Abstract
Copy gains involving chromosome 7p represent one of the most common genomic alterations found in melanomas, suggesting the presence of “driver” cancer genes. We identified several tumor samples that harbored focal amplifications situated at the peak of common chromosome 7p gains, in which the minimal common overlapping region spanned the ETV1 oncogene. Fluorescence in situ hybridization (FISH) analysis revealed copy gains spanning the ETV1 locus in >40% of cases, with ETV1 amplification present in 13% of primary and 18% of metastatic melanomas. Melanoma cell lines, including those with ETV1 amplification, exhibited dependency on ETV1 expression for proliferation and anchorage-independent growth. Moreover, over-expression of ETV1 in combination with oncogenic NRASG12D transformed primary melanocytes and promoted tumor formation in mice. ETV1 overexpression elevated MITF expression in immortalized melanocytes, which was necessary for ETV1-dependent oncogenicity. These observations implicate deregulated ETV1 in melanoma genesis and suggest a pivotal lineage dependency mediated by oncogenic ETS transcription factors in this malignancy.
Keywords: melanoma, amplification, ETV1, oncogene, ETS factors
INTRODUCTION
Recurrent tumor genomic alterations may pinpoint key drivers of tumorigenesis and offer avenues for rational therapeutic development. In some cancers, the importance of a gene to tumor maintenance is tightly linked to the presence of specific genetic alterations affecting that gene. In other cases, recurrent genomic changes illuminate mechanisms that are broadly operant, even in cancers where specific genetic alterations are not present at the “index” locus. Thus, some cancer gene mutations may serve as signifiers for cardinal pathways that are commonly altered by multiple genetic and epigenetic mechanisms across many cancers.
Chromosomal copy number changes represent highly prevalent genomic aberrations in cancer. In principle, genes targeted by such events might be expected to underlie fundamental tumorigenic mechanisms that are deregulated by both genetic and other means in many tumor types, as described above. However, many chromosomal alterations (e.g., copy gains, losses, and loss-of-heterozygosity (LOH)) involve broad, low-amplitude changes that may encompass an entire chromosome or chromosome arm. Dozens to hundreds of genes are altered by such genetic events; thus, characterizing the relevant effectors poses a significant challenge.
In melanoma, copy gains involving chromosome 7 are exceedingly common (up to 40% of cases) (1–3). Recent melanoma genomic studies suggest that the “peaks” of statistically significant chromosome 7 copy gains localize to independent loci on 7p and 7q (2, 3). Chromosome 7q contains the BRAF oncogene, which commonly undergoes activating point mutations in melanoma (4). However, the target(s) of 7p gains in melanoma—though equally common—remain uncharacterized. We therefore sought to identify candidate target oncogenes of chromosome 7p in melanoma. The results herein suggest that ETV1, an ETS transcription factor known to undergo genetic deregulation in several cancer types (5–7), is targeted by 7p21 amplification events and exerts a MAP kinase- and MITF-dependent tumor-promoting function in many melanomas. Thus, ETV1 may represent a critical effector within an oncogenic module that is broadly operant in melanoma.
MATERIALS AND METHODS
Microarray studies
High-density SNP array and gene expression microarray data generation were performed as described previously(3). A brief description is provided in Supplementary Methods.
Tissue microarray construction
The melanoma tumor progression array was generated through the collaborative efforts of three Skin SPORES (Harvard, MD Anderson and U Penn), and contains 480 0.6mm cores of tissues from 170 distinct clinical specimens, including 132 cores from 36 benign melanocytic nevi, 196 cores from 59 primary melanomas and 150 cores from 75 metastatic lesions (8).
Fluorescence in situ hybridization (FISH)
All bacterial artificial chromosome (BAC) clones were selected using the UCSC Genome Browser and obtained from the BACPAC Resource Center (CHORI). BAC probes preparation, labeling and hybridization was performed as described previously (9). To assess for ETV1 amplification, a dual-color FISH assay was designed using an ETV1 and a reference probe. To assess for ETV1 rearrangement, a dual color break-apart FISH assay was designed for the locus. Probes used and protocols followed are described in Supplementary Methods.
Genomic quantitative PCR (Q-PCR)
ETV1-locus genomic Q-PCR was performed following standard protocols and using two ETV1-specific primers. Protocols and primer sequences are provided in Supplementary Metohds.
Analysis of mRNA expression by quantitative RT-PCR
Quantitative RT-PCR was performed following standard protocols. Primer sequences and protocols are provided in Supplementary Methods.
Molecular cloning and expression of ETV1 and MITF
Human ETV1 was cloned from total fetal brain RNA. Total cDNA was generated with the SuperScript III first strand synthesis kit and random hexamers (Invitrogen). ETV1 cDNA was generated by PCR from the pool of fetal brain cDNA using primers containing ETV1-specific sequence flanked by a 5′ EcoRI site and a 3′-XhoI site to allow for cloning in to the retrovirus-blast vector pWZL (see Supplementary Methods for primer sequences). The pWZL-blast-HA-MITF vector used to overexpress melanocytic MITF was generated as described previously(10). All cDNAs were verified by Sanger sequencing.
Immunoblot analysis
Cell lysis and immunoblot analysis were performed as described in Supplementary Methods.
