Abstract
Purpose
The interferon regulatory factor 6 (IRF6), the gene that causes van der Woude syndrome has been shown to be associated with nonsyndromic cleft lip with or without palate in several populations. This study aimed to confirm the contribution of IRF6 to cleft lip with or without palate risk in additional Asian populations.
Methods
A set of 13 single nucleotide polymorphisms was tested for association with cleft lip with or without palate in 77 European American, 146 Taiwanese, 34 Singaporean, and 40 Korean case-parent trios using both the transmission disequilibrium test and conditional logistic regression models.
Results
Evidence of linkage and association was observed among all four populations; and two specific haplotypes [GC composed of rs2235373-rs2235371 (p.V274I) and AAG of rs599021-rs2235373-rs595918] showed the most significant over- and undertransmission among Taiwanese cases (P = 9 × 10−6 and P = 5 × 10−6, respectively). The AGC/CGC diplotype composed of rs599021-rs2235373-rs2013162 showed almost a 7-fold increase in risk among the Taiwanese sample (P < 10−3). These results confirmed the contribution of this gene to susceptibility of oral clefts across different populations; however, the specific single nucleotide polymorphisms showing statistical significance differed among ethnic groups.
Conclusion
The high-risk genotypes and diplotypes identified here may provide a better understanding of the etiological role of this gene in oral clefts and potential options for genetic counseling.
Keywords: Association, ethnicity, interferon regulatory factor 6, oral cleft, risk estimation, single nucleotide polymorphism
Nonsyndromic cleft lip with or without cleft palate (CL/P) is one of the most common birth defects with the birth prevalence being highest in Asian (2/1000 live births), intermediate in European (1/1000 live births), and lowest in African populations (0.4/1000 live births). CL/P is a complex disease with both genetic and environmental risk factors.1 Mutations in the interferon regulatory factor 6 gene (IRF6) located on chromosome 1q32.3-q41 are responsible for a majority of van der Woude syndrome (VWS) cases. VWS is an autosomal dominant syndrome that includes an oral cleft and pits on the lower lip in approximately 85% of cases. Fifteen percent of VWS cases have an isolated cleft with no lip pits and are clinically indistinguishable from nonsyndromic CL/P.2 The GATA124F08 marker located 1Mb from IRF6 has shown a significant heterogeneity LOD (1.15) with α = 0.45,3 and an anonymous marker (D1S205) in IRF6 has yielded significant evidence of linkage and linkage disequilibrium (LD) in 106 nonsyndromic CL/P trios.4 Recently, strong evidence of overtransmission of the G allele at the IRF6 c.820G>A marker (rs2235371) was found in CL/P case-parent trios from Asia and South America,5 and a significantly higher frequency of the GG genotype was observed among 192 Thai CL/P cases compared with controls (odds ratio = 1.67).6 This variant creates a valine→isoleucine substitution at amino acid 274 (p.V274I) in the protein-binding domain [the Smad-interferon regulatory factor binding domain (SMIR)] of IRF6, but the A allele is rare in white populations. Analysis of seven other single nucleotide polymorphisms (SNPs) in and around IRF6 has shown several distinct haplotypes demonstrating altered transmission in Io-wan and Danish trios.5 Confirmatory studies using Italian, European-American, and Belgian CL/P families, respectively, have strengthened the evidence that IRF6 is important in the etiology of nonsyndromic oral clefts.7–9 Risk of CL/P associated with particular variants in IRF6 may differ among ethnic groups, however. Here, we evaluated 13 SNPs in and around IRF6 to test for association with nonsyndromic CL/P in 77 European-American (including five incomplete trios), 146 (three incomplete trios) and 34 (11 incomplete trios) Han Chinese trios from Taiwan and Singapore, respectively, plus 40 (two incomplete trios) Korean CL/P trios. Expression of IRF6 in human craniofacial structures was also determined using publicly available data.
MATERIALS AND METHODS
Subjects
Infants born with isolated, nonsyndromic CL/P and their parents were ascertained through treatment centers in Maryland (Johns Hopkins University and University of Maryland), Taiwan (Chang Gung Memorial Hospital), Singapore (KK Women’s and Children’s Hospital), and Korea (Yonsei University Medical Center), respectively, under a protocol approved by the institutional review board at each institution as part of an international study of oral clefts. After informed consent was obtained from parents, ethnicity and other data were obtained through structured interviews.10 Both cases and parents provided blood samples.
SNP selection and genotyping
SNP markers in and around IRF6 were identified from the literature and the dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP/) using a NorthStar Searchlet program (Genetic Software Innovations, Inc., Cicero, NY). A final set of 13 SNPs were chosen based on the criteria of high “design scores” as provided by Illumina, Inc. (San Diego, CA), heterozygosity >0.1, and HapMap validation (www.hapmap.org/index.html.en). The final marker set included the rs2235371 and rs2013162, which previously showed significant associations in Asians and Europeans, respectively (Fig. 1). Primers for each SNP were synthesized using the Oligator technology by Illumina, Inc. as part of an oligo pool for the BeadLab 1000 system. Genomic DNA samples were prepared from peripheral blood lymphocytes by the protein precipitation method described previously11 and genotyped for SNP markers using the Golden Gate chemistry on Sentrix Array Matrices (Illumina, Inc.) at the Johns Hopkins SNP Center.12 The average distance between neighboring markers was 1.53 kb (based on the Build 36.1 of dbSNP). Two duplicates and four CEPH control DNA samples were included to evaluate genotyping consistency.
