Abstract
Reactive metabolites formed from benzene include benzene oxide, trans, trans muconaldehyde, quinones, thiol adducts, phenolic metabolites and oxygen radicals. Susceptibility to the toxic effects of benzene has been suggested to occur partly because of polymorphisms in enzymes involved in benzene metabolism which include cytochrome P450 2E1, epoxide hydrolases, myeloperoxidase, glutathione-S-transferases and quinone reductases. However, susceptibility factors not directly linked to benzene metabolism have also been associated with its toxicity and include p53, proteins involved in DNA repair, genomic stability and expression of cytokines and/or cell adhesion molecules. In this work, we examine potential relationships between metabolic and non-metabolic susceptibility factors using the enzyme NAD(P)H:quinone oxidoreductase (NQO1) as an example. NQO1 may also impact pathways in addition to metabolism of quinones due to protein-protein interactions or other mechanisms related to NQO1 activity. NQO1 has been implicated in stabilizing p53 and in maintaining microtubule integrity. Inhibition or knockdown of NQO1 in bone marrow endothelial cells has been found to lead to deficiencies of E-selectin, ICAM-1 and VCAM-1 adhesion molecule expression after TNFα stimulation. These examples illustrate how the metabolic susceptibility factor NQO1 may influence non-metabolic susceptibility pathways for benzene toxicity.
Keywords: Benzene, metabolic susceptibility factors, p53, NQO1, adhesion molecules, bone marrow endothelial cells
Metabolic factors in susceptibility to benzene toxicity
Benzene induces hematopoietic toxicity and can induce aplastic anemia, myelodysplasia and acute myeloid leukemia after chronic exposure [1;2]. The metabolism of benzene has been investigated extensively and previous reviews have characterized benzene metabolism in a comprehensive manner [3-7]. Consequently, this work is not intended to be a review of benzene metabolism but will focus on metabolic susceptibility factors for benzene toxicity which have been identified in both cell and animal studies and in studies of occupationally-exposed populations. Other susceptibility factors not directly linked to metabolism have also been identified in benzene toxicity and relationships between metabolic and non-metabolic susceptibility factors have not been previously considered. We will therefore discuss potential relationships between these two groups of susceptibility factors using the enzyme NAD(P)H:quinone oxidoreductase 1 (NQO1) as an example and highlight recent studies focusing on NQO1 in human bone marrow endothelial cells.
Benzene metabolism
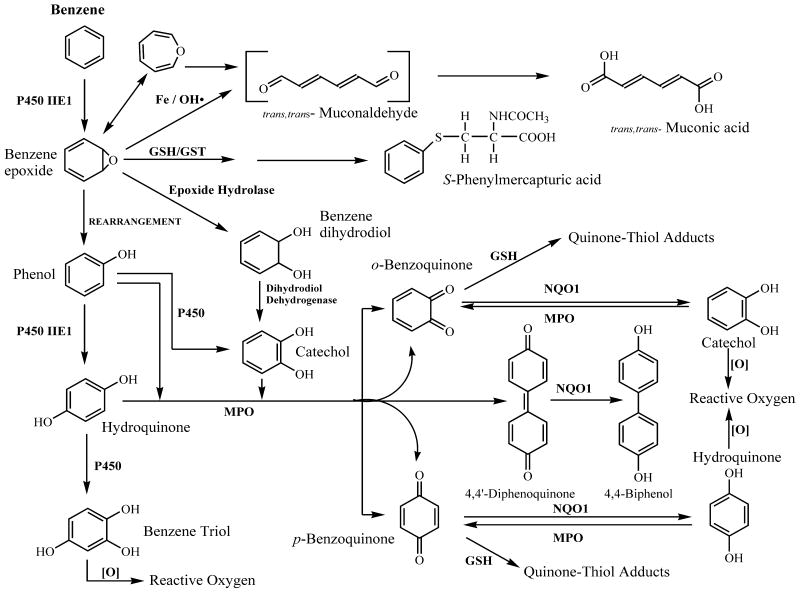
Metabolism of benzene is considered necessary for benzene toxicity and the evidence supporting this conclusion has been previously summarized [8-10]. A key finding in animal studies was that knockout of the first step in benzene metabolism mediated by cytochrome P450 2E1 totally abrogated benzene-induced myeloid toxicity and cytotoxicity [11]. Benzene metabolism in liver and in-situ in bone marrow could both conceivably contribute to benzene induced myeloid toxicity [10]. A simplified version of benzene metabolism is shown in Figure 1 where the majority of Phase II metabolic pathways including sulfation and glucuronidation have been omitted. It is important to note however that some phase II metabolites such as sulfate conjugates have been suggested as carrier forms of phenolic metabolites which are released in-situ in bone marrow due to a high concentration of sulfatase enzymes and a low content of sulfotransferases [12].
Figure 1. Benzene metabolic scheme.
Most Phase II pathways have been omitted. For potential reactive metabolites, see Table 1. For metabolic susceptibility factors, see Table 2. Adapted from [7],[10].
Reactive metabolites and metabolic susceptibility factors
Reactive metabolites formed from benzene include benzene epoxide [13-15], trans, trans muconaldehyde [16-19], phenolic metabolites of benzene [20-22] which can give rise to oxygen radicals upon autoxidation [23;24], reactive quinones and semiquinones formed from polyphenolic metabolites of benzene [25-28] [29] and quinone thiol adducts [30;31] (Table 1). Consequently, the metabolism of benzene is complex and gives rise to a large number of potentially reactive products which have been suggested to be important in benzene toxicity. Metabolic susceptibility factors (Table 2) have been identified in cellular studies, animals and in studies of occupationally-exposed human populations. Such susceptibility factors predictably encompass the wide range of benzene metabolic pathways and both phenotypic and genotypic variants of enzymes in these pathways have been investigated in epidemiological studies of benzene toxicity. The first step in benzene metabolism mediated by CYP2E1 represents a key metabolic susceptibility factor [11]. The involvement of other cytochrome P450s in benzene metabolism is also possible and recent work has shown that CYP4F3 was upregulated in peripheral white blood cells in 7 patients who had occupational benzene poisoning [32]. In the same study, phenol was found to be capable of inducing CYP4F3 in myeloid cell lines and in human neutrophils [32]. These observations may be significant and could provide a novel metabolic mechanism for benzene-induced myeloid toxicity if CYP4F3 is found to be capable of metabolizing benzene or phenol.