Cell culture
Culture conditions for short-term cultures and cell lines used are described in Supplementary Methods
Retroviral infections
Retroviruses were obtained by triple co-transfection of HEK293-ebna cells (Invitrogen), with the pWZL-blast vector (empty or containing the cDNA of interest) and the packaging and envelope plasmids pN8e-VSV-G and pN8e-GagPolΔS (kind gift from Dr Jay Morgenstern, Millenium Pharmaceuticals). P’mel* cells were infected with the retrovirus pBABE-zeo-NRASG12D or –BRAFV600E and selected by growth-factor deprivation in Ham’s F10 media supplemented with 10%FBS, L-Glutamine and P/S. p’mels*-BRAFV600E and -NRASG12D expressing cells were infected with the pWZL-blast -empty, -ETV1 or –HA-MITF retroviruses followed by blasticidin selection.
Lentivirally delivered short hairpin RNA (shRNA)
pLKO1-based lentiviral vector knockdown assays were performed following standard protocols described in Supplementary Methods.
Cell Proliferation assays
Cells were plated in triplicate in 12-well plates at 20,000 cells/well in a final volume of 2 ml of media. At each time point, cells were washed twice with PBS, trypsinized and counted with a cell counter (Beckman Coulter, Fullerton, CA).
Assessment of growth factor autonomy
Cells were plated on 24-well plates (7000 cells/well) in Ham’s F10 media containing 50ng/ml TPA, 1mM cAMP, L-Glutamine, 0.1mM IBMX, 1uM NA3VO4, 7% FBS and P/S (referred to as + Growth factors (+GF) media), or in Ham’s F10 media containing L-Glutamine, 10% FBS and P/S (referred to as - Growth factors (−GF) media). Growth factor dependence of cells was determined by counting surviving cells in +GF media compared to −GF media. At each time point, cells were washed with PBS, trypsinized and counted (hemacytometer).
Anchorage independent assays
Cells (10,000 or 20,000/well) suspended in media (2 ml) containing 0.7% noble agar, 0.5% fungizone (Sigma-Aldrich) and marker drug (when applicable) were seeded into 6 well plates containing a 0.5% noble agar layer (2ml). Colony formation in soft agar was assayed in triplicate.
Xenograft tumor experiments
Animal experiments were conducted according to Dana-Farber Cancer Institute Animal Care and Use Committee guidelines. Isogenic sets of transformed melanocytes were injected (5 ×10e5 cells per site in 100uL Hank’s buffered saline) at three flank positions (in cohorts of 5 animals) in 6 week old female NCR-Nu and tumor growth was monitored for 5–7 weeks post-injections. Animals were sacrificed before behavioral malaise followed by excision and analysis of resulting tumors.
RESULTS
Amplification of the ETV1 locus in melanoma
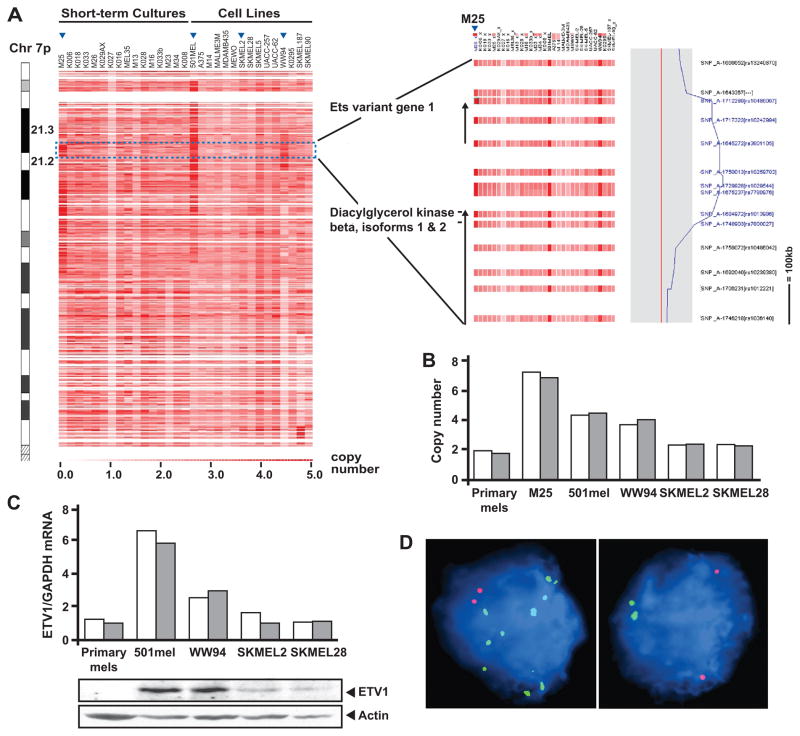
To identify candidate effector genes targeted by chromosome 7p gains in melanoma, we analyzed chromosomal copy number data from single nucleotide polymorphism (SNP) arrays performed on melanoma short-term cultures (STCs) and cell lines. As shown in Fig. 1A, many samples contained broad regions of chromosome 7p copy gain, as expected; however focal 7p21 amplification events were apparent in a few samples. Notably, the M25 short-term culture and the 501mel and WW94 cell lines harbored overlapping focal amplifications at the 7p21.3 locus (Fig. 1A; arrowheads). Inspection of genes common to the amplified regions identified ETV1 (Fig. 1A), a known oncogene and a member of the ETS transcription factor family (5–7). Quantitative genomic PCR confirmed ETV1 amplification in these samples compared to primary melanocytes and to cell lines without focal amplification (SKMEL2 and SKMEL28) (Fig. 1B). In addition, FISH analysis demonstrated higher numbers of ETV1-probe signals in 501mel and WW94, relative to SKMEL2 and SKMEL28 (Supplementary Fig. 1). Moreover, quantitative RT-PCR and immunoblot analyses also showed increased ETV1 mRNA and protein levels in 501mel and WW94, respectively (Fig. 1C), suggesting that the DNA amplification events resulted in increased ETV1 expression.