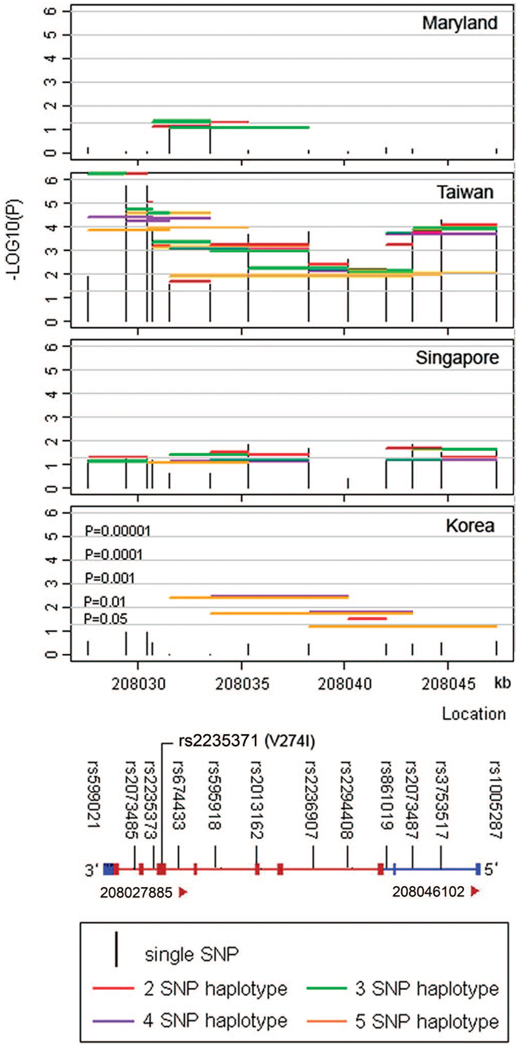
Fig. 1.
Significance of individual single nucleotide polymorphisms (SNPs) and sliding window haplotypes for the interferon regulatory factor 6 (IRF6) gene in four groups of nonsyndromic cleft lip with or without cleft palate trios. The −log10 (empirical P value) for the overall χ2 test for an individual SNP (vertical line) and for sliding windows of haplotypes of two to five SNPs (horizontal lines) is presented. Nominal significance levels are denoted by gray lines (5%, 1%, 0.1%, 0.01%, and 0.001%).
Statistical analysis
Within each population, the minor allele frequency (MAF), heterozygosity, and a χ2 test for Hardy-Weinberg equilibrium (HWE) at each SNP were computed among parents. Pairwise LD was computed as both D′ and r2 for all SNPs using the Haploview program (http://www.broad.mit.edu/mpg/haploview/index.php/).13,14 Individual SNPs and sliding windows of haplotypes consisting of two, three, four, and five SNPs were tested using the family-based association test program (http://www.biostat.harvard.edu/fbat/default.html).15 Empirical P values for observed versus expected transmission were obtained using the permutation option and these are presented as −log10 P values.16 A Web interface (SNPSpD) was used to perform the spectral decomposition correction for multiple comparisons (http://genepi.qimr.edu.au/general/daleN/SNPSpD/).17 Genotypic odds ratios (GORs) for heterozygotes and homozygotes were calculated separately for individual SNPs as well as for diplotypes consisting of two or three SNP haplotypes yielding statistical significance. GORs were obtained from conditional logistic regression models for matched sets consisting of the case and three “pseudosib” controls derived from the parental mating type using publicly available subroutines in the STATA software package (http://www-gene.cimr.cam.ac.uk/clayton/software/stata/).
Gene expression analysis
Expression of IRF6 in human craniofacial structures relevant to normal palate and lip development was determined using data obtained from the Craniofacial and Oral Gene Expression Network (COGENE) consortium (http://hg.wustl.edu/COGENE/). Data from seven serial analysis of gene expression (SAGE) libraries were used to assess gene expression patterns in different human embryonic tissues (i.e., 26-day-old human embryonic tissue, 4-week anterior rhombomere, 4-week posterior rhombomere, 4-week frontonasal prominence, 5-week frontonasal prominence, 6-week mandible, and 8.5-week upper lip).18
RESULTS
Proband gender for the four groups of case-parent trios is shown in Appendix Table A1 (available online at www.geneticsinmedicine.org). Examining duplicated samples revealed a very high reproducibility for genotypes (99.98%). Minor allele frequencies for rs2235371 and rs3753517 were too low to be informative in the Maryland samples (MAF <0.005 for both), and only 60% of genotypes were called at rs2294408 in the Singapore and Korean samples. All remaining SNPs gave no evidence of deviating from HWE (data not shown). Among the 13 SNPs, five groups of markers (rs599021-rs861019, rs2073485-rs2235373, rs2235371-rs3753517, rs674433-rs595918, and rs2013162-rs2236907-rs2294408-rs2073487-rs1005287) showed virtually complete LD (D′ = 1 and r2 > 0.8) in all four populations, so markers within each block became redundant (see Appendix Table A2 available online at www.geneticsinmedicine.org). Consequently, one tagging marker was chosen from each of these five groups (rs599021, rs2235373, rs2235371, rs595918, and rs2013162) to represent haplotypes showing significant transmission distortion and estimate GORs.
TDT analyses for individual markers and haplotypes
In Figure 1, only empirical P values <0.10 from the transmission disequilibrium test (TDT) are presented for haplotypes, whereas all empirical P values for the 13 individual SNPs in each of the four populations are presented. TDT results for individual SNPs showing significant evidence of linkage and LD among the four samples of CL/P trios are summarized in Table 1. Two SNPs, rs2073485 and rs2235373, which were in complete LD with one another (both D′ and r 2 = 1), yielded highly significant P values for both single marker and haplotype analyses among the 146 Taiwanese CL/P trios (P = 2 × 10−6 and lowest P < 10−6, respectively). In the 34 Singaporean trios, seven SNPs and their haplotypes yielded nominal significance (lowest P=0.014). Haplotypes consisting of three SNPs (rs2235371, rs674433, and rs595918) yielded nominal significance in the Maryland trios. In the Korean trios, most haplotypes, including rs2294408, were statistically significant; however, only nine families were informative for this marker. Experiment-wide significance thresholds required to keep the type I error rate at 5% for samples from Maryland, Taiwan, Singapore, and Korea are P < 0.010, 0.013, 0.017, and 0.013, respectively, based on the spectral decomposition correction. Therefore, statistical evidence observed among the Taiwanese, Singaporean, and Korean trios remained significant after correcting for multiple comparisons.
Table 1.
