Abstract
Species that pass repeatedly through narrow population bottlenecks (<100 individuals) are likely to have lost a large proportion of their genetic variation. Having genotyped 92 Raso larks Alauda razae, a Critically Endangered single-island endemic whose world population in the Cape Verdes over the last 100 years has fluctuated between about 15 and 130 pairs, we found variation at 7 of 21 microsatellite loci that successfully amplified, the remaining loci being monomorphic. At 6 of the polymorphic loci variation was sex-linked, despite the fact that these microsatellites were not sex-linked in the other passerine birds where they were developed. Comparative analysis strongly suggests that material from several different autosomes has been recently transferred to the sex chromosomes in larks. Sex-linkage might plausibly allow some level of heterozygosity to be maintained, even in the face of persistently small population sizes.
Electronic supplementary material
The online version of this article (doi:10.1007/s00239-010-9333-3) contains supplementary material, which is available to authorized users.
Keywords: Microsatellite polymorphism, Genetic bottlenecks, Raso lark, Alauda razae, Sex-linkage
Introduction
The study of small population genetics is of clear relevance to conservation, where low genetic diversity may be associated with reduced fitness (Spielman et al. 2004). However, the consequences of having low genetic diversity for species with persistently small populations, such as isolated endemics living in restricted habitats, are less clear. In particular, we need a better understanding of the extent to which low diversity may limit a species’ ability to adapt to new challenges, such as novel pathogens or climate change.
Genetic variation can be maintained in small populations in several ways. Lower organisms appear able to evolve higher mutation rates in response to need (Sniegowski et al. 1997). For higher organisms, a more likely mechanism involves balancing selection. Many studies report a link between particular genetic markers and some measure of fitness, particularly associated with resistance to pathogens (Acevedo-Whitehouse et al. 2005). Wherever such selection acts, neighbouring chromosomal regions will tend to have deeper gene trees and hence carry more variability (Charlesworth et al. 1997).
The Raso lark Alauda razae provides an interesting test case to ask how much variation can be maintained in a species with a tiny long-term effective population size. Since its description in 1898, this Cape Verdes endemic has been confined to the 7 km2 island of Raso where intermittent counts have recorded population fluctuations between about 15 and 130 pairs, with the population sex ratio often showing a strong male bias (Donald et al. 2003, 2005; M. de L. Brooke, pers. obs.). When, during Pleistocene sea level minima, Raso was connected to adjacent islands, the lark may have had a wider range and larger population. Under this scenario, recently confirmed by sub-fossil remains (Mateo et al. 2009), the lark would have been present on other islands (Branco, Santo Antão, Santa Luzia, São Vicente) when the Cape Verdes were first discovered and populated about 550 years ago. If these other lark populations disappeared because of habitat changes and/or the introduction of alien predators, the species would have become confined to Raso (Donald and Brooke 2006). Whatever the precise history, we can be confident the species has persisted for over 100 years at a population size of less than 300 animals, and often far fewer. For these reasons, it is classified as Critically Endangered (http://www.birdlife.org/datazone).
The Raso lark should harbour very little variability. With an effective population size (N
e) of ~50 and a microsatellite mutation rate, μ, of 10−3 per generation, the upper end of the range of rates reported (Brinkmann et al. 1998; Kayser et al. 2000; Xu et al. 2000), at mutation-drift equilibrium the expected heterozygosity, h, would be only ~16%, compared with 60–80% typical of most other species (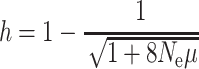 : Ohta and Kimura 1973). Actual levels of diversity will depend on how close the population is to equilibrium, and the severity of past bottlenecks. To determine the level of diversity persisting in this population we conducted an extensive survey of microsatellite markers in samples collected between 2004 and 2006 from 92 larks, roughly half the total number of individuals in the world available to be sampled in this period (M. de L. Brooke, unpubl. data).