Figure 1.
Amplification of the ETV1 locus in melanoma. A, SNP array-based copy number analysis of chromosome 7p (left panel; cytobands indicated) is shown for a collection of melanoma short-term cultures and cell lines. Increasing red intensity denotes increasing copy number (legend; bottom). The region containing the ETV1 locus is indicated (blue box), and a closer view is shown (right panel). ETV1 and the neighboring diacylglycerol kinase beta genes are indicated. The copy number at the ETV1 locus in sample M25 is shown graphically (right). B, quantitative genomic PCR assessment of ETV1 amplification is shown for M25, 501mel, WW94, SKMEL2 and SKMEL28 melanoma lines. Two sets of ETV1 locus-specific primers were used (white and gray bars). C, relative ETV1 mRNA and protein expression of the melanoma cell lines analyzed in B and in primary melanocytes were determined by quantitative RT-PCR using two different ETV1-specific primers (white and gray bars), and by immunoblot analysis using an α-ETV1 antibody. D, FISH analysis is shown for illustrative melanoma samples from a tissue microarray (TMA). Green- and red- labeled BAC probes detect the ETV1 and reference loci, respectively. A diploid case (left) and a case of ETV1 amplification (right) are shown.
To confirm the presence of ETV1 amplification in clinical specimens, we performed FISH analysis on an assembled melanoma tissue microarray (TMA) containing 170 evaluable nevi, primary and metastatic melanoma specimens (Table 1). The FISH results were segregated based on the quantity of ETV1 probe signals detected relative to the reference probe (see Materials and Methods). Detection of 2 ETV1 copies per nuclei was considered to indicate no amplification; between >2 and ≥6 ETV1 copies indicated “low-level” amplification; and >6 ETV1 copies represented “high-level” amplification. While no ETV1 copy gains were detected in any of the nevi examined, low-level ETV1 gains were detected at ~40% frequency in all melanomas examined, regardless of stage. These results were consistent with SNP array and CGH results reported here and elsewhere (2, 3). Notably, high-level ETV1 copy gains occurred in 13% of primary samples and in 18% of metastatic melanomas present on the TMA (Fig. 1D; Table 1). These results suggest that DNA amplifications involving the ETV1 locus occurred frequently in melanoma.
Table 1.
ETV1 FISH analysis of melanoma tissue samples
| % of samples |
|||||
|---|---|---|---|---|---|
| Sample type | Samples on array | Analyzable Samples* | No Amp | Low Amp† (<6) | High Amp§ (>6) |
| Thin Nevus | 21 | 10 | 100 | 0 | 0 |
| Thick Nevus | 15 | 10 | 100 | 0 | 0 |
| Thin Primary | 39 | 16 | 56.2 | 31.3 | 12.5 |
| Thick Primary | 20 | 15 | 46.7 | 40.0 | 13.3 |
| Lymph Node Mets | 39 | 17 | 23.6 | 58.8 | 17.6 |
| Visceral Mets | 46 | 23 | 43.5 | 39.1 | 17.4 |
Samples that rendered an informative result
Samples presenting 3 to 6 ETV1-probe signals
Samples presenting more than 6 ETV1-probe signals
Targeted ETV1 gene disruption in melanoma
As ETV1 undergoes translocation in some tumor types (6, 7, 11), we sought to determine if ETV1 gene rearrangements were evident in melanoma. Accordingly, we performed a FISH break-apart assay on the melanoma TMA using flanking telomeric and a centromeric ETV1 probes (Supplementary Fig. 2). All ETV1 translocations described to date show 5′ coding exons replaced by an ectopic promoter, and in some cases by 5′end exons of the partner gene. These rearrangements result in a truncated product that retains the ETS DNA binding domain, but whose expression is controlled by the fusion partner upstream regulatory promoter elements. Interestingly, the ETV1 FISH assay exhibited a “break-apart” pattern in two lymph node metastases (from a total of 41 lymph- and –visceral-metastases analyzed), suggestive of targeted gene disruption or possible translocation of the ETV1 locus (Supplementary Fig. 2). The ETV1 locus remained intact in the antecedent primary tumors (not shown). Interestingly, one of the lymph node metastases contained more signals corresponding to the telomeric probe (green) than to the centromeric probe (red) (Supplementary Fig. 2; right panel). Since ETV1 is positioned with its 5′ end nearest to the centromere this pattern could reflect loss of the centromeric region after translocation, or of selective amplification of a translocation product containing the (telomeric) 3′ end of ETV1. These results raised the possibility that ETV1 might undergo rare chromosomal translocation events in addition to the more common amplification events in melanoma.