Marker information and TDT results for SNPs in IRF6 showing significant evidence of linkage and LD in four groups of CL/P trios
| Allelesa | Haplotype size (P)c | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Population | SNP | Allele 1 frequency | 1 | 2 | T/NTb | SNP (P)c | 2 | 3 | 4 | 5 |
| Maryland | rs2235371 | 0.005 | T | C | 0/0 | NAd | 0.077 | 0.045 | ||
| rs674433 | 0.803 | T | G | 19/10 | 0.095 | 0.072 | 0.086 | 0.088 | ||
| rs595918 | 0.194 | A | G | 19/9 | 0.059 | 0.050 | 0.084 | |||
| rs2013162 | 0.345 | A | C | 28/25 | 0.782 | |||||
| rs2236907 | 0.350 | A | C | 29/27 | 0.789 | |||||
| Taiwan | rs599021 | 0.787 | A | C | 59/35 | 0.013 | <10−6 | <10−6 | 4 × 10−5 | 1 × 10−4 |
| rs2073485 | 0.482 | A | G | 98/42 | 2 × 10−6 | <10−6 | 2 × 10−5 | 6 × 10−5 | 3 × 10−5 | |
| rs2235373 | 0.482 | A | G | 97/41 | 2 × 10−6 | 1 × 10−5 | 3 × 10−5 | 5 × 10−5 | 1 × 10−4 | |
| rs2235371 | 0.423 | T | C | 87/44 | 2 × 10−4 | 7 × 10−4 | 5 × 10−4 | 8 × 10−4 | 7 × 10−4 | |
| rs674433 | 0.780 | T | G | 55/34 | 0.026 | 0.022 | 8 × 10−4 | 0.001 | 0.013 | |
| rs595918 | 0.220 | A | G | 55/34 | 0.026 | 6 × 10−4 | 0.001 | 0.012 | 0.011 | |
| rs2013162 | 0.566 | A | C | 93/49 | 2 × 10−4 | 6 × 10−4 | 0.006 | 0.012 | 0.013 | |
| rs2236907 | 0.567 | A | C | 94/49 | 2 × 10−4 | 0.004 | 0.007 | 0.008 | 0.011 | |
| rs2294408 | 0.566 | A | G | 68/37 | 0.002 | 0.007 | 0.007 | 0.010 | 0.009 | |
| rs861019 | 0.787 | A | G | 59/35 | 0.013 | 6 × 10−4 | 2 × 10−4 | 2 × 10−4 | ||
| rs2073487 | 0.433 | T | C | 94/49 | 2 × 10−4 | 1 × 10−4 | 1 × 10−4 | |||
| rs3753517 | 0.425 | A | G | 91/44 | 5 × 10−5 | 9 × 10−5 | ||||
| rs1005287 | 0.571 | T | C | 95/48 | 8 × 10−5 | |||||
| Singapore | rs599021 | 0.710 | A | C | 11/4 | 0.071 | 0.054 | 0.079 | ||
| rs2073485 | 0.394 | A | G | 15/6 | 0.049 | 0.051 | ||||
| rs2235373 | 0.394 | A | G | 15/6 | 0.049 | 0.084 | ||||
| rs2235371 | 0.317 | T | C | 11/4 | 0.071 | |||||
| rs674433 | 0.810 | T | G | 11/6 | 0.225 | 0.039 | 0.072 | |||
| rs595918 | 0.190 | A | G | 11/6 | 0.225 | 0.031 | 0.059 | |||
| rs2013162 | 0.509 | A | C | 18/6 | 0.014 | 0.039 | ||||
| rs2236907 | 0.514 | A | C | 17/6 | 0.022 | |||||
| rs2294408 | 0.476 | A | G | 8/5 | 0.405 | |||||
| rs861019 | 0.710 | A | G | 11/4 | 0.071 | 0.020 | 0.059 | 0.065 | ||
| rs2073487 | 0.486 | T | C | 18/6 | 0.014 | 0.022 | 0.023 | |||
| rs3753517 | 0.320 | A | G | 13/4 | 0.029 | 0.049 | ||||
| rs1005287 | 0.530 | T | C | 17/6 | 0.022 | |||||
| Korea | rs674433 | 0.748 | T | G | 14/14 | 1.000 | 0.004 | |||
| rs595918 | 0.252 | A | G | 14/14 | 1.000 | 0.004 | 0.018 | |||
| rs2013162 | 0.541 | A | C | 22/16 | 0.330 | 0.018 | 0.018 | |||
| rs2236907 | 0.539 | A | C | 22/16 | 0.330 | 0.019 | 0.017 | 0.067 | ||
| rs2294408 | 0.559 | A | G | 8/3 | NAd | 0.032 | 0.018 | 0.065 | 0.068 | |
| rs861019 | 0.783 | A | G | 17/12 | 0.353 | |||||
| rs2073487 | 0.461 | T | C | 22/16 | 0.330 | |||||
| rs3753517 | 0.382 | A | G | 21/15 | 0.317 | |||||
| rs1005287 | 0.533 | T | C | 23/16 | 0.262 | |||||
Overtransmitted alleles are in bold type.
Transmission/nontransmission counts from heterozygous parents.
Significant P values for individual SNP and global P values for sliding windows of haplotypes of two to five SNPs from TDT analyses. Tests significant after an SNPSpD correction are shown in bold.
Test statistic was not computed when <10 informative families were available for an individual marker.
TDT, transmission disequilibrium test; SNP, single nucleotide polymorphism; LD, linkage disequilibrium; CL/P, cleft lip with or without cleft palate; NA, not available.
In Table 2, the most common haplotype (AATGA) across five SNPs (rs599021, rs2235373, rs2235371, rs595918, and rs2013162) showed significant undertransmission (P = 0.00051), whereas two haplotypes, (A/C)GC(A/G)C, were significantly overtransmitted among Taiwanese CL/P children. Interestingly, two-SNP haplotypes [e.g., AA for rs599021 and rs2235373 (P=5 × 10−6) or GC composed of rs2235373 and rs2235371 (P = 9 × 10−6)] were more informative than the three- or four-SNP haplotypes. Here, all alleles are reported on the forward strand of the chromosome (NCBI build 36.1), although the gene is transcribed from the reverse strand. Allele designations need to be reversed when compared with published reports that used the transcription strand as the reference.
