Abstract
AIM: To study the characteristics of APC (adenomatous polyposis coli) gene germline mutation in Chinese patients with familial adenomatous polyposis (FAP).
METHODS: APC gene from 14 FAP families was amplified by polymerase chain reaction (PCR) and underwent direct sequencing to determine the micromutation type. For the samples without micromutation, the large fragment deletion of APC gene was examined by multiplex ligation-dependent probe amplification (MLPA).
RESULTS: There were gene micromutations in 9 families with a micromutation detection rate of 64.3% (9/14), including 6 frameshift mutations (66.7%), 1 nonsense mutation (11.1%) and 2 splicing mutations (22.2%). Large fragment deletions were detected by MLPA in 2 families. The total mutation detection rate of micromutations and large fragment deletions was 78.6% (11/14).
CONCLUSION: The detection rate of APC gene germline mutation can be improved by direct sequencing combined with MLPA large fragment deletion detection.
Keywords: Adenomatous polyposis coli gene, Familial adenomatous polyposis, Large fragment deletion, Multiplex ligation-dependent probe amplification, Mutation
INTRODUCTION
Familial adenomatous polyposis (FAP) is a rare autosomal dominant genetic disease, with an approximate incidence rate of 1/10 000. Clinical manifestations are mainly multiple adenomatous polyps in the large intestine (more than 100 polypi in total), and most of these patients fall ill in adolescence. Adenomatous polyps are a type of precancerous lesions. Thereby, cancerization will occur before the age of 40 years in almost 100% of patients without treatment. It was shown that the occurrence of FAP was related to APC (adenomatous polyposis coli) gene mutations located at 5q21-q22[1]. APC gene micromutations were identified in about 60%-70% of FAP patients[2], while large fragment deletion mutations of APC gene were identified in 10%-15%[3,4]. APC gene mutation screening in FAP patients and their family members cannot only further explore the pathogenesis of FAP and understand the APC gene mutation spectrum of Chinese FAP, but also predict the risk of FAP in “healthy members” of their families. It is also helpful for monitoring and in the clinical treatment of high-risk individuals with mutant genes, and can effectively decrease the incidence and mortality of FAP[5]. At present, only a few studies of Chinese FAP have been reported, and smaller sample size and lower detection rate (mostly at 50%) were the main problems in these studies. In order to further understand APC gene mutations in Chinese patients with FAP, a total of 14 FAP families were detected by direct sequencing combined with large fragment deletion detection in this study.
MATERIALS AND METHODS
Patients
From 2002 to 2008, 14 patients from FAP families diagnosed and treated in the General Hospital of Beijing Military Region were enrolled in this study. Diagnostic criteria were as follows: (1) more than 100 adenomatous polypi in total; (2) more than 20 adenomatous polypi in patients with a family history of FAP. The patients which included 7 males and 7 females were from Beijing, Hebei, Henan, Anhui, Inner Mongolia, Shan’xi, Fujian Provinces and other regions, and were aged 12-57 years (mean 35.21 years) with an onset age of 8-57 years (mean 28.14 years). All patients gave written informed consent.
Genomic DNA extraction
Ten milliliter peripheral venous blood was drawn from FAP patients and genomic DNA was extracted by the phenol/chloroform/isoamyl alcohol method.
Primer synthesis
Primer sequences for APC gene exons 1-15 were synthesized by Shanghai Sangon Biological Engineering Technology & Services Co. Ltd, as previously described[6].
PCR
Twenty microlitre PCR amplification reaction system contained 100 ng template DNA, 0.2 mmol/L of dNTP, 1.5 mmol/L of Mg2+, 0.1-0.2 μmol/L of upstream and downstream exon primers and 1-1.5 U TaqDNA polymerase. PCR reaction conditions were as follows: initial denaturation at 94°C for 5 min, followed by 35-40 cycles of denaturation at 94°C for 30 s, annealing for 30 s and extension at 72°C for 30 s, and a final extension at 72°C for 5 min, preservation was carried out at 4°C. After 1.5% agarose gel electrophoresis (containing EB dye), PCR products were observed by a gel imaging instrument. Fragment sizes of PCR products were indicated with DL2000 DNA Marker.
