Abstract
Although mammalian hearts show virtually no ability to regenerate, there is a growing initiative to determine whether existing cardiomyocytes or progenitor cells can be coaxed into eliciting a regenerative response. In contrast to mammals, a number of non-mammalian vertebrate species are able to regenerate their hearts1–3, including the zebrafish4,5, which can fully regenerate its heart following amputation of up to 20% of the ventricle. To directly address the source of newly formed cardiomyocytes during zebrafish heart regeneration, we first established a genetic strategy to lineage-trace cardiomyocytes in the adult fish, based on the Cre/lox system widely used in the mouse6. Using this system, we show here that regenerated heart muscle cells are derived from the proliferation of differentiated cardiomyocytes. Furthermore, we show that proliferating cardiomyocytes undergo limited dedifferentiation characterized by the disassembly of their sarcomeric structure, detachment from one another and expression of regulators of cell cycle progression. Specifically, we show that polo-like kinase1 (plk1) is an essential component of cardiomyocyte proliferation during heart regeneration. Our data provides the first direct evidence for the source of proliferating cardiomyocytes during zebrafish heart regeneration and indicates that stem/progenitor cells are not significantly involved in this process.
During heart regeneration in zebrafish, lost ventricular tissue is rapidly replaced. After as little as 1 month, most of the missing tissue has been regenerated by cardiomyocytes. The exact source of these new cardiomyocytes is a debated and unanswered question. To address this we developed and successfully implemented the tamoxifen (4-OHT) inducible Cre/Lox approach in zebrafish to genetically label regenerating cardiomyocytes (for a detailed description of the lines generated and/or methodologies, see Methods and Supplementary Figs. 1–9).
To examine the contribution made by differentiated cardiomyoctes to regenerated cardiac tissue, we performed a series of amputation experiments on adult zebrafish in which cardiomyocytes had been genetically labelled at 48 h post fertilization (hpf). Approximately 20% of the ventricle was removed and cardiac regeneration was subsequently assessed at 7, 14 and 30 days post amputation (dpa). At 7 dpa, the remaining cardiac tissue was uniformly GFPpos (Fig. 1a, b) with much of the missing tissue now replaced by a fibrin/collagen clot (n=5 hearts) (Fig. 1c). Strikingly, at 14 dpa, when a significant amount of regeneration had occurred, cardiomyocytes within the regenerating tissue were uniformly GFPpos (Fig. 1d, e) with only a small fibrin clot remaining (n=7 hearts) (Fig. 1f). These results suggest that the regenerated cardiomyocytes arise from differentiated GFPpos cardiomyocytes. These findings were further substantiated at 30 dpa, when regeneration is nearly complete; all of the cardiomyocytes within the regenerated cardiac tissue were clearly GFPpos (n=9 hearts) (Fig. 1g, h).
Fig. 1. Regenerated cardiomyocytes are derived from differentiated cardiomyocytes.
