Abstract
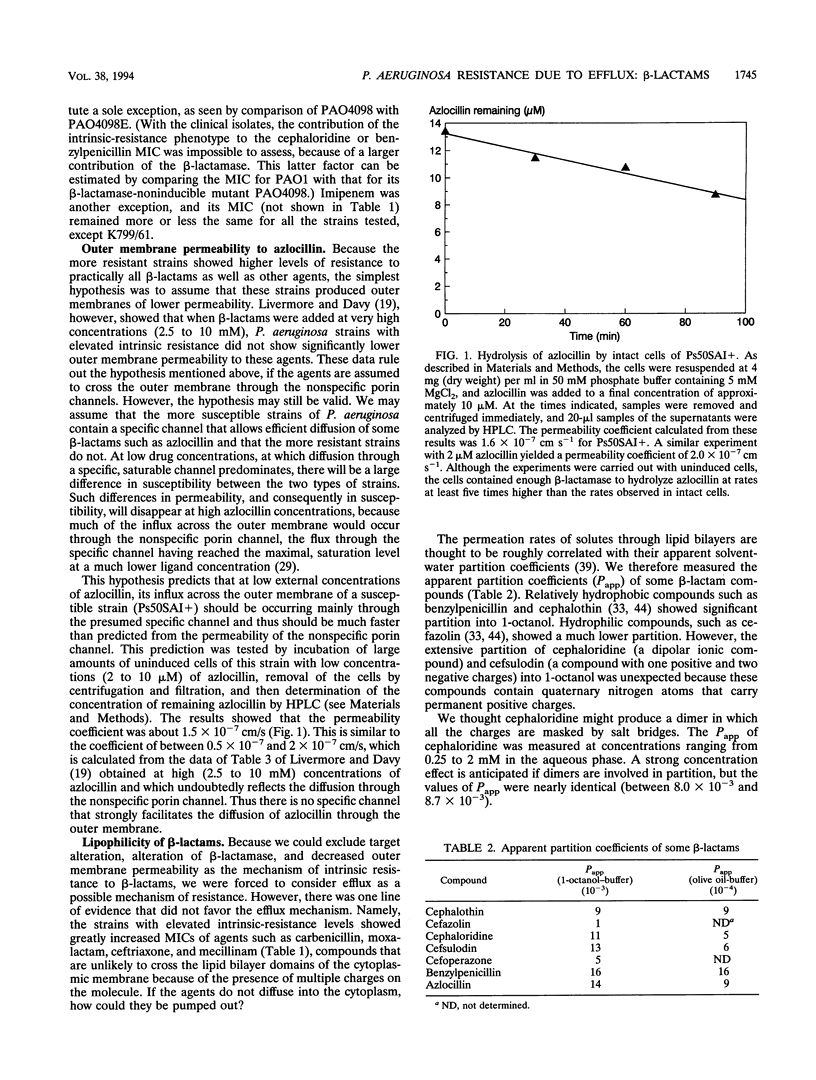
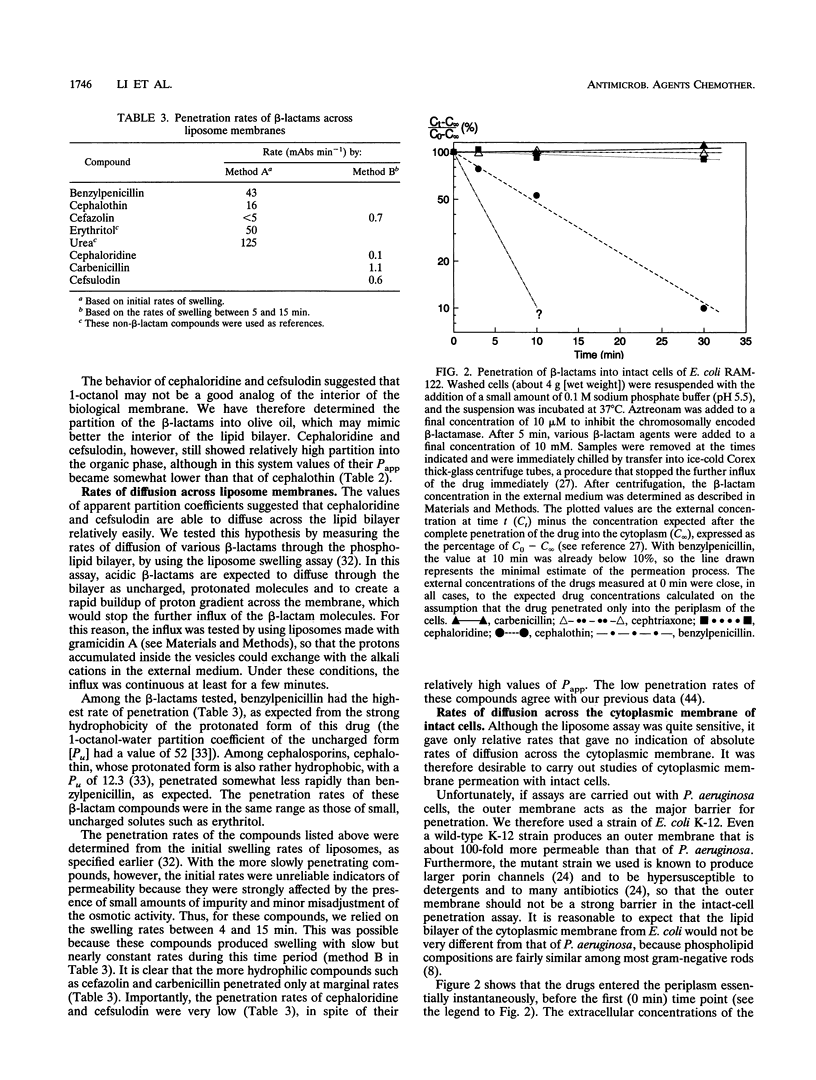
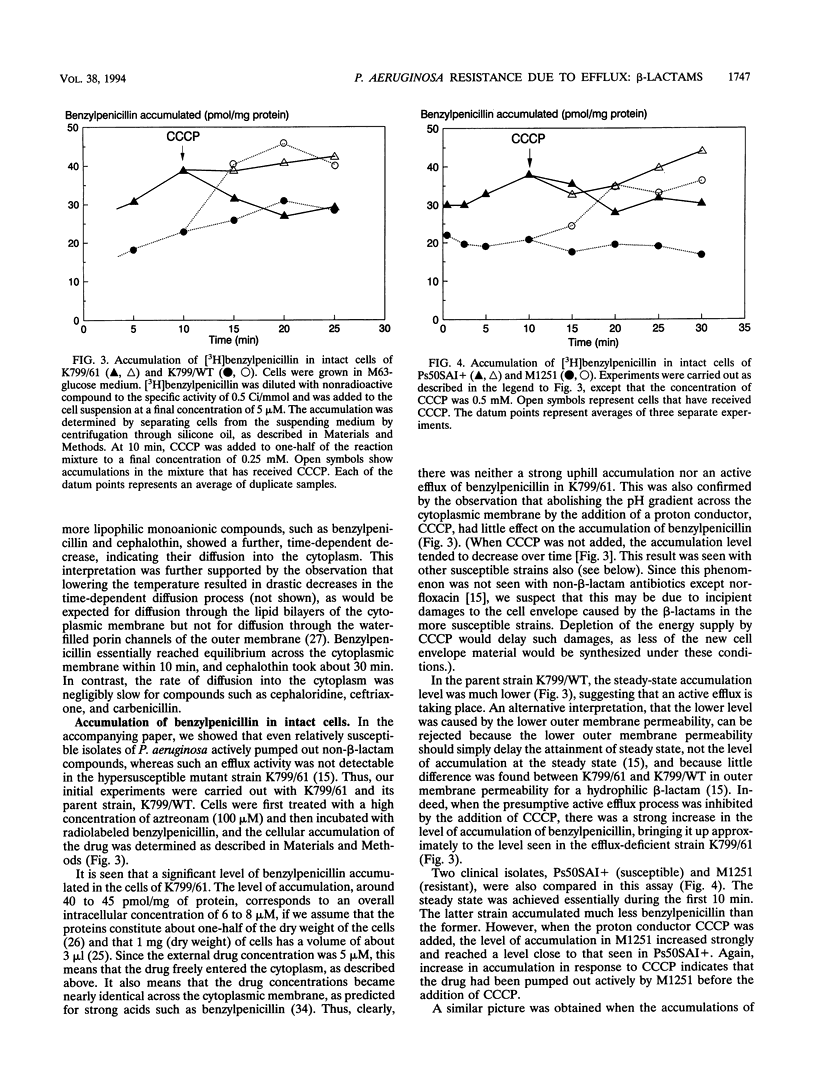
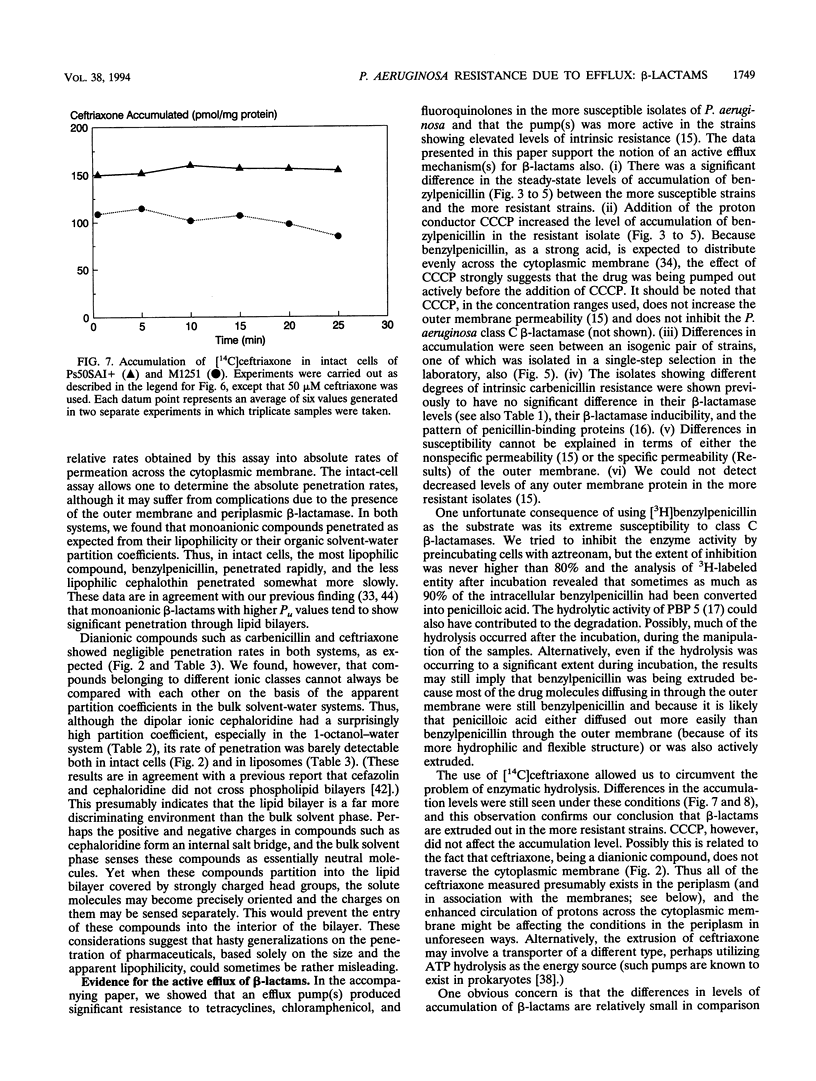
Wild-type strains of Pseudomonas aeruginosa are more resistant to various beta-lactam antibiotics as well as other agents than most enteric bacteria. Although resistance to compounds of earlier generations is explained by the synergism between the outer membrane barrier and the inducible beta-lactamase, it was puzzling to see significant levels of resistance to compounds that do not act as inducers or are not hydrolyzed rapidly by the chromosomally encoded enzyme. This intrinsic-resistance phenotype becomes enhanced in those strains with the so-called intrinsic carbenicillin resistance. In the accompanying paper (X.-Z. Li, D. M. Livermore, and H. Nikaido, Antimicrob. Agents Chemother. 38:1732-1741, 1994), we showed that active efflux played a role in the resistance, to various non-beta-lactam agents, of P. aeruginosa strains in general and that the efflux was enhanced in intrinsically carbenicillin-resistant strains. We show in this paper that, in comparison with the drug-hypersusceptible mutant K799/61, less benzylpenicillin was accumulated in wild-type strains of P. aeruginosa and that the accumulation levels were even lower in intrinsically carbenicillin-resistant strains. Deenergization by the addition of a proton conductor increased the accumulation level to that expected for equilibration across the cytoplasmic membrane. In intrinsically carbenicillin-resistant isolates, there was no evidence that either nonspecific or specific permeation rates of beta-lactams across the outer membrane were lowered in comparison with those of the more susceptible isolates. Furthermore, these carbenicillin-resistant isolates were previously shown to have no alteration in the level or the inducibility of beta-lactamase and in the affinity of penicillin-binding proteins. These data together suggest the involvement of an active efflux mechanism also in the resistance to beta-lactams. Hydrophilic beta-lactams with more than one charged group did not cross the cytoplasmic membrane readily. Yet one such compound, ceftriaxone, appeared