Table 1. Potential Reactive Metabolites of Benzene.
| Potential reactive metabolites of benzene |
|---|
| Benzene oxide |
| Trans,trans muconaldehyde |
| Quinones |
| Thiol adducts |
| Phenolics |
| Reactive oxygen species |
For citations see text
Table 2. Metabolic Susceptibility Factors in Benzene Toxicity.
| Factors | Susceptibility pathway |
|---|---|
| CYP2E1 | Rapid metabolizer phenotype and SNP's |
| CYP4F3 | ? |
| MPO | G463A promoter polymorphism leading to decreased transcription |
| GSH | Enzymes regulating levels |
| Reactive oxygen | Enzymes regulating levels |
| EH | Rapid metabolizer genotype, other variants |
| GST | Null variants in GSTT1 and GSTM1. GSTPi variants with decreased activity |
| NQO1*2 | Heterozygous (decreased activity) and homozygous (null) C609T variants |
| NQO1*3 | Reduced NQO1 activity |
For citations see text
Other metabolic susceptibility factors include epoxide hydrolase which is known to have genotypic variants with a range of activities [33] and glutathione, a key defense system against reactive metabolites [34]. Myeloperoxidase (MPO) can oxidize polyphenolic metabolites of benzene to electrophilic quinones. A promoter polymorphism in MPO (G463A) leads to decreased transcription and decreased enzymatic activity [35] and has been examined in epidemiological studies of benzene poisoning. Glutathione-S-transferases can impact benzene metabolism at a number of steps and null polymorphisms in GSTT1 and GSTM1 are well-characterized. GSTPi variants with altered enzyme activity have also been characterized [36]. Finally, the quinone reductases have attracted considerable attention as metabolic susceptibility factors. There are two major polymorphisms in NQO1. The NQO1*2 allele (C609T) has profound implications for phenotype and leads to the synthesis of a mutant protein which is rapidly degraded via the proteasome. The NQO1*2 allele is essentially, a null polymorphism in homozygous individuals [37;38] and exhibits a gene-dose effect with heterozygous carriers exhibiting intermediate enzyme activity [39]. The NQO1*2 allele is widespread in the population with the prevalence of the homozygous NQO1*2 genotype being between 4-34% depending on ethnic group [40-43]. The NQO1*3 allele is a relatively low frequency allele which has diminished activity for certain substrates [44]. Although up to 21 additional variants in the promoter region of NQO1 and additional variants in the coding region of the gene are known [45], the phenotypic implications of these variants are unclear. Furthermore, the low or sometimes unknown frequency of these alleles in the population has generally precluded their inclusion in epidemiological studies. NQO2 is an additional quinone reductase [46] but the role of the enzyme in benzene metabolism and the potential impact of any polymorphic variants in NQO2 on benzene toxicity is currently unknown.
Oxidative stress has been implicated in the toxic effects of benzene and its metabolites [22;47]. Oxygen radicals are produced during benzene metabolism and can induce direct toxic effects but at lower levels oxygen radicals can also influence signaling pathways and recent data suggests this may be critical in stem cell signaling. Cytokines which regulate hematopoiesis have been found to be able to generate ROS and TNFα-mediated inhibition of HSC self-renewal was shown to result from excessive ROS [48]. Dysregulation of reactive oxygen species production has also been implicated in abnormal hematopoiesis and potentially in functioning of the hematopoietic stem cell niche [49]. Enzymes modulating oxygen radical levels, particularly in stem cells, may therefore represent additional susceptibility factors for benzene toxicity.
Cell-specific metabolism and toxicity of the polyphenolic metabolites of benzene in cellular systems
Early studies of cell-specific metabolism [50] showed that the phenolic metabolites of benzene were more toxic in bone marrow cultures than benzene itself. One interesting metabolic balance is between MPO and NQO1. MPO can catalyze oxidation of hydroquinone or catechol to para or ortho-benzoquinone respectively while NQO1 can reduce quinones to their hydroquinone derivatives which are more readily excreted, are not electrophilic and do not undergo redox cycling. Consequently, NQO1 is viewed as a detoxification enzyme with respect to benzene metabolism and studies in both NQO1 knockout animals [51-53] and in humans occupationally exposed to benzene [54] have confirmed this view. Interestingly, Bauer et al [52] demonstrated that NQO1 knockout animals of both genders had greater sensitivity to benzene induced hematotoxicity than wild type controls. However, increased genotoxicity, as indicated by the frequency of micronucleated reticulocytes, only occurred in female mice. The authors suggested that different benzene metabolites may be responsible for hematotoxicity and genotoxicity [52;55].
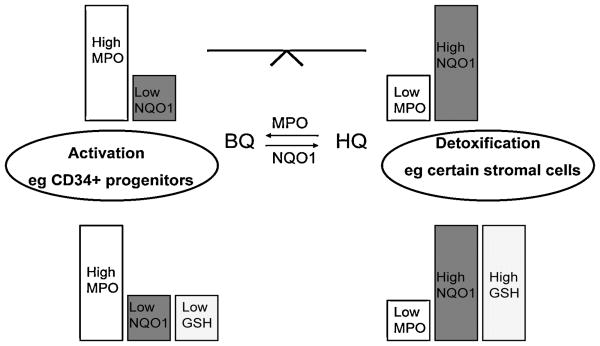
In cellular studies, the levels of MPO and NQO1 have been suggested to modulate the toxicity of phenolic metabolites of benzene particularly in stromal cells where multiple cell types exist with varying enzyme activities [56] [57]. For example, fibroblastoid cells in stroma tend to be less susceptible to hydroquinone due to an elevated NQO1 and decreased MPO content relative to macrophages. Another important determinant of stromal cell susceptibility to the phenolic metabolites of benzene is cellular glutathione levels [58;59]. A combination of NQO1, MPO, and GSH levels may therefore represent an approach to predicting the relative susceptibility of different cell types to the phenolic metabolites of benzene (Figure 2). Interestingly, CD34+ cells isolated from human bone marrow contain significant MPO [60] but no detectable levels of NQO1 [61] suggesting they would be susceptible to phenolic metabolites of benzene. However, NQO1 could be induced in isolated human bone marrow mononuclear or CD34+ progenitor cells after exposure to hydroquinone and the level of induction was dependent on NQO1 genotype. NQO1 was markedly induced by hydroquinone or catechol in isolated human bone marrow mononuclear cells genotyped as NQO1 wild type, intermediate induction occurred in heterozygous individuals whereas induction of NQO1 could not be detected in cells homozygous for the NQO1*2 polymorphism. These data provided a potential explanation of the protective role of the wild type NQO1 genotype in bone marrow cells that had no detectable resting levels of NQO1 [61]. CD34+ bone marrow cells have been shown to be a sensitive target for 1,4-benzoquinone-induced toxicity [62] and hydroquinone-induced apoptosis [61].
Figure 2. Metabolic susceptibility factors in stroma (MPO/NQO1/GSH).
Metabolic factors such as MPO, NQO1 and GSH can influence cell specific toxicity of benzene metabolites.
Metabolic susceptibility factors in epidemiological studies of benzene exposure
One of the first studies of metabolic susceptibility factors in workers occupationally exposed to benzene demonstrated that both a rapid metabolizer CYP2E1 phenotype and the NQO1*2 polymorphism were associated with an increased risk of benzene poisoning as defined by decreased white blood cell and platelet counts [54]. The combination of a rapid CYP2E1 phenotype and the NQO1 null genotype led to a 7.8 fold increased risk of benzene poisoning [54]. Subsequent studies have investigated the role of metabolic susceptibility factors in benzene poisoning and have confirmed a potential role for the NQO1*2 polymorphism in benzene poisoning [63-65]. Additional enzyme systems implicated in benzene poisoning included CYP2E1, GSTT1 and GSTM1 [63-65]. The NQO1*2 polymorphism together with epoxide hydrolase and GSTT1 polymorphisms were associated with induction of DNA single strand breaks in Bulgarian petrochemical workers while the MPO463 polymorphism and the NQO1*3 polymorphism influenced susceptibility to benzene hematotoxicity in Chinese workers exposed to low levels of benzene [66]. Epoxide hydrolase polymorphisms have also been recently implicated in susceptibility to benzene poisoning [67]. Not all of these studies were consistent in the metabolic susceptibility factors identified which probably reflects the often small numbers of cases studied and the different populations utilized. A recent review has summarized the literature on polymorphisms and biological effects of benzene exposure [68].