DNA sequencing
PCR reaction products were purified, and sequencing was performed by a DNA automatic sequencer (ABI PRISM 3730XL, USA). Changes in base sequence were confirmed by reverse sequencing.
Biological analyses
Sequence analyses were performed by BioEdit software. For the changed base sequences, the mutational site and type were determined in NCBI, and then the mutations were identified as new by referring to the mutations reported in the Human Gene Mutation Database (http://www.hgmd.cf.ac.uk/ac/gene.phpgene=APC) and the UMD-APC mutations database (http://www.umd.be/APC/).
Detection of large fragment deletions
Large fragment deletions were detected by multiplex ligation-dependent probe amplification (MLPA) only in samples without micromutations, and the experimental procedures followed the instruction manual of the MLPA kit (SALSA MLPA kit P043 APC, MRC-Holland, Amsterdam, the Netherlands). One hundred nanogram template DNA was denatured at 98°C for 5 min and hybridized with the probe liquid at 60°C overnight, subsequently, with thermal stability enzyme ligase 65, and spliced with long and short probes with the same sequences at 54°C for 15 min. Finally, the spliced probes were amplified by PCR with universal primers. Electrophoresis and collection of PCR amplification products were finished in an ABI 3700 sequenator, and the results were analyzed by GeneMapper software to obtain a peak map and peak area.
RESULTS
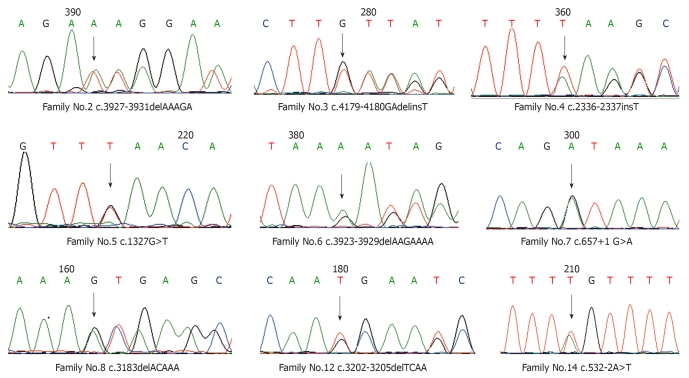
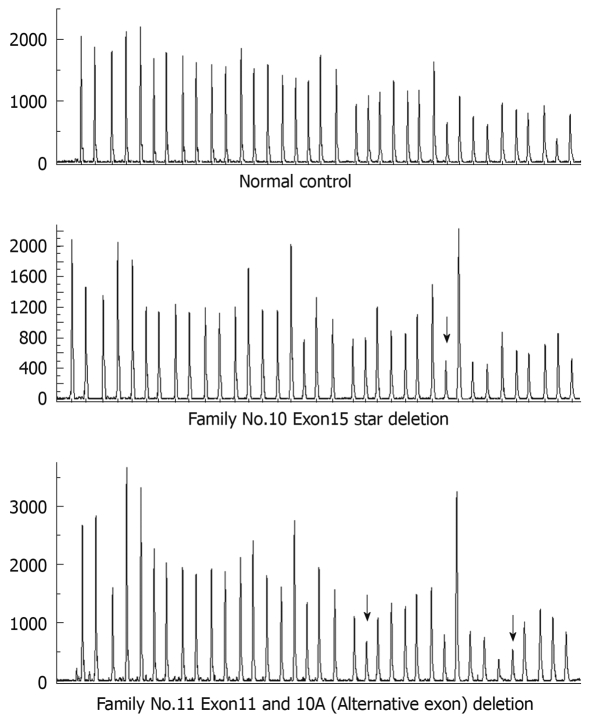
A total of 9 micromutations were identified by direct sequencing among these 14 unrelated FAP families with a mutation rate of 64.3%, including 6 frameshift mutations (66.7%), 2 splicing mutations (22.2%) and 1 nonsense mutation (11.1%), of which 4 mutations including c.2336-2337insT, c.3923-3929delAAGAAAA, c.532-2A>T and c.4179-4180GAdelinsT have not previously been reported (Table 1 and Figure 1). Among the 5 patients without micromutations identified by direct sequencing, large fragment deletions were identified by the MLPA method in 2 patients, including large fragment deletions of exon 11 and 10A (Alternative exon) in one patient and a large fragment deletion of exon 15 start in another patient (three probes included start, middle and end in the MLPA detection of exon 15), and these two large fragment deletions have not previously been reported (Table 1 and Figure 2).