to be extruded from the cells of more-resistant strains, although with this compound effects of proton conductors could not be shown. We postulate that wild-type strains of P. aeruginosa pump out such hydrophilic beta-lactams either from the periplasm or from the outer leaflet of the lipid bilayer of the cytoplasmic membrane, in a manner analogous to that hypothesized for multidrug resistance protein of human cancer cells (M.M. Gottesman and I. Pastan, Annu. Rev. Biochem. 62:385-427, 1993).
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Angus B. L., Carey A. M., Caron D. A., Kropinski A. M., Hancock R. E. Outer membrane permeability in Pseudomonas aeruginosa: comparison of a wild-type with an antibiotic-supersusceptible mutant. Antimicrob Agents Chemother. 1982 Feb;21(2):299–309. doi: 10.1128/aac.21.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellido F., Pechère J. C., Hancock R. E. Novel method for measurement of outer membrane permeability to new beta-lactams in intact Enterobacter cloacae cells. Antimicrob Agents Chemother. 1991 Jan;35(1):68–72. doi: 10.1128/aac.35.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K., Freudenberger J. S., Sykes R. B. Interaction of azthreonam and related monobactams with beta-lactamases from gram-negative bacteria. Antimicrob Agents Chemother. 1982 Sep;22(3):414–420. doi: 10.1128/aac.22.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. P., Hooper D. C., Wolfson J. S., Souza K. S., McMurry L. M., Levy S. B. Endogenous active efflux of norfloxacin in susceptible Escherichia coli. Antimicrob Agents Chemother. 1988 Aug;32(8):1187–1191. doi: 10.1128/aac.32.8.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan S. W., Schirmer T., Rummel G., Steiert M., Ghosh R., Pauptit R. A., Jansonius J. N., Rosenbusch J. P. Crystal structures explain functional properties of two E. coli porins. Nature. 1992 Aug 27;358(6389):727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- Fojo A., Akiyama S., Gottesman M. M., Pastan I. Reduced drug accumulation in multiply drug-resistant human KB carcinoma cell lines. Cancer Res. 1985 Jul;45(7):3002–3007. [PubMed] [Google Scholar]
