Abstract
Several studies have suggested that disrupting interactions of the G protein βγ subunits with downstream binding partners might be a valuable study for pharmaceutical development. Recently, small molecules have been found which bind to Gβγ with high apparent affinity in an enzyme-linked immunosorbent assay (ELISA), have demonstrated selective inhibition of interactions of Gβγ with downstream signaling partners, and have been shown to increase antinociceptive effects of morphine and inhibit inflammation in vivo. In this paper we examine several docking and scoring protocols for estimating binding affinities for a set of 830 ligands from the NCI diversity set to the Gβ1γ2 subunit and compared these with IC50s measured in a competition ELISA with a high-affinity peptidic ligand. The best-performing docking protocol used a consensus score and ensemble docking and resulted in a 6-fold enrichment of high-affinity compounds in the top-ranked 5% of the ligand data set.
INTRODUCTION
G protein-coupled receptors (GPCRs) trigger cellular signaling cascades through interactions with heterotrimeric guanine nucleotide binding proteins (G proteins) and are involved in many diverse physiological processes such as sensory perception, modulation of cardiac rhythm, neurotransmission, attraction of motile cells by chemotaxis, and regulation of mitosis.1,2 GPCRs are common targets for pharmaceutical intervention; around half of all drugs in clinical use are GPCR agonists or antagonists.3 In the standard model for signaling, the receptor acts as a catalyst for the exchange of GDP for GTP on the Gα subunit, which leads to dissociation from the Gβγ subunits. Gα and Gβγ are then free to interact with downstream signaling partners. Five distinct β subunits and twelve distinct γ subunits have been identified in the human and mouse genomes; these subunits can pair to form many Gβγ different combinations. Evidence suggests that the different Gβγ isoforms interact with different GPCRs, although this selectivity is not completely understood.4
Several studies have suggested that disruption of interactions of Gβγ with downstream binding partners might be a valuable strategy for pharmaceutical development.4–21 Recently, small molecules have been found which bind to Gβ1γ2, as demonstrated by the ability to compete with a high-affinity peptide ligand (SIGK) in an enzyme-linked immunosorbent assay (ELISA). An X-ray crystal structure of SIGK bound to Gβ1γ2 has been determined and shows that SIGK binds to the same region of the β subunit as the switch II region of Gα, in the center of the β-propeller.22 This region has been postulated to be a hotspot or a small area on a protein–protein interaction surface particularly important for mediating binding. The small molecules were discovered with the help of a small virtual screen of ~2000 compounds from the National Cancer Institute (NCI) diversity library, using the FlexX software package.23 One compound, M119 (NSC119910), was subsequently shown to inhibit Gβγ-dependent activation of phospholipase C and phosphoinositide 3-kinase γ (PI3K) and to increase the antinociceptive effects of morphine in mice.21 A very similar compound, gallein (which is commercially available at high purity), was found by surface plasmon resonance to bind Gβ1γ2 with Kd ~ 400 nM and was shown to inhibit PI3K activation and chemoattractant-dependent neutrophil migration and lessen carrageenan-induced inflammation in mice.20 In general, disruption of protein–protein interactions has the potential to be an important strategy for developing therapeutics. Although contact surfaces are often large in size and relatively flat,24–26 studies involving point mutations show that only a small subset of residues contribute most of the free energy of binding; these “hotspots” are often located at the center of the interface.27–33 Recent reviews of the principles of protein–protein interactions and their significance for drug design have been published.34,35
Virtual screening by docking compounds from chemical libraries has become widely used in drug discovery, for finding new lead compounds when a high-resolution structure of the target is available.24–26,36–40 Many docking programs have been developed over the past few decades, and good success in screening applications has been reported in some cases. However, accurate prediction of binding affinities remains challenging.41–46 All recently available programs provide false negatives (active compounds that are not given a high-ranking score by docking programs) and false positives (inactive compounds that are given a high-ranking score). Individual programs often show better ability to predict high-affinity compounds for particular receptors or compound classes. It is difficult to say in advance whether a particular program or scoring function will be successful for a certain case without any supporting experimental knowledge about the target.
In this paper, we examined several docking and scoring protocols for estimating binding affinities for a subset of 830 ligands from the NCI diversity set to the Gβ1γ2 subunit and compared these with IC50s measured in competition ELISA with the high-affinity SIGK peptidic ligand. The DOCK6 and GLIDE software packages were used, and the influences of the assignment of protonation states and partial charges of ligands and receptors were evaluated. Enrichment factors from both software programs (the number of high-affinity ligands found as a fraction of the amount of the database screened) were compared depending on variations. The influences of the assignment of protonation states and partial charges of ligands and protein receptor were evaluated. Solvation effects were introduced using the Poisson–Boltzmann continuum method. The effects of structural flexibility were explored in detail by molecular dynamics (MD) simulation. The aim of this study is to obtain the best-optimized docking method to apply to a larger database.
