes (αεθ), one DnaX clamp loader complex (τ2γδδ’ψχ), and two dimeric β sliding clamps [146–150]. Each core complex consists of one α catalytic subunit, which synthesizes nascent DNA, one ε 3’-to-5’ exonuclease proofreading subunit, which excises replication error catalyzed by α, and one θ subunit, which stimulates proofreading activity of ε. In addition to forming part of the ring structure of the DnaX clamp loader complex, the DnaX τ dimer also act to bridge two Pol III core complexes via physical interaction with the α subunits. This interaction involves the C-terminal tails of τ that extend out and away from DnaX. In contrast to E. coli, certain bacteria, such as the gram-positive Bacillus subtilis, contain two distinct Pol III enzymes (named DnaE and PolC), and may utilize one for replication of leading strand, and the other for lagging strand [151].
Replication priming in E. coli is performed by DnaG primase. During initiation of DNA replication, two hexameric DnaB helicases are loaded at opposite ends of the unwound origin [152, 153]. These helicases unwind the dsDNA, translocating 5’-to-3’ in an ATP-dependent fashion. While doing so, they recruit DnaG primase for synthesis of a short RNA primer, which will ultimately be elongated by Pol III [154, 155]. Following its synthesis, DnaG binds to the primer to protect it from nucleases such as RNAse HI that could degrade it [156]. DnaG also interacts with ssDNA binding protein (SSB) that binds to the ssDNA flanking the RNA primer. Interactions of the χ subunit of the DnaX clamp loader complex of Pol III HE with SSB serves to displace DnaG [156]. This exposes the 3’-OH end of the RNA primer, providing a site for DnaX to load a β clamp. Since DnaX is in association with two Pol III cores, this Pol is optimally positioned for preferential recruitment to the nascent replication fork. Thus, E. coli ensures that Pol III, and not one of its other four Pols is recruited for chromosomal replication through the use of a series of sophisticated and carefully orchestrated protein-protein and protein-nucleic acid interactions.
Once recruited, Pol III HE elongates the RNA primer in the 5’-to-3’ direction. Due to the anti-parallel structure of duplex DNA, one strand, referred to as leading, is in theory elongated in a continuous manner (but not in practice, due to frequent perturbations in the DNA template [157]), while the other, termed lagging, is synthesized discontinuously in ~1 kb pieces known as Okazaki fragments [158–160]. Since Pol III HE contains two core complexes, a single Pol III HE is able to simultaneously replicate both leading and lagging strands. In contrast to the leading strand, which in theory requires a single priming event, DnaG must repetitively prime the lagging strand. Since the leading and lagging strand Pol III core complexes are tethered via the τ2 subunits of DnaX, but the two DNA strands are anti-parallel, the lagging strand must loop around in order to accommodate simultaneous extension of both the leading and lagging strands, resulting in what Bruce Alberts termed a ‘trombone’ structure [161]. Upon completing an Okazaki fragment, the lagging strand Pol III core disengages from the DNA template in order to cycle to the next RNA primer. This cycling occurs roughly once every second. Interactions of the τ and χ subunits of DnaX with Pol IIIα and SSB, respectively, play pivotal roles in signaling to the lagging strand Pol III core that synthesis of the Okazaki fragment is complete, and help facilitate its cycling to the next primer [146, 162, 163].
Although Pol III performs the bulk of the DNA replication, Pol I serves a critically important role in lagging strand synthesis. In addition, it likely plays important roles in filling ssDNA gaps that arise as a result of obstructions, including collisions of the replicating Pol III HE with transcribing RNA polymerase [164]. With respect to its role on lagging strand, the 5’-to-3’ exonuclease activity of Pol I acts to remove RNA primers at the 5’-end of each Okazaki fragment, while the associated polymerase activity acts to extend the 3’-OH end of the adjacent Okazaki fragment (Fig. 1A). Following action of Pol I, E. coli DNA ligase seals the nick between adjacent Okazaki fragments, completing the maturation process. Both E. coli Pol I and DNA ligase interact with the β clamp [74, 76], consistent with a model in which the clamp acts to coordinate their functions with that of Pol III HE.
Although the roles of Pol III and Pol I in replication are well defined, possible roles for Pol II and Pol IV are not. Despite the fact that Pol II and Pol IV are SOS-regulated, they are both nonetheless present at significant levels in the absence of SOS induction (Table 2). Recent findings suggest that both Pol II and Pol IV gain access to the DNA during replication, particularly on lagging strand [69, 74]. Although Pol II possesses an intrinsic proofreading activity, Pol IV does not. Thus, unregulated access of Pol IV could result in gratuitous mutations. At least two distinct mechanisms act to limit the ability of Pol IV to catalyze gratuitous mutations during replication of undamaged DNA. First, Pol I competes directly with Pol IV, as well as other specialized Pols for access to ssDNA nicks or gaps located between immature Okazaki fragments [74]. Second, Pol II plays an antimutator role by acting as a proofreader for Pol IV, effectively removing misinsertions before they result in mutations [70, 165]. Thus, dynamic switching between Pol I, Pol II, Pol IV, and possibly Pol III [74] serves to limit access of Pol IV to the replication fork, as well as the number of gratuitous mutations catalyzed by Pol IV during replication of undamaged DNA.
The replisome frequently encounters lesions or other impediments. Differences in the mechanisms by which the leading and lagging strands are replicated have important implications with respect to how Pol III responds to these blocks, and thus influences Pol switching. For example, if the lagging strand Pol III core encounters a lesion, it could simply terminate that Okazaki fragment prematurely, leaving a ssDNA gap downstream of the lesion, which could be dealt with postreplicatively. This would likely have a minimal effect on replication. In contrast, when the leading strand Pol III core stalls at a lesion, it will become uncoupled from the helicase, blocking further synthesis. In this case, it is proposed that the replisome must be reassembled at a newly primed site downstream of the lesion to allow for continued synthesis [157, 166]. It is established that ssDNA gaps are generated following replication of damaged DNA in E. coli [167]. However, it remains to be determined whether specialized Pols have access to the fork during Pol III replication to enable TLS, irrespective of SOS induction. One advantage of a capacity for TLS is that it would reduce the frequency with which replication forks collapse. The recent findings that several specialized Pols catalyze bypass of certain classes of lesions in a largely accurate manner helps to make this type of model more palatable.
COORDINATION OF MULTPLE POLS DURING HIGH FIDELITY REPLICATION IN EUKARYOTES
Like E. coli, eukaryotes must also manage the actions of multiple Pols during high fidelity replication. This section briefly outlines the mechanisms by which eukaryotes coordinate the actions of Pol α, Pol ε, and Pol δ during DNA replication. Pol α, together with its associated primase activity, is responsible for primer synthesis [168]. As such, it is involved in both the initiation of the elongation process, as well as the repeated priming events required for lagging strand synthesis. Pol ε and Pol δ are currently thought to act as the leading and lagging strand Pols, respectively [169, 170]. The way in which eukaryotic proteins involved in DNA replication associate with each other to form the replisome is poorly understood. However, the finding that distinct Pols appear to have well-defined roles in replication strongly argues that the assembly mechanism somehow ensures that the right Pol is recruited to the right place at the right time.
Due to the involvement of at least three Pols, high fidelity replication in eukaryotes relies on at least three switching events. The first two switches involve a hand-off from Pol α to Pol ε on leading strand, or to Pol δ on lagging strand [171]. These switches are functionally analogous to the E. coli DnaG primase-Pol III switch, and similarly rely on a series of competitive protein-protein and protein-nucleic acid interactions involving the three Pols, the eukaryotic sliding clamp protein (PCNA), the eukaryotic clamp loader complex (RFC), and the heterotrimeric ssDNA binding protein RPA [172–174]. The main purpose of these switches is to displace Pol α from the DNA template, thereby exposing the 3’-OH end of the primer/template for RFC-mediated loading of PCNA and subsequent recruitment of Pol ε and Pol δ. As for lagging strand in E. coli, the Pol α-Pol δ switch is required for synthesis of each Okazaki fragment. In addition to these switching events, eukaryotes must also manage the actions of FEN1 and DNA ligase I during Okazaki fragment maturation. FEN1 is a 5’-to-3’ structure-specific DNA endonuclease that, together with the Dna2 helicase/nuclease, plays pivotal roles in removing RNA primers on lagging strand, analogous to the 3’-to-5’ exonuclease function of Pol I in E. coli [175, 176]. Subsequent to primer removal, Pol δ (or possibly Pol ε) fills in the ssDNA gap. Finally, the nick between adjacent Okazaki fragments is sealed by DNA ligase I [177]. Interactions of Pol δ, FEN1, and DNA ligase I with PCNA are crucial for the proper temporal regulation of Okazaki fragment maturation [178]. Thus, like E. coli, eukaryotes also utilize a sophisticated series of competitive protein-protein and protein-nucleic acid interactions to manage Pol-Pol and Pol-protein switching during chromosomal replication.
POLS ASSOCIATE DYNAMICALLY WITH THE REPLICATION FORK
Although we have known for many years now that the lagging strand Pol is dynamically associated with the replication fork, it was only recently determined that the leading strand Pol is similarly dynamically associated. This association has been termed ‘dynamic processivity,’ meaning that the leading strand Pol undergoes a rapid solution-to-replication fork exchange without impairing continued synthesis. Yang et al. [179] were the first to propose the idea of dynamic processivity, based on their study of DNA replication in bacteriophage T4. They determined that a catalytically inactive T4 gp43 Pol mutant replaced an actively replicating wild type gp43 Pol with a half-life of <1 min, despite the fact that actively replicating gp43 had a half-life of ≥9 min on primed DNA. The authors suggested that binding of multiple copies of gp43 to the homotrimeric T4 gp45 sliding clamp, and subsequent rapid exchanges of these gp43 Pols with the DNA template underlie the dynamic processivity. More recently, Hamdan et al. [180] described an analogous situation for bacteriophage T7. Instead of a clamp, the T7 gp5 Pol utilizes E. coli thioredoxin to anchor it on the DNA. Thus, in contrast to T4, rapid exchanges between ‘spare’ copies of the T7 gp5 Pol associated with the T7 helicase and the DNA template underlie its dynamic processivity.