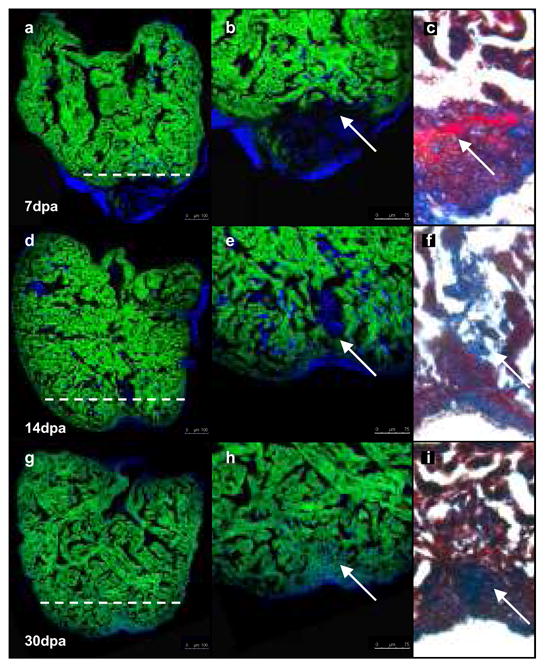
Cardiomyocytes in transgenic zebrafish (tg-cmlc2a-Cre-Ert2: tg-cmlc2a-LnL-GFP) were genetically labelled at 48 hpf by inducing Cre activity with tamoxifen. These embryos were then grown to adulthood (3months/sexually mature) at which point the heart was amputated and allowed to regenerate for 7 (a, b, c), 14 (d, e, f) or 30 days (g, h, i). Dashed white line represents the plane of amputation. At 7 dpa (a, b) relatively little regeneration has occurred. Trichromic staining indicates a fibrin clot has formed adjacent to the wound (c). By 14 dpa, GFPpos cardiomyocytes have regenerated a substantial amount of new cardiac tissue (d, e) and the fibrin clot is reduced in size (f). At 30 dpa, heart regeneration is virtually complete (g, h) and all of the regenerated tissue is comprised of GFPpos cardiomyocytes. The clot has been replaced by a small scar (h). Scale bars represent 100 μm in a, d, g, and 75 μm in b, e, h.
To determine the overall contribution made by GFPpos cardiomyocytes to the regenerated heart, we immunostained 7, 14 and 30 dpa regenerating heart sections with anti-GFP and the anti-sarcomeric myosin antibody MF20 (Supplementary Fig. 9a–d). We were unable to detect any cardiomyocytes labelled with MF20 or GFP alone (n=4836). Together, our results demonstrate that after amputation, the vast majority, if not all, of newly formed cardiomyocytes arise from pre-existing cardiomyocytes.
We next sought to determine whether GFPpos cardiomyocytes had re-entered the cell cycle. Adult GFPpos transgenic zebrafish were treated with bromodeoxyuridine (BrdU) for 7 days following amputation (Fig. 2a–f). Subsequently, at 14 dpa, we found a significant increase in the number of BrdUpos/GFPpos cardiomyocytes in regenerating hearts versus non-amputated controls (Fig. 2g). We can conclude from this that differentiated GFPpos cardiomyocytes have re-entered the cell cycle and engaged in DNA replication. We also analysed the position of BrdU-labelled GFPpos cardiomyocytes within the regenerating heart (Fig. 2h and inset). While the majority of BrdUpos/GFPpos labelled cardiomyocytes are concentrated around the wound, a proportion can also be found in regions far from the site of amputation. This suggests that the response to the injury affects the heart in a global manner.
Fig. 2. Differentiated cardiomyocytes re-enter the cell cycle.
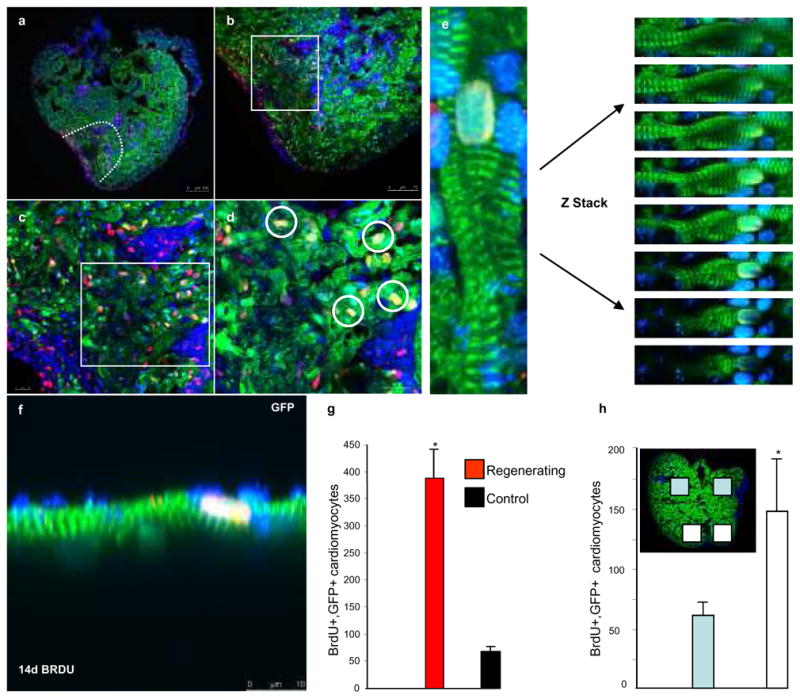
Transgenic zebrafish (tg-cmlc2a-Cre-Ert2: tg-cmlc2a-LnL-GFP) genetically labelled at 48 hpf and grown to adulthood were amputated then treated with BrdU at 7 dpa, hearts were then isolated and processed at 14 dpa (a–f). Green= GFPpos cardiomyocytes; Red=BrdUpos cells; Blue= DAPI; Yellow= BrdUpos/GFPpos cardiomyocytes (white rings in d).(g) Indicates the average number of BrdUpos/GFPpos cardiomyocytes/section +/− SEM, t-test * p<0.01; amputated n=17 sections from 7 different animals, control n=9 sections from 3 different animals. (h and inset) Indicates the distribution of BrdUpos/GFPpos cardiomyocytes (n=5 sections from 5 different animals). Scale bars represent 100 μm in a, 75 μm in b, and 10 μm in c, d, f.