ETV1 expression is elevated in melanoma
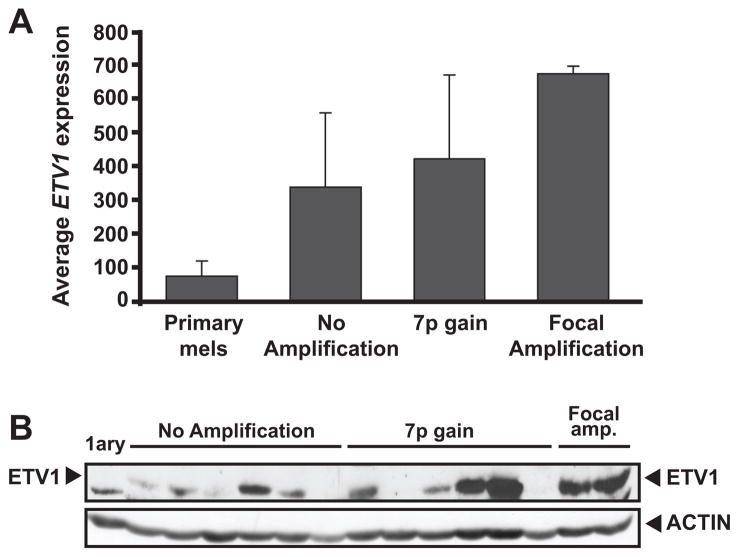
In some cancers, the oncogenicity of ETS transcription factors derives primarily from deregulated gene expression(5, 7, 11). To examine ETV1 expression in melanoma, we analyzed microarray data from a collection of metastatic short-term melanoma cultures and cell lines. ETV1 mRNA levels were elevated in most of the samples relative to primary melanocytes (Fig. 2A). As expected, two cell lines harboring the focal amplification (501mel and WW94) were among the highest ETV1-expressing samples (Figs. 2A–B). On average, the presence of chromosome 7p copy gains correlated with a slightly higher ETV1 expression compared to samples with no 7p gains (Fig. 2A), although this correlation did not reach statistical significance. Immunoblot analysis confirmed the increased levels of ETV1 protein compared to normal melanocytes in melanoma samples with and without 7p gains (Fig. 2B). Thus, elevated ETV1 expression was observed in melanoma samples relative to normal melanocytes regardless of 7p copy gains. Although we identified ETV1 as a melanoma oncogene based on genetic criteria, these results raised the possibility that ETV1 dysregulation might be achieved in melanoma by both genetic and non-genetic mechanisms.
Figure 2.
ETV1 expression in relation to chromosome 7p copy gains. A, normalized ETV1 expression levels in cultured primary melanocytes and melanoma lines are shown. Average expression values from subgroups stratified by ETV1 locus amplification are indicated (focal amplification: 501mel and WW94 lines). B, immunoblot analysis using α-ETV1 and α-actin antibodies is shown for a subset of melanoma cell lines and short-term cultures analyzed in (A).
ETV1 oncogene dependency in melanoma
To determine whether melanoma samples are dependent on ETV1 and whether ETV1 dependency correlates with ETV1 amplification, we studied the consequences of RNAi-mediated suppression of ETV1 expression on melanoma cell proliferation and colony formation. Here, we utilized two independent lentivirally delivered short hairpin RNA (shRNA) constructs that effectively reduced ETV1 mRNA and protein levels (Fig. 3A), as well as a control hairpin against the green fluorescent protein (shGFP). We tested the effects of ETV1 knockdown in 501mel and WW94, two cell lines with focal ETV1 amplification, as well as in SKMEL2 and SKMEL28 (Fig. 1B). To eliminate the possibility that the effects we observed where the consequence of off-target effects of the ETV1-specific shRNAs, we also tested HeLa cells, which do not express ETV1 (Supplementary Fig 3).
Figure 3.
Phenotypic effects of suppression of ETV1 expression in melanoma cell lines. A, ETV1 mRNA levels following infection with lentivirus containing ETV1-specific shRNAs (shETV1-3 and −5) relative to infection with control GFP-specific shRNAs (shGFP) in representative melanoma cell lines with (501mel and WW94) or without (SKMEL2 and SKMEL28) ETV1 amplification (top panels). Corresponding immunoblot studies of ETV1 protein are also shown (bottom panels). B, proliferation curves are shown for the melanoma cell lines in (A) after introduction of shRNAs against ETV1 (solid and dashed black lines) or GFP (control; gray lines). C, anchorage-independent growth of 501mel cells (ETV1-amplified) infected with lentivirus containing ETV1-specific or GFP-specific shRNAs.
Suppression of ETV1 resulted in a marked reduction of proliferation in both 501mel and WW94 cells (ETV1-amplified; Fig. 3B). Moreover, silencing of ETV1 suppressed anchorage independent growth in 501mel cells, while the control shRNA had no effect (Fig. 3C). The effect of ETV1 knockdown was more variable in melanoma cell lines that lacked chromosome 7p copy gains. The proliferation of SKMEL28 cells was substantially inhibited following ETV1 suppression; however, SKMEL2 cells were largely unaffected (Fig. 3B). These results suggested that ETV1 amplification induces an ETV1 dependency in melanoma, while supporting the hypothesis that ETV1 dependency also exists in some (but not all) melanoma cells lacking ETV1 amplification.