Certain features of the bacteriophage dynamic processivity extend to E. coli. The precise composition of the E. coli DnaX clamp loader has been somewhat controversial. Although Pritchard et al. convincingly demonstrated in 2000 that the reconstituted form of DnaX was comprised of τ2γδδ’ψχ [181], the question as to the actual composition of DnaX in vivo has recently reemerged. In reference to this, McInerney et al. [182] reconstituted four different forms of the DnaX clamp loader varying in their τ/γ composition, including γ3δδ’ψχ, τ1γ2δδ’ψχ, τ2γ1δδ’ψχ, and τ3δδ’ψχ. Characterization of these different forms of clamp loader revealed that the τ3 form was able to recruit three separate Pol III core complexes to the replication fork. As a result, this form of Pol III supported ~60% more total synthesis relative to that observed with τ2γ1δδ’ψχ that associates with just two Pol III cores, possibly due to more efficient recognition of and/or binding to the DNA template [182]. The finding that Okazaki fragments were short when using τ3δδ’ψχ was interpreted to suggest that multiple Pol III core complexes were acting to replicate lagging strand. Nossal et al. [183] have reported similar findings with the T4 system. Taken together, these findings illustrate the highly dynamic behavior of Pols associated with the replication fork, consistent with the idea that they switch more frequently than previously acknowledged.
THE SLIDING CLAMP TOOLBELT: A MODEL FOR COORDINATING HIGH FIDELITY REPLICATION WITH ERROR-PRONE TLS
One very popular model currently discussed in the literature for how organisms manage the actions of their different Pols is termed the toolbelt model. This model, as initially proposed by Pages & Fuchs in 2002 [184], postulates that two different Pols simultaneously bind to the same bacterial sliding clamp protein, with each Pol contacting a separate cleft on the clamp (Fig. 2B). Since the eukaryotic clamp is a homotrimer, it follows that it could simultaneously bind three different Pols. Despite the attractiveness of the toolbelt model, it is important to stress that it has not yet been rigorously tested. Thus, it is worthwhile discussing published results that pertain to this model. In support of the toolbelt model, Indiani et al. [132] demonstrate that a single β clamp could simultaneously bind Pol IV and the α catalytic subunit of Pol III in solution. It is currently unclear whether both Pols simultaneously interact with a clamp assembled on DNA, and whether both clefts are required for these interactions. Furthermore, using an in vitro replication assay, Indiani et al. demonstrated that Pol IV switched with a stalled Pol III in vitro, but was less proficient at switching with a Pol III that was actively replicating the DNA template. As noted above, Heltzel et al. recently reproduced these results [133]. Importantly, they additionally determined that a clamp bearing a single cleft was capable of supporting a switch between a stalled Pol III and Pol IV: these results challenge the toolbelt model as originally proposed [133]. Furukohri et al. [131] also reported that Pol IV switched with a stalled Pol III in vitro. However, these authors suggested that Pol IV actively displaced Pol III from the clamp during the switch, as discussed earlier in this review. This active displacement was suggested to involve interactions of Pol IV with both the clamp and Pol III. Thus, it is unclear at the present time whether both Pols remain bound to the clamp throughout the switching, or whether the replicative Pol dissociates.
As outlined above, the toolbelt model fails to fully explain the molecular mechanisms underlying the decision as to which of the cell’s different Pols will bind the clamp, and when each will gain access to the DNA template. Three recent findings shed some new light on the mechanism by which Pol switch, and demonstrate that this process is far more complex and dynamic than previously acknowledged. Firstly, it has become increasingly clear that Pols interact with surfaces of the clamp in addition to the cleft region [27, 91, 94, 99, 100, 185]. Importantly, these non-cleft interactions appear to be crucial for proper Pol function, both in vitro [94, 185] and in vivo [27, 28, 74, 99, 185], and are suggested to play important roles in Pol selection/switching [26, 185], as discussed in the following section. Second, interactions of the clamp with its partner proteins can involve cooperativity. For example, two DnaX δ subunits can simultaneously bind a single β clamp, and they do so with negative cooperativity, presumably with each δ contacting a separate cleft on the clamp [100]. The mechanistic basis for this negative cooperativity is unknown. However, it was suggested to result from conformational changes induced in the peptide backbone of the clamp upon binding of the first δ [100]. Thus, binding of a single Pol to the clamp may act to alter the clamps structure, thus imposing a plastic hierarchy on subsequent clamp-Pol interactions, effectively dictating both the identity and the order in which subsequent Pols are recruited. Finally, the clamp interacts with the DNA template that it encircles [92]. The implications of these clamp-DNA interactions for Pol switching are discussed after the following section.
INTERACTIONS OF POLS WITH NON-CLEFT SURFACES OF THE CLAMP ARE CRUCIAL FOR THEIR PROPER FUNCTION: IMPLICATIONS FOR THE TOOLBELT MODEL
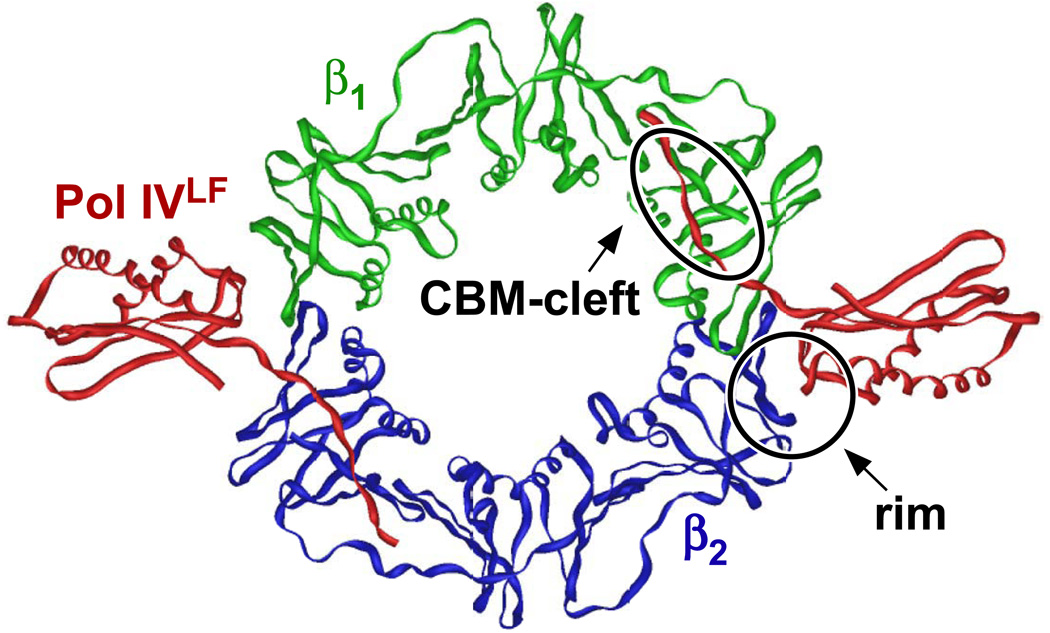
Interactions of Pols with surfaces of the clamp in addition to the cleft are crucial for their biological functions. E. coli Pol IV was the first Pol demonstrated to contacted surfaces of the clamp in addition to the cleft. Based on a crystal structure of the β clamp in complex with the Pol IV little finger domain (Pol IVLF; consisting of residues 243–351 of Pol IV) solved by Bunting et al. [91], Pol IV contacts at least two distinct surfaces of the clamp: the Pol IV CBM (residues 346–351) interacts with the hydrophobic cleft of the clamp, while residues V303, W304, and P305 of Pol IV (among others) reach over the dimer interface to contact E93 and L98 on the rim of the adjacent clamp protomer (Fig. 3). The authors suggested that Pol IV, when bound in this manner, would be unable to contact DNA, and therefore proposed that this structure represented an ‘inactive’ or ‘locked down’ conformation [91]. Thus, Pol IV could potentially reside at a replication fork in this inactive conformation so that is was available for TLS if needed, but would be unable to gain access to the DNA template until such time as it released the rim of the clamp. Consistent with this model, Xing et al. described multiple conformations for S. sulfataricus Dpo4 (an E. coli Pol IV ortholog) in complex with PCNA [186]. Their structures identified two hinge regions in Dpo4, suggesting that Dpo4 can adopt both ‘active’ and ‘inactive’ conformations on PCNA at the replication fork. In the active conformation, Dpo4 is bound to the DNA template and the cleft of PCNA. In contrast, the inactive conformation involves contact of Dpo4 with both cleft and non-cleft surfaces of PCNA, precluding Dpo4 from contacting the DNA template.
FIGURE 3. Crystal structure of the Pol IV little finger domain in complex with the E. coli β sliding clamp.
This structure was solved by Bunting et al. [91]. The Pol IV little finger (Pol IVLF) domain is comprised of residues 243–351 of Pol IV. Relative positions of the Pol IV CBM-clamp cleft (CBM-cleft), and the Pol IVLF-clamp rim (rim) contacts are indicated. This image was produced using imol and PDB coordinates 1UNN.