Recent studies have indicated that cardiomyocytes dedifferentiate in order to facilitate proliferation, yet it is unclear how far within the cardiac lineage they regress7,8. An increase in the expression of the cardiac progenitor associated genes nkx2.5 and hand2 during zebrafish heart regeneration has been reported9. However, our own in situ hybridisation analyses failed to detect any significant upregulation of either transcript (data not shown), confirming previous results by our laboratory5. Furthermore, genome-wide transcriptome data10,11 also failed to detect significant changes in the expression of either transcript during zebrafish heart regeneration. These results argue against an extensive dedifferentiation of cardiomyocytes as a pre-requisite for their proliferation in the context of heart regeneration.
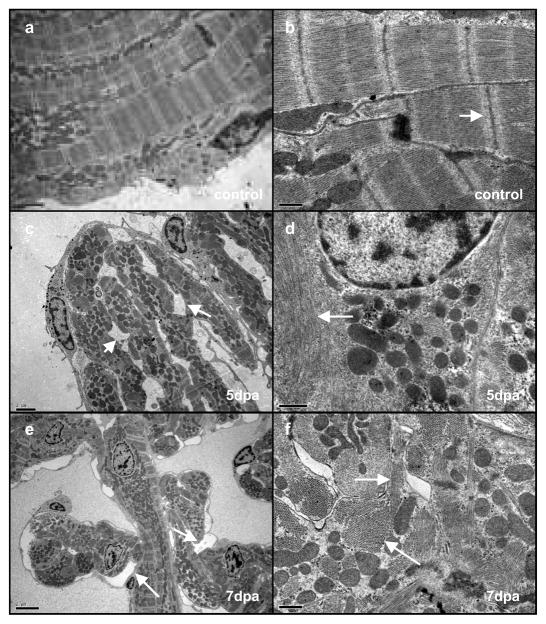
To address the extent to which cardiomyocytes dedifferentiate during zebrafish heart regeneration, we analysed regenerating hearts using transmission electron microscopy (TEM). In uninjured hearts (Fig. 3a, b) cardiomyocytes displayed an ordered arrangement of sarcomeres and mitochondria with clearly defined z-lines (Fig. 3b). Following amputation, cardiomyocytes began to detach from one another, creating large intercellular spaces (Fig. 3c, d). The aligned array of actin and myosin filaments visible in uninjured hearts (Fig. 3b) became disorganised (Fig. 3d), associated with a loss of z-line structure. Later, at 7 dpa (Fig. 3e, f), intercellular spaces were also readily visualised as cardiomyocytes detached from one another (Fig. 3e). Furthermore, although sarcomeric filaments were visible, there was a lack of organisation leading to the presence of both transverse and longitudinal sarcomeric structures within individual cardiomyocytes (Fig. 3f). Using these criteria, we were able to count the number of cardiomyocytes undergoing phenotypic changes (Supplementary Fig. 10a–c). Following amputation, the proportion of these structurally/morphologically altered cardiomyocytes increased, peaking at around day 7 (Supplementary Fig. 10c). Furthermore, their distribution within the regenerating heart closely resembled that of BrdU-labelled cardiomyocytes (Supplementary Fig. 10d and inset). Despite these changes, cardiomyocytes appeared healthy and did not display any of the hallmarks associated with cell death12,13 (Supplementary Fig. 11a–f). TUNEL labelling regenerating hearts further confirmed these observations (Supplementary Fig. 12a, b).