- George A. M., Levy S. B. Amplifiable resistance to tetracycline, chloramphenicol, and other antibiotics in Escherichia coli: involvement of a non-plasmid-determined efflux of tetracycline. J Bacteriol. 1983 Aug;155(2):531–540. doi: 10.1128/jb.155.2.531-540.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfine H. Comparative aspects of bacterial lipids. Adv Microb Physiol. 1972;8:1–58. doi: 10.1016/s0065-2911(08)60187-3. [DOI] [PubMed] [Google Scholar]
- Gottesman M. M., Pastan I. Biochemistry of multidrug resistance mediated by the multidrug transporter. Annu Rev Biochem. 1993;62:385–427. doi: 10.1146/annurev.bi.62.070193.002125. [DOI] [PubMed] [Google Scholar]
- Hooper D. C., Wolfson J. S., Souza K. S., Ng E. Y., McHugh G. L., Swartz M. N. Mechanisms of quinolone resistance in Escherichia coli: characterization of nfxB and cfxB, two mutant resistance loci decreasing norfloxacin accumulation. Antimicrob Agents Chemother. 1989 Mar;33(3):283–290. doi: 10.1128/aac.33.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashket E. R. Stoichiometry of the H+-ATPase of growing and resting, aerobic Escherichia coli. Biochemistry. 1982 Oct 26;21(22):5534–5538. doi: 10.1021/bi00265a024. [DOI] [PubMed] [Google Scholar]
- Lam J. S., Graham L. L., Lightfoot J., Dasgupta T., Beveridge T. J. Ultrastructural examination of the lipopolysaccharides of Pseudomonas aeruginosa strains and their isogenic rough mutants by freeze-substitution. J Bacteriol. 1992 Nov;174(22):7159–7167. doi: 10.1128/jb.174.22.7159-7167.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S. B. Active efflux mechanisms for antimicrobial resistance. Antimicrob Agents Chemother. 1992 Apr;36(4):695–703. doi: 10.1128/aac.36.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Z., Livermore D. M., Nikaido H. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: resistance to tetracycline, chloramphenicol, and norfloxacin. Antimicrob Agents Chemother. 1994 Aug;38(8):1732–1741. doi: 10.1128/aac.38.8.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore D. M., Davy K. W. Invalidity for Pseudomonas aeruginosa of an accepted model of bacterial permeability to beta-lactam antibiotics. Antimicrob Agents Chemother. 