Table 1.
Micromutations and large fragment deletions of APC gene germline mutation in Chinese patients with familial adenomatous polyposis detected in this study
| Family No. | Exons/introns | Base changes | Protein changes | Mutation types |
| 2 | Exon15 | c.3927-3931delAAAGA | p.Glu1309AspfsX4 | Frameshift mutation |
| 3 | Exon15 | c.4179-4180GAdelinsT1 | p.Asp1394LeufsX21 | Frameshift mutation |
| 4 | Exon15 | c.2336-2337insT1 | p.Leu779PhefsX9 | Frameshift mutation |
| 5 | Exon10 | c.1327G>T | p.Glu443X | Nonsense mutation |
| 6 | Exon15 | c.3923-3929delAAGAAAA1 | p.Lys1308ArgfsX11 | Frameshift mutation |
| 7 | Intron7 | c.657+1 G>A | Splicing mutation | |
| 8 | Exon15 | c.3183delACAAA | p.Gln1062X | Frameshift mutation |
| 10 | Exon15 start1 | Large fragment deletion | ||
| 11 | 10A (Alternative exon) and Exon111 | Large fragment deletion | ||
| 12 | Exon15 | c.3202_3205delTCAA | p.Ser1068GlyfsX57 | Frameshift mutation |
| 14 | Intron4 | c.532-2A>T1 | Splicing mutation |
Figure 1.
DNA sequencing of micromutations.
Figure 2.
Peak of large fragment deletion detected by multiplex ligation-dependent probe amplification (MLPA). Arrows show the reduced relative peak area of the amplification product of that probe which means heterozygous deletions of corresponding exons.
The detection rate of large fragment deletions in APC mutation-negative patients was 40%, while the total mutation rate of micromutations and large fragment deletions was 78.6%. Meanwhile, 7 Snp sites were detected in 14 families (Table 2). Missense mutations including c.2753C>A, c.4007G>C and c.3964C>T were found in families 3, 11 and 14 besides frameshift mutations, large fragment deletions and splicing mutations, respectively. Due to difficulty in assessing pathopoiesis of missense mutations, these 3 missense mutations were excluded in the calculation of the mutation detection rate during analyses of the results. Moreover, one base substitution of intron 14 (c.653+8T>C) was found in family 13. Due to unknown effects of intron mutation, this was also excluded in the calculation of the mutation detection rate.
Table 2.
Snp sites of APC gene germline mutation in Chinese patients with familial adenomatous polyposis detected by DNA sequencing
| Exons | APC gene loci | Base changes | Ncbi dbsnp ID | Positive families |
| Exon11 | 89271 | C/T | refSNP ID: rs2229992 | 1-8; 10; 12-14 |
| Exon13 | 90978 | A/G | refSNP ID: rs351771 | 1-12 |
| Exon15 | 102187 | A/G | refSNP ID: rs41115 | 1-7; 9-14 |
| Exon15 | 102742 | A/G | refSNP ID: rs42427 | 1-14 |
| Exon15 | 102976 | G/T | refSNP ID: rs866006 | 1-13 |
| Exon15 | 103173 | A/T | refSNP ID: rs459552 | 1-14 |
| Exon15 | 103588 | A/G | refSNP ID: rs465899 | 1-14 |
DISCUSSION
APC gene, a tumor suppressor gene, is the key gene in FAP. It is located at 5q21-q22 and contains an 8538 bp open reading-frame and a total of 15 exons. The APC gene product, APC protein, is a multi-regional binding protein containing 2843 amino acids. The majority of APC gene mutations will result in the earlier formation of terminal codons in downstream. APC protein will loose biological activities due to its truncated change. Although there are numerous methods for detecting APC gene micromutations, direct sequencing is the most direct and accurate method. MLPA is a new method for detecting large fragment deletions which is rapid, sensitive, specific and reliable and has other advantages. In order to improve the detection rate of APC gene mutations, direct sequencing and the MLPA method for detecting large fragment deletions were combined in this study.