Table 2.
Markers and haplotypes showing significant evidence of linkage and LD in 79 Taiwanese CL/P case-parent trios
| Marker/haplotype | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| rs599021 | rs2235373 | rs2235371 | rs595918 | rs2013162 | Allele freq | Trios(N)a | Obsb | Expb | P | Global Pc |
| Overtransmitted haplotypes | ||||||||||
| A | G | — | — | — | 0.304 | 76 | 88 | 72 | 0.002 | 2.7 × 10−5 |
| — | G | C | — | — | 0.513 | 77 | 111 | 85 | 9 × 10−6 | 3.1 × 10−5 |
| — | G | — | G | — | 0.294 | 73 | 87 | 70 | 0.001 | 2.9 × 10−5 |
| — | G | — | — | C | 0.429 | 91 | 107 | 86 | 9 × 10−4 | 4.1 × 10−5 |
| — | — | C | — | C | 0.431 | 92 | 116 | 95 | 7 × 10−4 | 0.001 |
| — | G | C | — | C | 0.429 | 91 | 114 | 93 | 9 × 10−4 | 8.7 × 10−5 |
| A | G | C | — | C | 0.217 | 65 | 62 | 53 | 0.037 | 2.6 × 10−4 |
| C | G | C | — | C | 0.212 | 65 | 62 | 51 | 0.022 | 2.6 × 10−4 |
| — | G | C | A | C | 0.217 | 65 | 63 | 54 | 0.037 | 2.6 × 10−4 |
| — | G | C | G | C | 0.212 | 65 | 63 | 52 | 0.022 | 2.6 × 10−4 |
| Undertransmitted haplotypes | ||||||||||
| A | A | — | — | — | 0.488 | 79 | 71 | 99 | 5 × 10−6 | 2.7 × 10−5 |
| A | — | v | G | — | 0.575 | 93 | 103 | 125 | 5.1 × 10−4 | 0.003 |
| — | A | — | G | — | 0.488 | 79 | 78 | 106 | 5 × 10−6 | 2.9 × 10−5 |
| — | A | T | — | 0.427 | 77 | 56 | 77 | 5.1 × 10−4 | 3.1 × 10−5 | |
| — | A | — | — | A | 0.488 | 77 | 67 | 94 | 9 × 10−6 | 4.1 × 10−5 |
| A | A | T | — | — | 0.427 | 77 | 65 | 86 | 5.1 × 10−4 | 1.1 × 10−4 |
| A | A | — | G | — | 0.488 | 79 | 75 | 103 | 5 × 10−6 | 8.7 × 10−5 |
| A | A | — | v | A | 0.488 | 77 | 74 | 101 | 9 × 10−6 | 1.6 × 10−4 |
| — | A | — | G | A | 0.488 | 77 | 74 | 101 | 9 × 10−6 | 1.6 × 10−4 |
| — | A | T | G | — | 0.427 | 77 | 67 | 88 | 5.1 × 10−4 | 1.1 × 10−4 |
| A | A | — | G | A | 0.488 | 77 | 74 | 101 | 9 × 10−6 | 1.6 × 10−4 |
| Haplotypes consisting of 4 SNPs | ||||||||||
| A | A | T | G | A | 0.427 | 77 | 72 | 93 | 5.1 × 10−4 | 2.6 × 10−4 |
| A | G | C | A | C | 0.217 | 65 | 62 | 53 | 0.037 | |
| C | G | C | G | C | 0.212 | 65 | 62 | 51 | 0.022 | |
| A | G | C | G | A | 0.084 | 40 | 28 | 23 | 0.116 | |
| A | A | C | G | A | 0.060 | 31 | 10 | 16 | 0.048 | |
Number of informative case-parent trios.
Observed and expected frequencies of alleles defining haplotype transmitted.
The P value for a statistical model with whole haplotypes consisted of two, three, or four SNPs.
LD, linkage disequilibrium; CL/P, cleft lip with or without cleft palate; freq, frequency; Obs, observed; Exp, expected.
Genotypic and diplotypic odds ratios
As shown in Table 3, G/G and C/C homozygotes at rs2235373 and rs2013162 had a significantly increased risk of being CL/P cases [GOR = 4.94 and 3.78, 95% confidence interval (CI) = 2.43–10.04 and 1.93–7.41, respectively], whereas C/C, C/C, and A/A homozygotes at rs599021, rs2235371, and rs595918 were more likely to be CL/P cases compared with reference homozygotes among Taiwanese trios (GOR = 2.92, 2.75, and 3.20, respectively, and 95% CI: 1.15–7.42, 1.37–5.52, and 1.15–8.90, respectively), although the global P value for the conditional logistic model of rs595918 was not significant. The C/C genotype at rs2013162 increased the risk of being a CL/P case among Singaporean trios (GOR = 6.88, 95% CI: 1.17–40.34).
Table 3.