Non-metabolic susceptibility factors in benzene toxicity
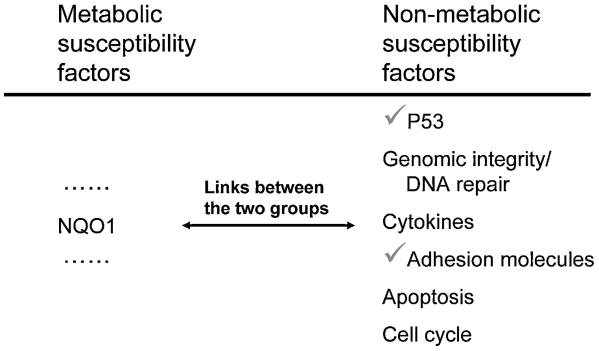
The adverse effects of benzene and its metabolites are widespread and consequently a wide variety of cellular systems are affected. A summary of non-metabolic susceptibility factors which are potentially important in benzene toxicity is shown in Table 3. p53 has been demonstrated to be important in benzene toxicity in animal models [69-72]. Benzene has been recognized as inducing chromosomal damage [73-76] and genes involved in DNA repair/genomic integrity have been shown to influence benzene toxicity in occupationally exposed populations [77]. Additional susceptibility factors associated with cytokines [78;79], adhesion molecules [78] and multiple gene pathways associated with cell cycle and apoptosis [70;71;80-82] have also been characterized. Thus, both metabolic and non-metabolic susceptibility factors have been described for benzene toxicity.
Table 3. Non-Metabolic Susceptibility Factors in Benzene Toxicity.
| Non-metabolic susceptibility factors in benzene toxicity |
|---|
| p53 |
| Genomic integrity and DNA repair |
| Cytokines |
| Adhesion Molecules |
| Apoptosis |
| Cell Cycle |
For citations see text
Are there links between metabolic and non-metabolic susceptibility factors in benzene toxicity?
Since both metabolic and non-metabolic susceptibility factors for benzene toxicity have been characterized, we have examined potential links between the two groups using NQO1 as an example.
The metabolic capability of NQO1 with respect to quinone reduction is well recognized [83;84]. Many quinones of different structural classes can be reduced via NQO1 to their hydroquinone derivatives via a mechanism proposed to involve hydride transfer [85;86]. Because metabolic enzymes are often expressed and/or induced to high levels in cellular systems, other functions for theses enzymes might be predicted. In the case of NQO1, the enzyme has been found to scavenge superoxide directly and function as a superoxide reductase [87]. However, the very poor rate constant of this reaction suggests that unless NQO1 is expressed or induced to very high levels of expression, which occurs in certain tumor cells or after marked cellular stress, this mechanism is unlikely to be of physiological relevance. Another interesting function of NQO1 which was discovered in Xenopus is stabilization of microtubules [88]. Considering the potential key role of electrophilic reactive metabolites at the level of sulfhydryl-rich microtubules in the mechanism of toxicity of benzene [89], NQO1-mediated modulation of microtubule stability in human bone marrow cells is deserving of investigation.
An additional function that has been proposed for NQO1 is stabilization of p53 against proteasomal degradation [90;91]. NQO1 interacts with p53 in a protein-protein interaction [92] which may explain this effect. It has been demonstrated that NQO1-knockout animals had no detectable NQO1 in bone marrow but importantly markedly decreased levels of p53 and consequently lower levels of apoptosis and increased bone marrow cellularity [51]. The protective effect of NQO1 has been proposed to occur at the level of the 20S proteasome [91].
In summary, it is recognized that NQO1 can have multiple roles and may influence oxygen radical levels, stabilize microtubules and stabilize p53. Given the influence of NQO1 in modulating p53 stability, the protective effects of NQO1 in benzene toxicity may not solely reflect its metabolic ability to reduce quinones to hydroquinones. The effect of NQO1 at the level of p53 may also indirectly affect other downstream susceptibility factors such as proteins involved in apoptosis and cell cycle.
NQO1 expression in bone marrow stroma. Studies using transformed human bone marrow endothelial cells (HBMEC)
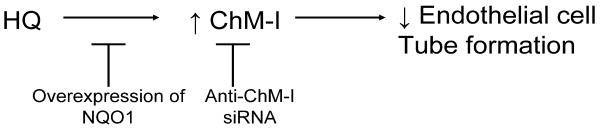
Within bone marrow, it is known that stromal cells express relatively high levels of NQO1 and expression occurs in a cell specific manner [57;93]. Bone marrow endothelial cells express elevated levels of NQO1 and we have used a transformed human bone marrow endothelial cell line [94] to examine both potential mechanisms of toxicity of benzene metabolites and the protective role of NQO1 [95;96]. Although endothelial cells have relatively high levels of NQO1, they are still susceptible to polyphenolic metabolites of benzene such as hydroquinone albeit at higher concentrations than stromal cells expressing low NQO1 levels. In a recent study we examined gene expression changes in HBMEC cells after treatment with 10 μM hydroquinone [96]. One of these genes, chondromodulin 1 (ChM-I), was upregulated by HQ treatment and we found the inhibitory effects of HQ on endothelial cell tube formation could be partially abrogated by ChM-I knockdown [96]. These data suggested a role for ChM-I in the inhibitory effects of HQ on HBMEC and cellular transfection of NQO1 blocked the effects of HQ. The potential role of ChM-I in the effects of HQ in HBMEC is summarized in Figure 3.
Figure 3. Mechanisms of HQ induced inhibition of tube formation in HBMEC.
An important role for ChM-I.
Using HBMEC to probe the effects of loss of NQO1
The homozygous NQO1 *2 polymorphism is essentially a null polymorphism with only trace levels of the mutant NQO1*2 protein detectable in homozygous individuals [37-39]. We have utilized HBMEC to model the effects of a loss of NQO1 in two different ways. We have utilized either mechanism-based inhibitors of NQO1 to irreversibly block NQO1 activity or we have knocked down NQO1 expression in HBMEC using anti-NQO1 siRNA. Treatment of HBMEC with ES936, a mechanism-based inhibitor of NQO1 [97], led to modulation of multiple genes in a microarray study and one of the downregulated genes was an adhesion molecule VCAM-1 [98]. The discovery of an adhesion molecule altered in this model was noteworthy since the HBMEC cellular system was originally developed and characterized based on its adhesive properties towards human progenitor cells [94]. Blocking antibodies to E-selectin, VCAM-1 and ICAM-1 were found to markedly inhibit CD34+ cell adhesion to HBMEC [94].
Adhesion of progenitor cells to endothelial cells can modulate progenitor cell signaling or differentiation [99;100]. In addition, endothelial cells in bone marrow form the vascular niche, one of the two major stem cell niches which regulate progenitor/stem cell signaling, recruitment and trafficking [101]. The other recognized stem cell niche is the osteoblastic niche [101]. Adhesion molecules are important in niche function and play key roles in stem cell mobilization and homing in the vascular niche. Adhesion molecules implicated in the function of the vascular niche include N- cadherin, VCAM-1, and osteopontin [101]. Multiple chemokines are also likely to play important roles [102].