In addition to these elegant structures, recent functional studies have clearly demonstrated the importance of non-cleft Pol-clamp contacts to proper Pol usage. For example, deletion of the C-terminal five residues of the E. coli β clamp served to abolish its interaction with the DnaX clamp loader complex, indicating that this mutation severely impaired the CBM-cleft interaction [100]. Nevertheless, this mutant clamp retained a significant affinity for the α catalytic subunit of Pol III [100], demonstrating the importance that non-cleft contacts make in this interaction. Furthermore, based on the x-ray crystal structure, substitution of residues H148-R152 in the clamp with poly-alanine failed to influence the structure of the cleft region, but nevertheless severely impaired physical and functional interactions of the clamp with both Pol II and Pol IV [94]. Taken together, these findings illustrate that clamp-Pol interactions involve multiple surfaces in addition to the CBM-cleft contact.
Given the importance of non-cleft surfaces on the clamp to proper Pol function/utilization in E. coli, it seems likely that these surfaces contribute to switching [26–28, 74, 94, 99, 185]. Coincidently, the dynamic processivity models discussed above suggest one mechanism by which these non-cleft contacts could contribute to Pol switching in a toolbelt-like model. Specifically, multiple Pols may associate simultaneously with a single clamp via a combination of both CBM-cleft and non-cleft contacts. The Pol contacting the cleft would have the advantage in gaining access to the DNA template relative to the Pol bound to non-cleft surfaces (Fig. 2C). Interaction of E. coli Pol IV with the rim of the β clamp represents an example of a possible inactive conformation involving non-cleft contacts. Since different non-cleft contacts could act to retain different Pols, it may be possible for a single clamp to simultaneously recruit multiple Pols via this mechanism. Thus, the E. coli clamp may simultaneously bind more than two Pols (or partners), while PCNA may bind to more than three. Switching between these Pols could involve exchange of the Pol bound in the active conformation with Pols bound in inactive conformations, similar to mechanisms discussed above for dynamic processivity [179, 180]. Consistent with this model, Heltzel et al. recently determined that interaction of Pol IV with the rim of the clamp is dispensable for DNA replication activity, but is nevertheless required for switching with a stalled Pol III [133]. Thus, Pols may make certain contacts with the clamp that are specific to switching, and others that are dispensable for switching, but are required for stimulation of DNA replication (Fig. 2C). This model represents a radically different view than that portrayed by the traditional toolbelt model, invoking far more dynamic roles for both the clamp and Pols involved in the switching process, and explains how a single cleft on the clamp could support a Pol switch [133].
SLIDING CLAMP-DNA INTERACTIONS CONTRIBUTE TO POL SWITCHING
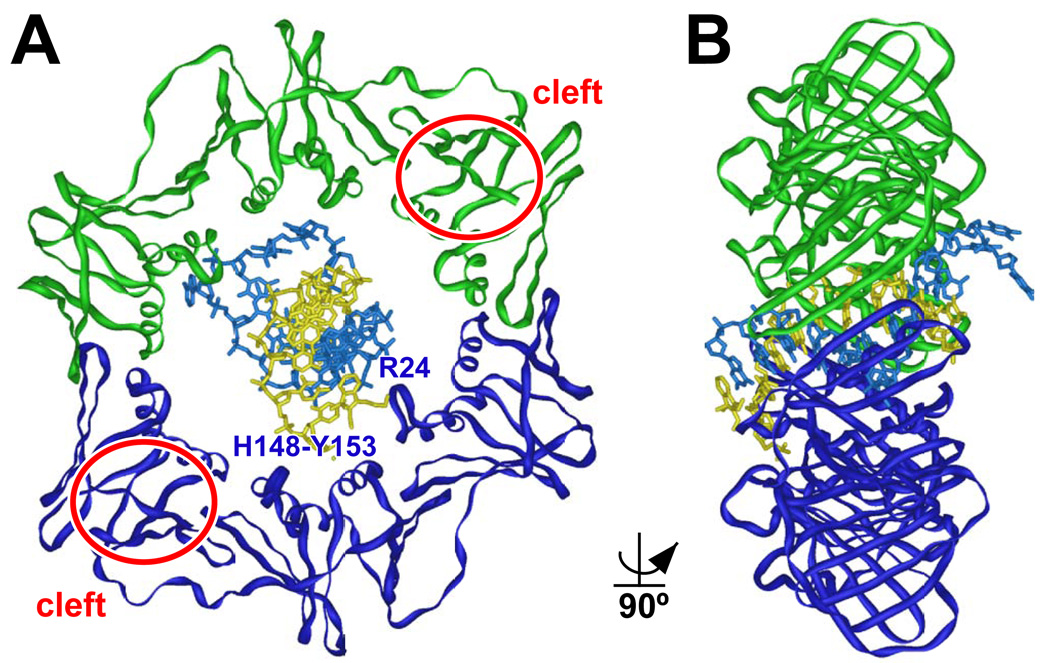
Recently, Georgescu et al. [92] described a crystal structure of the E. coli β sliding clamp on a short primed DNA. In this structure, three different surfaces of the clamp made specific contacts with the DNA template, including residues R24, H148-Q149-Y152-Y153, and the hydrophobic cleft. Residues R24, H148, Q149, Y152, and Y153 (among others) interacted with the dsDNA template, while the cleft contacted the ssDNA located downstream of the 3’-OH end of the primer (Fig. 4). As a result of these interactions, which appear to be conserved in PCNA [92], the DNA template passes through the clamp at an ~22° angle relative to the vertical plane of the clamp. Incidentally, one of the two clefts on β is closer to the DNA template. Hence, the Pol associated with this cleft would have the advantage in controlling the 3’-OH end of the DNA template relative to the Pol attached to the other, more distant cleft (Fig. 2B). In light of these results, Georgescu et al. [92] suggested that clamp-DNA interactions contributed to Pol switching in a toolbelt-like model by facilitating toggling of the clamp on the DNA template, granting the different Pols sequential access to the 3’-OH end of the DNA template.
FIGURE 4. Crystal structure of the E. coli β clamp on DNA.
This structure was solved by Georgescu et al. [92]. Positions of residues R24 and H148-Y153, which interact with dsDNA, are indicated, as are the locations of the hydrophobic clefts (red rings). (A) Front (C-side) and (B) side (90° rotation) views of the clamp-DNA complex are shown. The β clamp homodimer is depicted in green and blue, while dsDNA is gold and blue. This image was produced using imol and PDB coordinates 3BEP.
Heltzel et al. [94] recently characterized a mutant form of the β clamp bearing poly-alanine substitutions of residues H148-R152, which in the crystal made contact with the dsDNA template. Their results demonstrated that, in addition to contacting dsDNA, these residues were also required for physical and functional interactions with Pol II and Pol IV, implying that specialized Pols may compete with the DNA template for access to the clamp [94]. In contrast, these same residues did not contribute in a significant way to interaction of the clamp with Pol III, nor did they appear to be essential for Pol III replication in vivo or in vitro [94]. Thus, clamp-DNA interactions may regulate the ability of specialized Pols to contact the clamp. For example, the combination of Pol III and the DNA template (dsDNA and/or ssDNA) may act to effectively block sites on the clamp that specialized Pols must bind to in order to gains access to the replication fork and/or DNA template (Fig. 2D). In this case, conformational changes induced in Pol III and/or the clamp by contact with a DNA lesion may unmask one or more of these sites, enabling access of the specialized Pol to the DNA template. Thus, viewed in this way, clamp-DNA interactions may represent an additional level of regulation that helps to control the transition between active and inactive conformations of Pols bound to the clamp in a toolbelt-like model.
CONCLUDING REMARKS AND FUTURE PERSPECTIVES
As outlined in this review, remarkable progress has been made over the last ten years with respect to our understanding of how organisms manage the actions of their different Pols to enable switching between them during DNA transactions. Despite this progress, many important questions remain to be answered. For example, it is still unclear exactly where and when TLS take place. Are specialized Pols present at the fork during DNA replication to provide a capacity for TLS, or does the replisome prefer to ‘jump’ over lesions in the DNA, leaving ssDNA gaps that are repaired postreplicatively, as first suggested by Rupp & Howard-Flanders [167] more than 40 years ago? With respect to Pol switching, do combinations of different Pols actually switch via one or more ordered mechanisms, or does switching rely on maximizing the number of different Pols available at a replication fork to allow for stochastic sampling? Regardless of the answer to this question, it will also be important to define the contribution of non-cleft interactions to both Pol function and Pol switching. Another important question pertains to the roles played by clamp-DNA interactions: do certain Pols compete with the DNA template for interaction with the clamp, and if so, what effect do these competitive interactions have on Pol switching? Finally, it is important that we begin to define what happens to the structure of the clamp when different Pols bind to it. For example, do certain Pols interact cooperatively with the clamp, and if so, does this serve to impose a plastic hierarchy in clamp-Pol interactions? Based on the remarkable progress that has been made over the past ten years, it seems likely that many exciting and unexpected discoveries will be made over the next decade.