METHODS
Measurement of Binding Affinities
830 compounds taken from the NCI diversity set (comprising a total of 1990 compounds) were tested for competition with the SIGK peptide in ELISA as described in refs 16 and 21. Compounds with less than 300 μM of median inhibitory concentration (IC50) value were assumed to be high-affinity compounds; a total of 36 such high-affinity compounds were found in this assay (see Table 1).
Table 1.
Median Inhibitory Concentration (IC50) Values of 36 Binding Inhibitors of SIGK with Maximum Value of Less than 300 μM and the Molecular Weight for Each Inhibitor
| compound | MW | IC50 (μM) | compound | MW | IC50 (μM) |
|---|---|---|---|---|---|
| 119910 | 370 | 0.2 | 45382 | 440 | 35.4 |
| 119911 | 314 | 0.2 | 34238 | 402 | 42 |
| 119913 | 394 | 0.7 | 150289 | 464 | 42.5 |
| 51535 | 547 | 2.4 | 88915 | 550 | 47.7 |
| 119889 | 650 | 4.3 | 19630 | 183 | 53.6 |
| 14163 | 414 | 5.9 | 117079 | 473 | 56 |
| 2805 | 246 | 7.1 | 83217 | 407 | 56.9 |
| 118176 | 471 | 8.1 | 17770 | 543 | 59.1 |
| 39207 | 787 | 10.7 | 45126 | 424 | 69.7 |
| 22225 | 387 | 11.4 | 668394 | 452 | 77.7 |
| 12155 | 445 | 24 | 90831 | 560 | 92.9 |
| 9608 | 378 | 16.5 | 117199 | 362 | 161.6 |
| 69343 | 546 | 16.8 | 170423 | 497 | 166.6 |
| 125910 | 566 | 18 | 95676 | 536 | 182.4 |
| 128437 | 477 | 24 | 59275 | 523 | 197.1 |
| 83224 | 136 | 24.9 | 121771 | 260 | 198.1 |
| 14555 | 281 | 26 | 3284 | 512 | 261.9 |
| 23128 | 899 | 27 | 24048 | 442 | 292.1 |
Preparation of Receptors
The crystal structure of Gβγ complexed with the SIGK peptidic ligand (PDB ID 1XHM) was separated into protein and ligand PDB files. The protein was processed to add hydrogen atoms and remove water molecules and minimized to alleviate bad steric contacts with the Protein Preparation Wizard utility in Maestro (Schrödinger, Inc.). Two different sets of partial charges were employed for the receptor: AMBER charges,47 assigned using the CHIMERA program,48 and OPLS/AA charges,49 which were automatically assigned by Glide version 4.5 (Schrödinger, Inc.) in the receptor grid calculation. The protein–ligand file was subsequently used to select spheres in the hotspot of the receptor in the process of receptor preparation by the DOCK6 program.50
Preparation of Ligands
Coordinates for molecules in the NCI diversity set were downloaded from NCI. Metal ions were removed and hydrogens were added using SYBYL version 6.8 (Tripos, Inc.). Tautomers were generated using ionization states estimated for pH 7 with the LigPrep program (Schrödinger, Inc.). Four different sets of partial charges (CM1,51 CM2,52 AM1-BCC,53 and Mulliken54) were assigned to each ligand using the Antechamber program in the AMBER software package. OPLS ligand charges were also obtained using Glide by the default setting.
Docking
Using the DMS program in the DOCK6 software package, the solvent-accessible molecular surface of the protein binding site was calculated using a probe radius of 1.4 Å. Receptor spheres were generated using the program SPHGEN. Spheres covering the hotspot were selected within 10 Å from the positions of the heavy atoms of the SIGK peptide ligand. The grid box enclosing the selected spheres was generated with an extra 5 Å added in each dimension. Ligand flexibility was employed during the docking process. When performing docking with Glide, the inner box, which defines the range of motion for the center of each ligand, was set to 10 Å on each side. The outer (enclosing) box, which includes the entire hole at the center of Gβγ, was set to 11 Å longer than the inner box on each side. Docking calculations were performed using both standard-precision (SP) and extraprecision (XP) modes.
Introducing Solvation Effects
The Poisson–Boltzmann continuum method implemented in MEAD55 was used to introduce solvation effects. The best poses of ligand molecules from DOCK6 were used for solvation free energy calculations. A grid spacing of 1.0 Å was applied, and a focused grid of 0.25 Å was utilized. The differences of solvation free energies of ligands in protein and in water were calculated.