1991 May;35(5):916–921. doi: 10.1128/aac.35.5.916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore D. M. Interplay of impermeability and chromosomal beta-lactamase activity in imipenem-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992 Sep;36(9):2046–2048. doi: 10.1128/aac.36.9.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore D. M. Penicillin-binding proteins, porins and outer-membrane permeability of carbenicillin-resistant and -susceptible strains of Pseudomonas aeruginosa. J Med Microbiol. 1984 Oct;18(2):261–270. doi: 10.1099/00222615-18-2-261. [DOI] [PubMed] [Google Scholar]
- Livermore D. M. Radiolabelling of penicillin-binding proteins (PBPs) in intact Pseudomonas aeruginosa cells: consequences of beta-lactamase activity by PBP-5. J Antimicrob Chemother. 1987 Jun;19(6):733–742. doi: 10.1093/jac/19.6.733. [DOI] [PubMed] [Google Scholar]
- Livermore D. M., Yang Y. J. Beta-lactamase lability and inducer power of newer beta-lactam antibiotics in relation to their activity against beta-lactamase-inducibility mutants of Pseudomonas aeruginosa. J Infect Dis. 1987 Apr;155(4):775–782. doi: 10.1093/infdis/155.4.775. [DOI] [PubMed] [Google Scholar]
- Ma D., Cook D. N., Alberti M., Pon N. G., Nikaido H., Hearst J. E. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J Bacteriol. 1993 Oct;175(19):6299–6313. doi: 10.1128/jb.175.19.6299-6313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda N., Ohya S. Cross-resistance to meropenem, cephems, and quinolones in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992 Sep;36(9):1847–1851. doi: 10.1128/aac.36.9.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzanke B. F., Ecker D. J., Yang T. S., Huynh B. H., Müller G., Raymond K. N. Escherichia coli iron enterobactin uptake monitored by Mössbauer spectroscopy. J Bacteriol. 1986 Aug;167(2):674–680. doi: 10.1128/jb.167.2.674-680.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra R., Benson S. A. Genetic identification of the pore domain of the OmpC porin of Escherichia coli K-12. J Bacteriol. 1988 Aug;170(8):3611–3617. doi: 10.1128/jb.170.8.3611-3617.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlradt P. F., Menzel J., Golecki J. R., Speth V. Lateral mobility and surface density of lipopolysaccharide in the outer membrane of Salmonella typhimurium. Eur J Biochem. 1974 Apr 16;43(3):533–539. doi: 10.1111/j.1432-1033.1974.tb03440.x. [DOI] [PubMed] [Google Scholar]