ACKNOWLEDGEMENTS
I thank Justin Heltzel for help with preparing Figure 3 and Figure 4, and both expert reviewers for their insightful comments. Work in the author’s lab is supported by grant GM066094 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Kenyon CJ, Walker GC. DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc Natl Acad Sci U S A. 1980;77:2819–2823. doi: 10.1073/pnas.77.5.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kannouche P, Fernandez de Henestrosa AR, Coull B, Vidal AE, Gray C, Zicha D, Woodgate R, Lehmann AR. Localization of DNA polymerases eta and iota to the replication machinery is tightly co-ordinated in human cells. EMBO J. 2003;22:1223–1233. doi: 10.1093/emboj/cdf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sutton MD, Opperman T, Walker GC. The Escherichia coli SOS mutagenesis proteins UmuD and UmuD' interact physically with the replicative DNA polymerase. Proc Natl Acad Sci U S A. 1999;96:12373–12378. doi: 10.1073/pnas.96.22.12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 5.McCulloch SD, Kokoska RJ, Masutani C, Iwai S, Hanaoka F, Kunkel TA. Preferential cis-syn thymine dimer bypass by DNA polymerase eta occurs with biased fidelity. Nature. 2004;428:97–100. doi: 10.1038/nature02352. [DOI] [PubMed] [Google Scholar]
- 6.McCulloch SD, Kokoska RJ, Kunkel TA. Efficiency, fidelity and enzymatic switching during translesion DNA synthesis. Cell Cycle. 2004;3:580–583. [PubMed] [Google Scholar]
- 7.Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, Prakash L, Prakash S, Todo T, Walker GC, Wang Z, Woodgate R. The Y-family of DNA polymerases. Mol Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 8.Godoy VG, Jarosz DF, Walker FL, Simmons LA, Walker GC. Y-family DNA polymerases respond to DNA damage-independent inhibition of replication fork progression. EMBO J. 2006;25:868–879. doi: 10.1038/sj.emboj.7600986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogi T, Lehmann AR. The Y-family DNA polymerase kappa (pol kappa) functions in mammalian nucleotide-excision repair. Nat Cell Biol. 2006;8:640–642. doi: 10.1038/ncb1417. [DOI] [PubMed] [Google Scholar]
- 10.McIlwraith MJ, Vaisman A, Liu Y, Fanning E, Woodgate R, West SC. Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol Cell. 2005;20:783–792. doi: 10.1016/j.molcel.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 11.Kawamoto T, Araki K, Sonoda E, Yamashita YM, Harada K, Kikuchi K, Masutani C, Hanaoka F, Nozaki K, Hashimoto N, Takeda S. Dual roles for DNA polymerase eta in homologous DNA recombination and translesion DNA synthesis. Mol Cell. 2005;20:793–799. doi: 10.1016/j.molcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 12.McDonald JP, Tissier A, Frank EG, Iwai S, Hanaoka F, Woodgate R. DNA polymerase iota and related rad30-like enzymes. Philos Trans R Soc Lond B Biol Sci. 2001;356:53–60. doi: 10.1098/rstb.2000.0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bebenek K, Tissier A, Frank EG, McDonald JP, Prasad R, Wilson SH, Woodgate R, Kunkel TA. 5’-Deoxyribose phosphate lyase activity of human DNA polymerase iota in vitro. Science. 2001;291:2156–2159. doi: 10.1126/science.1058386. [DOI] [PubMed] [Google Scholar]
- 14.Fersht AR. Fidelity of replication of phage phi X174 DNA by DNA polymerase III holoenzyme: spontaneous mutation by misincorporation. Proc Natl Acad Sci U S A. 1979;76:4946–4950. doi: 10.1073/pnas.76.10.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sloane DL, Goodman MF, Echols H. The fidelity of base selection by the polymerase subunit of DNA polymerase III holoenzyme. Nucleic Acids Res. 1988;16:6465–6475. doi: 10.1093/nar/16.14.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas DC, Roberts JD, Sabatino RD, Myers TW, Tan CK, Downey KM, So AG, Bambara RA, Kunkel TA. Fidelity of mammalian DNA replication and replicative DNA polymerases. Biochemistry. 1991;30:11751–11759. doi: 10.1021/bi00115a003. [DOI] [PubMed] [Google Scholar]
- 17.Jarosz DF, Godoy VG, Delaney JC, Essigmann JM, Walker GC. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature. 2006;439:225–228. doi: 10.1038/nature04318. [DOI] [PubMed] [Google Scholar]
- 18.Neeley WL, Delaney S, Alekseyev YO, Jarosz DF, Delaney JC, Walker GC, Essigmann JM. DNA polymerase V allows bypass of toxic guanine oxidation products in vivo. J Biol Chem. 2007;282:12741–12748. doi: 10.1074/jbc.M700575200. [DOI] [PubMed] [Google Scholar]
- 19.Abdulovic AL, Jinks-Robertson S. The in vivo characterization of translesion synthesis across UV-induced lesions in Saccharomyces cerevisiae: insights into Pol zeta- and Pol eta-dependent frameshift mutagenesis. Genetics. 2006;172:1487–1498. doi: 10.1534/genetics.105.052480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robbins JH, Kraemer KH, Flaxman BA. DNA repair in tumor cells from the variant form of xeroderma pigmentosum. J Invest Dermatol. 1975;64:150–155. doi: 10.1111/1523-1747.ep12533310. [DOI] [PubMed] [Google Scholar]
- 21.Robbins JH, Kraemer KH, Lutzner MA, Festoff BW, Coon HG. Xeroderma pigmentosum.An inherited diseases with sun sensitivity, multiple cutaneous neoplasms, and abnormal DNA repair. Ann Intern Med. 1974;80:221–248. doi: 10.7326/0003-4819-80-2-221. [DOI] [PubMed] [Google Scholar]
- 22.Masutani C, Araki M, Yamada A, Kusumoto R, Nogimori T, Maekawa T, Iwai S, Hanaoka F. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. Embo J. 1999;18:3491–3501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 24.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase,Poleta. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 25.Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 26.Sutton MD. The Escherichia coli dnaN159 mutant displays altered DNA polymerase usage and chronic SOS induction. J Bacteriol. 2004;186:6738–6748. doi: 10.1128/JB.186.20.6738-6748.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sutton MD, Duzen JM. Specific amino acid residues in the beta sliding clamp establish a DNA polymerase usage hierarchy in Escherichia coli. DNA Repair (Amst) 2006;5:312–323. doi: 10.1016/j.dnarep.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 28.Maul RW, Sutton MD. Roles of the Escherichia coli RecA protein and the global SOS response in effecting DNA polymerase selection in vivo. J Bacteriol. 2005;187:7607–7618. doi: 10.1128/JB.187.22.7607-7618.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Viguera E, Petranovic M, Zahradka D, Germain K, Ehrlich DS, Michel B. Lethality of bypass polymerases in Escherichia coli cells with a defective clamp loader complex of DNA polymerase III. Mol Microbiol. 2003;50:193–204. doi: 10.1046/j.1365-2958.2003.03658.x. [DOI] [PubMed] [Google Scholar]
- 30.Little JW, Edmiston SH, Pacelli LZ, Mount DW. Cleavage of the Escherichia coli lexA protein by the recA protease. Proc Natl Acad Sci U S A. 1980;77:3225–3229. doi: 10.1073/pnas.77.6.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics. 2001;158:41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo Y, Pfuetzner RA, Mosimann S, Paetzel M, Frey EA, Cherney M, Kim B, Little JW, Strynadka NC. Crystal structure of LexA: a conformational switch for regulation of self-cleavage. Cell. 2001;106:585–594. doi: 10.1016/s0092-8674(01)00479-2. [DOI] [PubMed] [Google Scholar]
- 33.Sassanfar M, Roberts JW. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J Mol Biol. 1990;212:79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- 34.Neher SB, Flynn JM, Sauer RT, Baker TA. Latent ClpX-recognition signals ensure LexA destruction after DNA damage. Genes Dev. 2003;17:1084–1089. doi: 10.1101/gad.1078003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Little JW. The SOS regulatory system: control of its state by the level of RecA protease. J Mol Biol. 1983;167:791–808. doi: 10.1016/s0022-2836(83)80111-9. [DOI] [PubMed] [Google Scholar]
- 36.Qiu Z, Goodman MF. The Escherichia coli polB locus is identical to dinA, the structural gene for DNA polymerase II. Characterization of Pol II purified from a polB mutant. J Biol Chem. 1997;272:8611–8617. doi: 10.1074/jbc.272.13.8611. [DOI] [PubMed] [Google Scholar]
- 37.Bonner CA, Hays S, McEntee K, Goodman MF. DNA polymerase II is encoded by the DNA damage-inducible dinA gene of Escherichia coli. Proc Natl Acad Sci U S A. 1990;87:7663–7667. doi: 10.1073/pnas.87.19.7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim SR, Matsui K, Yamada M, Gruz P, Nohmi T. Roles of chromosomal and episomal dinB genes encoding DNA pol IV in targeted and untargeted mutagenesis in Escherichia coli. Mol Genet Genomics. 2001;266:207–215. doi: 10.1007/s004380100541. [DOI] [PubMed] [Google Scholar]
- 39.Woodgate R, Ennis DG. Levels of chromosomally encoded Umu proteins and requirements for in vivo UmuD cleavage. Mol Gen Genet. 1991;229:10–16. doi: 10.1007/BF00264207. [DOI] [PubMed] [Google Scholar]