GORs for heterozygotes and homozygotes for individual SNPs showing significant evidence of linkage and LD in four CL/P groups
| Population | Maryland | Taiwan | Singapore | Korea | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Genotype | Na | GOR | 95% CI | N | GOR | 95% CI | N | GOR | 95% CI | N | GOR | 95% CI |
| rs599021 | na = 58 (P = 0.806)b | n = 141 (P = 0.045) | n = 23 (P = 0.033) | n = 38 (P = 0.377) | |||||||||
| A/A | 63 | 1.00 | Reference | 258 | 1.00 | Reference | 47 | 1.00 | Reference | 67 | 1.00 | Reference | |
| C/A | 100 | 0.95 | 0.44–2.05 | 150 | 1.65 | 0.98–2.78 | 32 | 0.86 | 0.17–4.36 | 43 | 1.95 | 0.74–5.13 | |
| C/C | 35 | 0.72 | 0.24–2.16 | 22 | 2.92 | 1.15–7.42 | 13 | 10.16 | 0.87–118.38 | 4 | 1.68 | 0.27–10.68 | |
| rs2235373 | n = 63 (P = 0.750) | n = 139 (P < 1 × 10−4) | n = 23 (P = 0.018) | n = 38 (P = 0.254) | |||||||||
| A/A | 7 | 1.00 | Reference | 94 | 1.00 | Reference | 13 | 1.00 | Reference | 16 | 1.00 | Reference | |
| G/A | 52 | 2.16 | 0.24–19.33 | 199 | 1.48 | 0.82–2.67 | 42 | 0.54 | 0.10–2.96 | 59 | 2.16 | 0.59–7.90 | |
| G/G | 142 | 1.99 | 0.20–19.41 | 136 | 4.94 | 2.43–10.04 | 37 | 3.58 | 0.61–21.24 | 38 | 3.15 | 0.75–13.34 | |
| rs2235371 | n = 48 (P = N.A.) | n = 137 (P < 1 × 10−4) | n = 22 (P = 0.170) | n = 31 (P = 0.604) | |||||||||
| T/T | 0 | — | — | 77 | 1.00 | Reference | 8 | 1.00 | Reference | 14 | 1.00 | Reference | |
| C/T | 1 | — | — | 182 | 0.81 | 0.43–1.54 | 37 | 1.81 | 0.18–18.65 | 49 | 1.53 | 0.38–6.24 | |
| C/C | 147 | — | — | 163 | 2.75 | 1.37–5.52 | 45 | 5.79 | 0.45–74.31 | 44 | 2.27 | 0.43–12.12 | |
| rs595918 | n = 59 (P = 0.119) | n = 139 (P = 0.065) | n = 23 (P = 0.281) | n = 37 (P = 1.000) | |||||||||
| G/G | 127 | 1.00 | Reference | 256 | 1.00 | Reference | 57 | 1.00 | Reference | 64 | 1.00 | Reference | |
| A/G | 62 | 2.53 | 0.99–6.48 | 147 | 1.47 | 0.89–2.44 | 32 | 1.33 | 0.42–4.20 | 41 | 1.00 | 0.37–2.71 | |
| A/A | 9 | 2.81 | 0.37–21.12 | 26 | 3.20 | 1.15–8.90 | 3 | 7.31 | 0.58–92.88 | 8 | 1.00 | 0.21–4.83 | |
| rs2013162 | n = 54 (P = 0.900) | n = 137 (P = 1 × 10−4) | n = 23 (P = 0.015) | n = 38 (P = 0.617) | |||||||||
| A/A | 20 | 1.00 | Reference | 127 | 1.00 | Reference | 20 | 1.00 | Reference | 32 | 1.00 | Reference | |
| C/A | 96 | 1.02 | 0.37–2.79 | 210 | 1.28 | 0.78–2.10 | 47 | 1.16 | 0.27–5.08 | 61 | 1.44 | 0.53–3.89 | |
| C/C | 83 | 1.21 | 0.39–3.72 | 92 | 3.78 | 1.93–7.41 | 25 | 6.88 | 1.17–40.34 | 24 | 1.89 | 0.52–6.91 | |
N and n refer to the number of subjects carrying the genotype (including both cases and pseudosibs) and the number of case/pseudocontrol sets generated, respectively.
P values of χ2 tests for the conditional logistic regression model for each SNP.
GORs, genotypic odds ratios; SNPs, single nucleotide polymorphisms; LD, linkage disequilibrium; CL/P, cleft lip with or without cleft palate; CI, confidence interval.
To determine diplotype specific risks in the Taiwanese trios, two-, three- and four-marker models were tested as shown in Table 4. Diplotypes with frequency <4% were analyzed as a single group. The AG/CG diplotype for rs599021 and rs2235373 showed the most increased risk of being a CL/P case among all two-SNP diplotypes composed from the four SNPs identified in Table 3 (i.e., 5.95 times higher than AA/AA, the reference diplotype group; 95% CI: 2.53–13.99). Interestingly, the GOR for diplotypes that included A/C heterozygotes at rs599021 and homozygotes for the high-risk allele at other loci showed the greatest risk. For instance, the AGC/CGC diplotype showed a higher risk of being a CL/P (GOR = 6.99, 95% CI: 2.70–18.06) than did the CGC/CGC diplotype (GOR = 3.70, 95% CI: 1.31–10.46). Using data from the COGENE consortium, the IRF6 gene was found to be expressed in the 4-week frontonasal prominence among seven SAGE libraries.
Table 4.