Docking Using a Conformational Ensemble
A relatively simple way to take in to account the effects of receptor flexibility is to dock to an ensemble, rather than a single structure.56–58 To create an ensemble of receptor conformations for docking, molecular dynamics (MD) simulations of the unbound receptor were performed with the SANDER module of the AMBER9.0 software package. The parm99 force field parameters were used, and solvent effects were incorporated using the pairwise generalized Born model of Case and co-workers.59–61 Prior to a production run, the receptor was first relaxed by steepest descent energy minimization, followed by conjugate gradient energy minimization. The system was then heated over 25 ps from 0 to 300 K. An MD simulation of 1 ns was performed to produce a conformational ensemble of the receptor at 300 K. A nonbonded cutoff distance of 12 Å and a time step of 2 fs was used. Constant temperature was maintained using the Langevin thermostat, and all bonds involving hydrogen atoms were constrained using the SHAKE algorithm. Coordinates were saved every 1000 steps, for a total of 500 snapshots. The snapshots were classified into ten groups by the fuzzy c-means (FCM) clustering algorithm using the GA Fuzzy Clustering program.62 From each cluster, the structure closest to the center was chosen for docking. A docking score was obtained for each of the ten conformations. These scores were then averaged to rank compounds.
RESULTS
Influence of Protonation State of Ligands
Enrichment factors (i.e., the number of high-affinity compounds found as a function of the amount of the ranked database examined) were calculated using both unrefined original forms of NCI database ligands and ligands with protonation states corrected for pH 7, using LigPrep. AM1-BCC charges were used for the ligand, with AMBER charges for the receptor. When the structures in the unrefined original ligand database were inspected, most of the titratable functional groups such as carboxyls were found to be protonated. The charges on the acid functional groups were also significantly different from those on the refined ligands. However, most of the basic functional groups such as amines were deprotonated in both the refined and unrefined ligand structures. Docking was performed with DOCK6 using the Grid scoring function as described above. Enrichment factors are shown in Figure 1. With unrefined, original protonation states, DOCK6 could not effectively identify active ligand compounds, since the enrichment curve was only slightly better than random selection. After the ligand protonation state was corrected, the percent known active ligands found in the top tenth of the ranked database was increased from 13% to 33%, suggesting that the protonation state of ligands has a significant effect on molecular recognition by the Gβγ active site.
Figure 1.
Docking enrichment plot with unrefined ligand protonation state and refined ligand protonation state at pH 7.0, using the Grid scoring function of DOCK6. AM1-BCC and AMBER charges were chosen for ligands and receptor, respectively. The percent of the ranked database screened (x-axis) is plotted against the percent of known ligands found (y-axis). The black line represents the values expected if all known ligands were ranked in the top 10% of the list. The red line shows results expected if ligands were ranked completely randomly.
Influence of Partial Charges
To investigate the role of partial charges in docking and scoring accuracy, charges for Gβγ were assigned using both AMBER99 and OPLS/AA force fields; four different sets of charges (CM1, CM2, AM1-BCC, and Mulliken) were assigned to the ligands. The DOCK6 program was used to generate poses and obtain scores. Results are shown in Figure 2 and suggest that overall, more accurate results are obtained using AMBER receptor charges. Using AM1-BCC, 28% of the known high-affinity ligands were found in the top 10% of the database using OPLS receptor charges, while 33% of known ligands were found using AMBER receptor charges. In general, AM1-BCC charges resulted in the greatest enrichment, followed by Mulliken, CM2, and CM1. This trend held for both sets of receptor charges. The influence of the receptor charges was less significant than that from the ligand charges.
Figure 2.
Docking enrichment plots for comparing different receptor and ligand partial charges using DOCK6. Receptor charges and ligand charges used for docking are a) AMBER/(BCC, CM2, CM1, Mulliken, and OPLS) and b) OPLS/(BCC, CM2, CM1, and Mulliken).
Comparing Docking Methods
Success in enrichment and in accurately reproducing correct binding poses has been reported for the Glide docking program.63–65 We computed enrichment factors with Glide; results are shown in Figure 3. The best enrichment (33% of high-affinity ligands found in the top 10% of the database) was slightly higher with DOCK6 than with Glide (21% of high-affinity ligands found in the top 10% of the database). Combinations of AMBER and OPLS receptor charges and AM1-BCC and OPLS ligand charges were used. Similar to results with DOCK6, the AMBER receptor charges were generally superior to the OPLS receptor charges when using Glide. When OPLS charges were used for the ligands, Glide SP with AMBER receptor charges found 21% of known ligands in the top 10% of the ranked database, while Glide SP with OPLS receptor charges found only 12% of known ligands. When AM1-BCC charges were used for the ligand and AMBER charges were used for the receptor, the fraction of actives found in the top 10% dropped from 21% to 9%. Results indicate that the AMBER/OPLS receptor/ligand charge combination is the most effective to identify known active compounds for this system with Glide. However, when the AMBER/OPLS receptor/ligand charge combination was applied to DOCK6, the enrichment in the top 10% of the database was less than that from the AMBER/BCC receptor/ligand charge combination, as shown in Figure 2a.
Figure 3.
Performance of different combinations of receptor and ligand charges using Glide. Yellow, purple, brown, and turqoise lines show enrichment factors calculated using Glide SP and different charge sets. Each line shows a different combination of receptor and ligand charges. For example, the yellow line represents the combination of AMBER receptor charge and OPLS ligand charge. The blue line represents enrichment from redocking using ligand poses from the AMBER-OPLS charge combination, under Glide XP mode.