Our work using HBMEC demonstrated that either pharmacological inhibition of NQO1 using ES936 or knockdown of NQO1 expression using anti-NQO1 siRNA led to decreased resting levels of VCAM-1 [98]. Since adhesion molecule levels are markedly stimulated by TNFα in HBMEC [94], we continued our studies investigating the effects of inhibition or knockdown of NQO1 on levels of adhesion molecules by incorporating TNFα into the experimental design. When adhesion molecule expression in HBMEC was stimulated by TNFα, expression of E-selectin, ICAM-1 and VCAM-1 could be inhibited either by genetic knockdown or pharmacological inhibition of NQO1 (Zhou et al., unpublished data). Importantly, NQO1 inhibition also led to impaired adhesion of KG1a CD34+ cells to HBMEC imparting functional significance to downregulation of adhesion molecules. TNFα-induced adhesion molecule expression occurs via the transcription factor NF-κB and TNFα-induced NF-κB expression has recently been found to be inhibited in NQO1-knockout animals [103]. Inhibition of NF-κB activation as a result of inhibition or knockdown of NQO1 could therefore provide a potential mechanism for inhibited adhesion molecule expression. These findings may have implications for progenitor/stem cell adhesion and vascular niche function under conditions where NQO1 activity or protein level is depleted such as in the case of individuals expressing the homozygous NQO1*2 polymorphism. Interestingly, polymorphisms in both VCAM-1 [78] and TNFα [79] have been associated with susceptibility to benzene-induced hematotoxicity. In addition, hydroquinone is known to inhibit NF-κB activation in multiple cell lines [104-106] and the activity of other transcription factors such as PU.1 and AP-1 in progenitor cells [107;108].
Summary and future work
In summary, metabolic susceptibility factors such as NQO1 can influence non-metabolic susceptibility factors associated with benzene-induced hematotoxicity (Figure 4). NQO1 can influence p53 stability in bone marrow and can modulate adhesion molecule production and downstream CD34+ cell adhesion to HBMEC. The effects of NQO1 at the level of adhesion molecule expression may be of relevance to the functioning of the vascular stem cell niche and is deserving of further investigation.
Figure 4. The metabolic susceptibility factor NQO1 influences other non-metabolic susceptibility factors.
NQO1 modulates p53 stability and adhesion molecule expression.
Interactions between metabolic and non-metabolic susceptibility factors are unlikely to be restricted to NQO1. Glutathione-S-transferase Pi, for example, is known to regulate JNK signaling and cell proliferation [109;110] and its effects in modulating benzene toxicity may include both metabolic and non-metabolic mechanisms. Potential effects of metabolic susceptibility factors at other levels of the benzene toxicity cascade should be considered in future studies.
Acknowledgments
Supported by NIH grant ES09554
Abbreviations
- HBMEC
transformed human bone marrow endothelial cells
- MPO
myeloperoxidase
- NQO1, NAD(P)H
quinone oxidoreductase 1
- GST
glutathione-S-transferase
- ChM-I
chondromodulin 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Aksoy M. Benzene hematotoxicity. In: Aksoy M, editor. Benzene Carcinogenicity. CRC Press; Boca Raton: 1988. pp. 59–112. [Google Scholar]
- 2.Aksoy M. Benzene Carcinogenicity. In: Aksoy M, editor. Benzene Carcinogenicity. CRC Press; Boca raton: 1988. pp. 113–151. [Google Scholar]
- 3.Snyder R, Longacre SL, Witmer CM, Kocsis JJ. Biochemical toxicology of benzene. Rev Biochem Tox. 1980;3:123–154. [Google Scholar]
- 4.Kalf GF. Recent advances in the metabolism and toxicity of benzene. Crit Rev Toxicol. 1987;18:141–159. doi: 10.3109/10408448709089859. [DOI] [PubMed] [Google Scholar]
- 5.Yardley-Jones A, Anderson D, Parke DV. The toxicity of benzene and its metabolism and molecular pathology in human risk assessment. Br J Ind Med. 1991;48:437–444. doi: 10.1136/oem.48.7.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Snyder R, Hedli CC. An overview of benzene metabolism. Environ Health Perspect. 1996;104 6:1165–1171. doi: 10.1289/ehp.961041165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ross D. Metabolic basis of benzene toxicity. Eur J Haematol. 1996;57 60:111–118. doi: 10.1111/j.1600-0609.1996.tb01656.x. [DOI] [PubMed] [Google Scholar]
- 8.Irons RD, Greenlee WF, Wierda D, Bus JS. Relationship between benzene metabolism and toxicity: a proposed mechanism for the formation of reactive intermediates from polyphenol metabolites. Adv Exp Med Biol. 1981;136(Pt A):229–243. doi: 10.1007/978-1-4757-0674-1_13. [DOI] [PubMed] [Google Scholar]
- 9.Cooper KR, Snyder R. Benzene Metabolism. In: Aksoy M, editor. Benzene Carcinogenicity. CRC Press; Boca Raton: 1988. pp. 33–58. [Google Scholar]
- 10.Ross D. The role of metabolism and specific metabolites in benzene-induced toxicity: evidence and issues. J Toxicol Environ Health A. 2000;61:357–372. doi: 10.1080/00984100050166361. [DOI] [PubMed] [Google Scholar]
- 11.Valentine JL, Lee SST, Seaton MJ, Asgharian B, Farris G, Corton JC, Gonzalez FJ, Medinsky MA. Reduction of benzene metabolism and toxicity in mice that lack CYP2E1 expression. Toxicol Appl Pharmacol. 1996;141:205–213. doi: 10.1006/taap.1996.0277. [DOI] [PubMed] [Google Scholar]
- 12.Low LK, Lambert CE, Meeks JR, Naro PA, Mackerer CR. Tissue specific metabolism of benzene in Zymbal gland and other solid tumor target tissues in rats. J Am Coll Toxicol. 1995;14:40–60. [Google Scholar]
- 13.Tunek A, Platt K, Bentley P, Oesch F. Microsomal metabolism of benzene to species irreversibly binding to microsomal protein and effects of modifications of this metabolism. Mol Pharmacol. 1978;14:920–929. [PubMed] [Google Scholar]
- 14.Lindstrom AB, Yeowell-O'Connell K, Waidyanatha S, McDonald TA, Golding BT, Rappaport SM. Formation of hemoglobin and albumin adducts of benzene oxide in mouse, rat, and human blood. Chem Res Toxicol. 1998;11:302–310. doi: 10.1021/tx9701788. [DOI] [PubMed] [Google Scholar]
- 15.McDonald TA, Yeowell-O'Connell K, Rappaport SM. Comparison of protein adducts of benzene oxide and benzoquinone in the blood and bone marrow of rats and mice exposed to [14C/13C6[benzene. Cancer Res. 1994;54:4907–4914. [PubMed] [Google Scholar]