ETV1 overexpression cooperates with oncogenic NRAS and BRAF to transform immortalized human melanocytes
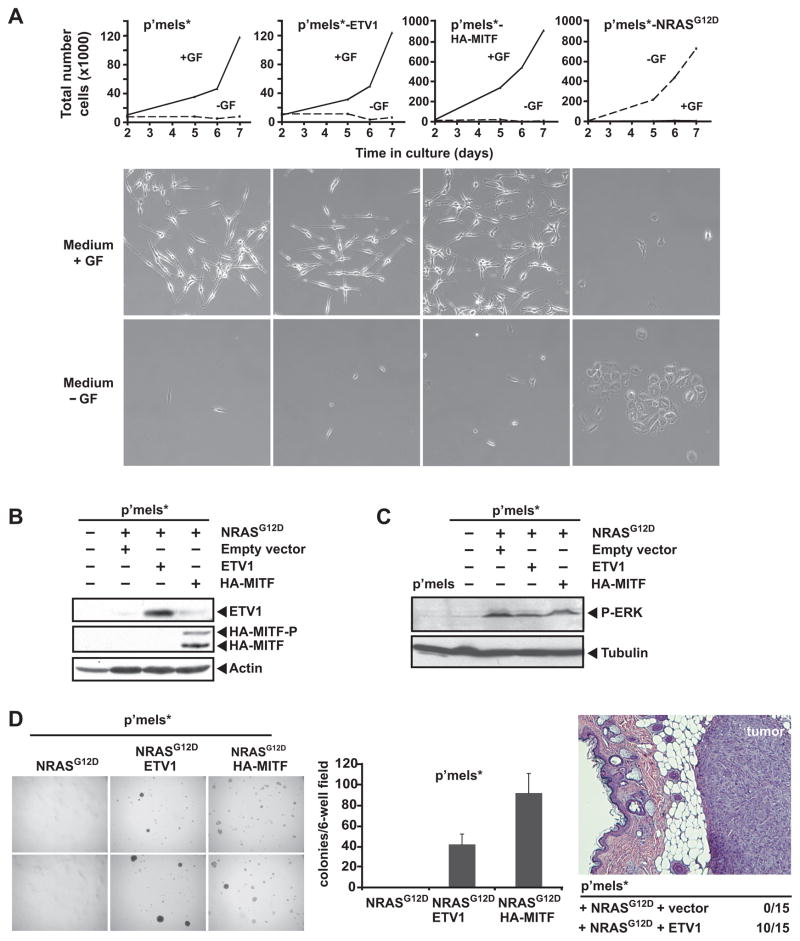
To test the hypothesis that ETV1 functions as an oncogene in melanoma, we overexpressed ETV1 in genetically-modified primary melanocytes (hereafter referred to as p’mel* cells). As described previously (10), p’mel* cells were immortalized through ectopic expression of hTERT, p53DD (dominant-negative p53) and CDK4(R24C) (INK-resistant CDK4). These factors facilitated telomere stabilization as well as retinoblastoma (RB) and p53 pathway inactivation, which occur commonly in this malignancy. Although immortalized, p’mel* cells require TPA and cyclic AMP (cAMP) agonists for survival, a hallmark of non-transformed melanocytes.
Overexpression of ectopic ETV1 alone did not overcome the TPA and cAMP growth factor requirement in p’mel* cells (Fig. 4A). As expected (10), similar results were obtained following ectopic expression of MITF, a known oncogenic transcription factor in melanoma. However, most melanomas also harbor activating NRAS or BRAF mutations that result in aberrant MAP kinase pathway activation (4, 12, 13). Toward this end, we expressed NRASG12D oncoprotein in p’mel* cells (Fig. 4B). When expressed alone, NRASG12D activated MAP kinase signaling (Fig 4C), but failed to confer robust anchorage-independent growth (Fig. 4D). This result is reminiscent of prior studies using the BRAFV600E oncogene in these cells (10). In contrast, combined expression of both ETV1 and NRASG12D strongly induced anchorage independent growth in p’mel* cells, with a phenotype that resembled the effect of ectopic MITF expression (Fig. 4D). Combined ETV1 and BRAFV600E expression also conferred soft agar growth to p’mel* cells, albeit with lower efficiency (Supplementary Fig. 4). Thus, ETV1 overexpression is able to cooperate with both NRASG12D and BRAFV600E to transform immortalized human melanocytes.
Figure 4.
Effects of ETV1 overexpression on melanocyte transformation. A, proliferation curves are shown for p’mel* cells (immortalized human melanocytes described in the text) and p’mel* cells expressing ETV1, HA-MITF or NRASG12D cultured in F10 media in the presence (+GF) or absence (−GF) of TPA and cAMP growth factors. Cells were photographed after 5 days in culture. B, C, immunoblot analyses of p’mel* and p’mel*-NRASG12D cells infected with empty vector, ETV1 or HA-MITF retroviruses are shown. Antibodies recognizing ETV1, HA epitope, actin, phospho-ERK and tubulin were used. HA-MITF-P denotes the phosphorylated form of HA-MITF. D, (left)anchorage-independent growth of p’mel* cells expressing NRASG12D alone or in combination with ETV1 or HA-MITF is shown (photographs from two independent assays are shown for each cell type). (middle) Colony counts from these experiments are also indicated. (right) p’mel*-NRASG12D cells infected with empty- or ETV1-retroviruses were injected subcutaneously in mice. Tumor formation following injection is indicated numerically, and a histological section of a representative ETV1-dependent tumor is shown (stained with hematoxylin and eosin).