- Nikaido H., Nikaido K., Harayama S. Identification and characterization of porins in Pseudomonas aeruginosa. J Biol Chem. 1991 Jan 15;266(2):770–779. [PubMed] [Google Scholar]
- Nikaido H., Normark S. Sensitivity of Escherichia coli to various beta-lactams is determined by the interplay of outer membrane permeability and degradation by periplasmic beta-lactamases: a quantitative predictive treatment. Mol Microbiol. 1987 Jul;1(1):29–36. doi: 10.1111/j.1365-2958.1987.tb00523.x. [DOI] [PubMed] [Google Scholar]
- Nikaido H. Outer membrane barrier as a mechanism of antimicrobial resistance. Antimicrob Agents Chemother. 1989 Nov;33(11):1831–1836. doi: 10.1128/aac.33.11.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. Outer membrane of Salmonella typhimurium. Transmembrane diffusion of some hydrophobic substances. Biochim Biophys Acta. 1976 Apr 16;433(1):118–132. doi: 10.1016/0005-2736(76)90182-6. [DOI] [PubMed] [Google Scholar]
- Nikaido H., Rosenberg E. Y., Foulds J. Porin channels in Escherichia coli: studies with beta-lactams in intact cells. J Bacteriol. 1983 Jan;153(1):232–240. doi: 10.1128/jb.153.1.232-240.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H., Rosenberg E. Y. Porin channels in Escherichia coli: studies with liposomes reconstituted from purified proteins. J Bacteriol. 1983 Jan;153(1):241–252. doi: 10.1128/jb.153.1.241-252.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H., Thanassi D. G. Penetration of lipophilic agents with multiple protonation sites into bacterial cells: tetracyclines and fluoroquinolones as examples. Antimicrob Agents Chemother. 1993 Jul;37(7):1393–1399. doi: 10.1128/aac.37.7.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W., Spratt B. G. Regulation of the permeability of the gonococcal cell envelope by the mtr system. Mol Microbiol. 1994 Feb;11(4):769–775. doi: 10.1111/j.1365-2958.1994.tb00354.x. [DOI] [PubMed] [Google Scholar]