- 16.Fuchs D, von Soo A. Hoppe-Seylers Z.Physiologische Chemie. 1916;98:11–13. [Google Scholar]
- 17.Parke DV, Williams RT. Studies in detoxication 49. The metabolism of benzene containing 14C-benzene. Biochem J. 1953;54:231–238. doi: 10.1042/bj0540231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Witz G, Latriano L, Goldstein BD. Metabolism and toxicity of trans,trans-muconaldehyde, an open- ring microsomal metabolite of benzene. Environ Health Perspect. 1989;82:19–22. doi: 10.1289/ehp.898219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bleasdale C, Kennedy G, MacGregor JO, Nieschalk J, Pearce K, Watson WP, Golding BT. Chemistry of muconaldehydes of possible relevance to the toxicology of benzene. Environ Health Perspect. 1996;104 6:1201–1209. doi: 10.1289/ehp.961041201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rickert DE, Baker TS, Bus JS, Barrow CS, Irons RD. Benzene disposition in the rat after exposure by inhalation. Toxicol Appl Pharmacol. 1979;49:417–423. doi: 10.1016/0041-008x(79)90441-1. [DOI] [PubMed] [Google Scholar]
- 21.Eastmond DA, Smith MT, Irons RD. An interaction of benzene metabolites reproduces the myelotoxicity observed with benzene exposure. Toxicol Appl Pharmacol. 1987;91:85–95. doi: 10.1016/0041-008x(87)90196-7. [DOI] [PubMed] [Google Scholar]
- 22.Kolachana P, Subrahmanyam VV, Meyer KB, Zhang L, Smith MT. Benzene and its phenolic metabolites produce oxidative DNA damage in HL-60 cells in vitro and in the bone marrow in-vivo. Cancer Res. 1993;53:1023–1026. [PubMed] [Google Scholar]
- 23.Hiraku Y, Kawanishi S. Oxidative DNA damage and apoptosis induced by benzene metabolites. Cancer Res. 1996;56:5172–5178. [PubMed] [Google Scholar]
- 24.Subrahmanyam VV, Ross D, Eastmond DA, Smith MT. Potential role of free radicals in benzene-induced myelotoxicity and leukemia. Free Rad Biol Med. 1991;11:495–515. doi: 10.1016/0891-5849(91)90063-9. [DOI] [PubMed] [Google Scholar]
- 25.Sawahata T, Rickert DE, Greenlee WF. Metabolism of benzene and its metabolites in bone marrow. In: Irons RD, editor. Toxicology of the Blood and Bone Marrow. Raven Press; New York: 1985. pp. 141–148. [Google Scholar]
- 26.Sadler A, Subrahmanyam VV, Ross D. Oxidation of catechol by horseradish peroxidase and human leukocyte peroxidase: reactions of o-benzoquinone and o- benzosemiquinone. Toxicol Appl Pharmacol. 1988;93:62–71. doi: 10.1016/0041-008x(88)90025-7. [DOI] [PubMed] [Google Scholar]
- 27.McDonald TA, Waidyanatha S, Rappaport SM. Production of benzoquinone adducts with hemoglobin and bone- marrow proteins following administration of [13C6]benzene to rats. Carcinogenesis. 1993;14:1921–1925. doi: 10.1093/carcin/14.9.1921. [DOI] [PubMed] [Google Scholar]
- 28.Pfeiffer E, Metzler M. Interaction of p-benzoquinone and p-biphenoquinone with microtubule proteins in vitro. Chem Biol Interact. 1996;102:37–53. doi: 10.1016/0009-2797(96)03730-1. [DOI] [PubMed] [Google Scholar]
- 29.Levay G, Ross D, Bodell WJ. 32P-postlabeling detection of benzene-DNA adducts in HL-60 cells, mouse bone marrow macrophages and human bone marrow. Carcinogenesis. 1993;14:2329–2334. doi: 10.1093/carcin/14.11.2329. [DOI] [PubMed] [Google Scholar]
- 30.Monks TJ, Lau SS. Toxicology of quinone-thioethers. Crit Rev Toxicol. 1992;22:243–270. doi: 10.3109/10408449209146309. [DOI] [PubMed] [Google Scholar]
- 31.Bratton SB, Lau SS, Monks TJ. Identification of quinol thioethers in bone marrow of hydroquinone/phenol- treated rats and mice and their potential role in benzene/mediated hematotoxicity. Chem Res Toxicol. 1997;10:859–865. doi: 10.1021/tx960208r. [DOI] [PubMed] [Google Scholar]
- 32.Zhao Z, He X, Bi Y, Xia Y, Tao N, Li L, Ma Q. Induction of CYP4F3 by benzene metabolites in human white blood cells in vivo in human promyelocytic leukemic cell lines and ex vivo in human blood neutrophils. Drug Metab Dispos. 2009;37:282–291. doi: 10.1124/dmd.108.023192. [DOI] [PubMed] [Google Scholar]
- 33.Przybyla-Zawislak BD, Srivastava PK, Vazquez-Matias J, Mohrenweiser HW, Maxwell JE, Hammock BD, Bradbury JA, Enayetallah AE, Zeldin DC, Grant DF. Polymorphisms in human soluble epoxide hydrolase. Mol Pharmacol. 2003;64:482–490. doi: 10.1124/mol.64.2.482. [DOI] [PubMed] [Google Scholar]
- 34.Twerdok LE, Rembish SJ, Trush MA. Induction of quinone reductase and glutathione in bone marrow cells by 1,2-dithiole-3-thione. Effect on hydroquinone-induced cytotoxicity. Toxicol Appl Pharmacol. 1992;112:273–281. doi: 10.1016/0041-008x(92)90197-z. [DOI] [PubMed] [Google Scholar]
- 35.London SJ, Lehman TA, Taylor JA. Myeloperoxidase genetic polymorphism and lung cancer risk. Cancer Research. 1997;57:5001–5003. [PubMed] [Google Scholar]
- 36.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 37.Traver RD, Horikoshi T, Danenberg KD, Stadlbauer THW, Danenberg PV, Ross D, Gibson NW. NAD(P)H:quinone oxidoreductase gene expression in human colon carcinoma cells: Characterization of a mutation which modulates DT-diaphorase activity and mitomycin sensitivity. Cancer Res. 1992;52:797–802. [PubMed] [Google Scholar]
- 38.Traver RD, Siegel D, Beall HD, Phillips RM, Gibson NW, Franklin WA, Ross D. Characterization of a polymorphism in NAD(P)H: Quinone oxidoreductase (DT-diaphorase) Br J Cancer. 1997;75:69–75. doi: 10.1038/bjc.1997.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siegel D, McGuinness SM, Winski S, Ross D. Genotype-phenotype relationships in studies of a polymorphism in NAD(P)H:quinone oxidoreductase 1. Pharmacogenetics. 1999;9:113–121. doi: 10.1097/00008571-199902000-00015. [DOI] [PubMed] [Google Scholar]
- 40.Kelsey KT, Wiencke JK, Christiani DC, Zuo Z, Spitz MR, Xu X, Lee BK, Schwartz BS, Traver RD, Ross D. Ethnic variation in the prevalence of a common NAD(P)H:quinone oxidoreductase polymorphism and its implications for anticancer chemotherapy. Br J Cancer. 1997;76:852–854. doi: 10.1038/bjc.1997.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaedigk A. Interethnic differences of drug-metabolizing enzymes. Int J Clin Pharmacol Ther. 2000;38:61–68. doi: 10.5414/cpp38061. [DOI] [PubMed] [Google Scholar]
- 42.Kiffmeyer WR, Langer E, Davies SM, Envall J, Robison LL, Ross JA. Genetic polymorphisms in the Hmong population: implications for cancer etiology and survival. Cancer. 2004;100:411–417. doi: 10.1002/cncr.11913. [DOI] [PubMed] [Google Scholar]