The Glide extended-precision (XP) mode has previously been reported to be more effective for finding known active compounds for a large and diverse set of ligands and receptors than Glide SP.66 However, it was designed as a refinement tool for use with relatively good ligand poses from Glide SP because it demands more intensive computational expense. We took the best-scoring poses from Glide SP mode using the AMBER/OPLS combination of charges and docked again using Glide XP mode. This actually provided degradation of the fraction of actives found in the top 10% of the database from 21% to 12%. However, in the top 30% of the database, enrichment was essentially the same.
Solvation Free Energy
To account for solvation, we used the Poisson–Boltzmann continuum method implemented in MEAD. Because PB energies are known to be very sensitive to receptor–ligand geometry, only the best poses from previous docking with DOCK6 were considered. The ligand charge set used in this method was AM1-BCC, which gave the best enrichment out of the charge sets we considered. The solvation free energy differences of the ligand in protein and in water were added to the DOCK6 Grid scoring function. (Solvation free energy calculations were not included for Glide, because the Glidescore already includes a solvation term.) Calculated solvation free energy differences were small and had little effect on enrichment factors.
Consensus Scoring
We constructed a simple consensus score by adding together the grid score from DOCK6 using AMBER/AM1-BCC receptor/ligand charges and the Glidescore from Glide SP with AMBER/OPLS charges. In this case, the fraction of actives in the top 5% of the database increased to 28%, and the fraction of actives found in the top 10% was increased to 37%. When the correction for solvation free energies was added to the consensus scoring function, the enrichment factors in the top 5% and 10% of the database were the same (Figure 4).
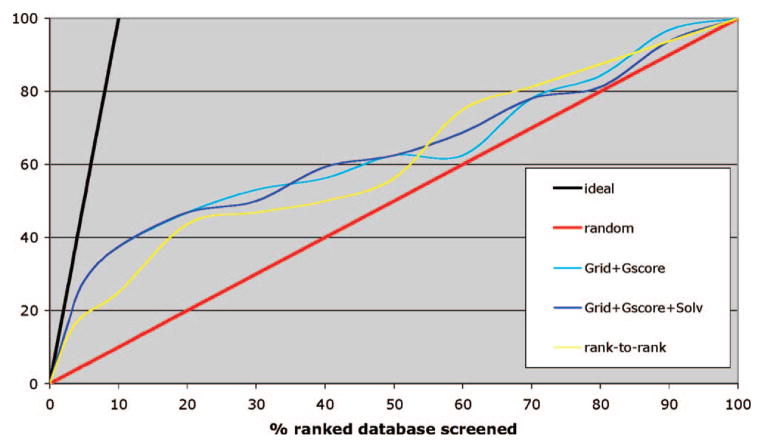
Figure 4.
Docking enrichment plots presenting consensus scoring function of Grid score from DOCK6 (AMBER-BCC charges) and Glidescore from Glide SP (AMBER-OPLS charges). The blue line represents a simple combination of Grid score, solvation energy, and Glidescore. The yellow line represents a “rank-to-rank” consensus scoring function of Glidescore and Grid score that includes solvation energy: that is, compounds are ranked using the Glidescore and the DOCK Grid score including solvation energy. A consensus rank is given as the average of these two ranks.
Accounting for Receptor Flexibility by Ensemble Docking
Ensemble docking—that is, docking to ensembles of conformations of receptors taken from molecular dynamics simulations—was performed to account for receptor flexibility. MD simulations did not significantly change the overall receptor structure but produced fluctuations (Figure 5). The receptor and ligand charge combinations used were AMBER/AM1-BCC for DOCK6 and AMBER/OPLS for GlideSP, which resulted in the best enrichment as described above. In general ensemble docking improved enrichment but only by a slight amount (Figure 6). For DOCK6, the fraction of actives found in the top 10% of the database was slightly improved (from 33% to 38%). For Glide SP, ensemble docking resulted in fewer actives found in the top 10%. However, consensus scoring with a combination of the DOCK6 grid score, solvation correction from MEAD, and the Glidescore resulted in an increase of the number of actives found in the top 10% of the database to 38%. The result shows that the performance with ensemble docking is better for the top fraction of the database, which is most valuable as it is this fraction that is likely to be tested in practice.67 Several studies have shown that docking scores are often highly correlated with molecular weight, indicating that nonspecific interactions are the most significant contributors to scores.67 Therefore, for comparison, we calculated enrichment simply ranking compounds by molecular weight. Results indicate that a consensus score with ensemble docking provides superior enrichment in the top fraction of the database; however, in considering larger fractions (greater than the top fifth or so), a simple ranking by molecular weight does just as well.
Figure 5.
A superposition of the ten receptor structures used for ensemble docking, taken from MD simulation. The ten conformations are represented by blue wire with only backbone traces. The X-ray crystal structure of the receptor without bound SIGK is represented by white wire.
Figure 6.