ETV1 overexpression promotes melanoma tumor formation in vivo
We next investigated the effect of ETV1 overexpression on melanoma genesis in a xenograft model. Here, p’mel* cells expressing NRASG12D alone or in combination with ETV1 were injected subcutaneously into immunodeficient mice and monitored for tumor formation. P’mel* cells expressing NRASG12D alone failed to form tumors (0/15 mice examined). In contrast, p’mel* cells expressing both NRASG12D and ETV1 formed tumors in 67% of cases (10 tumors/15 injections) (Fig. 4D). Together with the soft agar findings above, these results suggested that the ETV1 transcription factor can function as an oncogene in melanoma.
ETV1 overexpression induces MITF up-regulation in p’mel*-NRASG12D cells
In previous work, our group showed that the MITF transcription factor, a master regulator of melanocyte lineage development and survival, cooperates with BRAFV600E to function as a melanoma oncogene (10). Consistent with these earlier observations, overexpression of HA-tagged melanocytic MITF in p’mel*-NRASG12D cells also resulted in robust colony formation (Fig. 4D). These results indicated that MAP kinase pathway activation by either NRAS or BRAF mutation could cooperate with MITF to transform p’mel* cells. Together with the observation that ETV1 transforms p*mel cells in the same genetic context, these findings also raised the possibility that a mechanistic relationship might exist between ETV1 and MITF with regards to melanocyte transformation.
To test this hypothesis, we performed immunoblot analysis of lysates derived from p’mel*-NRASG12D cells expressing empty vector, ETV1, or HA-MITF. Interestingly, ETV1 overexpression strongly induced endogenous MITF protein up-regulation in p’mel*-NRASG12D cells (Fig. 5A, left). Quantitative RT-PCR using MITF primers revealed a concomitant increase in MITF mRNA levels (Fig. 5A, right). The melanocytic MITF promoter contains several four-base GGAA/T sequences characteristic of the ETS factor consensus binding site; however, ETV1 did not augment MITF promoter activity in luciferase reporter assays (data not shown). Thus, ETV1 expression induced MITF mRNA and protein expression markedly but indirectly in p’mel*-NRASG12D cells.
Figure 5.
ETV1 regulation of MITF expression. A, (left) immunoblot analyses of p’mel*-NRASG12D cells infected with empty vector, ETV1 or HA-MITF retroviruses are shown. Antibodies recognizing ETV1, HA epitope, and actin were used. MITF-P and HA-MITF-P denote the phosphorylated forms of endogenous MITF and HA-MITF, respectively. (right) Relative mRNA expression levels of endogenous melanocytic MITF are shown for the cell types analyzed at left as determined by RT-PCR. B, (left) suppression of MITF expression is shown for p’mel*-NRASG12D cells overexpressing HA-MITF or ETV1 following infection with lentivirus containing shRNAs against MITF (shMITF) or GFP (shGFP). (right) Anchorage-independent growth of cells analyzed at left. C, (left) immunoblot analysis of MITF levels following suppression of ETV1 expression using ETV1-specific shRNAs in 501mel cells. (right) ETV1 and MITF mRNA levels are shown for the cells analyzed at left, as determined by RT-PCR (in triplicate) and normalized to GAPDH.
MITF is necessary for ETV1 oncogenicity in melanoma
Given its oncogenic properties in MAP kinase-driven melanomas, we next examined whether MITF is required for ETV1-mediated transformation in p’mel*-NRASG12D cells. Here, we suppressed MITF expression using a MITF-specific shRNA in p’mel*-NRASG12D cells that overexpressed ETV1. As a control, we performed similar experiments in p’mel*-NRASG12D cells overexpressing MITF (Fig. 5B, left). Silencing of MITF greatly diminished the anchorage-dependent growth phenotype induced by both ETV1 and MITF overexpression in these cells (Fig. 5B, right). To eliminate the possibility that the effects we observed where the consequence of off-target effects of the MITF-specific shRNA, we also tested MCF7 cells, which do not express MITF (Supplementary Fig. 5). Thus, MITF is required for ETV1-dependent oncogenicity in this setting.
Since ETV1 overexpression augmented MITF expression (at least indirectly) as described above, we next investigated whether reduction of ETV1 expression in established melanoma cell lines might down-regulate endogenous MITF levels. Indeed, RNAi-mediated suppression of ETV1 resulted in a measurable albeit modest reduction in endogenous MITF in 501mel cells, which harbor ETV1 amplification (Fig. 5C, left). Moreover, shRNAs directed against MITF reduced both proliferation (data not shown) and colony formation in soft agar in 501mel cells (Supplementary Fig. 6). In contrast to the studies of p’mel*-NRASG12D cells above, we did not detect a corresponding decrease in MITF mRNA following ETV1 knockdown (Fig. 5C, right). These results suggested that the oncogenic function of ETV1 may derive at least in part through regulation of MITF protein levels in human melanoma cells.