Fig. 3. Cardiomyocytes dedifferentiate resulting in the disassembly of sarcomeric structure and detachment.
Electron microscopy of a control heart (a, b), a 5-dpa regenerating heart (c, d) and a 7-dpa regenerating heart (e, f). Cardiomyocytes in unamputated control samples show a tightly organised sarcomeric structure (a), at higher magnification (b) the Z-lines are clearly visible (white arrow). At 5 dpa many of the cardiomyocytes display a disorganised sarcomeric structure (c) along with the appearance of intercellular spaces (white arrows). Closer examination reveals a loss of Z-lines (d, white arrow). At 7 dpa there is a similar loss of structure and appearance of intercellular spaces (e white arrows). At higher magnification (f) myosin fibres are visible (arrows) however both longitudinal (upper arrow) and transverse (lower arrow) fibres are present within the same cardiomyocyte indicating disorganised sarcomeric structure. Scale bars represent 0.5 μm in a, b, d, and 2 μm in c, e, f.
The sequence of events described thus far would indicate that during regeneration, cardiomyocytes detach from one another and disassemble their sarcomeric structure presumably to facilitate cell cycle re-entry. If this is correct, then cardiomyocytes in the process of cell division should not have any discernable sarcomeric structure. To test this hypothesis we labelled dividing cardiomyocytes with both phosphorylated histone 3 (PH3), a well established marker of cells undergoing mitosis, and either GFP or MF20. Despite the rarity of these events (an average of 3 cells per section, n=12 sections from 4 hearts) none of the PH3-labelled cells showed any discernable organisation of their sarcomeric structure (Supplementary Fig. 13a–d). Furthermore we also immunolabelled GFPpos cardiomyocytes for proliferating cell nuclear antigen (PCNA)(Supplementary Fig. 14a–c). Similar to PH3-labelled cardiomyocytes, PCNApos/GFPpos cardiomyocytes showed disorganized sarcomeric structure (Supplementary Fig. 14b, c) These results indicate that the sarcomere is disassembled in the process of cardiomyocyte divison, similar to the set of events described recently in the mouse8.
Next we sought to determine whether cardiomyocyte dedifferentiation during heart regeneration was associated with specific changes in gene expression. Previous reports have indicated an increase in expression of the mitotic checkpoint kinase mps1 in cardiomyocytes during regeneration and that perturbation of this effectively inhibits regeneration4. Thus, we examined the expression of polo like kinase 1(plk1), a regulator of cell cycle progression that was detected as upregulated in previous microarray analyses of zebrafish heart regeneration11. plk1 transcripts were markedly upregulated in regenerating hearts at 1, 3, and 7 dpa, as detected by RT-PCR, closely resembling those of mps1 (Fig. 4a). In addition, in situ hybridisation analyses revealed that plk1 expression increased in cardiomyocytes during regeneration (Fig. 4b, c).
Fig. 4. plk1 is necessary for cardiac regeneration.