Direct sequencing showed that the APC gene mutation rate in Chinese FAP patients (Mainland) was 64.3%, which was significantly higher than that in Taiwan (50%)[6], Hong Kong (50%)[6] and other reports in the Mainland (48.39%[7] and 50%[8]). This difference in Chinese APC gene mutation rates might result from different human subjects and detection methods and small sample sizes. The detection rate in this study was close to that in Japan (67%[9] and 64.67%[10]), lower than that in Greece (83%)[6] and Chile (87.5%)[11], and slightly higher than that in the Czech Republic (59.3%)[12], Slovakia (61.5%)[12] and South Korea (61%)[13]. Combined with the MLPA method for detecting large fragment deletions, the total detection rate of APC gene mutations was up to 78.6%, which was higher than that reported in Czechoslovakia (67.6%)[9].
Among the 9 micromutations detected, the majority were frameshift mutations (66.7%) located at exon 15 (66.7%). Eventually, 77.8% of the mutations including nonsense mutations and frameshift mutations resulted in the earlier formation of terminal codons, and truncated changes were found in APC proteins. Substitution of bases in site +1 of exon 7 splicing district in family 7 and in site -2 of exon 5 splicing district in family 14 might lead to abnormal mRNA splicing. Therefore, the synthesized proteins were different from the wild-type. Four new micromutations found in this study were located at codon 779, 1308, 1394 and c.532-2, respectively.
With the exception of the above-mentioned 9 micromutations, 7 Snp sites were identified in this study, which was consistent with NCBI reports. Base substitution located at c.653+8 in intron 14 was found in family 13. However, the mechanisms of intron mutation are not yet understood. Therefore, the relationship between the mutation at c.653+8 and the onset of FAP should be studied further.
We also found two mutations in one family, including: (1) a frameshift mutation c.4179-4180GAdelinsT and a missense mutation c.2753C>A in family 3; (2) large fragment deletions of exon 10A and exon 11 and a missense mutation c.4007G>C in family 11; and (3) splicing mutations c.532-2A>T and a missense mutation c.3964C>T in family 14. Because the frameshift mutation in family 3, large fragment deletion in family 11 and splicing mutation in family 14 were definite pathogenic mutations, it was difficult to identify the pathopoiesis of simultaneous missense mutations, indicating that undetected definite pathogenic mutations would decrease after comprehensive screening for all exons of APC genes combined with large fragment deletion detection. Therefore, functional experiments should be carried out to identify the pathopoiesis of missense mutations and understand the causal relation with occurrence of diseases.
Two large fragment deletions were found by the MLPA method in patients without micromutations detected by direct sequencing. Deletions of 10A and exon 11 were found in one case. Because 10A was located at 1.6 kb in the downstream of exon 10, this indicated that deletions of exon 10A and exon 11 including some parts of intron 10 existed in this case. Deletion of exon 15 start was found in another case. These two large fragment deletions have not been reported in the Human Gene Mutation Database (http://www.hgmd.cf.ac.uk/ac/gene.phpgene=APC). MLPA showed that the detection rate of large fragment deletions in patients without APC gene mutations was 40% (2/5), which was higher than that in Belgium (15%)[3] and the Netherlands (6.4%)[14]. Because of the small sample size in the research on large fragment deletions (5 cases) and few reports on APC gene large fragment deletions in Chinese patients, studies on APC gene large fragment deletions in Chinese FAP need to be carried out with a larger sample size.
In addition, 10A, an additional APC gene exon, was found recently located at 1.6 kb in the downstream of exon 10. 10A consisted of 54 bases encoding 18 amino acids[15]. One case of 10A deletion was detected in this study. Therefore, APC gene screening should include exon 10A. Recently, the studies on MYH genes and MYH associated polyposis (MAP) revealed that MAP accounted for 5%-7.5 % of FAP. It was found by Sieber et al[16] that diallelic MYH mutations accounted for 6.6 % of polyposis families without APC gene mutations, and accounted for almost one third of FAP in families with attenuation-type familial adenomatous polyposis. Detection of MYH gene mutations should be performed in patients who have no mutations shown by mutation detection and large fragment deletion detection of APC genes.