DISCUSSION
The systematic characterization of target or “driver” genes enacted by recurrent genomic aberrations offers a means to elucidate mechanisms of tumorigenesis. However, many prevalent chromosomal copy number aberrations consist of large, low-amplitude events that do not provide sufficient resolution to pinpoint driver genes definitively. Rare focal genetic alterations may enrich for “driver” cancer genes that are also enacted by highly recurrent, non-focal events spanning the same locus. In this study, we identified three focal amplifications encompassing 7p21.3, which nominated the ETS transcription factor ETV1 as a candidate driver gene targeted by chromosome 7p gains in melanoma. Tissue microarray analyses also identified ETV1 amplification in 13%–18% of melanomas, with occasional evidence of targeted gene disruption (e.g., translocation) involving this locus. Subsequent studies indicated that ETV1 expression is required in several melanoma contexts, including cell lines harboring ETV1 amplification. Ectopic ETV1 overexpression in the context of aberrant MAP kinase pathway activation transformed immortalized human melanocytes. While these findings do not exclude the possibility that other genes located at chromosome 7p21 or elsewhere on 7p may be targeted by these same copy gains, our results strongly suggest that ETV1 functions as a melanoma oncogene.
Recent findings have drawn increasing attention to the role of ETS transcription factors in cancer. Since the original ETS sequence was discovered in an avian erythroblastosis virus (14), ~40 additional ETS factors have been identified (reviewed in (15)). Multiple members of the ETS family undergo oncogenic dysregulation in cancer, often through chromosomal translocation (reviewed in (15) and in (16)). In Ewing Sarcoma (EWS) (6), EWS:ETV1 translocations result in highly transforming chimeric ETS fusion proteins (17, 18). More recently, chromosomal translocations involving ETV1 and other ETS genes were found in over 40% of prostate cancers (7, 19). Most commonly, these translocations interpose the promoter and 5′ coding exons of the TMPRSS2 gene upstream of an ETS factor gene (ERG, ETV1, ETV4, or ETV5), resulting in androgen-dependent regulation and elevated expression of these genes (7, 19) results described herein add malignant melanoma to the growing list of cancers where ETS transcription factors in general—and ETV1 in particular—exert critical tumor dependencies.
In melanoma, ETV1 dependency may also extend beyond cases of 7p21 amplification/copy gain. Analyses of gene expression microarray data showed significantly higher ETV1 mRNA levels in melanoma relative to primary melanocytes, yet we did not observe a clear correlation between ETV1 mRNA expression and chromosome 7p copy gains per se. Thus, chromosomal copy changes represent only one mechanism of ETV1 up-regulation in melanoma. Moreover, melanoma cell viability was reduced following ETV1 knockdown in two cell lines with ETV1 amplification (and high ETV1 expression), but at least one melanoma cell line lacking 7p copy gain also exhibited a reliance on ETV1 for viability. These results are reminiscent to studies of MITF dependency in melanoma: focal amplifications targeting MITF are found in only 10–15% of melanomas (10) yet MITF dependency is broadly operant and not exclusive to MITF-amplified tumors. Thus, ETV1 amplifications and copy gains may highlight a widely relevant ETV1 dependency in melanoma.
In our hands, the ability of deregulated ETV1 to transform immortalized human melanocytes is dependent on constitutive MAP kinase pathway activation by gain-of-function BRAF or NRAS mutations. This result is reminiscent of studies in prostate and mammary transgenic mouse models, where ERG or ETV1 expression alone was insufficient to drive tumorigenesis (19–23). Our findings also accord well with studies indicating that ETS factors become activated by MAP kinase-mediated phosphorylation ((24, 25) and reviewed in (15)). Interestingly, a recent study of oncogenic MAP kinase output in melanoma showed that ERK also regulates the transcriptional expression of several ETS factors, including ETV1 (26). In this report, ETV1 mRNA levels in BRAFV600E melanoma cell lines were dramatically reduced following pharmacologic MAP kinase inhibition using a MEK inhibitor (26). Together with published evidence, our findings support a model wherein deregulated ETV1 expression in melanoma (by genomic amplification or other means) elaborates an oncogenic signal that is both enabled and potentiated by concomitant MAP kinase pathway activation.
Ectopic ETV1 expression in immortalized human melanocytes results in up-regulation of MITF mRNA and protein levels. This effect, though indirect, is necessary for ETV1 mediated melanoycte transformation. These results indicate that MITF expression may be necessary to elaborate a full ETV1 dependency in melanoma. ETV1 also regulates the expression of other proteins with tumorigenic functions, including hTERT (27). Our observations imply that MITF, which itself is activated by MAP kinase (28), may participate in a melanocytic transcriptional program that includes ETV1 and possibly other factors. This program may become deregulated at multiple points during melanoma genesis or progression.
In conclusion, this study suggests that deregulated ETV1 underpins a melanoma oncogene dependency involving both MAP kinase and MITF activation. The high prevalence of chromosome 7p gains together with activating MAP kinase mutations (e.g., BRAF or NRAS mutation) in melanoma may offer an optimal setting for ETV1-driven oncogenicity. Given the abundance of genetically characterized in vitro models for this malignancy, melanoma may offer a robust context for elucidating downstream oncogenic mechanisms linked to ETV1 and possibly other ETS factors in many human cancers.
Supplementary Material
Acknowledgments
We thank M. Bachrach for technical assistance. J.J.V. and L.A.G acknowledge financial support from the Department of Defense Prostate Cancer Program (Postdoctoral Traineeship Award), National Institutes of Health, National Cancer Institute, Robert Wood Johnson Foundation, Prostate Cancer Foundation, and Burroughs-Wellcome Fund. D.E.F is Distinguished Clinical Scholar of the Doris Duke Medical Foundation and acknowledges grant support from NIH.
