Abstract
Genomic imprinting is widely conserved amongst placental mammals. Imprinted expression of IGF2R, however, differs between mice and humans. In mice, Igf2r imprinted expression is seen in all fetal and adult tissues. In humans, adult tissues lack IGF2R imprinted expression, but it is found in fetal tissues and Wilms' tumors where it is polymorphic and only seen in a small proportion of tested samples. Mouse Igf2r imprinted expression is controlled by the Air (Airn) ncRNA whose promoter lies in an intronic maternally-methylated CpG island. The human IGF2R gene carries a homologous intronic maternally-methylated CpG island of unknown function. Here, we use transfection and transgenic studies to show that the human IGF2R intronic CpG island is a ncRNA promoter. We also identify the same ncRNA at the endogenous human locus in 16–40% of Wilms' tumors. Thus, the human IGF2R gene shows evolutionary conservation of key features that control imprinted expression in the mouse.
Keywords: Genomic imprinting, IGF2R, AIR (AIRN) ncRNA, Wilms' tumor
Introduction
Genomic imprinting is an epigenetic process that leads to parental-specific expression in diploid cells [1,2]. In mammals, genomic imprinting affects approximately 80 autosomal genes that are mostly grouped into small clusters [3,4]. Imprinted gene expression has been identified in diverse mammals and, with the exception of egg-laying mammals, may be conserved in all mammalian classes – reinforcing a proposed link between genomic imprinting and maternal control of fetal growth [5]. Despite this general conservation amongst mammals, many genes have been identified that lack conserved imprinted expression between mice and humans [6]. This has been suggested to be an adaptation to different reproductive strategies in human monotoccus (i.e., single offspring) and mouse polytoccus (i.e., multiple offspring) pregnancies [7].
The mouse Igf2r (insulin-like growth factor type 2 receptor) gene is part of an imprinted cluster spanning 500 kb on chromosome 17, that contains three maternally-expressed protein-coding genes (Igf2r, Slc22a2 and Slc22a3), and, one paternally-expressed ncRNA (Air, named from Antisense to Igf2r RNA Noncoding) [8]. Despite the extensive synteny of this region between mice and humans (http://www.ensembl.org), imprinted expression differs. In mice, Igf2r and Air show ubiquitous imprinted expression in fetal, extra-embryonic and adult tissues with the exception of post-mitotic neurons [9], while Slc22a2 and Slc22a3 show imprinted expression only in placenta in a temporally-regulated manner [10]. In humans, imprinted expression of IGF2R appears to be absent from all tested adult tissues [11,12]. However, human IGF2R, SLC22A2 and SLC22A3 all show a polymorphic and concordant form of imprinted expression in placenta in approximately 40% of informative samples [7]. Polymorphic imprinting of IGF2R has also been seen in early fetal tissue, cultured amniotic cells, lymphoblastoid cells and Wilms' tumors [13-16]. However, the imprinted status of human IGF2R remains controversial as some studies show a complete lack of imprinted IGF2R expression in fetal tissues [17] and some publications refer to human IGF2R as a non-imprinted gene [18].
Imprinted expression of the mouse Igf2r, Slc22a2 and Slc22a3 genes is caused by paternal-specific expression of the Air ncRNA. Imprinted expression of Air itself, is caused by a DNA methylation imprint acquired during oocyte development that silences the maternal Air promoter [19]. Mouse Air expression is driven by a CpG island promoter in Igf2r intron 2, that is antisense to the Igf2r promoter thus the Air ncRNA overlaps the 5′ part of Igf2r. The Air ncRNA does not overlap the two other genes it silences [20]. The mouse Air ncRNA is an unusual RNAPII transcript that evades cotranscriptional splicing, resulting in a mature 108 kb ncRNA that is nuclear localized and relatively unstable [21].
The human IGF2R gene also contains a CpG island in intron 2 that carries a similar maternal-specific DNA-methylation imprint in all tested fetal and adult tissues [15,22]. However, no evidence of a human AIR transcript has been found [13]. Since CpG islands strongly correlate with promoter predictions [23], we test here the ability of the human IGF2R intron 2 CpG island to act as a promoter in vitro in transfection assays, and, in vivo in transgenic mice. The results show that the human IGF2R intron 2 CpG island is a promoter that produces a ncRNA orientated antisense to human IGF2R. Moreover, we use the transgenic human cDNAs to design RT-PCR assays that identified the same ncRNA at the endogenous human locus in 16–40% of human Wilms' tumor samples. The transcriptional start site of the endogenous human AIR ncRNA was mapped in Wilms' tumors samples and shown to coincide with a DNase1 hypersensitive site present only on the unmethylated paternal intron 2 CpG island. The presence of a DNase1 site at the endogenous human locus that indicates open chromatin, together with the demonstration that the human IGF2R intron 2 CpG island can be an active promoter in transfection and transgene assays, indicates that lack of AIR expression in human tissues may result from the absence of essential transcription factors for this ncRNA promoter.
Results
Promoter activity of the human IGF2R intron 2 CpG island in transfection assays
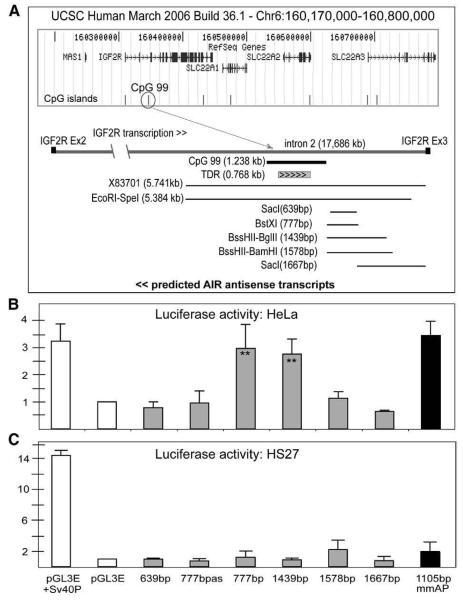
Transient transfection assays were performed in two human cell lines using fragments from the human IGF2R intron 2 CpG island (here called CGI-2) subcloned into the pGL3 expression vector. Six reporter constructs (Fig. 1A), containing a SV40 enhancer plus different lengths of CGI-2 sequences were tested for promoter activity. Of all human fragments, only a 777 bp BstXI fragment and an overlapping 1439 bp BssHII–BglII fragment (X83701/3344–4121 bp and X83701/3359–4798 bp) showed significant Luciferase activity in HeLa cells (Fig. 1B). Similar activity was shown by a control mouse Air promoter (mmAP). No promoter activity was detected from a 1578 bp BssHII–BamHI fragment also spanning the promoter region. No fragment had promoter activity in HS27 human foreskin fibroblasts (Fig. 1C), or in mouse NIH3T3 fibroblasts (data not shown). These results show that sequences immediately upstream of human CGI-2 have promoter activity in some cell types.
Fig. 1.