- 40.Elledge SJ, Walker GC. Proteins required for ultraviolet light and chemical mutagenesis. Identification of the products of the umuC locus of Escherichia coli. J Mol Biol. 1983;164:175–192. doi: 10.1016/0022-2836(83)90074-8. [DOI] [PubMed] [Google Scholar]
- 41.Frank EG, Ennis DG, Gonzalez M, Levine AS, Woodgate R. Regulation of SOS mutagenesis by proteolysis. Proc Natl Acad Sci U S A. 1996;93:10291–10296. doi: 10.1073/pnas.93.19.10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perry KL, Elledge SJ, Mitchell BB, Marsh L, Walker GC. umuDC and mucAB operons whose products are required for UV light- and chemical-induced mutagenesis: UmuD, MucA,and LexA proteins share homology. Proc Natl Acad Sci U S A. 1985;82:4331–4335. doi: 10.1073/pnas.82.13.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferentz AE, Walker GC, Wagner G. Converting a DNA damage checkpoint effector (UmuD2C) into a lesion bypass polymerase (UmuD'2C) Embo J. 2001;20:4287–4298. doi: 10.1093/emboj/20.15.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peat TS, Frank EG, McDonald JP, Levine AS, Woodgate R, Hendrickson WA. Structure of the UmuD' protein and its regulation in response to DNA damage. Nature. 1996;380:727–730. doi: 10.1038/380727a0. [DOI] [PubMed] [Google Scholar]
- 45.Shinagawa H, Iwasaki H, Kato T, Nakata A. RecA protein-dependent cleavage of UmuD protein and SOS mutagenesis. Proc Natl Acad Sci U S A. 1988;85:1806–1810. doi: 10.1073/pnas.85.6.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nohmi T, Battista JR, Dodson LA, Walker GC. RecA-mediated cleavage activates UmuD for mutagenesis: mechanistic relationship between transcriptional derepression and posttranslational activation. Proc Natl Acad Sci U S A. 1988;85:1816–1820. doi: 10.1073/pnas.85.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burckhardt SE, Woodgate R, Scheuermann RH, Echols H. UmuD mutagenesis protein of Escherichia coli: overproduction, purification, and cleavage by RecA. Proc Natl Acad Sci U S A. 1988;85:1811–1815. doi: 10.1073/pnas.85.6.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Battista JR, Ohta T, Nohmi T, Sun W, Walker GC. Dominant negative umuD mutations decreasing RecA-mediated cleavage suggest roles for intact UmuD in modulation of SOS mutagenesis. Proc Natl Acad Sci U S A. 1990;87:7190–7194. doi: 10.1073/pnas.87.18.7190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reuven NB, Tomer G, Livneh Z. The mutagenesis proteins UmuD' and UmuC prevent lethal frameshifts while increasing base substitution mutations. Mol Cell. 1998;2:191–199. doi: 10.1016/s1097-2765(00)80129-x. [DOI] [PubMed] [Google Scholar]
- 50.Tang M, Shen X, Frank EG, O'Donnell M, Woodgate R, Goodman MF. UmuD'(2)C is an error-prone DNA polymerase,Escherichia coli pol V. Proc Natl Acad Sci U S A. 1999;96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reuven NB, Arad G, Maor-Shoshani A, Livneh Z. The mutagenesis protein UmuC is a DNA polymerase activated by UmuD' RecA, and SSB and is specialized for translesion replication. J Biol Chem. 1999;274:31763–31766. doi: 10.1074/jbc.274.45.31763. [DOI] [PubMed] [Google Scholar]
- 52.Dutreix M, Moreau PL, Bailone A, Galibert F, Battista JR, Walker GC, Devoret R. New recA mutations that dissociate the various RecA protein activities in Escherichia coli provide evidence for an additional role for RecA protein in UV mutagenesis. J Bacteriol. 1989;171:2415–2423. doi: 10.1128/jb.171.5.2415-2423.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sweasy JB, Witkin EM, Sinha N, Roegner-Maniscalco V. RecA protein of Escherichia coli has a third essential role in SOS mutator activity. J Bacteriol. 1990;172:3030–3036. doi: 10.1128/jb.172.6.3030-3036.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pham P, Seitz EM, Saveliev S, Shen X, Woodgate R, Cox MM, Goodman MF. Two distinct modes of RecA action are required for DNA polymerase V-catalyzed translesion synthesis. Proc Natl Acad Sci U S A. 2002;99:11061–11066. doi: 10.1073/pnas.172197099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schlacher K, Leslie K, Wyman C, Woodgate R, Cox MM, Goodman MF. Goodman, DNA polymerase V and RecA protein, a minimal mutasome. Mol Cell. 2005;17:561–572. doi: 10.1016/j.molcel.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 56.Schlacher K, Cox MM, Woodgate R, Goodman MF. RecA acts in trans to allow replication of damaged DNA by DNA polymerase V. Nature. 2006;442:883–887. doi: 10.1038/nature05042. [DOI] [PubMed] [Google Scholar]
- 57.Fujii S, Gasser V, Fuchs RP. The biochemical requirements of DNA polymerase V-mediated translesion synthesis revisited. J Mol Biol. 2004;341:405–417. doi: 10.1016/j.jmb.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 58.Fujii S, Isogawa A, Fuchs RP. RecFOR proteins are essential for Pol V-mediated translesion synthesis and mutagenesis. EMBO J. 2006;25:5754–5763. doi: 10.1038/sj.emboj.7601474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kato T, Shinoura Y. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol Gen Genet. 1977;156:121–131. doi: 10.1007/BF00283484. [DOI] [PubMed] [Google Scholar]
- 60.Steinborn G. Uvm mutants of Escherichia coli K12 deficient in UV mutagenesis. I. Isolation of uvm mutants and their phenotypical characterization in DNA repair and mutagenesis. Mol Gen Genet. 1978;165:87–93. doi: 10.1007/BF00270380. [DOI] [PubMed] [Google Scholar]
- 61.Steinborn G. Uvm mutants of Escherichia coli K12 deficient in UV mutagenesis. II. Further evidence for a novel function in error-prone repair. Mol Gen Genet. 1979;175:203–208. doi: 10.1007/BF00425537. [DOI] [PubMed] [Google Scholar]
- 62.Napolitano R, Janel-Bintz R, Wagner J, Fuchs RP. All three SOS-inducible DNA polymerases Pol II, Pol IV and Pol V) are involved in induced mutagenesis. EMBO J. 2000;19:6259–6265. doi: 10.1093/emboj/19.22.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pham P, Rangarajan S, Woodgate R, Goodman MF. Roles of DNA polymerases V and II in SOS-induced error-prone and error-free repair in Escherichia coli. Proc Natl Acad Sci U S A. 2001;98:8350–8354. doi: 10.1073/pnas.111007198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berdichevsky A, Izhar L, Livneh Z. Error-free recombinational repair predominates over mutagenic translesion replication in E. coli. Mol Cell. 2002;10:917–924. doi: 10.1016/s1097-2765(02)00679-2. [DOI] [PubMed] [Google Scholar]
- 65.Wagner J, Etienne H, Janel-Bintz R, Fuchs RP. Genetics of mutagenesis in E. coli: various combinations of translesion polymerases (Pol II, IV and V) deal with lesion/sequence context diversity. DNA Repair (Amst) 2002;1:159–167. doi: 10.1016/s1568-7864(01)00012-x. [DOI] [PubMed] [Google Scholar]
- 66.Nowosielska A, Janion C, Grzesiuk E. Effect of deletion of SOS-induced polymerases, pol II, IV, V on spontaneous mutagenesis in Escherichia coli mutD5. Environ Mol Mutagen. 2004;43:226–234. doi: 10.1002/em.20019. [DOI] [PubMed] [Google Scholar]
- 67.Kuban W, Banach-Orlowska M, Schaaper RM, Jonczyk P, Fijalkowska IJ. Role of DNA polymerase IV in Escherichia coli SOS mutator activity. J Bacteriol. 2006;188:7977–7980. doi: 10.1128/JB.01088-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Al Mamun AA. Elevated expression of DNA polymerase II increases spontaneous mutagenesis in Escherichia coli. Mutat Res. 2007;625:29–39. doi: 10.1016/j.mrfmmm.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 69.Kuban W, Banach-Orlowska M, Bialoskorska M, Lipowska A, Schaaper RM, Jonczyk P, Fijalkowska IJ. Mutator phenotype resulting from DNA polymerase IV overproduction in Escherichia coli: preferential mutagenesis on the lagging strand. J Bacteriol. 2005;187:6862–6866. doi: 10.1128/JB.187.19.6862-6866.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Curti E, McDonald JP, Mead S, Woodgate R. DNA polymerase switching: effects on spontaneous mutagenesis in Escherichia coli. Mol Microbiol. 2009;71:315–331. doi: 10.1111/j.1365-2958.2008.06526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wolff E, Kim M, Hu K, Yang H, Miller JH. Polymerases leave fingerprints: analysis of the mutational spectrum in Escherichia coli rpoB to assess the role of polymerase IV in spontaneous mutation. J Bacteriol. 2004;186:2900–2905. doi: 10.1128/JB.186.9.2900-2905.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kai M, Wang TS. Checkpoint activation regulates mutagenic translesion synthesis. Genes Dev. 2003;17:64–76. doi: 10.1101/gad.1043203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Burgers PM, Kornberg A, Sakakibara Y. The dnaN gene codes for the beta subunit of DNA polymerase III holoenzyme of escherichia coli. Proc Natl Acad Sci U S A. 1981;78:5391–5395. doi: 10.1073/pnas.78.9.5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maul RW, Sanders LH, Lim JB, Benitez R, Sutton MD. Role of Escherichia coli DNA polymerase I in conferring viability upon the dnaN159 mutant strain. J Bacteriol. 2007;189:4688–4695. doi: 10.1128/JB.00476-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lopez de Saro FJ, Marinus MG, Modrich P, O'Donnell M. The beta sliding clamp binds to multiple sites within MutL and MutS. J Biol Chem. 2006;281:14340–14349. doi: 10.1074/jbc.M601264200. [DOI] [PubMed] [Google Scholar]