- 43.Ross D, Siegel D. NQO1. Functions and Pharmacogenetics. Methods in Enzymology. 2004;382B:115–144. doi: 10.1016/S0076-6879(04)82008-1. [DOI] [PubMed] [Google Scholar]
- 44.Pan SS, Forrest GL, Akman SA, Hu LT. NAD(P)H:quinone oxidoreductase expression and mitomycin C resistance developed by human colon cancer HCT 116 cells. Cancer Res. 1995;55:330–335. [PubMed] [Google Scholar]
- 45.Nebert DW, Roe AL, Vandale SE, Bingham E, Oakley GG. NAD(P)H:quinone oxidoreductase (NQO1) polymorphism, exposure to benzene, and predisposition to disease: A HuGE review. Genet Med. 2002;4:62–70. doi: 10.1097/00125817-200203000-00003. [DOI] [PubMed] [Google Scholar]
- 46.Jaiswal AK, Burnett P, Adesnik M, McBride OW. Nucleotide and deduced amino acid sequence of a human cDNA (NQO2) corresponding to a second member of the NAD(P)H:quinone oxidoreductase gene family. Extensive polymorphism at the NQO2 gene locus on chromosome 6. Biochemistry. 1990;29:1899–1906. doi: 10.1021/bi00459a034. [DOI] [PubMed] [Google Scholar]
- 47.Winn LM. Homologous recombination initiated by benzene metabolites: a potential role of oxidative stress. Toxicol Sci. 2003;72:143–149. doi: 10.1093/toxsci/kfg008. [DOI] [PubMed] [Google Scholar]
- 48.Naka K, Muraguchi T, Hoshii T, Hirao A. Regulation of reactive oxygen species and genomic stability in hematopoietic stem cells. Antioxid Redox Signal. 2008;10:1883–1894. doi: 10.1089/ars.2008.2114. [DOI] [PubMed] [Google Scholar]
- 49.Ghaffari S. Oxidative stress in the regulation of normal and neoplastic hematopoiesis. Antioxid Redox Signal. 2008;10:1923–1940. doi: 10.1089/ars.2008.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harrison K, Randoll FW. An application of bone marrow cultures to toxicology and therapeutics. J Exp Phys. 1947;34:141–152. doi: 10.1113/expphysiol.1948.sp000921. [DOI] [PubMed] [Google Scholar]
- 51.Long DJ, Gaikwad A, Multani A, Pathak S, Montgomery CA, Gonzalez FJ, Jaiswal AK. Disruption of the NAD(P)H:Quinone Oxidoreductase 1 (NQO1) Gene in Mice Causes Myelogenous Hyperplasia. Cancer Res. 2002;62:3030–3036. [PubMed] [Google Scholar]
- 52.Bauer AK, Faiola B, Abernethy DJ, Marchan R, Pluta LJ, Wong VA, Roberts K, Jaiswal AK, Gonzalez FJ, Butterworth BE, Borghoff S, Parkinson H, Everitt J, Recio L. Genetic Susceptibility to Benzene-induced Toxicity: Role of NADPH: Quinone Oxidoreductase-1. Cancer Res. 2003;63:929–935. [PubMed] [Google Scholar]
- 53.Iskander K, Jaiswal AK. Quinone oxidoreductases in protection against myelogenous hyperplasia and benzene toxicity. Chem Biol Interact. 2005;153-154:147–157. doi: 10.1016/j.cbi.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 54.Rothman N, Smith MT, Hayes RB, Traver RD, Hoener BA, Campleman S, Li GL, Dosemeci M, Linet M, Zhang LP, Xi LQ, Wacholder S, Lu W, Meyer KB, Titenko-Holland N, Stewart JT, Yin SN, Ross D. Benzene poisoning, a risk factor for hematological malignancy, is associated with the NQ01609C-->T mutation and rapid fractional excretion of chlorzoxazone. Cancer Res. 1997;57:2839–2842. [PubMed] [Google Scholar]
- 55.Recio L, Bauer A, Faiola B. Use of genetically modified mouse models to assess pathways of benzene-induced bone marrow cytotoxicity and genotoxicity. Chem Biol Interact. 2005;153-154:159–164. doi: 10.1016/j.cbi.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 56.Smart RC, Zannoni VG. DT-Diaphorase and peroxidase influence the covalent binding of the metabolites of phenol, the major metabolite of benzene. Mol Pharmacol. 1984;26:105–111. [PubMed] [Google Scholar]
- 57.Thomas DJ, Sadler A, Subrahmanyam VV, Siegel D, Reasor MJ, Wierda D, Ross D. Bone marrow stromal cell bioactivation and detoxification of the benzene metabolite hydroquinone: comparison of macrophages and fibroblastoid cells. Mol Pharmacol. 1990;37:255–262. [PubMed] [Google Scholar]
- 58.Zhu H, Li YB, Trush MA. Differences in xenobiotic detoxifying activities between bone marrow stromal cells from mice and rats: Implications for benzene-induced hematotoxicity. J Toxicol Environ Health. 1995;46:183–201. doi: 10.1080/15287399509532028. [DOI] [PubMed] [Google Scholar]
- 59.Trush MA, Twerdok LE, Rembish SJ, Zhu H, Li YB. Analysis of target cell susceptibility as a basis for the development of a chemoprotective strategy against benzene-induced hematotoxicities. Environ Health Perspect. 1996;104 6:1227–1234. doi: 10.1289/ehp.961041227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schattenberg DG, Stillman WS, Gruntmeir JJ, Helm KM, Irons RD, Ross D. Peroxidase activity in murine and human hematopoietic progenitor cells: Potential relevance to benzene-induced toxicity. Mol Pharmacol. 1994;46:346–351. [PubMed] [Google Scholar]
- 61.Moran JL, Siegel D, Ross D. A potential mechanism underlying the increased susceptibility of individuals with a polymorphism in NAD(P)H:quinone oxidoreductase 1 (NQO1) to benzene toxicity. Proc Natl Acad Sci U S A. 1999;96:8150–8155. doi: 10.1073/pnas.96.14.8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abernethy DJ, Kleymenova EV, Rose J, Recio L, Faiola B. Human CD34+ hematopoietic progenitor cells are sensitive targets for toxicity induced by 1,4-benzoquinone. Toxicol Sci. 2004;79:82–89. doi: 10.1093/toxsci/kfh095. [DOI] [PubMed] [Google Scholar]
- 63.Wan J, Shi J, Guan J, Ye R, Gao X, Liu W, Hui L, Cao D, Jin X, Hu G, Xia Z. Relation of genetic polymorphism of NQO1 and GSTT1 with risks of chronic benzene poisoning. Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi. 2002;20:340–343. [PubMed] [Google Scholar]
- 64.Wan J, Shi J, Hui L, Wu D, Jin X, Zhao N, Huang W, Xia Z, Hu G. Association of genetic polymorphisms in CYP2E1, MPO, NQO1, GSTM1, and GSTT1 genes with benzene poisoning. Environ Health Perspect. 2002;110:1213–1218. doi: 10.1289/ehp.021101213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen Y, Li G, Yin S, Xu J, Ji Z, Xiu X, Liu L, Ma D. Genetic polymorphisms involved in toxicant-metabolizing enzymes and the risk of chronic benzene poisoning in Chinese occupationally exposed populations. Xenobiotica. 2007;37:103–112. doi: 10.1080/00498250601001662. [DOI] [PubMed] [Google Scholar]