Human CGI-2 promoter activity in transient transfection assays. (A) 630 kb map from human Chr.6 including IGF2R (http://genome.ucsc.edu, Human Build 36.1 assembly March 2006). CpG islands indicated by bars – CpG-99 (circled) is a 1238 bp CpG island located in IGF2R intron 2 (160,346,255–160,347,492) that contains 99 CpG dinucleotides with a 0.66 CpG:GpC observed:expected ratio. A map of IGF2R intron 2 is shown underneath, the direction of the IGF2R mRNA is indicated and exons 2 and 3 by black boxes. The position of CpG-99 (identified using CpG plot, http://www.ebi.ac.uk/servicestmp/cpgplot) within IGF2R intron 2, is indicated by a black line. A 768 bp region within CpG-99 that contains a series of Tandem Direct Repeats (TDR) is indicated by the grey box marked with arrowheads. The lines underneath show the fragments used in transient transfection analyses. Clone pE3up (X83701) contains a 5.741 kb fragment from intron 2 of IGF2R and was used to prepare five fragments to test for transient expression after subcloning into the pGLE3 vector (Sac1/639 bp, BstXI/777 bp, BssHII–BglII/1439 bp, BssHII–BamHI/1578 bp, SacI/1667 bp). The EcoRI–SpeI fragment was used to prepare transgenic constructs shown in Fig. 2. The predicted orientation of AIR antisense transcripts is shown underneath. (B–C) Luciferase activity after transfection in HeLa and HS27 human fibroblast cells. Activity of the control pGL3E vector without promoter was set to 1. Results are shown for the pGL3E vector plus control promoters (Sv40 and mouse Air promoters: Sv40P and mmAP) or plus the test human IGF2R intron 2 CpG island fragments shown above. Note that fragment BstXI (777 bp) was assayed in sense (777 bp) and antisense (777 bpas) orientation. **Statistically significant (p < 0.001) promoter activity. The mean values of two biological replicates, each performed in five technical replicates, are shown.
Promoter activity of the human IGF2R intron 2 CpG island in mouse transgenes
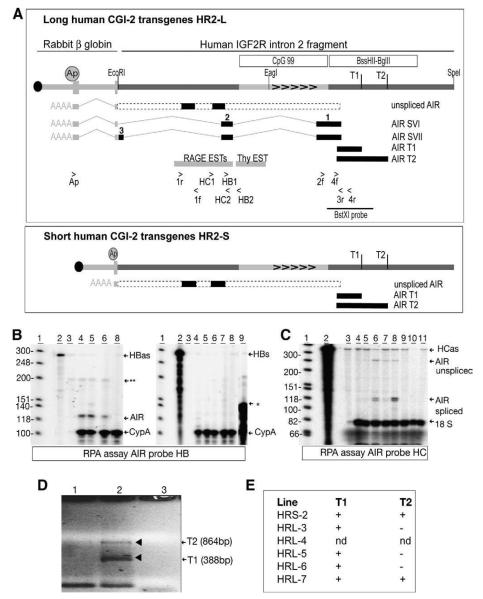
To test expression of human CGI-2 in a normal chromatin environment and validate the transient expression studies, mouse transgenes carrying human CGI-2 were generated. Two transgenic constructs: Human Region 2 Long (HR2-L) and Human Region 2 Short (HR2-S) were generated (Fig. 2A). HR2-L contained a 5383 bp human CGI-2 fragment that included the region identified above showing transient promoter activity, joined to a 1.2 kb polyadenylation signal orientated to terminate the predicted AIR transcript. HR2-S is identical except for a shorter polyadenylation cassette. Nine mouse transgenic lines representing nine independent integration sites with transgene copy numbers ranging from 4 to 25 were generated by oocyte injection (Fig. 2B). Two transgenes (HR2-L2 and HR2-L8) did not transmit through the germ line and expression was analyzed in founder (F0) mice. Of the remaining 7 transgenic lines, all were free of methylation after transmission through the maternal and paternal germ line and 8/9 transgenes expressed a human CGI-2 transcript (Fig. 2B, and data not shown).
Fig. 2.
Human CGI-2 transgenic mouse constructs. (A) Map of human CGI-2 constructs used to create transgenic mice. HR2-L: a single LoxP1 site (black circle), a 1.2 kb rabbit β-globin polyA cassette that contains exon 3, intron 2 and the 3′ end of exon 2 (grey bar with exons as grey boxes), 13 bp of vector sequence and 1–5384 bp (EcoRI–SpeI) from X83701. HR2-S differs in length of the polyA cassette that is reduced to 0.5 kb and only contains the 3′ end of exon 3 and thus lacks any splice acceptors. White box; the 1439 bp BssHII–BglII fragment with promoter activity identified in Fig. 1. SVI probe; the human spliced AIR transcript used as a Northern blot probe in C. BstXI; the fragment used for Southern blots in B. Grey bar with arrowheads; 768 bp region shown in Fig. 1A containing tandem direct repeats. Grey circle labeled Ap; polyA signal orientated to terminate the predicted AIR transcript. (B) Table showing methylation and expression status of human CGI-2 in 9 transgenic lines. TG-Line; transgenic mouse line, Copy No; transgene copy number (see methodsMaterials and methods), Mat-meth and Pat-meth; methylation upon maternal and paternal transmission (see Materials and methods), Mat-exp and Pat-exp; expression upon maternal and paternal transmission (C). nt; not transmitted through germline. F0; founder generation. (C) Northern blots showing expression of 3 human CGI-2 transcripts (4 kb, 1 kb and 0.5 kb) in adult brain from different transgenic lines, using probe SVI. Mouse Gapdh was used as a loading control. L7; HR2-L7, L5; HR2-L5, L4; HR2-L4, S1; HR2-S1, wt; wildtype brain, L3; HR2-L3, S2; HR2-S2. (D) Northern blot of adult S2 transgene organs showing tissue-specific expression in spleen and brain of a 4 kb human CGI-2 transcript. 1; liver, 2; lung, 3; kidney, 4; spleen, 5; uterus, 6; brain, 7; wild type brain. (E) Northern blot of paternal (HR2-L3-Pat) and maternal (HR2-L3-Mat) expression in adult HR2-L3 transgenic organs showing a lack of imprinted expression. Lung (lane 2, 4 kb transcript) and heart (lane 3, 1.5 kb transcript) showed variation between maternal and paternal blots that was not reproducible and reflects a loading variation. Lanes 1; liver, 2; lung, 3; heart, 4; kidney, 5; testis (4 kb transcript), 6; spleen (0.5 kb transcript), 7; brain (1.5 kb transcript), 8; wildtype brain. *non-specific band. (F) Northern blot of paternal (HR2-L5-Pat) and maternal (HR2-L5-Mat) expression in adult HR2-L5 transgenic organs showing a lack of imprinted expression. The weak expression in the kidney sample (lane 4, 0.5 kb transcript) showed variation between maternal and paternal blots that was not reproducible and reflects a loading variation. Lanes wt; wildtype brain, 1; liver, 2; lung, 3; heart, 4; kidney, 5; testis (0.5 kb transcript), 6; spleen, 7; brain (0.5 kb transcript).