- 76.Lopez de Saro FJ, O'Donnell M. Interaction of the beta sliding clamp with MutS, ligase, and DNA polymerase I. Proc Natl Acad Sci U S A. 2001;98:8376–8380. doi: 10.1073/pnas.121009498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Becherel OJ, Fuchs RP, Wagner J. Pivotal role of the beta-clamp in translesion DNA synthesis and mutagenesis in E. coli cells. DNA Repair (Amst) 2002;1:703–708. doi: 10.1016/s1568-7864(02)00106-4. [DOI] [PubMed] [Google Scholar]
- 78.Wagner J, Fujii S, Gruz P, Nohmi T, Fuchs RP. The beta clamp targets DNA polymerase IV to DNA and strongly increases its processivity. EMBO Rep. 2000;1:484–488. doi: 10.1093/embo-reports/kvd109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bonner CA, Stukenberg PT, Rajagopalan M, Eritja R, O'Donnell M, McEntee K, Echols H, Goodman MF. Processive DNA synthesis by DNA polymerase II mediated by DNA polymerase III accessory proteins. J Biol Chem. 1992;267:11431–11438. [PubMed] [Google Scholar]
- 80.Hughes AJ, Jr, Bryan SK, Chen H, Moses RE, McHenry CS. Escherichia coli DNA polymerase II is stimulated by DNA polymerase III holoenzyme auxiliary subunits. J Biol Chem. 1991;266:4568–4573. [PubMed] [Google Scholar]
- 81.Lenne-Samuel N, Wagner J, Etienne H, Fuchs RP. The processivity factor beta controls DNA polymerase IV traffic during spontaneous mutagenesis and translesion synthesis in vivo. EMBO Rep. 2002;3:45–49. doi: 10.1093/embo-reports/kvf007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 83.Dionne I, Nookala RK, Jackson SP, Doherty AJ, Bell SD. A Heterotrimeric PCNA in the Hyperthermophilic Archaeon Sulfolobus solfataricus. Mol Cell. 2003;11:275–282. doi: 10.1016/s1097-2765(02)00824-9. [DOI] [PubMed] [Google Scholar]
- 84.Kong XP, Onrust R, O'Donnell M, Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 85.Krishna TS, Kong XP, Gary S, Burgers PM, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 86.Moarefi I, Jeruzalmi D, Turner J, O'Donnell M, Kuriyan J. Crystal structure of the DNA polymerase processivity factor of T4 bacteriophage. J Mol Biol. 2000;296:1215–1223. doi: 10.1006/jmbi.1999.3511. [DOI] [PubMed] [Google Scholar]
- 87.Argiriadi MA, Goedken ER, Bruck I, O'Donnell M, Kuriyan J. Crystal structure of a DNA polymerase sliding clamp from a Gram-positive bacterium. BMC Struct Biol. 2006;6:2. doi: 10.1186/1472-6807-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dalrymple BP, Kongsuwan K, Wijffels G, Dixon NE, Jennings PA. A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. Proc Natl Acad Sci U S A. 2001;98:11627–11632. doi: 10.1073/pnas.191384398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jeruzalmi D, Yurieva O, Zhao Y, Young M, Stewart J, Hingorani M, O'Donnell M, Kuriyan J. Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E. coli DNA polymerase III. Cell. 2001;106:417–428. [PubMed] [Google Scholar]
- 90.Burnouf DY, Olieric V, Wagner J, Fujii S, Reinbolt J, Fuchs RP, Dumas P. Structural and biochemical analysis of sliding clamp/ligand interactions suggest a competition between replicative and translesion DNA polymerases. J Mol Biol. 2004;335:1187–1197. doi: 10.1016/j.jmb.2003.11.049. [DOI] [PubMed] [Google Scholar]
- 91.Bunting KA, Roe SM, Pearl LH. Structural basis for recruitment of translesion DNA polymerase Pol IV/DinB to the beta-clamp. EMBO J. 2003;22:5883–5892. doi: 10.1093/emboj/cdg568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Georgescu RE, Kim SS, Yurieva O, Kuriyan J, Kong XP, O'Donnell M. Structure of a sliding clamp on DNA. Cell. 2008;132:43–54. doi: 10.1016/j.cell.2007.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ivanov I, Chapados BR, McCammon JA, Tainer JA. Proliferating cell nuclear antigen loaded onto double-stranded DNA: dynamics, minor groove interactions and functional implications. Nucleic Acids Res. 2006;34:6023–6033. doi: 10.1093/nar/gkl744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Heltzel JMH, Scouten Ponticelli SK, Sanders LH, Duzen JM, Cody V, Pace J, Snell EH, Sutton MD. Sliding clamp-DNA interactions are required for viability and contribute to DNA polymerase management in Escherichia coli. Journal of Molecular Biology. 2009;387:74–91. doi: 10.1016/j.jmb.2009.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Indiani C, O'Donnell M. The replication clamp-loading machine at work in the three domains of life. Nat Rev Mol Cell Biol. 2006;7:751–761. doi: 10.1038/nrm2022. [DOI] [PubMed] [Google Scholar]
- 96.Jeruzalmi D, O'Donnell M, Kuriyan J. Crystal structure of the processivity clamp loader gamma (gamma) complex of E. coli DNA polymerase III. Cell. 2001;106:429–441. doi: 10.1016/s0092-8674(01)00463-9. [DOI] [PubMed] [Google Scholar]
- 97.Kazmirski SL, Zhao Y, Bowman GD, O'Donnell M, Kuriyan J. Out-of-plane motions in open sliding clamps: molecular dynamics simulations of eukaryotic and archaeal proliferating cell nuclear antigen. Proc Natl Acad Sci U S A. 2005;102:13801–13806. doi: 10.1073/pnas.0506430102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Leu FP, Hingorani MM, Turner J, O'Donnell M. The delta subunit of DNA polymerase III holoenzyme serves as a sliding clamp unloader in Escherichia coli. J Biol Chem. 2000;275:34609–34618. doi: 10.1074/jbc.M005495200. [DOI] [PubMed] [Google Scholar]
- 99.Sutton MD, Duzen JM, Maul RW. Mutant forms of the Escherichia coli beta sliding clamp that distinguish between its roles in replication and DNA polymerase V-dependent translesion DNA synthesis. Mol Microbiol. 2005;55:1751–1766. doi: 10.1111/j.1365-2958.2005.04500.x. [DOI] [PubMed] [Google Scholar]
- 100.Ponticelli SH, Ponticelli J, Duzen M, Sutton MD. Contributions of the individual hydrophobic clefts of the Escherichia coli beta sliding clamp to clamp loading, DNA rpelication, and clamp recycling. Nucleics Acid Research. 2009;37:2796–2809. doi: 10.1093/nar/gkp128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cai J, Gibbs E, Uhlmann F, Phillips B, Yao N, O'Donnell M, Hurwitz J. A complex consisting of human replication factor C p40, p37, and p36 subunits is a DNA-dependent ATPase and an intermediate in the assembly of the holoenzyme. J Biol Chem. 1997;272:18974–18981. doi: 10.1074/jbc.272.30.18974. [DOI] [PubMed] [Google Scholar]
- 102.Xu H, Zhang P, Liu L, Lee MY. A novel PCNA-binding motif identified by the panning of a random peptide display library. Biochemistry. 2001;40:4512–4520. doi: 10.1021/bi010103+. [DOI] [PubMed] [Google Scholar]
- 103.Dohrmann PR, McHenry CS. A bipartite polymerase-processivity factor interaction: only the internal beta binding site of the alpha subunit is required for processive replication by the DNA polymerase III holoenzyme. J Mol Biol. 2005;350:228–239. doi: 10.1016/j.jmb.2005.04.065. [DOI] [PubMed] [Google Scholar]
- 104.Haracska L, Acharya N, Unk I, Johnson RE, Hurwitz J, Prakash L, Prakash S. A single domain in human DNA polymerase iota mediates interaction with PCNA: implications for translesion DNA synthesis. Mol Cell Biol. 2005;25:1183–1190. doi: 10.1128/MCB.25.3.1183-1190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Guo C, Sonoda E, Tang TS, Parker JL, Bielen AB, Takeda S, Ulrich HD, Friedberg EC. REV1 protein interacts with PCNA: significance of the REV1 BRCT domain in vitro and in vivo. Mol Cell. 2006;23:265–271. doi: 10.1016/j.molcel.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 106.Vidal AE, Kannouche P, Podust VN, Yang W, Lehmann AR, Woodgate R. Proliferating cell nuclear antigen-dependent coordination of the biological functions of human DNA polymerase iota. J Biol Chem. 2004;279:48360–48368. doi: 10.1074/jbc.M406511200. [DOI] [PubMed] [Google Scholar]
- 107.Haracska L, Johnson RE, Unk I, Phillips B, Hurwitz J, Prakash L, Prakash S. Physical and functional interactions of human DNA polymerase eta with PCNA. Mol Cell Biol. 2001;21:7199–7206. doi: 10.1128/MCB.21.21.7199-7206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Haracska L, Kondratick CM, Unk I, Prakash S, Prakash L. Interaction with PCNA is essential for yeast DNA polymerase eta function. Mol Cell. 2001;8:407–415. doi: 10.1016/s1097-2765(01)00319-7. [DOI] [PubMed] [Google Scholar]
- 109.Bi X, Barkley LR, Slater DM, Tateishi S, Yamaizumi M, Ohmori H, Vaziri C. Rad18 regulates DNA polymerase kappa and is required for recovery from S-phase checkpoint-mediated arrest. Mol Cell Biol. 2006;26:3527–3540. doi: 10.1128/MCB.26.9.3527-3540.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Haracska L, Unk I, Johnson RE, Phillips BB, Hurwitz J, Prakash L, Prakash S. Stimulation of DNA synthesis activity of human DNA polymerase kappa by PCNA. Mol Cell Biol. 2002;22:784–791. doi: 10.1128/MCB.22.3.784-791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang SJ, Zeng XR, Zhang P, Toomey NL, Chuang RY, Chang LS, Lee MY. A conserved region in the amino terminus of DNA polymerase delta is involved in proliferating cell nuclear antigen binding. J Biol Chem. 1995;270:7988–7992. doi: 10.1074/jbc.270.14.7988. [DOI] [PubMed] [Google Scholar]