GORs for heterozygotes and homozygotes for two-, three-, and four-SNP haplotypes consisting of SNPs increasing risk in 146 Taiwanese CL/P case-parent trios
| Diplotype | Na (%) | GOR (95% CI) | Diplotype | Na (%) | GOR (95% CI) |
|---|---|---|---|---|---|
| rs599021-rs2235373 (P = 1 × 10−4)b n = 556 | rs2235373-rs2235371 (P < 10−4) n = 548 | ||||
| AA/AA | 140 (25.2) | Reference | AT/AT | 100 (18.3) | Reference |
| AA/AG | 153 (27.5) | 1.35 (0.69–2.66) | AT/AC | 31 (5.7) | 0.18 (0.04–0.90) |
| AA/CG | 103 (18.5) | 1.62 (0.75–3.50) | AT/GC | 221 (40.3) | 0.85 (0.44–1.66) |
| AG/AG | 61 (11.0) | 5.10 (2.08–12.48) | AC/GC | 31 (5.7) | 1.47 (0.52–4.16) |
| AG/CG | 68 (12.2) | 5.95 (2.53–13.99) | GC/ GC | 152 (27.7) | 3.51 (1.66–7.42) |
| CG/CG | 25 (4.5) | 3.96 (1.40–11.24) | 4 types (<4%) | 13 (2.4) | 0.61 (0.05–7.01) |
| 3 types (<4%) | 6 (3.3) | 1.91 (0.15–24.12) | |||
| rs599021-rs2235371 (P = 2 × 10−4) n = 548 | rs2235373-rs2013162 (P = 1 × 10−4) n = 548 | ||||
| AT/AT | 101 (18.4) | Reference | AA/AA | 137 (25.0) | Reference |
| AT/AC | 164 (29.9) | 0.70 (034–1.43) | AA/GA | 39 (7.1) | 1.64 (0.60–4.52) |
| AT/CC | 91 (16.6) | 0.97 (0.43–2.19) | AA/GC | 212 (38.7) | 1.34 (0.73–2.47) |
| AC/AC | 81 (14.8) | 2.29 (1.01–5.20) | GA/GC | 41 (7.5) | 3.94 (1.48–10.46) |
| AC/CC | 82 (15.0) | 3.27 (1.46–7.31) | GC/ GC | 103 (18.8) | 5.01 (2.35–10.66) |
| CC/CC | 25 (4.6) | 3.05 (1.08–8.63) | 4 types (<4%) | 16 (2.9) | 1.26 (0.18–8.61) |
| 3 types (<4%) | 4 (0.7) | 2.04 (0.11–36.83) | |||
| rs599021-rs2013162 (P = 3.7 × 10−3) n = 548 | rs2235371-rs2013162 (P < 10−4) n = 544 | ||||
| AA/AA | 182 (33.2) | Reference | TA/TA | 98 (18.0) | Reference |
| AA/AC | 135 (24.6) | 1.14 (0.63–2.08) | TA/CA | 68 (12.5) | 0.57 (0.23–1.38) |
| AA/CC | 121 (22.1) | 1.44 (0.74–2.79) | TA/CC | 182 (33.5) | 0.78 (0.40–1.55) |
| AC/AC | 29 (5.3) | 3.71 (1.27–10.85) | CA/CC | 68 (12.5) | 2.17 (0.95–5.00) |
| AC/CC | 50 (9.1) | 4.84 (2.05–11.44) | CC/CC | 102 (18.8) | 3.62 (1.64–7.97) |
| CC/CC | 25 (4.6) | 3.16 (1.18–8.47) | 4 types (<4%) | 26 (4.8) | 0.37 (0.08–1.79) |
| 3 types (<4%) | 6 (1.1) | 1.45 (0.11–18.71) | |||
| rs599021-rs2235373-rs2235371 (P < 10−4) n = 548 | rs599021-rs2235371-rs2013162 (P = 1 × 10−4) n = 544 | ||||
| AAT/AAT | 99 (18.1) | Reference | ATA/ATA | 97 (17.8) | Reference |
| AAT/AAC | 31 (5.7) | 0.19 (0.04–0.93) | ATA/ACA | 68 (12.5) | 0.57 (0.23–1.40) |
| AAT/AGC | 128 (23.4) | 0.79 (0.36–1.72) | ATA/ACC | 89 (16.4) | 0.61 (0.26–1.42) |
| AAT/CGC | 90 (16.4) | 0.88 (0.38–2.03) | ATA/CCC | 90 (16.5) | 0.95 (0.41–2.19) |
| AGC/AGC | 59 (10.8) | 3.98 (1.56–10.14) | ACA/ACC | 37 (6.8) | 2.13 (0.79–5.72) |
| AGC/CGC | 68 (12.4) | 4.14 (1.72–9.96) | ACA/CCC | 31 (5.7) | 2.34 (0.84–6.53) |
| CGC/CGC | 25 (4.6) | 2.56 (0.90–7.29) | ACC/ACC | 27 (5.0) | 4.12 (1.26–13.51) |
| 11 types (<4%) | 48 (8.8) | 1.48 (0.54–4.07) | ACC/CCC | 50 (9.2) | 4.59 (1.76–11.97) |
| CCC/CCC | 25 (4.6) | 2.80 (0.97–8.05) | |||
| 8 types (<4%) | 30 (5.5) | 0.53 (0.12–2.30) | |||
| rs599021-rs2235373-rs2013162 (P = 5 × 10−4) n = 548 | rs2235373-rs2235371-rs2013162 (P < 10−4) n = 548 | ||||
| AAA/AAA | 135 (24.6) | Reference | ATA/ATA | 98 (18.0) | Reference |
| AAA/AGA | 39 (7.1) | 1.72 (0.63–4.70) | ATA/ACA | 31 (5.7) | 0.17 (0.04–0.84) |
| AAA/AGC | 105 (19.2) | 1.15 (0.55–2.39) | ATA/GCA | 36 (6.6) | 1.15 (0.41–3.25) |
| AAA/CGC | 103 (18.8) | 1.57 (0.73–3.41) | ATA/GCC | 181 (33.3) | 0.79 (0.39–1.56) |
| AGA/AGC | 23 (4.2) | 5.18 (1.61–16.67) | ACA/GCC | 26 (4.8) | 2.03 (0.68–6.02) |
| AGC/AGC | 28 (5.1) | 5.76 (1.83–18.16) | GCA/GCC | 41 (7.5) | 2.65 (0.98–7.19) |
| AGC/CGC | 50 (9.1) | 6.99 (2.70–18.06) | GCC/GCC | 102 (18.8) | 3.71 (1.67–8.23) |
| CGC/CGC | 25 (4.6) | 3.70 (1.31–10.46) | 12 types (<4%) | 29 (5.3) | 0.40 (0.07–2.15) |
| 8 types (<4%) | 40 (7.3) | 2.57 (0.83–7.97) | |||
| rs599021-rs2235373-rs2235371-rs2013162 (P < 10−4) n = 544 | |||||
| AATA/AATA | 97 (17.8) | Reference | AGCA/AGCC | 23 (4.2) | 3.55 (1.09–11.59) |
| AATA/AACA | 31 (5.7) | 0.19 (0.04–0.93) | AGCC/AGCC | 27 (5.0) | 4.58 (1.39–15.09) |
| AATA/AGCA | 36 (6.6) | 1.31 (0.46–3.72) | AGCC/CGCC | 50 (9.2) | 4.90 (1.85–12.97) |
| AATA/AGCC | 88 (16.2) | 0.63 (0.26–1.50) | CGCC/CGCC | 25 (4.6) | 2.53 (0.89–7.20) |
| AATA/CGCC | 90 (16.5) | 0.91 (0.39–2.10) | 20 types (<4%) | 77 (14.2) | 1.68 (0.07–2.15) |
N and n refer to the number of subjects carrying the genotype (including both cases and pseudosibs) and the number of case/pseudocontrol sets.