Human CGI-2 expression analyzed by Northern blot of adult mouse brain showed expression of three different sized transcripts (Fig. 2C). A 0.5 kb transcript in lines HR2-L7, HR2-L5 and HR2-L4, a 1.0 kb transcript in line HR2-L3, and a 4.0 kb transcript in line HR2-S2. Line HR2-S2 also expressed the 4.0 kb transcript strongly in spleen and weakly in uterus (Fig. 2D). Six transgenic lines (HR2-S2, HR2-L3, HR2-L4, HR2-L5, HR2-L6, HR2-L7) were tested for imprinted human CGI-2 expression in 13.5dpc embryo, placenta and adult mouse tissues. All transgenes were equally expressed following maternal and paternal transmission and thus lacked imprinted expression, as shown for HR2-L3 and HR2-L5 (Fig. 2E, F). Line HR2-L3 expressed all three RNA isoforms (0.5, 1.0, 4.0 kb) in a tissue-specific fashion that were all similarly expressed upon maternal and paternal transmission (Fig. 2E). Line HR2-L5 expressed only the 1.0 kb isoform on maternal and paternal transmission. Notably, although the above transient expression studies showed no expression from a 1578 bp fragment, mouse transgenes that included this region were expressed when inserted in 6 independent integration sites. In summary, the results show that in embryonic and adult tissues in transgenic mice the human CGI-2 is an active non-imprinted promoter.
Transgenic human CGI-2 produces spliced and unspliced transcripts
Northern blots in Fig. 2 identified various sized human CGI-2 transcripts indicating alternative splicing. We have previously shown that the mouse Air ncRNA is an unusual RNAPII promoter that produces predominantly unspliced 108 kb transcripts and a small percentage of spliced transcripts that appear to be non-functional [21]. The processing of human CGI-2 transcripts was investigated by RT-PCR and sequencing using multiple primers shown in Fig. 3A. Two spliced variants (AIR SVI, Accession No. DQ220010 and AIR SVII, Accession No. DQ220011, Sup. Fig. 1) were identified in mice carrying the HR2-L transgene construct. Both spliced variants used the same human CGI-2 exons 1 and 2 as well as the splice donor/acceptor in the β-globin polyadenylation cassette, however, SVII contained an additional human CGI-2 exon (Fig. 3A and data not shown). Evidence for unspliced human CGI-2 transcripts was also obtained from primers 1r/1f that amplified a 281 bp fragment in both transgene types (data not shown). The presence of unspliced human CGI-2 transcripts is suggested from the abundant 4.0 kb signal in Northern blots shown in Fig. 2, that is similar to the length of transgene DNA downstream of sequences showing transient promoter activity. The unspliced 4.0 kb cDNA was the only isoform seen in HR2-S2 transgenes carrying the shorter polyA cassette.
Fig. 3.
Human CGI-2 transgenes express spliced and unspliced transcripts. (A) Human CGI-2 transcripts analyzed by RT-PCR for HR2-L (top box) and HR2-S (lower box) transgenes. A transgene map is shown above (details as in Fig. 2A) with primer locations. T1, T2 indicate the sequence identified by 5′RACE with primers 2f+4f, as transcriptional start sites (D). Grey bars indicate ESTs in this region (http://genome.ucsc.edu) see Sup. Fig. 2. The position of splice variants SVI and SVII identified by primers Ap+3r, is shown. Introns are indicated by grey lines, exons by numbered black bars. The dashed bar indicates the predicted unspliced AIR transcript corresponding to the 4 kb Northern blot signal in Fig. 2C. Black bars within this indicate unspliced AIR transcripts detected with primers 1r, 1f and HC RPA probe. HC1/HC2 and HB1/HB2: primers used to prepare RNase protection probes. (B) RNase protection assay with probe HB in brain samples from adult transgenic mice showing expression of spliced human CGI-2 transcripts. The probe HBas protects transcripts with the opposite orientation to IGF2R and identifies a 120 bp spliced SVI and SVII transcript (labeled AIR, present in lanes 4–6), and a 199 bp unspliced transcript (not distinguishable from a non-specific product**). Probe HBs protect a 120 bp product with the same orientation to IGF2R and fails to identify any transcripts from the human CGI-2. An in vitro transcribed 140 bp fragment from probe HBas is used as a positive control for the HBs probe (*lane 9). Mouse Cyclophilin A (protecting 105 bp) was used as a loading control. 1; marker, 2,3; yeast RNA controls minus/plus RNase treatment, 4; HR2-L4 brain, 5; HR2-L6 brain, 6; HR2-L3 brain, 7; HR2-L5 brain, 8; wild type brain. (C) RNase protection assay with probe HC in adult HR2-L3 transgenic organs showing relative abundance of spliced and unspliced human CGI-2 transcripts. The probe protects 121 bp spliced AIR transcript in lanes 5–8 (the band is weak in lane 5), and a 260 bp unspliced AIR transcript in lanes 5–8 (the band is weak in lane 5). Mouse 18S probe (protecting 80 bp) was used as a loading control. 1; marker, 2; HC probe, 3; yeast controls minus RNase, 4; liver, 5; lung, 6; brain, 7; heart, 8; testis, 9; kidney, 10; wild type testes control, 11; wild type kidney control. (D) Identification of human AIR transcriptional start sites in HR2-S2 transgenic mice by 5′RACE with gene specific primers 2f and 4f. Lane 1; minus phosphotase control, lane 2; plus phosphotase, lane 3; no template. T1, T2; transcription start site 1 and 2. (E) T1 and T2 start sites in human CGI-2 transgenic mice. nd; not done.
RNase Protection Assays (RPA) were used to identify the orientation of human CGI-2 transcripts. The HB antisense probe recognized a specific 120 bp fragment in transgenic RNA samples corresponding to the opposite strand encoding IGF2R (Fig. 3B, left). Transcription in the opposite direction was not seen (Fig. 3B, right). RPA was also used to show the relative abundance of the spliced to unspliced human CGI-2 transcripts, which varied from 4.3:1 to 2.1:1 (Fig. 3C). Note that in the mouse, spliced Air transcripts are more stable than unspliced transcripts [21].
Sequence analysis of the spliced variants and of unspliced human CGI-2 transcripts (http://repeatmasker.org, http://www.ncbi.nlm.nih.gov/projects/gorf/) shows they contain 30% interspersed repeats and lack a continuous open reading frame longer than 120 bp, indicating that all are non-coding (Sup. Fig. 1 and data not shown). These results show that human CGI-2 transgenes can express both unspliced and spliced non-coding transcripts from the DNA strand opposite to IGF2R. Human CGI-2 transcripts are here named AIR (Antisense IGF2R RNA) to correspond with the mouse name.
Transgenic human CGI-2 transcriptional start sites
Two transcriptional start sites were identified in mouse brain for line L7 (Fig. 3D). T1 (Accession No. DQ220013) is located 721 bp upstream of human CGI-2 at position 3904 bp in X83701. T2 (Accession No. DQ220014) is located further upstream at position 4377 bp in X83701 (Sup. Fig. 1). Both T1 and T2 lie within the 0.7 kb fragment with promoter activity identified in Fig. 1. T1 was identified in all tested transgenic lines (Fig. 3E) while T2 was only seen in L7 and S2. Active transcription originating upstream of T2 was also identified but this was shown not to arise from an upstream start site but from read-through of the upstream transgene copy (data not shown). In summary, the human AIR ncRNA has at least two transcriptional starts sites that lie upstream to the CpG island in CGI-2 transgenes.