References
- 1.Bastian BC, LeBoit PE, Hamm H, Brocker EB, Pinkel D. Chromosomal gains and losses in primary cutaneous melanomas detected by comparative genomic hybridization. Cancer Res. 1998;58:2170–5. [PubMed] [Google Scholar]
- 2.Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–47. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 3.Lin WM, Baker AC, Beroukhim R, et al. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res. 2008;68:664–73. doi: 10.1158/0008-5472.CAN-07-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 5.Baert JL, Monte D, Musgrove EA, et al. Expression of the PEA3 group of ETS-related transcription factors in human breast-cancer cells. Int J Cancer. 1997;70:590–7. doi: 10.1002/(sici)1097-0215(19970304)70:5<590::aid-ijc17>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 6.Jeon IS, Davis JN, Braun BS, et al. A variant Ewing’s sarcoma translocation (7;22) fuses the EWS gene to the ETS gene ETV1. Oncogene. 1995;10:1229–34. [PubMed] [Google Scholar]
- 7.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 8.Kim M, Gans JD, Nogueira C, et al. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell. 2006;125:1269–81. doi: 10.1016/j.cell.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 9.Perner S, Wagner PL, Demichelis F, et al. EML4-ALK fusion lung cancer: a rare acquired event. Neoplasia. 2008;10:298–302. doi: 10.1593/neo.07878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garraway LA, Widlund HR, Rubin MA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–22. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 11.Hermans KG, van der Korput HA, van Marion R, et al. Truncated ETV1, fused to novel tissue-specific genes, and full-length ETV1 in prostate cancer. Cancer Res. 2008;68:7541–9. doi: 10.1158/0008-5472.CAN-07-5930. [DOI] [PubMed] [Google Scholar]
- 12.Carr J, Mackie RM. Point mutations in the N-ras oncogene in malignant melanoma and congenital naevi. Br J Dermatol. 1994;131:72–7. doi: 10.1111/j.1365-2133.1994.tb08460.x. [DOI] [PubMed] [Google Scholar]
- 13.Thomas RK, Baker AC, Debiasi RM, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–51. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 14.Leprince D, Gegonne A, Coll J, et al. A putative second cell-derived oncogene of the avian leukaemia retrovirus E26. Nature. 1983;306:395–7. doi: 10.1038/306395a0. [DOI] [PubMed] [Google Scholar]
- 15.Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- 16.Hsu T, Trojanowska M, Watson DK. Ets proteins in biological control and cancer. J Cell Biochem. 2004;91:896–903. doi: 10.1002/jcb.20012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bailly RA, Bosselut R, Zucman J, et al. DNA-binding and transcriptional activation properties of the EWS-FLI-1 fusion protein resulting from the t(11;22) translocation in Ewing sarcoma. Mol Cell Biol. 1994;14:3230–41. doi: 10.1128/mcb.14.5.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ouchida M, Ohno T, Fujimura Y, Rao VN, Reddy ES. Loss of tumorigenicity of Ewing’s sarcoma cells expressing antisense RNA to EWS-fusion transcripts. Oncogene. 1995;11:1049–54. [PubMed] [Google Scholar]
- 19.Tomlins SA, Laxman B, Dhanasekaran SM, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–9. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 20.Carver BS, Tran J, Gopalan A, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–24. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.King JC, Xu J, Wongvipat J, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41:524–6. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Netzer S, Leenders F, Dumont P, Baert JL, de Launoit Y. Ectopic expression of the ets transcription factor ER81 in transgenic mouse mammary gland enhances both urokinase plasminogen activator and stromelysin-1 transcription. Transgenic Res. 2002;11:123–31. doi: 10.1023/a:1015248525364. [DOI] [PubMed] [Google Scholar]
- 23.Shin S, Kim TD, Jin F, et al. Induction of prostatic intraepithelial neoplasia and modulation of androgen receptor by ETS variant 1/ETS-related protein 81. Cancer Res. 2009;69:8102–10. doi: 10.1158/0008-5472.CAN-09-0941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bosc DG, Goueli BS, Janknecht R. HER2/Neu-mediated activation of the ETS transcription factor ER81 and its target gene MMP-1. Oncogene. 2001;20:6215–24. doi: 10.1038/sj.onc.1204820. [DOI] [PubMed] [Google Scholar]
- 25.Janknecht R. Analysis of the ERK-stimulated ETS transcription factor ER81. Mol Cell Biol. 1996;16:1550–6. doi: 10.1128/mcb.16.4.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pratilas CA, Taylor BS, Ye Q, et al. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci U S A. 2009;106:4519–24. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goueli BS, Janknecht R. Upregulation of the Catalytic Telomerase Subunit by the Transcription Factor ER81 and Oncogenic HER2/Neu, Ras, or Raf. Mol Cell Biol. 2004;24:25–35. doi: 10.1128/MCB.24.1.25-35.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hemesath TJ, Price ER, Takemoto C, Badalian T, Fisher DE. MAP kinase links the transcription factor Microphthalmia to c-Kit signalling in melanocytes. Nature. 1998;391:298–301. doi: 10.1038/34681. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.