: Ohta and Kimura 1973). Actual levels of diversity will depend on how close the population is to equilibrium, and the severity of past bottlenecks. To determine the level of diversity persisting in this population we conducted an extensive survey of microsatellite markers in samples collected between 2004 and 2006 from 92 larks, roughly half the total number of individuals in the world available to be sampled in this period (M. de L. Brooke, unpubl. data).
Methods
Raso larks were caught with mist nets, ringed to establish individual identity, and reliably sexed using the marked sexual dimorphism in wing and bill size (Donald et al. 2005). Our sample comprised 61 males and 31 females. After cleaning the bird’s skin with ethanol, a drop of 0.5 M EDTA was placed on the brachial vein, which was then pricked. The resultant drop of blood was transferred immediately to EDTA-moistened filter paper, which was then air dried in the field. The filter paper strip was kept dry and separated from other samples, on silica gel, until transfer to a −70°C freezer.
DNA was purified using a salt extraction method (Richardson et al. 2001). Thirty-five microsatellite loci were screened for amplification and polymorphism in eight randomly chosen adult Raso larks (Table 1). These loci were chosen either because they were available in our laboratory, or using information from the BIRDMARKER webpage (http://www.shef.ac.uk/misc/groups/molecol/deborah-dawson-birdmarkers.html). Loci showing good amplification and polymorphism (Tables 1, 2) were then used to genotype all 92 Raso lark samples. PCR reactions were carried out in a 10 μl volume containing 20–50 ng DNA, 0.2 mM of each dNTP, 0.5 μM of each primer, 10 mM Tris–HCl, 50 mM KCl, 1.5 mM MgCl2 and 0.15 U Taq DNA polymerase (Roche Diagnostics GmbH, Mannheim, Germany), using a program of: 94°C for 1 min, 40 cycles of 94°C for 30 s, Ta for 45 s and 72°C for 45 s, followed by 72°C for 2 min (For locus specific Ta’s see Table 2). Fluorescently labelled PCR products were separated on an ABI 377 automatic sequencer and allele lengths determined using Genescan 3.1 software.
Table 1.
Microsatellite primers tested for amplification and polymorphism in Raso lark (n = 8)
| Marker | PCR amplification |
|---|---|
| Aar2 | NA |
| Aar4 | M |
| Aar8 | M |
| Ase3 | M |
| Ase4 | NA |
| Ase6 | NA |
| Ase9 | P |
| Ase10 | M |
| Ase18 | P |
| Ase25 | NA |
| Ase27 | NA |
| Ase35 | NA |
| Ase37 | P |
| Ase42 | NA |
| Ase48 | NA |
| Ase56 | M |
| Ase58 | NA |
| Calex01 | M |
| Calex08 | P |
| Cuμ4 | P |
| Esc2 | NA |
| Esc6 | M |
| Fhu2 | M |
| Hru2 | P |
| Hru7 | NA |
| Lei160 | M |
| Lox1 | M |
| Mcy4 | M |
| Mjg1 | NA |
| Mme12 | P |
| Pca7 | M |
| Pca9 | NA |
| Pdo5 | M |
| Pocc6 | M |
| Ppi2 | NA |
NA no amplification, M amplification but monomorphic, P amplification and polymorphic
See Supplementary Material for information on studies originally describing these markers
Table 2.