Non-coding AIR transcripts at the endogenous human IGF2R intron 2 CpG island
The human CGI-2 mouse transgenic expression pattern raised the possibility that the endogenous human IGF2R intron 2 CpG island would also contain an active promoter for an antisense AIR ncRNA. We first examined the EST database for evidence of expression (www.ensemble.org, March 2006 human genome sequence build 36.1 Chr.6:160,230,000–160,340,000). Eight ESTs were found within the human IGF2R intron 2 CpG island just downstream to the AIR promoter identified in the above transgenic/transfection experiments (Sup. Fig. 2A). These comprised a single thymus EST (AW008515) corresponding to an unspliced AIR ncRNA transcript and seven RAGE (Random Activation of Gene Expression) ESTs (BG182740, BG196263, BG216625, BG209200, BG191601, BG186411, and BG216164) obtained from a promoter insertion screen [24]. All seven RAGE clones used the same splice acceptor site at the start of human AIR exon 2 identified in the transgenic experiments (position 1872 bp in X83701) which indicates that the exon 2 splice acceptor identified in the transgene experiments is functional in the human genome. Examination of the region between the human IGF2R intron 2 CpG island and the flanking MAS1 gene that corresponds to the mouse Air transcription unit, identified 15 human ESTs many of which are expressed in the nervous system, that are putative AIR partial cDNAs (Sup. Fig. 2B).
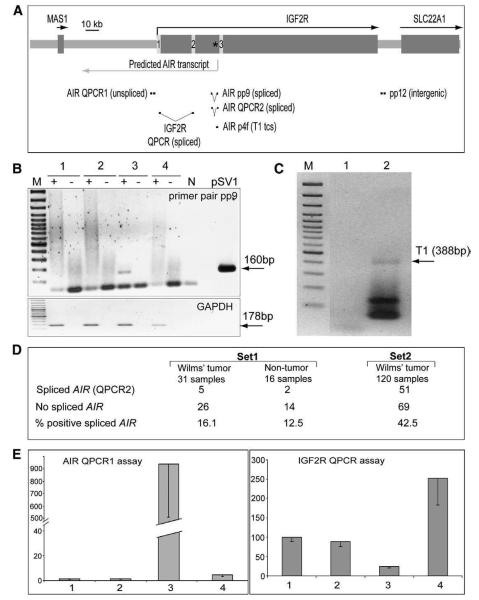
We next used RT-PCR to test for spliced non-coding AIR transcripts in Wilms' tumors, previously reported as showing polymorphic IGF2R imprinted expression [9]. Primer pair pp9 that spanned exons 1–2 of the transgenic human AIR ncRNA amplified the correct 160 bp fragment from 1 in 3 tested Wilms' tumors cell lines (Fig. 4B). This fragment was sequenced to show it arose from a correctly spliced human AIR ncRNA (data not shown). We used 5′RACE to identify the endogenous human AIR ncRNA transcriptional start in the STA-WT3ab Wilms' tumors cell line. A single transcriptional start site was identified, identical to the T1 site used in the human CGI-2 transgenes (Fig. 4C). Next, we assayed spliced human AIR ncRNA in two sets of Wilms' tumors tissue samples (Fig. 4D). In Set 1 comprising 31 Wilms' tumors and related non-tumor samples, respectively 16.1% and 12.5% were positive. In Set 2 comprising 120 Wilms' tumors, 42.5% were positive. In the majority of Wilms' tumors samples the amplified spliced AIR product was seen as a relatively weak band, only a few samples produced a strong PCR band (Sup. Figs. 3,4). This indicates that while the human IGF2R intron 2 CpG island can function as a promoter for the AIR ncRNA, its expression may be restricted to a subpopulation of cells in the tested samples.
Fig. 4.
Spliced and unspliced AIR transcripts at the endogenous human locus. (A) Map of part of human chromosome 6q27 showing IGF2R and flanking genes (dark grey boxes) with transcription orientation indicated by arrows and the human CGI-2 marked by an asterisk. Primers and QPCR assays are indicated underneath (details in Sup. Table 1). (B) Expression of spliced AIR by RT-PCR using primer pair pp9 in human Wilms' tumor cell lines (lane 1: Sk-Nep1-negative, lane 2: G-401-negative, lane 3:STA-WT3ab-positive for a 160 bp product) and lane 4: HS27 human foreskin fibroblasts. Plasmid SV1 containing the spliced human AIR product from mouse transgenes was used as a positive control. Human GAPDH was used as loading control. N; negative control PCR reaction without template. (C) Identification of the endogenous human AIR transcriptional start site in STA-WT3ab Wilms' tumor cells by 5′RACE with primers 2f and 4f. Lane 1; minus phosphotase control, lane 2; plus phosphotase, T1; transcription start site 1. (D) Summary of expression of spliced AIR by RT-PCR using primer pair pp9, in two sets of Wilms' tumor samples (see Sup. Figs. 3,4). Set 1 comprises successively accessioned cases of Wilms' tumor seen at Columbia University Medical Center that were not selected for any particular tumor stage or characteristics. The samples were from primary nephrectomy without pre-operative chemotherapy. Set 2 are Wilms' tumor samples from the German SIOP/GPOH 93-01 study and represent a largely unbiased series of tumors, predominantly with pre-operative chemotherapy as mandated by the European protocol. (E) Quantitative RT-PCR analysis of unspliced AIR (AIR QPCR1 assay) and IGF2R (IGF2R QPCR assay) expression in human Wilms' tumor cell lines – 1; Sk-Nep1, 2; G-401, 3; STA-WT3ab, and 4; HS27 cells, using assays shown in A. Values average three technical replicates normalized to GAPDH.
Since expression of the mouse Air ncRNA correlates with silencing of one parental Igf2r allele we used QPCR to test if there was a correlation between high expression of the human AIR ncRNA in Wilms' tumors and low expression of IGF2R. We were not able to perform an allele-specific assay of human IGF2R in these samples as matched genomic DNA was not available. We first assayed AIR and IGF2R in three Wilms' tumors cell lines. Fig. 4E shows that AIR is expressed only in the STA-WT3ab Wilms' tumor cell line, and that this cell line shows the lowest expression of IGF2R. We then applied the same assay to Wilms' tumor samples that expressed no AIR, or showed medium AIR expression or high AIR expression as judged from the non-quantitative PCR data shown in Sup. Fig. 4A. Samples with no or medium AIR expression showed no correlation between AIR expression and reduced IGF2R expression while 3/6 samples with high AIR expression showed reduced IGF2R expression. (Sup. Fig. 4B). Thus the data do not show a clear correlation between human AIR expression and reduced IGF2R expression as shown for the mouse Igf2r imprinted gene cluster [19]. However, since Wilms' tumor samples most likely contain mixtures of cells only some of which express AIR, the data do not exclude a correlation between expression of the AIR ncRNA and silencing of IGF2R in cis. Further experiments using homogenous cell populations or single cell assays will be needed to test if human AIR can silence IGF2R in cis.