In conclusion, there were a variety of APC gene mutations in Chinese FAP, and mutation detection rates were relatively high. Four new micromutations and 2 new large fragment deletions were found in this study. APC gene mutation detection rates could be improved effectively by direct sequencing combined with the MLPA method for large fragment deletions. Therefore, it was very necessary to add large fragment deletion detection to conventional detection of molecular genetics.
COMMENTS
Background
Familial adenomatous polyposis (FAP) is a rare autosomal dominant genetic disease. Clinical manifestations are mainly multiple adenomatous polyps in large intestine, and most patients fall ill in adolescence. Cancerization will occur before the age of 40 years in almost 100% of patients without treatment. FAP is related to APC (adenomatous polyposis coli) gene mutations located at 5q21-q22.
Research frontiers
APC gene micromutations were identified in about 60%-70% of FAP patients, while large fragment deletions were identified in 10%-15%. Direct sequencing is the most accurate method for detecting APC gene micromutations. Multiplex ligation-dependent probe amplification (MLPA) is a new method for detecting large fragment deletions.
Innovations and breakthroughs
In order to improve the detection rate of APC gene mutations, direct sequencing and the MLPA method for large fragment deletion detection were combined in this study. The total detection rate of APC gene mutations was up to 78.6%. Moreover, 4 novel micromutations and 2 novel large fragment deletions were found in this study.
Applications
APC gene mutation detection rates could be improved effectively by direct sequencing combined with the MLPA method for large fragment deletions. Therefore, it was very necessary to add large fragment deletion detection to conventional detection of molecular genetics.
Terminology
Multiplex ligation-dependent probe amplification (MLPA) is a variation of PCR that permits multiple targets to be amplified with only a single primer pair. Each probe consists of a two oligonucleotides which recognise adjacent target sites on the DNA. One probe oligonucleotide contains the sequence recognised by the forward primer, the other sequence is recognised by the reverse primer. Only when both probe oligonucleotides are hybridized to their respective targets, can they be ligated into a complete probe.
Peer review
The work described in this report is a service to the community concerned with APC gene mutations. The discovery of new mutations in an important gene and disease sometimes can qualify for a publication, especially in a relevant journal. This article is well written and the work done properly.
Footnotes
Supported by The National Natural Science Foundation of China, No. 30940086
Peer reviewer: Dr. Anthony T Yeung, BS, MS, PhD, Fox Chase Cancer Center, Room R404, 333 Cottman Avenue, Philadelphia, PA 19111, United States
S- Editor Wang JL L- Editor Webster JR E- Editor Ma WH
References
- 1.Leppert M, Dobbs M, Scambler P, O’Connell P, Nakamura Y, Stauffer D, Woodward S, Burt R, Hughes J, Gardner E. The gene for familial polyposis coli maps to the long arm of chromosome 5. Science. 1987;238:1411–1413. doi: 10.1126/science.3479843. [DOI] [PubMed] [Google Scholar]
- 2.González S, Blanco I, Campos O, Julià M, Reyes J, Llompart A, Cabeza E, Germà JR, Obrador A, Capellá G. Founder mutation in familial adenomatous polyposis (FAP) in the Balearic Islands. Cancer Genet Cytogenet. 2005;158:70–74. doi: 10.1016/j.cancergencyto.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Michils G, Tejpar S, Thoelen R, van Cutsem E, Vermeesch JR, Fryns JP, Legius E, Matthijs G. Large deletions of the APC gene in 15% of mutation-negative patients with classical polyposis (FAP): a Belgian study. Hum Mutat. 2005;25:125–134. doi: 10.1002/humu.20122. [DOI] [PubMed] [Google Scholar]