Docking enrichment plots showing ensemble docking against 10 different receptor conformations made by MD simulation. The consensus score (blue line) is the sum of Grid scores from DOCK (AMBER-BCC charges), solvation free energies, and Glidescores from Glide SP (AMBER-OPLS charges), averaged over all receptor conformations. The brown line is the enrichment plot calculated by ranking compounds by molecular weight, which ranges from 100 to 890 amu.
CONCLUSIONS
In this work we examined the ability of docking and scoring methods to predict the binding of small molecules to a protein–protein interface, the hotspot of the G protein β subunit. The influence of variations in the ligand protonation state, initial structures of proteins and ligands, partial charges, scoring functions, and docking protocols was investigated using the DOCK6 and Glide programs. Docking to a broad protein–protein interaction surface is a difficult problem, but it was possible to obtain significant enrichment in the number of active compounds found in the top-ranking portion of the database. The use of a consensus score and taking receptor flexibility into account by docking to an ensemble of configurations from molecular dynamics simulation resulted in greater enrichment. Limitations remain, such as the neglect of changes of conformational entropy upon binding. Furthermore, docking programs could not directly rank compounds according to affinities, and there was little correlation observed between scores and measured inhibition constants. For example, the highest-affinity compound M119 (IC50 = 200 nM) was filtered out of the top 10% of the database during this screening process, although it appeared in the top 30% for all methods. In agreement with previous studies, enrichment provided by docking is comparable to ranking compounds simply by molecular weight, although ensemble docking using a consensus score was better at finding high-affinity compounds in the top-ranked fraction of the database.
Somewhat surprisingly, the binding area of small molecules observed in docking does not always overlap with the binding of the high-affinity SIGK peptide ligand (Figure 7). In general, small compounds were observed to dock inside the hole of the Gβγ hotspot, rather than on the surface. While this might be an artifact of the modeling, if poses are reasonable accurate, it suggests that small molecules can disrupt the interaction without completely occluding the binding surface.
Figure 7.
Docking pose of a compound (NSC83217) bound to the Gβγ subunits, generated using DOCK6, superimposed with the bound high-affinity SIGK peptide. The SIGK peptide and compound are represented by green and orange colored sticks, respectively. The receptor is represented by gray wire, showing only the backbone structure.
Future work will involve applying the methods investigated here to larger databases of millions of compounds. It is expected that docking/scoring methods will be able to make a contribution toward discovery of new high-affinity ligands with the potential for selective inhibition of interactions of Gβγ with downstream signaling partners as well as to a better understanding of the structural basis for signaling specificity.
Acknowledgments
We thank Dr. Orr Ravitz and Cen Gao for their assistance with computational problems. This work was supported in part with funds from the Camille and Henry Dreyfus Foundation.
REFERENCES AND NOTES
- 1.Gilman AG. G-Proteins - Transducers of Receptor-Generated Signals. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- 2.Hamm HE. The many faces of G protein signaling. J Biol Chem. 1998;273(2):669–672. doi: 10.1074/jbc.273.2.669. [DOI] [PubMed] [Google Scholar]
- 3.Klabunde T, Hessler G. Drug design strategies for targeting G-protein-coupled receptors. ChemBioChem. 2002;3(10):929–944. doi: 10.1002/1439-7633(20021004)3:10<928::AID-CBIC928>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 4.Smrcka AV. G protein beta gamma subunits: Central mediators of G protein-coupled receptor signaling. Cell Mol Life Sci. 2008;65(14):2191–2214. doi: 10.1007/s00018-008-8006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eckhart AD, Ozaki T, Tevaearai H, Rockman HA, Koch WJ. Vascular-targeted overexpression of G protein-coupled receptor kinase-2 in transgenic mice attenuates beta-adrenergic receptor signaling and increases resting blood pressure. Mol Pharmacol. 2002;61(4):749–758. doi: 10.1124/mol.61.4.749. [DOI] [PubMed] [Google Scholar]
- 6.Li Z, Jiang HP, Xie W, Zhang ZC, Smrcka AV, Wu DQ. Roles of PLC-beta 2 and -beta 3 and PI3K gamma in chemoattractant-mediated signal transduction. Science. 2000;287(5455):1046–1049. doi: 10.1126/science.287.5455.1046. [DOI] [PubMed] [Google Scholar]