A DNase1 site marks the unmethylated human IGF2R intron 2 CpG island
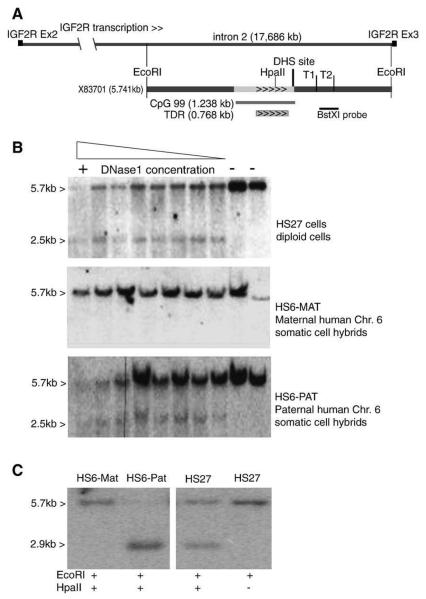
DNase1 hypersensitive (DHS) sites identify short regions sensitive to nucleases because of nucleosome displacement, which are often binding sites of transcription factors and thus likely to be cis-acting regulatory elements such as promoters or enhancers. We tested if the endogenous human IGF2R intron 2 CpG island contained a DHS site in HS27 normal diploid human fibroblasts, which lack AIR expression as shown in Fig. 4B. Fig. 5A shows a map of the tested region and the position of the human IGF2R intron 2 CpG island. A strong DHS site was identified in HS27 cells that mapped to a ~500 bp region between the CpG island and the major transcription start T1 identified in transgenic studies and in Wilms' tumors (Fig. 5B). Moreover, analysis of somatic-cell-hybrid cell lines containing either a paternal or a maternal human Chr.6 (that contains IGF2R) on a mouse background [25], shows that this DHS site is found only on the paternal chromosome. Fig. 5C shows that the paternal copy of human Chr.6 lacks a methylation imprint present over the human IGF2R intron 2 CpG island on the maternal chromosome. Thus, the paternal human IGF2R intron 2 CpG island lacks DNA methylation and contains a DNase1 hypersensitive site indicating a potential for expression.
Fig. 5.
The unmethylated human IGF2R intron 2 CpG island contains a DNase1 hypersensitive site. (A) Map of human IGF2R intron 2 (17.686 kb) showing the direction of IGF2R transcription. A map of the fragment used to generate the mouse transgenes is shown underneath. The length of the intronic CpG island (CpG 99) is indicated and the location of a cluster of tandem direct repeats (TDR) within this island is shown. The BstXI probe used in the Southern blots in B,C is shown. The transcription start sites T1, T2, mapped in Fig. 3A are indicated. The paternal-specific DHS site identified in B, is marked with a resolution of ±500 bp. (B) Identification of a paternal-specific DNase1 hypersensitive site. Top: Southern blot from HS27 human foreskin fibroblast cells treated with different concentrations of DNase1, digested with EcoRI and hybridized with probe BstXI. The last two lanes contain control nuclei treated either with incubation buffer at 37 °C or left on ice. Samples treated with DNase1 show a second 2.5 kb band in addition to the expected 5.7 kb EcoRI band. The position of this DNase1 hypersensitive site can be determined with a resolution of ±500 bp to be at the border between the 1440 bp fragment with promoter activity and the CpG island (indicated above in A). The middle and lower panel shows Southern blots from mouse A9 cells containing a full mouse genome plus a single human chromosome 6 from the maternal parent (HS6-MAT) or from the paternal parent (HS6-PAT) [25], treated as described for A. The 2.5 kb band produced by treatment with DNase1 is only present in somatic cell hybrids containing the paternal chromosome. (C) DNA methylation of the IGF2R intron 2 CpG island is maternal-specific. HS27 cells contain maternal and paternal chromosomes and produce 2 bands, one from the maternally-methylated allele (5.7 kb) and one from the unmethylated paternal allele (2.9 kb). HS6-MAT somatic cell hybrids retain the DNA-methylation imprint on the maternal copy of the IGF2R intron 2 CpG island (5.7 kb), while HS6-PAT have a methylation-free island (2.9 kb). The HS6-PAT somatic cell hybrid lacks AIR expression (data not shown and [25]).
Discussion
Here we have tested if the human IGF2R gene that shows polymorphic imprinted expression is able to express an antisense ncRNA from a CpG island located in intron 2. Studies from the mouse imprinted Igf2r gene has shown that the antisense Air ncRNA, which is expressed from a homologous intron 2 CpG island, is directly responsible for silencing Igf2r and the flanking Slc22a2 and Slc22a3 genes in cis [19]. We used transfection and mouse transgenic assays to demonstrate that the human IGF2R intron 2 CpG island has promoter activity. Moreover, this promoter can express an antisense ncRNA that we name AIR in agreement with the mouse nomenclature [20], in both mouse transgenes and in Wilms' tumors. The mouse transgenes were an essential tool to localize human AIR expression as previous studies based on random RT-PCR assays throughout the 99 kb region corresponding to the mouse Air transcript (from IGF2R intron 2 – MAS1), failed to identify AIR transcription [13]. Our demonstration that the human IGF2R intron 2 CpG island is an active promoter is supported by the identification of RAGE EST clones that use the splice acceptor of human AIR exon 2 [24], and by our identification of a DNase1 hypersensitive site that is often associated with cis-regulatory elements [26], which is only present on the unmethylated paternal allele.
The human IGF2R intron 2 CpG island was previously known to show three similarities to the homologous mouse Igf2r intron 2 CpG island that controls imprinted expression of the mouse Air ncRNA. These similar features are; that both lie in the second IGF2R/Igf2r intron close to exon 3, both are modified by a DNA-methylation imprint only on the maternal chromosome and both lie in a chromosomal region showing asynchronous behavior in S-phase cells [15,22]. The results presented here identify a fourth similarity in that both the human and mouse IGF2R/Igf2r intron 2 CpG islands, can act as promoters for the AIR/Air antisense ncRNA. We further show that the human AIR ncRNA is expressed in 16–40% of Wilms' tumors and that high expression of the AIR ncRNA correlates in 3/6 tumor samples, with reduced IGF2R expression. These data indicate that the human IGF2R imprinted gene cluster which also includes the SLC22A2 and SLC22A3 genes that only show imprinted expression in placenta [7], shows evolutionary conservation of the key features that control imprinted expression in the mouse.