Characteristics of seven polymorphic microsatellites in Raso lark (n = 92)
| Locus | Ta | # alleles | Allele length | Species where microsatellite first identified and found to be autosomal |
|---|---|---|---|---|
| Ase9 | 55 | 2 | 135, 139 | Acrocephalus sechellensis |
| Ase18 | 55 | 7 | 203, 205, 214, 218, 220, 222, 224 | Acrocephalus sechellensis |
| Ase37 | 50 | 2 | 214, 236 | Acrocephalus sechellensis |
| Calex08 | 55 | 2 | 198, 228 | Charadrius alexandrinus |
| Cuμ4 | 55 | 2 | 125, 127 | Catharus ustulatus |
| Hru2 | 50 | 2 | 119, 140 | Hirundo rustica |
| Mme12 | 50 | 2 | 165, 172 | Melospiza melodia |
Results
Thirty-three of the 35 microsatellite primers screened had been developed for other passerine species where cross-species and cross-family utility of primers is well established (Dawson et al. 2000), while the remaining two had been developed for non-passerine species, a plover Charadrius alexandrinus and the chicken Gallus gallus. Fourteen failed to amplify. Of the remainder, another 14 showed no variation but seven were polymorphic in Raso larks (Tables 1, 2). Of the polymorphic markers, six appear to be sex-linked. At four loci (Ase9, Ase37, Hru2, Mme12), all females were heterozygous while all males were homozygous for one, and only one, of the two female alleles. Despite our modest sample size, the probability of 61 males all being homozygous for an allele at frequency 0.83 [=(61 × 2 + 31)/184] is 1.6 × 10−10, while the probability that all 31 females are heterozygous is 7.4 × 10−18, giving a combined probability of 1.24 × 10−27. However, given the likely presence of many relatives in such a small population, a more conservative calculation would be of the probability, given the observed numbers of heterozygotes and homozygotes, that these correspond perfectly to the two sexes. The chance that all 31 heterozygotes are female is given by (31/92) * (30/91) ··· * (1/62) = 3.3 * 10−25, allowing extremely confident rejection of the null hypothesis that these are autosomal markers. The most parsimonious explanation of our data is that the W and Z chromosomes each carry one allele, this allele differing between the two chromosomes (in birds the female is the heterogametic sex). At one further locus (Ase 18), all females were heterozygous for allele 205 and another allele, while none of the males carried allele 205. Here, a similar calculation can be made (31 animals carry the 205 allele and these are all the 31 females), so it seems that the W chromosome is fixed for allele 205 while the Z chromosome is polymorphic, carrying one of six alleles, but not allele 205. This locus is the only one at which we found more than two alleles, perhaps hinting that it lies near a gene where selection favours diversity. At the last putatively sex-linked locus (Cuμ4), all 31 females were 125 homozygotes while, of the males, 39 were ‘125:125’ homozygotes, two ‘127:127’ homozygotes and 20 ‘125:127’ heterozygotes. This pattern is highly unlikely for a neutral autosomal locus (all 31 females have the same genotype that is present in 71 of 92 animals, P = (71/91) * (70/90) * ··· (40/62) = 0.000043). Arguably more likely is that the marker lies on the Z chromosome only. Then, by chance, all 31 females in our sample could have inherited a 125-carrying Z chromosome. This scenario is about 117.5 times as likely as an autosomal location, but nevertheless rather improbable (frequency of 125 allele = 0.843, P = 0.84331 = 0.005). However, it should be remembered that our sample contains many relatives that are not independent observations, increasing the chance of a significant deviation from random expectation. The final locus (Calex 08) appears to be a normal autosomal locus with two alleles.
Discussion
To find any appreciable microsatellite polymorphism in the Raso lark is rather unexpected, given the long-term small size of the population. That this preserved polymorphism is almost exclusively sex-linked is surprising for two reasons. First, none of these markers are reported to be sex-linked in the five species (Table 2) from which they were originally developed. This was confirmed by writing to the authors, asking them to re-test for evidence of sex-linkage; all replied in the negative. In addition, Ase9, Ase18 and Cuu4 have been mapped to chicken autosomes three, three and five, respectively (Dawson et al. 2006), confirming directly the lack of sex-linkage. Second, sex-linked loci are relatively rare. Surveying all issues of Molecular Ecology Resources in 2008, we found 413 new markers developed for 22 species of bird of which only 13 were sex-linked. Only a minority of studies explicitly test for sex-linkage, but of those that do, two large studies reveal 11 of 170 markers to be sex-linked (6.5%) (Jaari et al. 2008; Leder et al. 2008), significantly fewer (χ21 = 39.9, using Haber’s correction, P < 0.001), than the six of seven we observed in Raso larks.