Global P value for the conditional logistic model.
GORs, genotypic odds ratios; SNP, single nucleotide polymorphism; CL/P, cleft lip with or without cleft palate.
DISCUSSION
Zucchero et al.5 showed strong evidence of overtransmission of the valine (V) allele at p.V274I (rs2235371) in IRF6; however, the estimated attributable risk (11.6%) and the estimated 3-fold increased recurrence risk among Filipino CL/P case-parent trios must be interpreted carefully because it was assumed that carrying this allele was directly causal and uncorrelated with other risk factors.6 This SNP is not highly polymorphic in Europeans, although Asians have allele frequencies around 66% for the G allele (p.274V).9,19 Thus, subsequent studies in European-derived populations focused on four SNPs (rs1319435, rs2013162, rs2235375, and rs2235543) with higher heterozygosity levels. Scapoli et al.7 detected overtransmission of the G and C alleles for markers rs2013162 and rs2235375 (P = 0.004 and P = 0.002, respectively) and all haplotypes carrying these common alleles among 219 Italian CL/P trios (the GTGA haplotype showed significant undertransmission, P = 0.0003). Blanton et al.8 detected overtransmission of the C allele at rs2013162 (P = 0.05), and all haplotypes including the A allele at this marker were significantly undertransmitted to cases (the lowest P = 0.002 for CAXT haplotype among 216 European-American families with CL/P). Ghassibé et al.9 observed transmission distortion of the GG and TG haplotypes (P= 0.004 and P= 0.036, respectively) for two markers (rs2013162 and rs2235543) and confirmed overtransmission of the G allele at rs2013162 (P = 0.01) in 195 Belgium families (this sample included some immigrant families from other populations). Alleles were designated (T/G) for rs2013162 based on the 3′ to 5′ orientation of this gene in some studies, whereas here these alleles are designated A/C based on their 5′ to 3′ orientation on the chromosome. Reported significance levels from these published studies did not include corrections for multiple comparisons.
Initially, we analyzed 103 European-American trios, 171 and 66 Han Chinese trios from Taiwan and Singapore, respectively, and 42 Korean trios with either CL/P or isolated cleft plate (results not shown). When stratified by type of cleft, we found greater statistical significance among CL/P groups, despite the smaller numbers, confirming the possibility of etiologic heterogeneity (e.g., P = 3.6 × 10−5 vs. P = 2 × 10−6 at rs2235373 alone among all 171 Taiwanese trios vs. the 146 CL/P trios).
Two SNPs (rs2073485 and rs2235373), located next to, but not in LD with, the V274I variant, yielded statistical significance for individual SNPs and haplotypes among Taiwanese trios, even after correcting for multiple comparisons. In particular, the G allele at rs2235373 significantly increased the risk of being a CL/P case, whereas the A allele was underrepresented among Taiwanese cases. Overtransmission of the C allele at rs2013162 (located in the fifth exon of IRF6) was not confirmed here among European-American trios (from Maryland), in contrast to three previously reported studies of European-derived populations. In our Han Chinese populations (Taiwan and Singapore), the C allele at this synonymous SNP significantly increased the risk of being a CL/P case, whereas haplotypes including the A allele at the same marker were consistently undertransmitted to CL/P cases (Table 2).
This is the first study that has considered genotypic and diplotypic risks for specific SNP markers in IRF6 particularly in Taiwanese, Singaporean, and Korean samples. However, the number of Singaporean and Korean trios available may not be sufficient to detect SNPs with weak or moderate effects on risk. Although the C allele at rs599021 was overtransmitted to cases and C/C homozygotes showed an increased risk of being a CL/P case, C/A heterozygotes seemed to be at higher risk when diplotypes were considered with other SNPs (Table 4). The fact that mutations in IRF6 cause VWS, which usually includes CL/P, combined with significant evidence of linkage and association with CL/P in our study and other case-parent trio studies strongly suggests that IRF6 itself is a causal gene for CL/P, but not the only one.
IRF6 is one of nine members of a family of transcription factors (IRFs) that share a highly conserved helix-turn helix DNA-binding domain and a less conserved protein-binding domain. These domains exert diverse functions including regulating host defense pathways.20–22 The Irf6 gene was expressed in the ectoderm fusion forming the upper lip and primary palate in both mouse and chick, but only in the developing secondary palate of the mouse (which fuses as in humans).23 Similar expression patterns for IRF6 are also seen in human craniofacial structures, although the biological function of IRF6 during development of the lip and palate in humans remains uncertain.
Significant results observed from SNPs other than p.V274I (rs2235371) suggested that V274I itself is not causal, but rather in LD with some causal mutation in IRF6. Patterns of LD between SNPs and the SNPs that individually showed statistical significance differed across our sample populations (see Appendix Table A2). However, our data confirm that the IRF6 gene is associated with increased risk of CL/P, and thus the regions showing statistical evidence of association (e.g., rs599021, rs2235373, rs2235371, and rs2013162 for two Chinese groups; rs2294408 for Korean trios) should be searched further for causal variants. High-risk genotypes and diplotypes identified here may provide a better understanding of the etiological role that IRF6 plays in CL/P and could prove useful in genetic counseling, if these findings can be confirmed in subsequent studies.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by Grants P60-DE13078, R01-DE014581, and NO1-DE92630 from the National Institute of Dental and Craniofacial Research and Grant 2003-041-E00109 from the Korean Research Foundation. The authors acknowledge Smile Train, Inc. for support of research on clefts in China. The authors thank all participants who donated samples for the international study of oral clefts and all staff members who were involved in this research.