Polymorphic imprinted expression of the human IGF2R gene cluster
The human IGF2R–SLC22A2–SLC22A3 gene cluster shows a concordant but polymorphic form of genomic imprinting, in which only a subset of tested samples show imprinted expression and at this stage of our knowledge, this behavior appears to be limited to fetal tissues and Wilms' tumors [7,13,14,16]. In contrast to polymorphic imprinted expression, the DNA-methylation imprint is always present on the maternal copy of the human IGF2R intron 2 CpG island in all cells of an individual [15,22,27,28]. Thus, human cells carry a methylation imprint on the IGF2R intron 2 CpG island that in most cases does not lead to imprinted expression. In the mouse, the maternal-specific methylation imprint on the Igf2r intron 2 CpG island is used to repress the Air ncRNA that is the key silencer of this gene cluster [19,21]. However, the mouse Igf2r gene can also separate the presence of the methylation imprint from imprinted expression, by regulating Air ncRNA transcription. For example, mouse preimplantation embryos, ES cells and post-mitotic neurons show bi-allelic Igf2r expression in the presence of the maternal methylation imprint because they lack Air expression [9,29,30,31]. This indicates that polymorphic imprinted expression of the human IGF2R imprinted gene cluster could be explained by a differential ability to transcribe the AIR ncRNA promoter in the human population, most likely arising from regulation of essential trans-acting factors.
Human IGF2R intron 2 CpG island is a typical CpG island promoter
CpG islands, defined as regions with a higher CpG content than the genome average, are the most reliable feature for gene-prediction in the mammalian genome and are associated with 60% of promoters [32].The human IGF2R intron 2 CpG island contains consensus CG-rich transcription factor binding sites, such as SP1, AP1 and GATA that are mainly clustered between the 3′ end of the island and the T1 transcription start site (data not shown). The main human AIR transcription start site was mapped 721 bp upstream of the CpG island. We also precisely determined the start sites of the endogenous mouse Air promoter to be at the 5′ border of the CpG island (data not shown). Thus, for both the mouse and the human intron 2 promoters, the bulk of the CpG island is internal to the transcribed ncRNA and the mapped promoters lie upstream. Previously, promoters were considered to be part of the CpG island [23]. The internal position of the AIR/Air CpG islands may indicate they act as cis-regulators such as enhancers, rather than promoters. The expression pattern of human CGI-2 transgenes was different to that shown by the mouse Air ncRNA. Human CGI-2 constructs were expressed in most transgenic lines and were highly expressed in adult brain, testes and a few other tissues but lowly expressed in adult heart. Mouse Igf2r intron 2 CpG island transgenes have a similar ability to be expressed as a transgene, but they show highest expression in adult heart that is typical of the endogenous transcript [33]. The human CGI-2 transgenes did not acquire a maternal-specific methylation imprint, however, this feature is also absent from the similar-sized mouse transgenes [33,34]. The lack of a methylation imprint in human CGI-2 transgenes explains expression following maternal and paternal transmission, as the methylation imprint is necessary for imprinted expression in mice [21,35].
Functional consequences of polymorphic human IGF2R imprinted expression
The IGF2R gene is a non-transducing intracellular transport receptor with multiple growth-suppressor functions, that include activation of the TGFβ growth suppressor and degradation of the IGF2 growth factor [36]. There is now a growing list of human tumors that show reduced IGF2R function as a consequence of genetic changes arising from genomic instability of the FRA6E fragile site that spans a 3.6 Mb region containing IGF2R [37], mutations of mismatch repair enzymes [38], or, inactivating mutations in coding regions of the gene [39]. In many cases these genetic mutations are accompanied by loss of the other parental allele (known as loss of heterozygosity) and IGF2R is now designated as a tumor suppressor gene [40].
Polymorphic imprinted expression of the human IGF2R gene has only been observed in fetal and tumor tissues [7], while adult tissues would normally express IGF2R from both the maternal and paternal allele [17,18]. However, in view of our demonstration here that the human IGF2R intron 2 CpG island contains an active AIR promoter, a scenario could be envisaged whereby gain of paternal expression of the AIR ncRNA would lead to silencing of the paternal IGF2R allele, even in adult tissues. Thus, reduced IGF2R function could theoretically arise in tumors epigenetically, as a consequence of a gain of imprinted gene silencing. Changes in the imprinted status of the IGF2 growth factor have been described in many tumor types, however, in this case IGF2 shows a loss of imprinted expression with aberrant activation of the normally silent maternal allele [41]. Nonetheless, there are precedents for a form of epigenetic silencing in human disease and tumors that is dependent on expression of antisense ncRNA [42]. For example, a Thalassaemia has been identified that results from a de novo transcriptional overlap of the alpha globin gene [43], and silencing of the p15 (CDKN2B) tumor suppressor gene in leukemia correlates with gain of expression of an antisense ncRNA that overlaps the promoter [44]. We have shown here that the human IGF2R gene contains an intronic CpG island promoter that expresses the AIR ncRNA in mouse transgenes and in Wilms' tumors. In mice, experiments have shown that the Air ncRNA can silence all three gene in the imprinted Igf2r gene cluster [19]. Our results here do not show a clear correlation in Wilms' tumors between high AIR expression and reduced IGF2R expression. However, they do not exclude this possibility. Further experiments using homogenous cell populations or single cell assays will be needed to test the consequence of human AIR expression on the downstream IGF2R promoter to determine if gain of AIR expression could provide an epigenetic mimic of the genetic changes leading to loss IGF2R function in tumors.
Materials and methods
Transient transfection assays
DNA fragments were subcloned into pGL3-Enhancer vector (Promega) (from X83701: 639 bp SacI fragment bp3434-4073, 777 bp BstXI fragment bp3344–4121, 1439 bp BssHII–BglII fragment bp3359–4798, 1578 bp BssHII–BamHI fragment bp3359–4937, 1667 bp SacI fragment bp4074–5741, from AJ249895: 1105 bp PstI–EcoRI fragment bp126132–127237) and assays were performed as described [20]. Transfections were performed five times and repeated with two different DNA preparations.
Transgenic constructs and transgenic mice
Construct HR2-L contains a 5384 bp human fragment (X83701 1–5384 bp) upstream to a 1200 bp polyadenylation cassette (rabbit β-globin, 31392–32590 bp/M18818) and LoxP1 (oligonucleotides 5′-AGCTTATAACTTCGTATAATGTATGCTATACGAAGTTATG and 5′-TCGACATAACTTCGTATAGCATACATTATACGAAGTTATA). 13 bp of vector backbone remained between the human fragment and the polyA cassette including an EcoRV site. Construct HR2-S differs from construct HR2-L only in the length of the polyA cassette (32033–32590 bp/M18818) that includes the polyA signal but lacks the last intron. The polyA signal was orientated to terminate the predicted AIR antisense transcript. Transgenic mice were generated by injecting DNA into B6/CBA fertilized oocytes using standard procedures. Transgenes were bred onto a FVB background and identified by DNA analysis.
Methylation analysis
Tail DNA from transgenic mice was extracted, digested with methyl-sensitive enzymes HpaII or NotI in combination with HindIII using standard methods. Hybridization probe BstXI (577 bp BstXI fragment from X83701). Complete digestion was verified with HTF9c probe that contains an unmethylated CpG island (NM_008307). Transgene copy number was determined by hybridization of EcoRV digested DNA with probe BstXI.
DNase1 assay
Tissue culture cells (107–108) were trypsinized into a single cell suspension. The preparation of nuclei and the DNase1 (Roche) digest were as previously described using a concentration range from 90–1000 U/ml [45]. DNase1 treated DNA was prepared and analyzed by Southern blot using standard protocols.