- 112.Bertram JG, Bloom LB, O'Donnell M, Goodman MF. Increased dNTP binding affinity reveals a nonprocessive role for Escherichia coli beta clamp with DNA polymerase IV. J Biol Chem. 2004;279:33047–33050. doi: 10.1074/jbc.C400265200. [DOI] [PubMed] [Google Scholar]
- 113.Maga G, Hubscher U. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J Cell Sci. 2003;116:3051–3060. doi: 10.1242/jcs.00653. [DOI] [PubMed] [Google Scholar]
- 114.Naktinis V, Turner J, O'Donnell M. A molecular switch in a replication machine defined by an internal competition for protein rings. Cell. 1996;84:137–145. doi: 10.1016/s0092-8674(00)81000-4. [DOI] [PubMed] [Google Scholar]
- 115.Xiao W, Chow BL, Broomfield S, Hanna M. The Saccharomyces cerevisiae RAD6 group is composed of an error-prone and two error-free postreplication repair pathways. Genetics. 2000;155:1633–1641. doi: 10.1093/genetics/155.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 117.Chiu RK, Brun J, Ramaekers C, Theys J, Weng L, Lambin P, Gray DA, Wouters BG. Lysine 63-polyubiquitination guards against translesion synthesis-induced mutations. PLoS Genet. 2006;2:e116. doi: 10.1371/journal.pgen.0020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Solomon DA, Cardoso MC, Knudsen ES. Dynamic targeting of the replication machinery to sites of DNA damage. J Cell Biol. 2004;166:455–463. doi: 10.1083/jcb.200312048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Soria G, Podhajcer O, Prives C, Gottifredi V. P21Cip1/WAF1 downregulation is required for efficient PCNA ubiquitination after UV irradiation. Oncogene. 2006;25:2829–2838. doi: 10.1038/sj.onc.1209315. [DOI] [PubMed] [Google Scholar]
- 120.Frampton J, Irmisch A, Green CM, Neiss A, Trickey M, Ulrich HD, Furuya K, Watts FZ, Carr AM, Lehmann AR. Postreplication repair and PCNA modification in Schizosaccharomyces pombe. Mol Biol Cell. 2006;17:2976–2985. doi: 10.1091/mbc.E05-11-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Leach CA, Michael WM. Ubiquitin/SUMO modification of PCNA promotes replication fork progression in Xenopus laevis egg extracts. J Cell Biol. 2005;171:947–954. doi: 10.1083/jcb.200508100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Simpson LJ, Ross AL, Szuts D, Alviani CA, Oestergaard VH, Patel KJ, Sale JE. RAD18-independent ubiquitination of proliferating-cell nuclear antigen in the avian cell line DT40. EMBO Rep. 2006;7:927–932. doi: 10.1038/sj.embor.7400777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, Gygi SP, Ploegh HL, Bernards R, D'Andrea AD. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8:339–347. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 124.Avkin S, Sevilya Z, Toube L, Geacintov N, Chaney SG, Oren M, Livneh Z. p53 and p21 regulate error-prone DNA repair to yield a lower mutation load. Mol Cell. 2006;22:407–413. doi: 10.1016/j.molcel.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 125.Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, Hofmann K, Dikic I. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 126.Bailly V, Lauder S, Prakash S, Prakash L. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J Biol Chem. 1997;272:23360–23365. doi: 10.1074/jbc.272.37.23360. [DOI] [PubMed] [Google Scholar]
- 127.Opperman T, Murli S, Smith BT, Walker GC. A model for a umuDC-dependent prokaryotic DNA damage checkpoint. Proc Natl Acad Sci U S A. 1999;96:9218–9223. doi: 10.1073/pnas.96.16.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sutton MD, Farrow MF, Burton BM, Walker GC. Genetic interactions between the Escherichia coli umuDC gene products and the beta processivity clamp of the replicative DNA polymerase. J Bacteriol. 2001;183:2897–2909. doi: 10.1128/JB.183.9.2897-2909.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sutton MD, Murli S, Opperman T, Klein C, Walker GC. umuDC-dnaQ Interaction and its implications for cell cycle regulation and SOS mutagenesis in Escherichia coli. J Bacteriol. 2001;183:1085–1089. doi: 10.1128/JB.183.3.1085-1089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Uchida K, Furukohri A, Shinozaki Y, Mori T, Ogawara D, Kanaya S, Nohmi T, Maki H, Akiyama M. Overproduction of Escherichia coli DNA polymerase DinB (Pol IV) inhibits replication fork progression and is lethal. Mol Microbiol. 2008;70:608–622. doi: 10.1111/j.1365-2958.2008.06423.x. [DOI] [PubMed] [Google Scholar]
- 131.Furukohri A, Goodman MF, Maki H. A dynamic polymerase exchange with Escherichia coli DNA polymerase IV replacing DNA polymerase III on the sliding clamp. J Biol Chem. 2008;283:11260–11269. doi: 10.1074/jbc.M709689200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Indiani C, McInerney P, Georgescu R, Goodman MF, O'Donnell M. A sliding-clamp toolbelt binds high- and low-fidelity DNA polymerases simultaneously. Mol Cell. 2005;19:805–815. doi: 10.1016/j.molcel.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 133.Heltzel JMH, Maul RW, Scouten Ponticelli SH, Sutton MD. A model for DNA polymerase switching involving a single cleft and the rim of the sliding clamp. Proc Natl Acad Sci U S A. doi: 10.1073/pnas.0903460106. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kannouche P, Fernandez de Henestrosa AR, Coull B, Vidal AE, Gray C, Zicha D, Woodgate R, Lehmann AR. Localization of DNA polymerases eta and iota to the replication machinery is tightly co-ordinated in human cells. EMBO J. 2002;21:6246–6256. doi: 10.1093/emboj/cdf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.D'Souza S, Walker GC. Novel role for the C terminus of Saccharomyces cerevisiae Rev1 in mediating protein-protein interactions. Mol Cell Biol. 2006;26:8173–8182. doi: 10.1128/MCB.00202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Guo C, Fischhaber PL, Luk-Paszyc MJ, Masuda Y, Zhou J, Kamiya K, Kisker C, Friedberg EC. Mouse Rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J. 2003;22:6621–6630. doi: 10.1093/emboj/cdg626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lawrence CW, Christensen R. UV mutagenesis in radiation-sensitive strains of yeast. Genetics. 1976;82:207–232. doi: 10.1093/genetics/82.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lemontt JF. Genetic and physiological factors affecting repair and mutagenesis in yeast. Basic Life Sci. 1980;15:85–120. doi: 10.1007/978-1-4684-3842-0_7. [DOI] [PubMed] [Google Scholar]
- 139.Nelson JR, Gibbs PE, Nowicka AM, Hinkle DC, Lawrence CW. Evidence for a second function for Saccharomyces cerevisiae Rev1p. Mol Microbiol. 2000;37:549–554. doi: 10.1046/j.1365-2958.2000.01997.x. [DOI] [PubMed] [Google Scholar]
- 140.Becherel OJ, Fuchs RP. Mechanism of DNA polymerase II-mediated frameshift mutagenesis. Proc Natl Acad Sci U S A. 2001;98:8566–8571. doi: 10.1073/pnas.141113398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shachar S, Ziv O, Avkin S, Adar S, Wittschieben J, Reissner T, Chaney S, Friedberg EC, Wang Z, Carell T, Geacintov N, Livneh Z. Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J. 2009;28:383–393. doi: 10.1038/emboj.2008.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yuan F, Yanbin Zhang Y, Rajpal DK, Wu X, Guo D, Wang M, Taylor J-s, Wang Z. Specificity of DNA Lesion Bypass by the Yeast DNA Polymerase h. J Biol Chem. 2000;275:8233–8239. doi: 10.1074/jbc.275.11.8233. [DOI] [PubMed] [Google Scholar]
- 143.Caillet-Fauquet P, Maenhaut-Michel G. Nature of the SOS mutator activity: genetic characterization of untargeted mutagenesis in Escherichia coli. Mol Gen Genet. 1988;213:491–498. doi: 10.1007/BF00339621. [DOI] [PubMed] [Google Scholar]
- 144.Fujii S, Fuchs RP. Defining the position of the switches between replicative and bypass DNA polymerases. EMBO J. 2004;23:4342–4352. doi: 10.1038/sj.emboj.7600438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Johnson KA. Conformational coupling in DNA polymerase fidelity. Annu Rev Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- 146.McHenry CS. Chromosomal replicases as asymmetric dimers: studies of subunit arrangement and functional consequences. Mol Microbiol. 2003;49:1157–1165. doi: 10.1046/j.1365-2958.2003.03645.x. [DOI] [PubMed] [Google Scholar]
- 147.Naktinis V, Onrust R, Fang L, O'Donnell M. Assembly of a chromosomal replication machine:two DNA polymerases, a clamp loader, and sliding clamps in one holoenzyme particle. II. Intermediate complex between the clamp loader and its clamp. J Biol Chem. 1995;270:13358–13365. [PubMed] [Google Scholar]
- 148.Onrust R, Finkelstein J, Naktinis V, Turner J, Fang L, O'Donnell M. Assembly of a chromosomal replication machine: two DNA polymerases, a clamp loader, and sliding clamps in one holoenzyme particle. I. Organization of the clamp loader. J Biol Chem. 1995;270:13348–13357. doi: 10.1074/jbc.270.22.13348. [DOI] [PubMed] [Google Scholar]