- 66.Lan Q, Zhang L, Li G, Vermeulen R, Weinberg RS, Dosemeci M, Rappaport SM, Shen M, Alter BP, Wu Y, Kopp W, Waidyanatha S, Rabkin C, Guo W, Chanock S, Hayes RB, Linet M, Kim S, Yin S, Rothman N, Smith MT. Hematotoxicity in workers exposed to low levels of benzene. Science. 2004;306:1774–1776. doi: 10.1126/science.1102443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun P, Qian J, Zhang ZB, Wan JX, Wu F, Jin XP, Fan WW, Lu DR, Zhao NQ, Christiani DC, Xia ZL. Polymorphisms in phase I and phase II metabolism genes and risk of chronic benzene poisoning in a Chinese occupational population. Carcinogenesis. 2008;29:2325–2329. doi: 10.1093/carcin/bgn208. [DOI] [PubMed] [Google Scholar]
- 68.Dougherty D, Garte S, Barchowsky A, Zmuda J, Taioli E. NQO1, MPO, CYP2E1, GSTT1 and GSTM1 polymorphisms and biological effects of benzene exposure--a literature review. Toxicol Lett. 2008;182:7–17. doi: 10.1016/j.toxlet.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 69.Kawasaki Y, Hirabayashi Y, Kaneko T, Kanno J, Kodama Y, Matsushima Y, Ogawa Y, Saitoh M, Sekita K, Uchida O, Umemura T, Yoon BI, Inoue T. Benzene-induced hematopoietic neoplasms including myeloid leukemia in Trp53-deficient C57BL/6 and C3H/He mice. Toxicol Sci. 2009;110:293–306. doi: 10.1093/toxsci/kfp107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yoon BI, Li GX, Kitada K, Kawasaki Y, Igarashi K, Kodama Y, Inoue T, Kobayashi K, Kanno J, Kim DY, Inoue T, Hirabayashi Y. Mechanisms of benzene-induced hematotoxicity and leukemogenicity: cDNA microarray analyses using mouse bone marrow tissue. Environ Health Perspect. 2003;111:1411–1420. doi: 10.1289/ehp.6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boley SE, Wong VA, French JE, Recio L. p53 heterozygosity alters the mRNA expression of p53 target genes in the bone marrow in response to inhaled benzene. Toxicol Sci. 2002;66:209–215. doi: 10.1093/toxsci/66.2.209. [DOI] [PubMed] [Google Scholar]
- 72.Boley SE, Anderson EE, French JE, Donehower LA, Walker DB, Recio L. Loss of p53 in benzene-induced thymic lymphomas in p53+/- mice: evidence of chromosomal recombination. Cancer Res. 2000;60:2831–2835. [PubMed] [Google Scholar]
- 73.Zhang L, Rothman N, Wang Y, Hayes RB, Yin SN, Li GL, Smith MT. Aneuploidy of chromosomes 7,8 and 9 detected by fluorescence in-situ hybridization in workers exposed to benzene. Proc Amer Assoc Cancer Res. 1995;36:112. [Google Scholar]
- 74.Zhang L, Eastmond DA, Smith MT. The nature of chromosomal aberrations detected in humans exposed to benzene. Crit Rev Toxicol. 2002;32:1–42. doi: 10.1080/20024091064165. [DOI] [PubMed] [Google Scholar]
- 75.Eastmond DA, Mondrala ST, Hasegawa L. Topoisomerase II inhibition by myeloperoxidase-activated hydroquinone: a potential mechanism underlying the genotoxic and carcinogenic effects of benzene. Chem Biol Interact. 2005;153-154:207–216. doi: 10.1016/j.cbi.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 76.Lau A, Belanger CL, Winn LM. In utero and acute exposure to benzene: investigation of DNA double-strand breaks and DNA recombination in mice. Mutat Res. 2009;676:74–82. doi: 10.1016/j.mrgentox.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 77.Lan Q, Zhang L, Shen M, Jo WJ, Vermeulen R, Li G, Vulpe C, Lim S, Ren X, Rappaport SM, Berndt SI, Yeager M, Yuenger J, Hayes RB, Linet M, Yin S, Chanock S, Smith MT, Rothman N. Large-scale evaluation of candidate genes identifies associations between DNA repair and genomic maintenance and development of benzene hematotoxicity. Carcinogenesis. 2009;30:50–58. doi: 10.1093/carcin/bgn249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lan Q, Zhang L, Shen M, Smith MT, Li G, Vermeulen R, Rappaport SM, Forrest MS, Hayes RB, Linet M, Dosemeci M, Alter BP, Weinberg RS, Yin S, Yeager M, Welch R, Waidyanatha S, Kim S, Chanock S, Rothman N. Polymorphisms in cytokine and cellular adhesion molecule genes and susceptibility to hematotoxicity among workers exposed to benzene. Cancer Res. 2005;65:9574–9581. doi: 10.1158/0008-5472.CAN-05-1419. [DOI] [PubMed] [Google Scholar]
- 79.Lv L, Kerzic P, Lin G, Schnatter AR, Bao L, Yang Y, Zou H, Fu H, Ye X, Gross SA, Armstrong TW, Irons RD. The TNF-alpha 238A polymorphism is associated with susceptibility to persistent bone marrow dysplasia following chronic exposure to benzene. Leuk Res. 2007;31:1479–1485. doi: 10.1016/j.leukres.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 80.Hirabayashi Y. p53-dependent gene profiling for reactive oxygen species after benzene inhalation: special reference to genes associated with cell cycle regulation. Chem Biol Interact. 2005;153-154:165–170. doi: 10.1016/j.cbi.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 81.Faiola B, Fuller ES, Wong VA, Recio L. Gene expression profile in bone marrow and hematopoietic stem cells in mice exposed to inhaled benzene. Mutat Res. 2004;549:195–212. doi: 10.1016/j.mrfmmm.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 82.Sun P, Qiu Y, Zhang Z, Wan J, Wang T, Jin X, Lan Q, Rothman N, Xia ZL. Association of genetic polymorphisms, mRNA expression of p53 and p21 with chronic benzene poisoning in a chinese occupational population. Cancer Epidemiol Biomarkers Prev. 2009;18:1821–1828. doi: 10.1158/1055-9965.EPI-09-0140. [DOI] [PubMed] [Google Scholar]
- 83.Lind C, Hochstein P, Ernster L. DT-Diaphorase as a quinone reductase: A cellular control device against semiquinone and superoxide radical formation. Arch Biochem Biophys. 1982;216:178–185. doi: 10.1016/0003-9861(82)90202-8. [DOI] [PubMed] [Google Scholar]
- 84.Thor H, Smith MT, Hartzell P, Bellomo G, Jewell SA, Orrenius S. The metabolism of menadione (2-methyl-1,4-naphthoquinone) by isolated hepatocytes. A study of the implication of oxidative stress in intact cells. J Biol Chem. 1982;257:12419–12425. [PubMed] [Google Scholar]
- 85.Faig M, Bianchet MA, Talalay P, Chen S, Winski S, Ross D, Mario AL. Structures of recombinant human and mouse NAD(P)H:quinone oxidoreductases: Species comparison and structural changes with substrate binding and release. Proc Natl Acad Sci U S A. 2000;97:3177–3182. doi: 10.1073/pnas.050585797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bianchet MA, Faig M, Amzel LM. Structure and mechanism of NADPH:quinone acceptor oxidoreductases (NQO) Meth Enzymol. 2004;382:144–174. doi: 10.1016/S0076-6879(04)82009-3. [DOI] [PubMed] [Google Scholar]
- 87.Siegel D, Gustafson DL, Dehn DL, Han JY, Boonchoong P, Berliner LJ, Ross D. NAD(P)H:quinone oxidoreductase 1: Role as a superoxide scavenger. Molecular Pharmacology. 2004;65:1–10. doi: 10.1124/mol.65.5.1238. [DOI] [PubMed] [Google Scholar]