- 4.Nieuwenhuis MH, Vasen HF. Correlations between mutation site in APC and phenotype of familial adenomatous polyposis (FAP): a review of the literature. Crit Rev Oncol Hematol. 2007;61:153–161. doi: 10.1016/j.critrevonc.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Vasen HF, Möslein G, Alonso A, Aretz S, Bernstein I, Bertario L, Blanco I, Bülow S, Burn J, Capella G, et al. Guidelines for the clinical management of familial adenomatous polyposis (FAP) Gut. 2008;57:704–713. doi: 10.1136/gut.2007.136127. [DOI] [PubMed] [Google Scholar]
- 6.Wei SC, Su YN, Tsai-Wu JJ, Wu CH, Huang YL, Sheu JC, Wang CY, Wong JM. Genetic analysis of the APC gene in Taiwanese familial adenomatous polyposis. J Biomed Sci. 2004;11:260–265. doi: 10.1007/BF02256569. [DOI] [PubMed] [Google Scholar]
- 7.Cai SR, Zhang SZ, Zheng S. [Detection of adenomatous polyposis coli gene mutations in 31 familial adenomatous polyposis families by using denaturing high performance liquid chromatography] Zhonghua Yixue Yichuanxue Zazhi. 2008;25:164–167. [PubMed] [Google Scholar]
- 8.Lou Z, Yu ED, Meng RG, Fu CG, Liu LJ. Preliminary study on APC gene germline mutation in familial adenomatous polyposis patients. Dier Junyi Daxue Xuebao. 2006;27:358–361. [Google Scholar]
- 9.Miyoshi Y, Ando H, Nagase H, Nishisho I, Horii A, Miki Y, Mori T, Utsunomiya J, Baba S, Petersen G. Germ-line mutations of the APC gene in 53 familial adenomatous polyposis patients. Proc Natl Acad Sci USA. 1992;89:4452–4456. doi: 10.1073/pnas.89.10.4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagase H, Miyoshi Y, Horii A, Aoki T, Petersen GM, Vogelstein B, Maher E, Ogawa M, Maruyama M, Utsunomiya J. Screening for germ-line mutations in familial adenomatous polyposis patients: 61 new patients and a summary of 150 unrelated patients. Hum Mutat. 1992;1:467–473. doi: 10.1002/humu.1380010603. [DOI] [PubMed] [Google Scholar]
- 11.De la Fuente MK, Alvarez KP, Letelier AJ, Bellolio F, Acuña ML, León FS, Pinto E, Carvallo P, López-Köstner F. Mutational screening of the APC gene in Chilean families with familial adenomatous polyposis: nine novel truncating mutations. Dis Colon Rectum. 2007;50:2142–2148. doi: 10.1007/s10350-007-9044-z. [DOI] [PubMed] [Google Scholar]
- 12.Stekrova J, Sulova M, Kebrdlova V, Zidkova K, Kotlas J, Ilencikova D, Vesela K, Kohoutova M. Novel APC mutations in Czech and Slovak FAP families: clinical and genetic aspects. BMC Med Genet. 2007;8:16. doi: 10.1186/1471-2350-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Won YJ, Park KJ, Kwon HJ, Lee JH, Kim JH, Kim YJ, Chun SH, Han HJ, Park JG. Germline mutations of the APC gene in Korean familial adenomatous polyposis patients. J Hum Genet. 1999;44:103–108. doi: 10.1007/s100380050118. [DOI] [PubMed] [Google Scholar]
- 14.Nielsen M, Bik E, Hes FJ, Breuning MH, Vasen HF, Bakker E, Tops CM, Weiss MM. Genotype-phenotype correlations in 19 Dutch cases with APC gene deletions and a literature review. Eur J Hum Genet. 2007;15:1034–1042. doi: 10.1038/sj.ejhg.5201871. [DOI] [PubMed] [Google Scholar]
- 15.Suleková Z, Ballhausen WG. A novel coding exon of the human adenomatous polyposis coli gene. Hum Genet. 1995;96:469–471. doi: 10.1007/BF00191808. [DOI] [PubMed] [Google Scholar]
- 16.Sieber OM, Lipton L, Crabtree M, Heinimann K, Fidalgo P, Phillips RK, Bisgaard ML, Orntoft TF, Aaltonen LA, Hodgson SV, et al. Multiple colorectal adenomas, classic adenomatous polyposis, and germ-line mutations in MYH. N Engl J Med. 2003;348:791–799. doi: 10.1056/NEJMoa025283. [DOI] [PubMed] [Google Scholar]