- 7.Hirsch E, Katanaev VL, Garlanda C, Azzolino O, Pirola L, Silengo L, Sozzani S, Mantovani A, Altruda F, Wymann MP. Central role for G protein-coupled phosphoinositide 3-kinase gamma in inflammation. Science. 2000;287(5455):1049–1053. doi: 10.1126/science.287.5455.1049. [DOI] [PubMed] [Google Scholar]
- 8.Xie W, Samoriski GM, McLaughlin JP, Romoser VA, Smrcka A, Hinkle PM, Bidlack JM, Gross RA, Jiang HP, Wu DQ. Genetic alteration of phospholipase C beta 3 expression modulates behavioral and cellular responses to mu opioids. Proc Natl Acad Sci USA. 1999;96(18):10385–10390. doi: 10.1073/pnas.96.18.10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iaccarino G, Smithwick LA, Lefkowitz RJ, Koch WJ. Targeting G(beta gamma) signaling in arterial vascular smooth muscle proliferation: A novel strategy to limit restenosis. Proc Natl Acad Sci USA. 1999;96(7):3945–3950. doi: 10.1073/pnas.96.7.3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iaccarino G, Koch WJ. Transgenic mice targeting the heart unveil G protein-coupled receptor kinases as therapeutic targets. Assay Drug Dev Technol. 2003;1(2):347–355. doi: 10.1089/154065803321204484. [DOI] [PubMed] [Google Scholar]
- 11.Bookout AL, Finney AE, Guo RS, Peppel K, Koch WJ, Daaka Y. Targeting G beta gamma signaling to inhibit prostate tumor formation and growth. J Biol Chem. 2003;278(39):37569–37573. doi: 10.1074/jbc.M306276200. [DOI] [PubMed] [Google Scholar]
- 12.Rockman HA, Chien KR, Choi DJ, Iaccarino G, Hunter JJ, Ross J, Lefkowitz RJ, Koch WJ. Expression of a beta-adrenergic receptor kinase 1 inhibitor prevents the development of myocardial failure in gene-targeted mice. Proc Natl Acad Sci USA. 1998;95(12):7000–7005. doi: 10.1073/pnas.95.12.7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yao LN, Fan PD, Jiang Z, Mailliard WS, Gordon AS, Diamond I. Addicting drugs utilize a synergistic molecular mechanism in common requiring adenosine and Gi-beta gamma dimers. Proc Natl Acad Sci USA. 2003;100(24):14379–14384. doi: 10.1073/pnas.2336093100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neptune ER, Bourne HR. Receptors induce chemotaxis by releasing the beta gamma subunit of G(i), not by activating G(q) or G(s) Proc Natl Acad Sci USA. 1997;94(26):14489–14494. doi: 10.1073/pnas.94.26.14489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muller A, Homey B, Soto H, Ge NF, Catron D, Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, Barrera JL, Mohar A, Verastegui E, Zlotnik A. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410(6824):50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 16.Smrcka AV, Lehmann DM, Dessal AL. G protein beta gamma subunits as targets for small molecule therapeutic development. Comb Chem High Throughput Screening. 2008;11(5):382–395. doi: 10.2174/138620708784534761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pons T, Gomez R, Chinea G, Valencia A. Beta-propellers: Associated functions and their role in human diseases. Curr Med Chem. 2003;10(6):505–524. doi: 10.2174/0929867033368204. [DOI] [PubMed] [Google Scholar]
- 18.Li D, Roberts R. WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell Mol Life Sci. 2001;58(14):2085–2097. doi: 10.1007/PL00000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehmann DM, Yuan C, Smrcka AV. Lipidomics and Bioactive Lipids. Vol. 434. Elsevier; San Diego, CA: 2007. Analysis and pharmacological targeting of phospholipase C beta interactions with G proteins; pp. 29–48. [DOI] [PubMed] [Google Scholar]
- 20.Lehmann DM, Seneviratne A, Smrcka AV. Small molecule disruption of G protein beta gamma subunit signaling inhibits neutrophil chemotaxis and inflammation. Mol Pharmacol. 2008;73(2):410–418. doi: 10.1124/mol.107.041780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonacci TM, Mathews JL, Yuan CJ, Lehmann DM, Malik S, Wu DQ, Font JL, Bidlack JM, Smrcka AV. Differential targeting of G beta gamma-subunit signaling with small molecules. Science. 2006;312(5772):443–446. doi: 10.1126/science.1120378. [DOI] [PubMed] [Google Scholar]
- 22.Davis TL, Bonacci TM, Sprang SR, Smrcka AV. Structural and molecular characterization of a preferred protein interaction surface on G protein beta gamma subunits. Biochemistry. 2005;44(31):10593–10604. doi: 10.1021/bi050655i. [DOI] [PubMed] [Google Scholar]
- 23.Hindle SA, Rarey M, Buning C, Lengauer T. Flexible docking under pharmacophore type constraints. J Comput-Aided Mol Des. 2002;16(2):129–149. doi: 10.1023/a:1016399411208. [DOI] [PubMed] [Google Scholar]