Our data further contrast with most other avian studies, which report sex-linked alleles on the Z chromosome only (Ellegren 2000), with all females being phenotypically homozygous (= hemizygous). This reflects the generally much larger size of the Z relative to the W chromosome (75 Mb: 0.26 Mb in chickens) (Dawson et al. 2006). W-linked loci are rare, though appear occasionally (Küpper et al. 2007). Our data reveal a radically different pattern, with, in all but locus Cuμ4, homologous alleles occurring on both the Z and the W chromosomes. By implication, Raso lark sex chromosomes differ markedly from those of other birds. A clue as to what has happened is given by Bulatova (1973), who observed enlarged sex chromosomes in three species of lark, suggestive of either a fusion or a translocation of chromosomal material between the sex chromosomes and one or more autosomes. The three species, bimaculated lark Melanocorypha bimaculata, greater short-toed lark Calandrella brachydactyla (cinerea) and shore lark Eremophila alpestris, all have much larger geographic ranges than the Raso lark. Moreover, Ase18 has been genotyped in skylarks Alauda arvensis (Hutchinson and Griffith 2008), where our re-analysis of the raw data reveals it is sex-linked, and in sparrows (S. Griffith, unpublished data), where we find it is autosomal.
With enlarged sex chromosomes, multiple markers showing a switch from autosomal to sex-linked patterns and allele distributions indicating homologous W and Z alleles, larks, and the Raso lark in particular, appear to have experienced wholesale transfer of material from the autosomes to the sex chromosomes. This transfer appears to have been recent, evidenced both by the unusually large sex chromosomes of some but not all larks, and by the presence of alleles on both the W and Z chromosomes. In most birds, the W chromosome is small, making sex-linked loci hemizygous and reflecting a frequent evolutionary pattern where one of the sex chromosomes tends to degenerate by mutation and loss of material (Charlesworth 1991; Rice 1994). Larks might be unusual in not suffering a degeneration of the W chromosome analogous to the widespread degeneration of the Y chromosome often seen in other animals (Charlesworth and Charlesworth 2000), and also in experimental studies where autosomal material deliberately fused to the sex chromosomes suffers degeneration (Bachtrog and Charlesworth 2000). However, it seems more likely to us that material from a recent autosomal translocation or fusion has had too little time for significant loss to occur. Since three of the six sex-linked loci could be mapped in the chicken genome, we can be confident that at least two autosomes are involved, not just one (an unlikely alternative is that two autosomes fused before a translocation to the sex chromosomes). Just why this has occurred remains unclear. The phenomenon of addition of autosomal material to the sex chromosomes, not dissimilar to that reported in humans (Lahn and Page 1999), appears to be shared by several species of lark including species with large ranges and population sizes (Bulatova 1973).
While the reasons for the transfer are unresolved, we note that it might plausibly provide a mechanism by which some level of heterozygosity could be maintained, even in the face of persistently small population sizes, the two sex chromosomes maintaining two lineages for all markers that reside on them. Small populations usually harbour little variability due to genetic drift, and this is particularly so for the sex chromosomes which, in the absence of strong diversifying selection, are expected to have smaller effective sizes than the autosomes. However, even though each of the sex chromosomes would tend quickly to become monomorphic in a small population, because the W and Z carry different copies, overall greater diversity would be maintained, at least until the W chromosome degeneration began in earnest. This remarkable observation of conserved sex-linked microsatellite diversity in the Raso lark highlights how rapidly genomic reorganisation can occur and provides a fascinating opportunity to study the evolution of autosomal material that has very recently come to lie on a well-established pair of sex chromosomes.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