Footnotes
The authors declare no conflict of interest.
Supplementary Appendices are available via the ArticlePlus feature at www.geneticsinmedicine.org. Please go to the April issue and click on the ArticlePlus link posted with the article in the Table of Contents to view this material.
References
- 1.Lidral AC, Murray JC. Genetic approaches to identify disease genes for birth defects with cleft lip/palate as a model. Birth Defects Res A Clin Mol Teratol. 2004;70:893–901. doi: 10.1002/bdra.20096. [DOI] [PubMed] [Google Scholar]
- 2.Kondo S, Schutte BC, Richardson RJ, Bjork BC, et al. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat Genet. 2002;32:285–289. doi: 10.1038/ng985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Field LL, Ray AK, Cooper ME, Goldstein T, et al. Genome scan for loci involved in nonsyndromic cleft lip with or without cleft palate in families from West Bengal, India. Am J Med Genet A. 2004;130:265–271. doi: 10.1002/ajmg.a.30252. [DOI] [PubMed] [Google Scholar]
- 4.Houdayer C, Bonaiti-Pellie C, Erguy C, Soupre V, et al. Possible relationship between the van der Woude syndrome (vWS) locus and nonsyndromic cleft lip with or without cleft palate (NSCL/P) Am J Med Genet. 2001;104:86–92. doi: 10.1002/1096-8628(20011115)104:1<86::aid-ajmg10053>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 5.Zucchero TM, Cooper ME, Maher BS, Daack-Hirsch S, et al. Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N Engl J Med. 2004;351:769–780. doi: 10.1056/NEJMoa032909. [DOI] [PubMed] [Google Scholar]
- 6.Srichomthong C, Siriwan P, Shotelersuk V. Significant association between IRF6 820G->A and non-syndromic cleft lip with or without cleft palate in the Thai population. J Med Genet. 2005;42:e46. doi: 10.1136/jmg.2005.032235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scapoli L, Palmieri A, Martinelli M, Pezzetti F, et al. Strong evidence of linkage disequilibrium between polymorphisms at the IRF6 locus and nonsyndromic cleft lip with or without cleft palate, in an Italian population. Am J Hum Genet. 2005;76:180–183. doi: 10.1086/427344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanton SH, Cortez A, Stal S, Mulliken JB, et al. Variation in IRF6 contributes to nonsyndromic cleft lip and palate. Am J Med Genet A. 2005;137:259–262. doi: 10.1002/ajmg.a.30887. [DOI] [PubMed] [Google Scholar]
- 9.Ghassibé M, Bayet B, Revencu N, Verellen-Dumoulin C, et al. Interferon regulatory factor-6: a gene predisposing to isolated cleft lip with or without cleft palate in the Belgian population. Eur J Hum Genet. 2005;13:1239–1242. doi: 10.1038/sj.ejhg.5201486. [DOI] [PubMed] [Google Scholar]
- 10.Beaty TH, Maestri NE, Hetmanski JB, Wyszynski DF, et al. Testing for interaction between maternal smoking and TGFA genotype among oral cleft cases born in Maryland 1992–1996. Cleft Palate Craniofac J. 1997;34:447–454. doi: 10.1597/1545-1569_1997_034_0447_tfibms_2.3.co_2. [DOI] [PubMed] [Google Scholar]
- 11.Bellus GA, Hefferon TW, Luna RIO, Hecht JT, et al. Achondroplasia is defined by recurrent G380R mutations of FGFR3. Am J Hum Genet. 1995;56:368–373. [PMC free article] [PubMed] [Google Scholar]
- 12.Oliphant A, Barker DL, Stuelpnagel JR, Chee MS. BeadArray™ technology: enabling accurate, cost- effective approach to high-throughput genotyping. BioTechniques. 2002;32:56–61. [PubMed] [Google Scholar]
- 13.Ardlie KG, Kruglyak L, Seielstad M. Patterns of linkage disequilibrium in the human genome. Nat Rev Genet. 2002;3:299–309. doi: 10.1038/nrg777. [DOI] [PubMed] [Google Scholar]
- 14.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 15.Rabinowitz D, Laird NM. A unified approach to adjusting association tests for population admixture with arbitrary pedigree structure and arbitrary missing marker information. Hum Hered. 2000;50:211–223. doi: 10.1159/000022918. [DOI] [PubMed] [Google Scholar]
- 16.Horvath S, Xu X, Laird NM. The family based association test method: strategies for studying general genotype-phenotype associations. Eur J Hum Genet. 2001;9:301–306. doi: 10.1038/sj.ejhg.5200625. [DOI] [PubMed] [Google Scholar]
- 17.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai J, Ash D, Kotch LE, Jabs EW, et al. Gene expression in pharyngeal arch 1 during human embryonic development. Hum Mol Genet. 2005;14:903–912. doi: 10.1093/hmg/ddi083. [DOI] [PubMed] [Google Scholar]
- 19.Hering R, Grundmann K. The IRF6 p. 274V polymorphism is not a risk factor for isolated cleft lip. Genet Med. 2005;7:209. doi: 10.1097/01.gim.0000157128.95526.79. [Letter]. [DOI] [PubMed] [Google Scholar]
- 20.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 21.Kim Y, Park J, Lee T, Yoo H. Identification of two novel mutations of IRF6 in Korean families affected with Van der Woude syndrome. Int J Mol Med. 2003;12:465–468. [PubMed] [Google Scholar]
- 22.Ben J, Jabs EW, Chong SS. Genomic, cDNA and embryonic expression analysis of zebrafish IRF6, the gene mutated in the human oral clefting disorders Van der Woude and popliteal pterygium syndromes. Gene Expr Patterns. 2005;5:629–638. doi: 10.1016/j.modgep.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 23.Knight AS, Schutte BC, Jiang R, Dixon MJ. Developmental expression analysis of the mouse and chick orthologues of IRF6: the gene mutated in Van der Woude syndrome. Dev Dyn. 2006;235:1441–1447. doi: 10.1002/dvdy.20598. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.