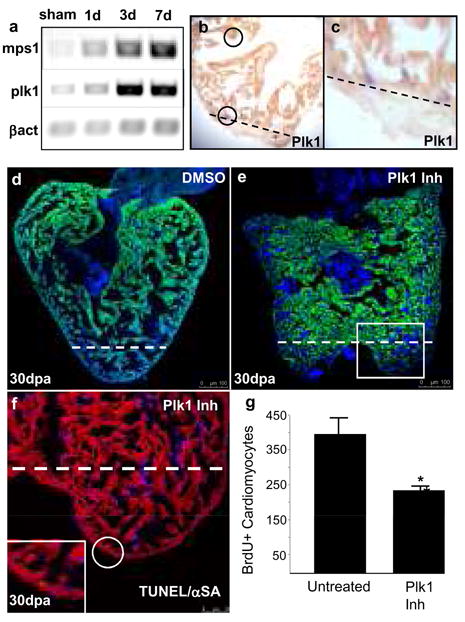
(a) Semi-quantitative RT-PCR of mps1 and plk1. (b, c) Double in-situ hybridization detecting plk1 mRNA (blue) and cmlc2a mRNA (red/brown) in a 7-dpa ventricle. The plane of amputation is indicated with a dashed line. (d) 30-dpa regenerating heart (DMSO treated). The plane of amputation is indicated with a dashed line. (e) 30-dpa regenerating heart (cyclapolin9 treated). The plane of amputation is indicated with a dashed line the white box indicates the area shown in (f). (f) Consecutive section taken from the same heart pictured in (e), labelled with TUNEL (green) and anti-α sarcomeric actin (red). The plane of amputation is indicated with a dashed line. Inset shows a TUNEL positive cardiomyocyte. (g) Plk1 inhibiton reduces the number of cardiomyocytes entering the cell cycle during heart regeneration. The graph indicates the average number of BrdUpos/GFPpos cardiomyocytes/section +/− SEM, t-test * p<0.01; Plk1 inhibitor (cyclapolin9) treated (n=30 sections from 10 different animals), untreated control (n=9 sections from 3 different animals). Scale bars represent 100 μm in d, e, and 50 μm in f.
To further assess the role of plk1 during heart regeneration, we utilised a recently described embryonic model of heart regeneration14. Using this model system we were able to specifically ablate cardiomyocytes in 48-hpf embryos. These were subsequently washed and allowed to recover with or without the Plk1 inhibitor cyclapolin 9. In the absence of the inhibitor, 67% of the embryos were able to regenerate their heart. However, in the presence of the inhibitor, this number fell substantially to 17% (the treatments used in these experiments have no effect on wildtype embryos) (Supplementary Fig. 15a–d). We confirmed these results in the adult setting by inhibiting Plk1 activity with cyclapolin 9 in regenerating zebrafish throughout the 30-day recovery period. Inhibition of Plk1 drastically inhibited the regenerative process (Fig. 4d, e). TUNEL labelling showed that this inhibition was not due to an increase in cardiomyocyte apoptosis (on average 1 cell per section, n=3 sections from 3 different hearts) (Fig. 4f). Furthermore we found a significant decrease in the number of BrdUpos/GFPpos cardiomyocytes in regenerating animals treated with the inhibitor (Fig. 4g). These results indicate that plk1 is essential for heart regeneration to proceed.