The molecular part of this work was assisted by the BIRDMARKER webpage maintained by D.A. Dawson at the NERC-funded Biomolecular Analysis Facility at Sheffield, UK and supported by a NWO-VICI grant to JK (86503003). Fieldwork was generously supported by Julian Francis and undertaken with the approval of the Direcção Geral do Ambiente of the Cape Verdes Government. Our thanks to Paul Donald and Jim Groombridge for comments.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Acevedo-Whitehouse K, Vicente J, Gortazar C, Höfle U, Fernández-de-Mera IG, Amos W. Genetic resistance to infection and severity of bovine tuberculosis in wild boar. Mol Ecol. 2005;14:3209–3217. doi: 10.1111/j.1365-294X.2005.02656.x. [DOI] [PubMed] [Google Scholar]
- Bachtrog D, Charlesworth B. Reduced levels of microsatellite variability on the neo-Y chromosome of Drosophila miranda. Curr Biol. 2000;10:1025–1031. doi: 10.1016/S0960-9822(00)00656-4. [DOI] [PubMed] [Google Scholar]
- Brinkmann B, Klintschar M, Neuhuber F, Hühne J, Rolf B. Mutation rate in human microsatellites: influence of the structure and length of the tandem repeat. Am J Hum Genet. 1998;62:1408–1415. doi: 10.1086/301869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulatova NS. Unusually large sex chromosomes in some larks (Aves: Alaudidae) Mamm Chrom Newsl. 1973;14:150–151. [Google Scholar]
- Charlesworth B. The evolution of sex chromosomes. Science. 1991;251:1030–1033. doi: 10.1126/science.1998119. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philos Trans R Soc Lond B. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Nordberg M, Charlesworth D. The effects of local selection, balanced polymorphism and background selection on equilibrium patterns of genetic diversity in subdivided populations. Genet Res. 1997;70:155–174. doi: 10.1017/S0016672397002954. [DOI] [PubMed] [Google Scholar]
- Dawson DA, Hanotte O, Greig C, Stewart IRK, Burke T. Polymorphic microsatellites in the blue tit Parus caeruleus and their cross-species utility in 20 songbird families. Mol Ecol. 2000;9:1941–1944. doi: 10.1046/j.1365-294x.2000.01094-14.x. [DOI] [PubMed] [Google Scholar]
- Dawson DA, Burke T, Hansson B, Pandhal J, Hale MC, Hinten GN, Slate J. A predicted microsatellite map of the passerine genome based on chicken-passerine sequence similarity. Mol Ecol. 2006;15:1299–1320. doi: 10.1111/j.1365-294X.2006.02803.x. [DOI] [PubMed] [Google Scholar]
- Donald PF, Brooke M de L (2006) An unlikely survivor: the peculiar natural history of the Raso Lark. Br Birds 99:420–430
- Donald PF, De Ponte M, Pitta Groz MJ, Taylor R. Status, ecology, behaviour and conservation of Raso Lark Alauda razae. Bird Conserv Int. 2003;13:13–28. doi: 10.1017/S0959270903003022. [DOI] [Google Scholar]
- Donald PF, Brooke M de L, Bolton MR, Taylor R, Wells CE, Marlow T, Hille SM (2005) Status of the Raso Lark Alauda razae in 2003, with further notes on sex ratio, behaviour and conservation. Bird Conserv Int 15:165–172
- Ellegren H. Evolution of the avian sex chromosomes and their role in sex determination. Trends Ecol Evol. 2000;15:188–192. doi: 10.1016/S0169-5347(00)01821-8. [DOI] [PubMed] [Google Scholar]
- Gibbs HL, Tabak LM, Hobson K. Characterization of microsatellite DNA loci for a neotropical migrant songbird, the Swainson’s thrush (Catharus ustulatus) Mol Ecol. 1999;8:1551–1552. doi: 10.1046/j.1365-294x.1999.00673.x. [DOI] [PubMed] [Google Scholar]