- 24.Cheng AC, Coleman RG, Smyth KT, Cao Q, Soulard P, Caffrey DR, Salzberg AC, Huang ES. Structure-based maximal affinity model predicts small-molecule druggability. Nat Biotechnol. 2007;25(1):71–75. doi: 10.1038/nbt1273. [DOI] [PubMed] [Google Scholar]
- 25.Janin J, Chothia C. The structure of protein-protein recognition sites. J Biol Chem. 1990;265(27):16027–16030. [PubMed] [Google Scholar]
- 26.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285(5):2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 27.Clackson T, Wells JA. A hot-spot of binding energy in a hormone-receptor interface. Science. 1995;267(5196):383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 28.Desrosiers DC, Peng ZY. A binding free energy hot spot in the ankyrin repeat protein GABP beta mediated protein-protein interaction. J Mol Biol. 2005;354(2):375–384. doi: 10.1016/j.jmb.2005.09.045. [DOI] [PubMed] [Google Scholar]
- 29.Keskin O, Ma BY, Nussinov R. Hot regions in protein-protein interactions: The organization and contribution of structurally conserved hot spot residues. J Mol Biol. 2005;345(5):1281–1294. doi: 10.1016/j.jmb.2004.10.077. [DOI] [PubMed] [Google Scholar]
- 30.Moreira IS, Fernandes PA, Ramos MJ. Hot spots-A review of the protein-protein interface determinant amino-acid residues. Proteins. 2007;68(4):803–812. doi: 10.1002/prot.21396. [DOI] [PubMed] [Google Scholar]
- 31.Moreira IS, Fernandes PA, Ramos MJ. Protein-protein recognition: a computational mutagenesis study of the MDM2-P53 complex. Theor Chem Acc. 2008;120:4–6. 533–542. [Google Scholar]
- 32.Scott JK, Huang SF, Gangadhar BP, Samoriski GM, Clapp P, Gross RA, Taussig R, Smrcka AV. Evidence that a protein-protein interaction ‘hot spot’ on heterotrimeric G protein beta gamma subunits is used for recognition of a subclass of effectors. EMBO J. 2001;20(4):767–776. doi: 10.1093/emboj/20.4.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shulman-Peleg A, Shatsky M, Nussinov R, Wolfson HJ. Spatial chemical conservation of hot spot interactions in protein-protein complexes. BMC Biol. 2007:5. doi: 10.1186/1741-7007-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keskin O, Gursoy A, Ma B, Nussinov R. Principles of protein-protein interactions: what are the preferred ways for proteins to interact. Chem Rev. 2008;108(4):1225–44. doi: 10.1021/cr040409x. [DOI] [PubMed] [Google Scholar]
- 35.Keskin O, Gursoy A, Ma B, Nussinov R. Towards drugs targeting multiple proteins in a systems biology approach. Curr Top Med Chem. 2007;7(10):943–51. doi: 10.2174/156802607780906690. [DOI] [PubMed] [Google Scholar]
- 36.Brooijmans N, Kuntz ID. Molecular recognition and docking algorithms. Annu Rev Biophys Biomol Struct. 2003;32:335–373. doi: 10.1146/annurev.biophys.32.110601.142532. [DOI] [PubMed] [Google Scholar]
- 37.Brown SP, Muchmore SW. High-throughput calculation of protein-ligand binding affinities: Modification and adaptation of the MM-PBSA protocol to enterprise grid computing. J Chem Inf Model. 2006;46(3):999–1005. doi: 10.1021/ci050488t. [DOI] [PubMed] [Google Scholar]
- 38.Jorgensen WL. The many roles of computation in drug discovery. Science. 2004;303(5665):1813–1818. doi: 10.1126/science.1096361. [DOI] [PubMed] [Google Scholar]
- 39.Shoichet BK. Virtual screening of chemical libraries. Nature. 2004;432(7019):862–865. doi: 10.1038/nature03197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walters WP, Stahl MT, Murcko MA. Virtual screening - an overview. Drug Discovery Today. 1998;3(4):160–178. [Google Scholar]
- 41.Cummings MD, DesJarlais RL, Gibbs AC, Mohan V, Jaeger EP. Comparison of virtual screening programs. Abstr Pap Am Chem Soc. 2003;226:U456–U456. [Google Scholar]
- 42.Cummings MD, DesJarlais RL, Gibbs AC, Mohan V, Jaeger EP. Comparison of automated docking programs as virtual screening tools. J Med Chem. 2005;48(4):962–976. doi: 10.1021/jm049798d. [DOI] [PubMed] [Google Scholar]
- 43.Kellenberger E, Rodrigo J, Muller P, Rognan D. Comparative evaluation of eight docking tools for docking and virtual screening accuracy. Proteins. 2004;57(2):225–242. doi: 10.1002/prot.20149. [DOI] [PubMed] [Google Scholar]
- 44.Mohan V, Gibbs AC, Cummings MD, Jaeger EP, DesJarlais RL. Docking: Successes and challenges. Curr Pharm Des. 2005;11(3):323–333. doi: 10.2174/1381612053382106. [DOI] [PubMed] [Google Scholar]
- 45.Onodera K, Satou K, Hirota H. Evaluations of molecular docking programs for virtual screening. J Chem Inf Model. 2007;47(4):1609–1618. doi: 10.1021/ci7000378. [DOI] [PubMed] [Google Scholar]