- 88.Wignall SM, Gray NS, Chang YT, Juarez L, Jacob R, Burlingame A, Schultz PG, Heald R. Identification of a novel protein regulating microtubule stability through a chemical approach. Chem Biol. 2004;11:135–146. [PubMed] [Google Scholar]
- 89.Irons RD, Neptun DA. Effects of the principal hydroxy metabolites of benzene on microtubule polymerization. Arch Toxicol. 1980;45:297–305. doi: 10.1007/BF00293810. [DOI] [PubMed] [Google Scholar]
- 90.Asher G, Lotem J, Cohen B, Sachs L, Shaul Y. Regulation of p53 stability and p53-dependent apoptosis by NADH quinone oxidoreductase 1. Proc Natl Acad Sci U S A. 2001;98:1188–1193. doi: 10.1073/pnas.021558898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gong X, Kole L, Iskander K, Jaiswal AK. NRH:quinone oxidoreductase 2 and NAD(P)H:quinone oxidoreductase 1 protect tumor suppressor p53 against 20s proteasomal degradation leading to stabilization and activation of p53. Cancer Res. 2007;67:5380–5388. doi: 10.1158/0008-5472.CAN-07-0323. [DOI] [PubMed] [Google Scholar]
- 92.Anwar A, Dehn D, Siegel D, Kepa JK, Tang LJ, Pietenpol JA, Ross D. Interaction of Human NAD(P)H:Quinone Oxidoreductase 1 (NQO1) with the Tumor Suppressor Protein p53 in Cells and Cell-free Systems. J Biol Chem. 2003;278:10368–10373. doi: 10.1074/jbc.M211981200. [DOI] [PubMed] [Google Scholar]
- 93.Siegel D, Ryder J, Ross D. NAD(P)H: quinone oxidoreductase 1 expression in human bone marrow endothelial cells. Toxicol Lett. 2001;125:93–98. doi: 10.1016/s0378-4274(01)00426-x. [DOI] [PubMed] [Google Scholar]
- 94.Schweitzer KM, Vicart P, Delouis C, Paulin D, Drager AM, Langenhuijsen MM, Weksler BB. Characterization of a newly established human bone marrow endothelial cell line: distinct adhesive properties for hematopoietic progenitors compared with human umbilical vein endothelial cells. Lab Invest. 1997;76:25–36. [PubMed] [Google Scholar]
- 95.Bironaite D, Siegel D, Moran JL, Weksler BB, Ross D. Stimulation of endothelial IL-8 (eIL-8) production and apoptosis by phenolic metabolites of benzene in HL-60 cells and human bone marrow endothelial cells. Chem Biol Interact. 2004;149:37–49. doi: 10.1016/j.cbi.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 96.Zhou H, Kepa JK, Siegel D, Shigenori M, Hiraki Y, Ross D. Benzene metabolite hydroquinone upregulates chondromodulin 1 and inhibits tube formation in human bone marrow endothelial cells. Mol Pharmacol. 2009 doi: 10.1124/mol.109.057323. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Winski SL, Faig M, Bianchet MA, Siegel D, Swann E, Fung K, Duncan MW, Moody CJ, Amzel LM, Ross D. Characterization of a Mechanism-Based Inhibitor of NAD(P)H:Quinone Oxidoreductase 1 by Biochemical, X-ray Crystallographic, and Mass Spectrometric Approaches. Biochemistry. 2001;40:15135–15142. doi: 10.1021/bi011324i. [DOI] [PubMed] [Google Scholar]
- 98.Zhou H, Dehn D, Kepa JK, Siegel D, Ross D. Downregulation of the cellular adhesion molecule VCAM-1 in human bone marrow endothelial cells after inhibition of NQO1 activity. Proc Amer Assoc Cancer Res. 2007;48(5371) [Google Scholar]
- 99.Dorshkind K. Regulation of hemopoiesis by bone marrow stromal cells and their products. Ann Rev Immunol. 1990;8:111–137. doi: 10.1146/annurev.iy.08.040190.000551. [DOI] [PubMed] [Google Scholar]
- 100.Eaves CJ, Cashman JD, Kay RJ, Dougherty GJ, Otsuka T, Gaboury LA, Hogge DE, Lansdorp PM, Eaves AC, Humphries RK. Mechanisms that regulate the cell cycle status of very primitive hematopoietic cells in long term human marrow cultures. II Analysis of positive and negative regulators produced by stromal cells within the adherent layer. Blood. 1991;78:110–117. [PubMed] [Google Scholar]
- 101.Yin T, Li L. The stem cell niches in bone. J Clin Invest. 2006;116:1195–1201. doi: 10.1172/JCI28568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang J, Li L. Stem cell niche: microenvironment and beyond. J Biol Chem. 2008;283:9499–9503. doi: 10.1074/jbc.R700043200. [DOI] [PubMed] [Google Scholar]
- 103.Ahn KS, Sethi G, Jain AK, Jaiswal AK, Aggarwal BB. Genetic deletion of NAD(P)H:quinone oxidoreductase 1 abrogates activation of nuclear factor-kappaB, IkappaBalpha kinase, c-Jun N-terminal kinase, Akt, p38, and p44/42 mitogen-activated protein kinases and potentiates apoptosis. J Biol Chem. 2006;281:19798–19808. doi: 10.1074/jbc.M601162200. [DOI] [PubMed] [Google Scholar]
- 104.Pyatt DW, Stillman WS, Irons RD. Hydroquinone, a reactive metabolite of benzene, inhibits NF-kappaB in primary human CD4+ T lymphocytes'. Toxicol Appl Pharmacol. 1998;149:178–184. doi: 10.1006/taap.1998.8369. [DOI] [PubMed] [Google Scholar]
- 105.Pyatt DW, Yang Y, Stillman WS, Cano LL, Irons RD. Hydroquinone inhibits PMA-induced activation of NFkappaB in primary human CD19+ B lymphocytes. Cell Biol Toxicol. 2000;16:41–51. doi: 10.1023/a:1007644620655. [DOI] [PubMed] [Google Scholar]
- 106.Kerzic PJ, Pyatt DW, Zheng JH, Gross SA, Le A, Irons RD. Inhibition of NF-kappaB by hydroquinone sensitizes human bone marrow progenitor cells to TNF-alpha-induced apoptosis. Toxicology. 2003;187:127–137. doi: 10.1016/s0300-483x(03)00064-7. [DOI] [PubMed] [Google Scholar]
- 107.Zheng JH, Pyatt DW, Gross SA, Le AT, Kerzic PJ, Irons RD. Hydroquinone modulates the GM-CSF signaling pathway in TF-1 cells. Leukemia. 2004;18:1296–1304. doi: 10.1038/sj.leu.2403389. [DOI] [PubMed] [Google Scholar]
- 108.Gross SA, Zheng JH, Le AT, Kerzic PJ, Irons RD. PU.1 phosphorylation correlates with hydroquinone-induced alterations in myeloid differentiation and cytokine-dependent clonogenic response in human CD34(+) hematopoietic progenitor cells. Cell Biol Toxicol. 2006;22:229–241. doi: 10.1007/s10565-006-0128-7. [DOI] [PubMed] [Google Scholar]
- 109.Adler V, Yin Z, Fuchs SY, Benezra M, Rosario L, Tew KD, Pincus MR, Sardana M, Henderson CJ, Wolf CR, Davis RJ, Ronai Z. Regulation of JNK signaling by GSTp. EMBO J. 1999;18:1321–1334. doi: 10.1093/emboj/18.5.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ruscoe JE, Rosario LA, Wang T, Gate L, Arifoglu P, Wolf CR, Henderson CJ, Ronai Z, Tew KD. Pharmacologic or genetic manipulation of glutathione S-transferase P1-1 (GSTpi) influences cell proliferation pathways. J Pharmacol Exp Ther. 2001;298:339–345. [PubMed] [Google Scholar]