RNA analysis
5′RACE
Total RNA and polyA+ RNA prepared by RNAwizTM (Ambion) was analyzed using the First-Choice® RLM-RACE kit (Ambion) and primer 2f (external) and 4f (internal) for 35 cycles. PCR products (T1, DQ220013 and T2, DQ220014) were subcloned and sequenced.
Northern blots
20 μg of total DNase1 (DNA-freeTM, Ambion) treated RNA was electrophoresed in 1% agarose/formaldehyde gels. Human specific SVI probe (EcoRI fragment DQ220010) and mouse specific Gapdh probes (BC083149) were used.
Ribonuclease Protection Assay (RPA)
The RPAIII (Ambion) kit was used. The 290 bp HB probe protects 120 bp from the spliced AIR transcript and 149 bp from unspliced AIR transcript. The 350 bp HC probe protects 121 bp from the spliced and 260 bp unspliced AIR transcript. Mouse Cyclophilin A probe that protected a 105 bp band and 18S ribosomal gene probe that protected an 80 bp band were used as loading controls.
Non-quantitative RT-PCR
Reverse transcription was performed on DNase1 treated RNA using random hexamer primers and RevertAid™ First Strand cDNA Synthesis kit (Fermentas). PCRs were performed using a PCR Reagent kit (Invitrogen) for 32 or 35 cycles. Different splice variants of the human CGI-2 transgene ncRNA were identified by RT-PCR using primers Ap located in exon 3 of the PolyA cassette and primer 3r or 1r. PCR products were subcloned or directly sequenced.
Quantitative RT-PCR
Assay primers are listed in Sup. Table 1.
Supplementary Material
Acknowledgments
We thank Benjamin Tycko for providing Wilms' tumor samples, the Barlow Lab for the ir help and support, this project was funded by: FWF-SFB (P1718-1312), FWF P15522(B08) and the Epigenome NoE (LSHG-CT-2004-053433).
Note added in proof
AIRN (Antisense IGF2R RNA Noncoding) is the new approved HGNC gene name.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ygeno.2008.08.004.
References
- 1.Barlow DP, Bartolomei MS. Genomic imprinting in mammals. In: David Allis C, Jenuwein Thomas, Reinberg Danny, editors. Epigenetics. Cold Spring Harbor Laboratory Press; New York: 2007. Chapter 19. [Google Scholar]
- 2.Solter D. Imprinting today: end of the beginning or beginning of the end? Cytogenet. Genome Res. 2006;113:12–16. doi: 10.1159/000090809. [DOI] [PubMed] [Google Scholar]
- 3.Beechey C, Peters J, Blake A. World Wide Web Site – Mouse Imprinting Data and References. 2005 [Google Scholar]
- 4.Thorvaldsen JL, Verona RI, Bartolomei MS. X-tra! X-tra! News from the mouse X chromosome. Dev. Biol. 2006;298:344–353. doi: 10.1016/j.ydbio.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 5.Haig D. Evolutionary conflicts in pregnancy and calcium metabolism–a review. Placenta. 2004;25(Suppl A):S10–S15. doi: 10.1016/j.placenta.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 6.Morison IM, Ramsay JP, Spencer HG. A census of mammalian imprinting. Trends Genet. 2005;21:457–465. doi: 10.1016/j.tig.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 7.Monk D, Arnaud P, Apostolidou S, Hills FA, Kelsey G, Stanier P, Feil R, Moore GE. Limited evolutionary conservation of imprinting in the human placenta. Proc. Natl. Acad. Sci. U. S. A. 2006;103:6623–6628. doi: 10.1073/pnas.0511031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Regha K, Latos PA, Spahn L. The imprinted mouse Igf2r/Air cluster‘a model maternal imprinting system. Cytogenet. Genome Res. 2006;113:165–177. doi: 10.1159/000090829. [DOI] [PubMed] [Google Scholar]
- 9.Yamasaki Y, Kayashima T, Soejima H, Kinoshita A, Yoshiura K, Matsumoto N, Ohta T, Urano T, Masuzaki H, Ishimaru T, Mukai T, Niikawa N, Kishino T. Neuron-specific relaxation of Igf2r imprinting is associated with neuron-specific histone modifications and lack of its antisense transcript Air. Hum. Mol. Genet. 2005;14:2511–2520. doi: 10.1093/hmg/ddi255. [DOI] [PubMed] [Google Scholar]
- 10.Zwart R, Sleutels F, Wutz A, Schinkel AH, Barlow DP. Bidirectional action of the Igf2r imprint control element on upstream and downstream imprinted genes. Genes Dev. 2001;15:2361–2366. doi: 10.1101/gad.206201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalscheuer VM, Mariman EC, Schepens MT, Rehder H, Ropers HH. The insulin-like growth factor type-2 receptor gene is imprinted in the mouse but not in humans. Nat. Genet. 1993;5:74–78. doi: 10.1038/ng0993-74. [DOI] [PubMed] [Google Scholar]
- 12.Ogawa O, McNoe LA, Eccles MR, Morison IM, Reeve AE. Human insulin-like growth factor type I and type II receptors are not imprinted. Hum. Mol. Genet. 1993;2:2163–2165. doi: 10.1093/hmg/2.12.2163. [DOI] [PubMed] [Google Scholar]
- 13.Oudejans CB, Westerman B, Wouters D, Gooyer S, Leegwater PA, van Wijk IJ, Sleutels F. Allelic IGF2R repression does not correlate with expression of antisense RNA in human extraembryonic tissues. Genomics. 2001;73:331–337. doi: 10.1006/geno.2001.6522. [DOI] [PubMed] [Google Scholar]
- 14.Xu Y, Goodyer CG, Deal C, Polychronakos C. Functional polymorphism in the parental imprinting of the human IGF2R gene. Biochem. Biophys. Res. Commun. 1993;197:747–754. doi: 10.1006/bbrc.1993.2542. [DOI] [PubMed] [Google Scholar]
- 15.Smrzka OW, Fae I, Stoger R, Kurzbauer R, Fischer GF, Henn T, Weith A, Barlow DP. Conservation of a maternal-specific methylation signal at the human IGF2R locus. Hum. Mol. Genet. 1995;4:1945–1952. doi: 10.1093/hmg/4.10.1945. [DOI] [PubMed] [Google Scholar]
- 16.Xu YQ, Grundy P, Polychronakos C. Aberrant imprinting of the insulin-like growth factor II receptor gene in Wilms' tumor. Oncogene. 1997;14:1041–1046. doi: 10.1038/sj.onc.1200926. [DOI] [PubMed] [Google Scholar]
- 17.Killian JKN, Wylie CM, Vu AA, Li TH, Hoffman T, Jirtle AR. Divergent evolution in M6P/IGF2R imprinting from the Jurassic to the Quaternary. Hum. Mol. Genet. 2001;10:1721–1728. doi: 10.1093/hmg/10.17.1721. R.J. [DOI] [PubMed] [Google Scholar]
- 18.Vu TH, Jirtle RL, Hoffman AR. Cross-species clues of an epigenetic imprinting regulatory code for the IGF2R gene. Cytogenet. Genome Res. 2006;113:202–208. doi: 10.1159/000090833. [DOI] [PubMed] [Google Scholar]