Overall, our studies show that zebrafish heart regeneration is primarily driven by pre-existing cardiomyocytes, rather than by progenitor cells, as has been previously suggested9. The tightness and specificity of our cardiomyocyte lineage-tracing system (Supplementary Figs. 1–9, see also Methods for more details) allow us to demonstrate that the vast majority, if not all, of heart muscle cells formed during regeneration arise from pre-existing cardiomyocytes. Even though we cannot formally exclude the possibility that stem/progenitor cells may give rise to cardiomyocytes during this process, in light of our results we can conclude that their contribution to newly formed myocardium would only be marginal. To facilitate proliferation, we found that pre-existing cardiomyocytes undergo limited dedifferentiation. Soon after amputation, cardiomyocytes close to the wound start to disassemble their sarcomeric structure and detach from one another. Furthermore, reduced sarcomeric structure is also observed in zebrafish cardiomyocytes re-entering cell cycle. Microarray analysis also confirms these findings with many sarcomeric genes being downregulated after amputation11. A comparable set of events has also been described in the newt. Here, the expression of cardiac sarcomeric genes is down regulated following amputation, then as regeneration proceeds, the expression returns to pre-amputation levels15. Similar structural changes are also associated with hibernating myocardium in humans following cardiac injury16. Hibernating cardiomyocytes typically show a depletion of sarcomeric structure and an expression pattern of structural proteins closely resembling foetal heart cells17. Although mammalian cardiomyocytes are unable to regenerate, it is tempting to speculate whether they can in fact complete one of the steps involved in this process. We finally show that, as a prelude to proliferation, zebrafish cardiomyocytes adjacent to the wound begin to express regulators of cell cycle progression, such as plk1 and mps14 which are necessary for the regenerative process to proceed. The fact that zebrafish cardiomyocytes dedifferentiate to a limited extent during heart regeneration, rather than undergoing dramatic changes in gene expression, offers hope that heart regeneration can also be induced in mammals. In this respect, our studies provide mechanistic insight into previous findings in mice indicating that forced expression of cell cycle regulators can induce regeneration following cardiac injury18.
Methods summary
Cardiomyocyte genetic labelling
Generation of transgenic lines is described in the supplementary methods. To induce recombination and subsequently genetically label cardiomyocytes, embryos/adults were treated with 4-OHT followed by a wash period of at least 1 week prior to amputation.
Zebrafish heart amputation
Adult fish were anesthetized in 0.4%Tricaine and secured ventral side up in a slotted sponge. Watchmaker forceps were used to remove the surface scales and penetrate the skin, muscle, and pericardial sac. Once exposed, the ventricle was gently pulled at the apex and cut with iridectomy scissors. After surgery, fish were immediately returned to system water. At the specified time points, hearts were removed and fixed in 4% paraformaldehyde overnight at 4°C, washed several times in PBS, equilibrated in 30% sucrose in PBS, and frozen for cryosectioning.
BrdU labelling
Fish were anesthetized in 0.4%Tricaine, and 0.5ml of a 2.5mg/ml solution of BrdU (in PBS) was injected into the abdominal cavity once every 24 h for 7 d. After 14 d, hearts were removed and fixed in 4% paraformaldehyde overnight at 4°C, washed several times in PBS, equilibrated in 30% sucrose in PBS, and frozen for cryosectioning.
Inhibitor treatment
Following amputation, adult fish were allowed to recover for 24hrs in system water. Cyclapolin 9 (Sigma C6493) was dissolved in DMSO and added to 400ml system water to a final concentration of 3μM. Water and inhibitor or DMSO alone were changed daily throughout the experimental procedures.
Supplementary Material
Acknowledgments
We would like to thank Maria C. Fabregat, Concepcion Rodriguez Esteban and Ilir Dubova for technical assistance and Adèle Faucherre for critique of the manuscript. ES was recipient of pre-doctoral fellowship from DIUE, Generalitat de Catalunya. This work was supported by grants from Fundacion Cellex, the Ipsen Foundation, the G. Harold and Leila Y. Mathers Charitable Foundation, MICINN, the National Institutes of Health and Sanofi-Aventis.
Footnotes
Author contributions
C.J., A.R., and J.C.I.B. conceived the project and designed the experiments. C.J. executed the molecular biology and established the transgenic lines. C.J., E.S. and M.R. executed the experiments. M.M. performed the immunohistochemistry and confocal/TEM imaging. C.J., A.R., and J.C.I.B. wrote the manuscript.