- 149.Onrust R, Finkelstein J, Turner J, Naktinis V, O'Donnell M. Assembly of a chromosomal replication machine: two DNA polymerases, a clamp loader, and sliding clamps in one holoenzyme particle. III. Interface between two polymerases and the clamp loader. J Biol Chem. 1995;270:13366–13377. doi: 10.1074/jbc.270.22.13366. [DOI] [PubMed] [Google Scholar]
- 150.Stukenberg PT, O'Donnell M. Assembly of a chromosomal replication machine: two DNA polymerases, a clamp loader, and sliding clamps in one holoenzyme particle. V. Four different polymerase-clamp complexes on DNA. J Biol Chem. 1995;270:13384–13391. doi: 10.1074/jbc.270.22.13384. [DOI] [PubMed] [Google Scholar]
- 151.Dervyn E, Suski C, Daniel R, Bruand C, Chapuis J, Errington J, Janniere L, Ehrlich SD. Two essential DNA polymerases at the bacterial replication fork. Science. 2001;294:1716–1719. doi: 10.1126/science.1066351. [DOI] [PubMed] [Google Scholar]
- 152.Carr KM, Kaguni JM. Stoichiometry of DnaA and DnaB protein in initiation at the Escherichia coli chromosomal origin. J Biol Chem. 2001;276:44919–44925. doi: 10.1074/jbc.M107463200. [DOI] [PubMed] [Google Scholar]
- 153.Fang L, Davey MJ, O'Donnell M. Replisome assembly at oriC, the replication origin of E. coli, reveals an explanation for initiation sites outside an origin. Mol Cell. 1999;4:541–553. doi: 10.1016/s1097-2765(00)80205-1. [DOI] [PubMed] [Google Scholar]
- 154.Tougu K, Marians KJ. The interaction between helicase and primase sets the replication fork clock. J Biol Chem. 1996;271:21398–21405. doi: 10.1074/jbc.271.35.21398. [DOI] [PubMed] [Google Scholar]
- 155.Tougu K, Marians KJ. The extreme C terminus of primase is required for interaction with DnaB at the replication fork. J Biol Chem. 1996;271:21391–21397. doi: 10.1074/jbc.271.35.21391. [DOI] [PubMed] [Google Scholar]
- 156.Yuzhakov A, Kelman Z, O'Donnell M. Trading places on DNA--a three-point switch underlies primer handoff from primase to the replicative DNA polymerase. Cell. 1999;96:153–163. doi: 10.1016/s0092-8674(00)80968-x. [DOI] [PubMed] [Google Scholar]
- 157.Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 158.Machida Y, Okazaki T, Okazaki R. Discontinuous replication of replicative form DNA from bacteriophage phiX174. Proc Natl Acad Sci U S A. 1977;74:2776–2779. doi: 10.1073/pnas.74.7.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sugino A, Hirose S, Okazaki R. RNA-linked nascent DNA fragments in Escherichia coli. Proc Natl Acad Sci U S A. 1972;69:1863–1867. doi: 10.1073/pnas.69.7.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sugino A, Okazaki R. RNA-linked DNA fragments in vitro. Proc Natl Acad Sci U S A. 1973;70:88–92. doi: 10.1073/pnas.70.1.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Alberts BM, Morris C, Mace D, Sinha N, Bittner M, Moran L. DNA Synthesis and Regulation. In: Goulian M, Hanawalt PC, Fox CF, editors. Menlo Park, CA: W. A. Benjamin; 1975. pp. 241–269. [Google Scholar]
- 162.Lopez de Saro FJ, Georgescu RE, O'Donnell M. A peptide switch regulates DNA polymerase processivity. Proc Natl Acad Sci U S A. 2003;100:14689–14694. doi: 10.1073/pnas.2435454100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Leu FP, Georgescu R, O'Donnell M. Mechanism of the E. coli tau processivity switch during lagging-strand synthesis. Mol Cell. 2003;11:315–327. doi: 10.1016/s1097-2765(03)00042-x. [DOI] [PubMed] [Google Scholar]
- 164.Pomerantz RT, O'Donnell M. The replisome uses mRNA as a primer after colliding with RNA polymerase. Nature. 2008;456:762–766. doi: 10.1038/nature07527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Banach-Orlowska M, Fijalkowska IJ, Schaaper RM, Jonczyk P. DNA polymerase II as a fidelity factor in chromosomal DNA synthesis in Escherichia coli. Mol Microbiol. 2005;58:61–70. doi: 10.1111/j.1365-2958.2005.04805.x. [DOI] [PubMed] [Google Scholar]
- 166.Heller RC, Marians KJ. Replisome assembly and the direct restart of stalled replication forks. Nat Rev Mol Cell Biol. 2006;7:932–943. doi: 10.1038/nrm2058. [DOI] [PubMed] [Google Scholar]
- 167.Rupp WD, Howard-Flanders P. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J Mol Biol. 1968;31:291–304. doi: 10.1016/0022-2836(68)90445-2. [DOI] [PubMed] [Google Scholar]
- 168.Grosse F, Krauss G. The primase activity of DNA polymerase alpha from calf thymus. J Biol Chem. 1985;260:1881–1888. [PubMed] [Google Scholar]
- 169.Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Tsurimoto T, Melendy T, Stillman B. Sequential initiation of lagging and leading strand synthesis by two different polymerase complexes at the SV40 DNA replication origin. Nature. 1990;346:534–539. doi: 10.1038/346534a0. [DOI] [PubMed] [Google Scholar]
- 172.Dornreiter I, Erdile LF, Gilbert IU, von Winkler D, Kelly TJ, Fanning E. Interaction of DNA polymerase alpha-primase with cellular replication protein A and SV40 T antigen. EMBO J. 1992;11:769–776. doi: 10.1002/j.1460-2075.1992.tb05110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Yuzhakov A, Kelman Z, Hurwitz J, O'Donnell M. Multiple competition reactions for RPA order the assembly of the DNA polymerase delta holoenzyme. EMBO J. 1999;18:6189–6199. doi: 10.1093/emboj/18.21.6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Tsurimoto T, Stillman B. Replication factors required for SV40 DNA replication in vitro. II. Switching of DNA polymerase alpha and delta during initiation of leading and lagging strand synthesis. J Biol Chem. 1991;266:1961–1968. [PubMed] [Google Scholar]
- 175.Rossi ML, Bambara RA. Reconstituted Okazaki fragment processing indicates two pathways of primer removal. J Biol Chem. 2006;281:26051–26061. doi: 10.1074/jbc.M604805200. [DOI] [PubMed] [Google Scholar]
- 176.Ayyagari R, Gomes XV, Gordenin DA, Burgers PM. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 AND DNA2. J Biol Chem. 2003;278:1618–1625. doi: 10.1074/jbc.M209801200. [DOI] [PubMed] [Google Scholar]
- 177.Barnes DE, Tomkinson AE, Lehmann AR, Webster AD, Lindahl T. Mutations in the DNA ligase I gene of an individual with immunodeficiencies and cellular hypersensitivity to DNA-damaging agents. Cell. 1992;69:495–503. doi: 10.1016/0092-8674(92)90450-q. [DOI] [PubMed] [Google Scholar]
- 178.Henricksen LA, Veeraraghavan J, Chafin DR, Bambara RA. DNA ligase I competes with FEN1 to expand repetitive DNA sequences in vitro. J Biol Chem. 2002;277:22361–22369. doi: 10.1074/jbc.M201765200. [DOI] [PubMed] [Google Scholar]
- 179.Yang J, Zhuang Z, Roccasecca RM, Trakselis MA, Benkovic SJ. The dynamic processivity of the T4 DNA polymerase during replication. Proc Natl Acad Sci U S A. 2004;101:8289–8294. doi: 10.1073/pnas.0402625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hamdan SM, Johnson DE, Tanner NA, Lee JB, Qimron U, Tabor S, van Oijen AM, Richardson CC. Dynamic DNA helicase-DNA polymerase interactions assure processive replication fork movement. Mol Cell. 2007;27:539–549. doi: 10.1016/j.molcel.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 181.Pritchard AE, Dallmann HG, Glover BP, McHenry CS. A novel assembly mechanism for the DNA polymerase III holoenzyme DnaX complex: association of deltadelta' with DnaX(4) forms DnaX(3)deltadelta'. EMBO J. 2000;19:6536–6545. doi: 10.1093/emboj/19.23.6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.McInerney P, Johnson A, Katz F, O'Donnell M. Characterization of a triple DNA polymerase replisome. Mol Cell. 2007;27:527–538. doi: 10.1016/j.molcel.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 183.Nossal NG, Makhov AM, Chastain PD, 2nd, Jones CE, Griffith JD. Architecture of the bacteriophage T4 replication complex revealed with nanoscale biopointers. J Biol Chem. 2007;282:1098–1108. doi: 10.1074/jbc.M606772200. [DOI] [PubMed] [Google Scholar]
- 184.Pages V, Fuchs RP. How DNA lesions are turned into mutations within cells? Oncogene. 2002;21:8957–8966. doi: 10.1038/sj.onc.1206006. [DOI] [PubMed] [Google Scholar]
- 185.Maul RW, Ponticelli SK, Duzen JM, Sutton MD. Differential binding of Escherichia coli DNA polymerases to the beta-sliding clamp. Mol Microbiol. 2007;65:811–827. doi: 10.1111/j.1365-2958.2007.05828.x. [DOI] [PubMed] [Google Scholar]
- 186.Xing G, Kirouac K, Shin YJ, Bell SD, Ling H. Structural insight into recruitment of translesion DNA polymerase Dpo4 to sliding clamp PCNA. Mol Microbiol. 2009;71:678–691. doi: 10.1111/j.1365-2958.2008.06553.x. [DOI] [PubMed] [Google Scholar]
- 187.Burgers PM, Koonin EV, Bruford E, Blanco L, Burtis KC, Christman MF, Copeland WC, Friedberg EC, Hanaoka F, Hinkle DC, Lawrence CW, Nakanishi M, Ohmori H, Prakash L, Prakash S, Reynaud CA, Sugino A, Todo T, Wang Z, Weill JC, Woodgate R. Eukaryotic DNA polymerases: proposal for a revised nomenclature. J Biol Chem. 2001;276:43487–43490. doi: 10.1074/jbc.R100056200. [DOI] [PubMed] [Google Scholar]
- 188.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. 2nd. Washington, D.C: ASM Press; 2006. DNA repair and mutagenesis. [Google Scholar]