- 19.Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 20.Lyle R, Watanabe D, te Vruchte D, Lerchner W, Smrzka OW, Wutz A, Schageman J, Hahner L, Davies C, Barlow DP. The imprinted antisense RNA at the Igf2r locus overlaps but does not imprint Mas1. Nat. Genet. 2000;25:19–21. doi: 10.1038/75546. [DOI] [PubMed] [Google Scholar]
- 21.Seidl CI, Stricker SH, Barlow DP. The imprinted Air ncRNA is an atypical RNAPII transcript that evades splicing and escapes nuclear export. Embo J. 2006;25:3565–3575. doi: 10.1038/sj.emboj.7601245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riesewijk AM, Schepens MT, Welch TR, van den Berg-Loonen EM, Mariman EM, Ropers HH, Kalscheuer VM. Maternal-specific methylation of the human IGF2R gene is not accompanied by allele-specific transcription. Genomics. 1996;31:158–166. doi: 10.1006/geno.1996.0027. [DOI] [PubMed] [Google Scholar]
- 23.Antequera F. Structure, function and evolution of CpG island promoters. Cell Mol. Life Sci. 2003;60:1647–1658. doi: 10.1007/s00018-003-3088-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrington JJ, Sherf B, Rundlett S, Jackson PD, Perry R, Cain S, Leventhal C, Thornton M, Ramachandran R, Whittington J, Lerner L, Costanzo D, McElligott K, Boozer S, Mays R, Smith E, Veloso N, Klika A, Hess J, Cothren K, Lo K, Offenbacher J, Danzig J, Ducar M. Creation of genome-wide protein expression libraries using random activation of gene expression. Nat. Biotechnol. 2001;19:440–445. doi: 10.1038/88107. [DOI] [PubMed] [Google Scholar]
- 25.Kugoh H, Mitsuya K, Meguro M, Shigenami K, Schulz TC, Oshimura M. Mouse A9 cells containing single human chromosomes for analysis of genomic imprinting. DNA Res. 1999;6:165–172. doi: 10.1093/dnares/6.3.165. [DOI] [PubMed] [Google Scholar]
- 26.Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sandovici I, Leppert M, Hawk PR, Suarez A, Linares Y, Sapienza C. Familial aggregation of abnormal methylation of parental alleles at the IGF2/H19 and IGF2R differentially methylated regions. Hum. Mol. Genet. 2003;12:1569–1578. doi: 10.1093/hmg/ddg167. [DOI] [PubMed] [Google Scholar]
- 28.Huang Z, Wen Y, Shandilya R, Marks JR, Berchuck A, Murphy SK. High throughput detection of M6P/IGF2R intronic hypermethylation and LOH in ovarian cancer. Nucleic Acids Res. 2006;34:555–563. doi: 10.1093/nar/gkj468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang ZQ, Fung MR, Barlow DP, Wagner EF. Regulation of embryonic growth and lysosomal targeting by the imprinted Igf2/Mpr gene. Nature. 1994;372:464–467. doi: 10.1038/372464a0. [DOI] [PubMed] [Google Scholar]
- 30.Braidotti G, Baubec T, Pauler F, Seidl C, Smrzka O, Stricker S, Yotova I, Barlow DP. The Air noncoding RNA: an imprinted cis-silencing transcript. Cold Spring Harb. Symp. Quant. Biol. 2004;69:55–66. doi: 10.1101/sqb.2004.69.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szabo PE, Mann JR. Biallelic expression of imprinted genes in the mouse germ line: implications for erasure, establishment, and mechanisms of genomic imprinting. Genes Dev. 1995;9:1857–1868. doi: 10.1101/gad.9.15.1857. [DOI] [PubMed] [Google Scholar]
- 32.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. U. S. A. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sleutels F, Barlow DP. Investigation of elements sufficient to imprint the mouse Air promoter. Mol. Cell Biol. 2001;21:5008–5017. doi: 10.1128/MCB.21.15.5008-5017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wutz A, Smrzka OW, Schweifer N, Schellander K, Wagner EF, Barlow DP. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature. 1997;389:745–749. doi: 10.1038/39631. [DOI] [PubMed] [Google Scholar]
- 35.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 36.Ghosh P, Dahms NM, Kornfeld S. Mannose 6-phosphate receptors: new twists in the tale. Nat. Rev. Mol. Cell Biol. 2003;4:202–212. doi: 10.1038/nrm1050. [DOI] [PubMed] [Google Scholar]
- 37.Denison SR, Callahan G, Becker NA, Phillips LA, Smith DI. Characterization of FRA6E and its potential role in autosomal recessive juvenile parkinsonism and ovarian cancer. Genes Chromosomes Cancer. 2003;38:40–52. doi: 10.1002/gcc.10236. [DOI] [PubMed] [Google Scholar]
- 38.Seitz S, Wassmuth P, Plaschke J, Schackert HK, Karsten U, Santibanez-Koref MF, Schlag PM, Scherneck S. Identification of microsatellite instability and mismatch repair gene mutations in breast cancer cell lines. Genes Chromosomes Cancer. 2003;37:29–35. doi: 10.1002/gcc.10196. [DOI] [PubMed] [Google Scholar]
- 39.Chappell SA, Walsh T, Walker RA, Shaw JA. Loss of heterozygosity at the mannose 6-phosphate insulin-like growth factor 2 receptor gene correlates with poor differentiation in early breast carcinomas. Br. J. Cancer. 1997;76:1558–1561. doi: 10.1038/bjc.1997.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oates AJ, Schumaker LM, Jenkins SB, Pearce AA, DaCosta SA, Arun B, Ellis MJ. The mannose 6-phosphate/insulin-like growth factor 2 receptor (M6P/IGF2R), a putative breast tumor suppressor gene. Breast Cancer Res. Treat. 1998;47:269–281. doi: 10.1023/a:1005959218524. [DOI] [PubMed] [Google Scholar]
- 41.Kaneda A, Feinberg AP. Loss of imprinting of IGF2: a common epigenetic modifier of intestinal tumor risk. Cancer Res. 2005;65:11236–11240. doi: 10.1158/0008-5472.CAN-05-2959. [DOI] [PubMed] [Google Scholar]
- 42.Yasuda J, Hayashizaki Y. The RNA continent. Adv. Cancer Res. 2008;99:77–112. doi: 10.1016/S0065-230X(07)99003-X. [DOI] [PubMed] [Google Scholar]
- 43.Tufarelli C, Stanley JA, Garrick D, Sharpe JA, Ayyub H, Wood WG, Higgs DR. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat. Genet. 2003;34:157–165. doi: 10.1038/ng1157. [DOI] [PubMed] [Google Scholar]
- 44.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pauler FM, Stricker SH, Warczok KE, Barlow DP. Long-range DNase I hypersensitivity mapping reveals the imprinted Igf2r and Air promoters share cis-regulatory elements. Genome Res. 2005;15:1379–1387. doi: 10.1101/gr.3783805. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.