- Hutchinson JMC, Griffith SC. Extra-pair paternity in the Skylark Alauda arvensis. Ibis. 2008;150:90–97. [Google Scholar]
- Jaari S, Välimäki K, Merilä J. Isolation and characterisation of 100 polymorphic microsatellite loci for the Siberian jay (Perisoreus infaustus) Mol Ecol Res. 2008;8:1469–1474. doi: 10.1111/j.1755-0998.2008.02223.x. [DOI] [PubMed] [Google Scholar]
- Jeffery KJ, Keller LF, Arcese P, Bruford MW. The development of microsatellite loci in the song sparrow, Melospiza melodia (Aves) and genotyping errors associated with good quality DNA. Mol Ecol Notes. 2001;1:11–13. doi: 10.1046/j.1471-8278.2000.00005.x. [DOI] [Google Scholar]
- Kayser M, Roewer L, Hedman M, Henke L, Henke J, Brauer S, Krüger C, Krawczak M, Nagy M, Dobosz T, Szibor R, de Knijff P, Stoneking M, Sajantila A. Characteristics and frequency of germline mutations at microsatellite loci from the human Y chromosome, as revealed by direct observation in father/son pairs. Am J Hum Genet. 2000;66:1580–1588. doi: 10.1086/302905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küpper C, Horsburgh GJ, Dawson DA, Ffrench-Constant R, Székely T, Burke T. Characterization of 36 polymorphic microsatellite loci in the Kentish plover (Charadrius alexandrinus) including two sex-linked loci and their amplification in four other Charadrius species. Mol Ecol Notes. 2007;7:35–39. doi: 10.1111/j.1471-8286.2006.01517.x. [DOI] [Google Scholar]
- Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- Leder EH, Karaiskou N, Primmer CR. Seventy new microsatellites for the pied flycatcher, Ficedula hypoleuca and amplification in other passerine birds. Mol Ecol Res. 2008;8:874–880. doi: 10.1111/j.1755-0998.2008.02096.x. [DOI] [PubMed] [Google Scholar]
- Mateo JL, López Jurado LF, Geniez P. Historical distribution of the Razo lark Alauda razae in the Cape Verdes Archipelago. Alauda. 2009;77:309–312. [Google Scholar]
- Ohta T, Kimura M. A model of mutation appropriate to estimate the number of electrophoretically detectable alleles in a finite population. Genet Res. 1973;22:201–204. doi: 10.1017/S0016672300012994. [DOI] [PubMed] [Google Scholar]
- Primmer CR, Møller AP, Ellegren H. Resolving genetic-relationships with microsatellite markers—a parentage testing system for the swallow Hirundo rustica. Mol Ecol. 1995;4:493–498. doi: 10.1111/j.1365-294X.1995.tb00243.x. [DOI] [PubMed] [Google Scholar]
- Rice WR. Degeneration of a nonrecombining chromosome. Science. 1994;263:230–232. doi: 10.1126/science.8284674. [DOI] [PubMed] [Google Scholar]
- Richardson DS, Jury FL, Dawson DA, et al. Fifty Seychelles warbler (Acrocephalus sechellensis) microsatellite loci polymorphic in Sylviidae species and their cross-species amplification in other passerine birds. Mol Ecol. 2000;9:2226–2231. doi: 10.1046/j.1365-294X.2000.105338.x. [DOI] [PubMed] [Google Scholar]
- Richardson DS, Jury FL, Blaakmeer J, Komdeur J, Burke T. Parentage assignment and extra-pair paternity in a cooperative breeder: the Seychelles warbler (Acrocephalus sechellensis) Mol Ecol. 2001;10:2263–2273. doi: 10.1046/j.0962-1083.2001.01355.x. [DOI] [PubMed] [Google Scholar]
- Sniegowski PD, Gerrish PJ, Lenski RE. Evolution of high mutation rates in experimental populations of E. coli. Nature. 1997;387:703–705. doi: 10.1038/42701. [DOI] [PubMed] [Google Scholar]
- Spielman D, Brook BW, Briscoe DA, Frankham R. Does inbreeding and loss of genetic diversity decrease disease resistance? Conserv Genet. 2004;5:439–448. doi: 10.1023/B:COGE.0000041030.76598.cd. [DOI] [Google Scholar]
- Xu X, Peng M, Fang Z, Xu X. The direction of microsatellite mutations is dependent upon allele length. Nat Genet. 2000;24:396–399. doi: 10.1038/74238. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