- 46.Warren GL, Andrews CW, Capelli AM, Clarke B, LaLonde J, Lambert MH, Lindvall M, Nevins N, Semus SF, Senger S, Tedesco G, Wall ID, Woolven JM, Peishoff CE, Head MS. A critical assessment of docking programs and scoring functions. J Med Chem. 2006;49(20):5912–5931. doi: 10.1021/jm050362n. [DOI] [PubMed] [Google Scholar]
- 47.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong GM, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang JM, Kollman P. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24(16):1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 48.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF chimera - A visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 49.Jorgensen WL, Maxwell DS, Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc. 1996;118(45):11225–11236. [Google Scholar]
- 50.Ewing TJA, Makino S, Skillman AG, Kuntz ID. DOCK 4.0: Search strategies for automated molecular docking of flexible molecule databases. J Comput-Aided Mol Des. 2001;15(5):411–428. doi: 10.1023/a:1011115820450. [DOI] [PubMed] [Google Scholar]
- 51.Storer JW, Giesen DJ, Cramer CJ, Truhlar DG. A new semiempirical approach in quantum chemistry. J Comput-Aided Mol Des. 1995;9(1):87–110. doi: 10.1007/BF00117280. [DOI] [PubMed] [Google Scholar]
- 52.Li JB, Zhu TH, Cramer CJ, Truhlar DG. New class IV charge model for extracting accurate partial charges from wave functions. J Phys Chem A. 1998;102(10):1820–1831. [Google Scholar]
- 53.Jakalian A, Jack DB, Bayly CI. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II Parameterization and validation. J Comput Chem. 2002;23(16):1623–1641. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 54.Mulliken RS. Electronic Population Analysis on LCAO/MO Molecular Wave Functions. J Chem Phys. 1955;23:1883–1840. [Google Scholar]
- 55.Tishmack PA, Bashford D, Harms E, VanEtten RL. Use of H-1 NMR spectroscopy and computer simulations to analyze histidine pK(a) changes in a protein tyrosine phosphatase: Experimental and theoretical determination of electrostatic properties in a small protein. Biochemistry. 1997;36(39):11984–11994. doi: 10.1021/bi9712448. [DOI] [PubMed] [Google Scholar]
- 56.Carlson HA, Masukawa KM, Rubins K, Bushman FD, Jorgensen WL, Lins RD, Briggs JM, McCammon JA. Developing a dynamic pharmacophore model for HIV-1 integrase. J Med Chem. 2000;43(11):2100–2114. doi: 10.1021/jm990322h. [DOI] [PubMed] [Google Scholar]
- 57.Huang SY, Zou XQ. Ensemble docking of multiple protein structures: Considering protein structural variations in molecular docking. Proteins. 2007;66(2):399–421. doi: 10.1002/prot.21214. [DOI] [PubMed] [Google Scholar]
- 58.Rao S, Sanschagrin PC, Greenwood JR, Repasky MP, Sherman W, Farid R. Improving database enrichment through ensemble docking. J Comput-Aided Mol Des. 2008;22(9):621–627. doi: 10.1007/s10822-008-9182-y. [DOI] [PubMed] [Google Scholar]
- 59.Hawkins GD, Cramer CJ, Truhlar DG. Pairwise solute descreening of solute charges from a dielectric medium. Chem Phys Lett. 1995;246(1–2):122–129. [Google Scholar]
- 60.Hawkins GD, Cramer CJ, Truhlar DG. Parametrized models of aqueous free energies of solvation based on pairwise descreening of solute atomic charges from a dielectric medium. J Phys Chem. 1996;100(51):19824–19839. [Google Scholar]
- 61.Tsui V, Case DA. Theory and applications of the generalized Born solvation model in macromolecular Simulations. Biopolymers. 2000;56(4):275–291. doi: 10.1002/1097-0282(2000)56:4<275::AID-BIP10024>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 62.Gordon HL, Somorjai RL. Fuzzy cluster - analysis of molecular-dynamics trajectories. Proteins. 1992;14(2):249–264. doi: 10.1002/prot.340140211. [DOI] [PubMed] [Google Scholar]
- 63.Kontoyianni M, McClellan LM, Sokol GS. Evaluation of docking performance: Comparative data on docking algorithms. J Med Chem. 2004;47(3):558–565. doi: 10.1021/jm0302997. [DOI] [PubMed] [Google Scholar]
- 64.Kontoyianni M, Sokol GS, McClellan LM. Evaluation of library ranking efficacy in virtual screening. J Comput Chem. 2005;26(1):11–22. doi: 10.1002/jcc.20141. [DOI] [PubMed] [Google Scholar]
- 65.Ray SS, Nowak RJ, Brown RH, Lansbury PT. Small-molecule-mediated stabilization of familial amyotrophic lateral sclerosis-linked superoxide dismutase mutants against unfolding and aggregation. Proc Natl Acad Sci USA. 2005;102(10):3639–3644. doi: 10.1073/pnas.0408277102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem. 2006;49(21):6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 67.Kim R, Skolnick J. Assessment of programs for ligand binding affinity prediction. J Comput Chem. 2008;29(8):1316–1331. doi: 10.1002/jcc.20893. [DOI] [PMC free article] [PubMed] [Google Scholar]