References
- 1.Oberpriller JO, Oberpriller JC. Response of the adult newt ventricle to injury. J Exp Zool. 1974;187:249–253. doi: 10.1002/jez.1401870208. [DOI] [PubMed] [Google Scholar]
- 2.Neff AW, Dent AE, Armstrong JB. Heart development and regeneration in urodeles. Int J Dev Biol. 1996;40:719–725. [PubMed] [Google Scholar]
- 3.Flink IL. Cell cycle reentry of ventricular and atrial cardiomyocytes and cells within the epicardium following amputation of the ventricular apex in the axolotl, Amblystoma mexicanum: confocal microscopic immunofluorescent image analysis of bromodeoxyuridine-labeled nuclei. Anat Embryol (Berl) 2002;205:235–244. doi: 10.1007/s00429-002-0249-6. [DOI] [PubMed] [Google Scholar]
- 4.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 5.Raya A, et al. Activation of Notch signaling pathway precedes heart regeneration in zebrafish. Proc Natl Acad Sci U S A. 2003;100 (Suppl 1):11889–11895. doi: 10.1073/pnas.1834204100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dor Y, Brown J, Martinez OI, Melton DA. Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature. 2004;429:41–46. doi: 10.1038/nature02520. [DOI] [PubMed] [Google Scholar]
- 7.Engel FB, et al. p38 MAP kinase inhibition enables proliferation of adult mammalian cardiomyocytes. Genes Dev. 2005;19:1175–1187. doi: 10.1101/gad.1306705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bersell K, Arab S, Haring B, Kühn B. Neuregulin1/ErbB4 Signaling Induces Cardiomyocyte Proliferation and Repair of Heart Injury. Cell. 2009;138:257–270. doi: 10.1016/j.cell.2009.04.060. [DOI] [PubMed] [Google Scholar]
- 9.Lepilina A, et al. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 10.Lien CL, Schebesta M, Makino S, Weber GJ, Keating MT. Gene expression analysis of zebrafish heart regeneration. PLoS Biol. 2006;4:e260. doi: 10.1371/journal.pbio.0040260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sleep E, et al. Transcriptomics approach to investigate zebrafish heart regeneration. J Cardiovasc Med (Hagerstown) 2010 doi: 10.2459/JCM.0b013e3283375900. In press. [DOI] [PubMed] [Google Scholar]
- 12.Abbate A, et al. Electron microscopy characterization of cardiomyocyte apoptosis in ischemic heart disease. Int J Cardiol. 2007;114:118–120. doi: 10.1016/j.ijcard.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 13.Cheville NF. ULTRASTRUCTURAL PATHOLOGY-AN INTRODUCTION TO INTERPRETATION. Iowa states: University Press; 1994. [Google Scholar]
- 14.Curado S, et al. Conditional targeted cell ablation in zebrafish: a new tool for regeneration studies. Dev Dyn. 2007;236:1025–1035. doi: 10.1002/dvdy.21100. [DOI] [PubMed] [Google Scholar]
- 15.Laube F, Heister M, Scholz C, Borchardt T, Braun T. Re-programming of newt cardiomyocytes is induced by tissue regeneration. J Cell Sci. 2006;119:4719–4729. doi: 10.1242/jcs.03252. [DOI] [PubMed] [Google Scholar]
- 16.Wijns W, Vatner SF, Camici PG. Hibernating myocardium. N Engl J Med. 1998;339:173–181. doi: 10.1056/NEJM199807163390307. [DOI] [PubMed] [Google Scholar]
- 17.Dispersyn GD, Geuens E, Ver Donck L, Ramaekers FC, Borgers M. Adult rabbit cardiomyocytes undergo hibernation-like dedifferentiation when co-cultured with cardiac fibroblasts. Cardiovasc Res. 2001;51:230–240. doi: 10.1016/s0008-6363(01)00326-1. [DOI] [PubMed] [Google Scholar]
- 18.Bicknell KA, Coxon CH, Brooks G. Can the cardiomyocyte cell cycle be reprogrammed? J Mol Cell Cardiol. 2007;42:706–721. doi: 10.1016/j.yjmcc.2007.01.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.