- Poole K., Krebes K., McNally C., Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol. 1993 Nov;175(22):7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds P. R., Mottur G. P., Bradbeer C. Transport of vitamin B12 in Escherichia coli. Some observations on the roles of the gene products of BtuC and TonB. J Biol Chem. 1980 May 10;255(9):4313–4319. [PubMed] [Google Scholar]
- Ross J. I., Eady E. A., Cove J. H., Cunliffe W. J., Baumberg S., Wootton J. C. Inducible erythromycin resistance in staphylococci is encoded by a member of the ATP-binding transport super-gene family. Mol Microbiol. 1990 Jul;4(7):1207–1214. doi: 10.1111/j.1365-2958.1990.tb00696.x. [DOI] [PubMed] [Google Scholar]
- Trias J., Dufresne J., Levesque R. C., Nikaido H. Decreased outer membrane permeability in imipenem-resistant mutants of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1989 Aug;33(8):1202–1206. doi: 10.1128/aac.33.8.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji A., Kubo O., Miyamoto E., Yamana T. Physicochemical properties of beta-lactam antibiotics: oil-water distribution. J Pharm Sci. 1977 Dec;66(12):1675–1679. doi: 10.1002/jps.2600661205. [DOI] [PubMed] [Google Scholar]
- Yamaguchi A., Hiruma R., Sawai T. Phospholipid bilayer permeability of beta-lactam antibiotics. J Antibiot (Tokyo) 1982 Dec;35(12):1692–1699. doi: 10.7164/antibiotics.35.1692. [DOI] [PubMed] [Google Scholar]
- Yoshimura F., Nikaido H. Diffusion of beta-lactam antibiotics through the porin channels of Escherichia coli K-12. Antimicrob Agents Chemother. 1985 Jan;27(1):84–92. doi: 10.1128/aac.27.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura F., Nikaido H. Permeability of Pseudomonas aeruginosa outer membrane to hydrophilic solutes. J Bacteriol. 1982 Nov;152(2):636–642. doi: 10.1128/jb.152.2.636-642.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann W. Penetration of beta-lactam antibiotics into their target enzymes in Pseudomonas aeruginosa: comparison of a highly sensitive mutant with its parent strain. Antimicrob Agents Chemother. 1980 Jul;18(1):94–100. doi: 10.1128/aac.18.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann W., Rosselet A. Function of the outer membrane of Escherichia coli as a permeability barrier to beta-lactam antibiotics. Antimicrob Agents Chemother. 1977 Sep;12(3):368–372. doi: 10.1128/aac.12.3.368. [DOI] [PMC free article] [PubMed] [Google Scholar]



