SUMMARY
Vaccinia virus (VACV) encodes DNA polymerase and additional proteins that enable cytoplasmic replication. We confirmed the ability of VACV DNA ligase mutants to replicate and tested the hypothesis that cellular ligases compensate for loss of viral gene expression. Knock-down of human DNA ligase I but not other ligases with siRNA or a specific inhibitor severely reduced replication of viral DNA in cells infected with VACV ligase-deficient mutants, indicating that the cellular enzyme plays a complementary role. Replication of ligase-deficient VACV was greatly reduced and delayed in resting primary cells, correlating with initial low levels of ligase I and subsequent viral induction and localization of ligase I in virus factories. These studies indicate that DNA ligation is essential for poxvirus replication and explain the ability of ligase deletion mutants to replicate in dividing cells but exhibit decreased pathogenicity in mice. Encoding of a ligase might allow VACV to “jump-start” DNA synthesis.
INTRODUCTION
Poxviruses are large DNA viruses notable for their replication in the cytoplasm of infected cells, wide distribution in nature, and ability to cause disease (Moss, 2007). Proteins encoded by vaccinia virus (VACV), the prototype poxvirus, that are essential for replication and processing of viral DNA include a DNA polymerase, primase/NTPase, uracil DNA glycosylase, processivity factor, protein kinase and Holliday junction resolvase (Moss and De Silva, 2006). Chordopoxviruses also encode an ATP-dependent DNA ligase that is expressed early in infection (Colinas et al., 1990; Kerr and Smith, 1989; Smith et al., 1989). The VACV DNA ligase, which can repair nicked duplex DNA substrates consisting of a 5’-phosphate terminated strand and a 3’-hydroxyl terminated strand, has been characterized extensively (Sekiguchi and Shuman, 1997).
Deletion of the DNA ligase gene from VACV and Shope fibroma virus had minor effects on replication (Colinas et al., 1990; Kerr and Smith, 1991; Parks et al., 1998), although the sensitivity of the mutant viruses to DNA damaging agents was increased (Kerr et al., 1991; Parks et al., 1998). The viability of the ligase mutant virus could be interpreted as support for an asymmetric DNA replication model, which posits only leading strand DNA synthesis (Moss and De Silva, 2006; Moyer and Graves, 1981). However, the recent discovery of a VACV DNA primase (De Silva et al., 2007; De Silva et al., 2009) has led to renewed interest in a DNA replication model that requires joining of Okazaki fragments on the lagging strand at the replication fork (Esteban and Holowczak, 1977; Olgiati et al., 1976). If the latter model is correct, then another unrecognized viral enzyme or a cellular DNA ligase must participate in DNA replication to compensate for loss of the viral ligase. Utilization of a cellular ligase was considered but evidence for this was not obtained (Kerr et al., 1991). Nevertheless, the availability of new methods, in particular RNA silencing, as well as better reagents encouraged us to reopen the question.
Vertebrates possess three homologous DNA ligases: I, III and IV (abbreviated Lig1, 3 and 4) (Ellenberger and Tomkinson, 2008). Lig1 participates in DNA replication by joining DNA fragments during lagging strand synthesis and also is involved in DNA repair. Lig3 (and its alternately spliced form Lig2) complexes with DNA repair protein XRCC1 to aid in sealing base excision mutations and recombinant fragments. Lig4 complexes with XRCC4 and catalyzes the final step in non-homologous DNA double-strand break repair. The VACV DNA ligase is homologous to the eukaryotic DNA ligases at the DNA binding and catalytic domains with the greatest similarity to Lig3 (Wang et al., 1994).
Here we show that replication of a VACV ligase deletion mutant in proliferating cells depends on cellular Lig1, which is recruited from the nucleus to cytoplasmic viral factories. Replication of ligase deficient VACV was greatly reduced and delayed in resting primary cells, correlating with initial low levels of Lig1 and subsequent viral induction and localization of that enzyme in virus factories. The defect in resting cells could explain the decreased pathogenicity of ligase-deficient VACV in a mouse model (Kerr et al., 1991). The synthesis of a viral ligase could give VACV a head start in replication and contribute to pathogenicity.
RESULTS
Lig1 Contributes to the Replication of DNA Ligase Deficient VACV
We constructed several recombinant VACV. First, we replaced the A50R open reading frame (ORF) encoding DNA ligase with that of enhanced green fluorescent protein (GFP) regulated by a VACV late promoter to form vΔA50gfp. Then, we made additional recombinants by replacing the GFP gene and promoter with an intact A50R ORF to form the revertant vA50Rev or with one containing a stop codon to form vA50Stop. The latter two constructs had the natural promoter upstream of the A50R ORF. The phenotypes of the revertant and stop codon viruses were similar to those of the wild-type parent and the deletion mutant, respectively (Fig. S1A). The mutant viruses replicated in a variety of cell lines with at most a half-log reduction in yield compared to the revertant (Fig. S1A). Similarly, DNA replication of mutant viruses determined by slot blot analysis and real-time PCR was unaffected or modestly reduced (Fig S1B). Overall, these results were consistent with previous reports (Kerr and Smith, 1991; Parks et al., 1998) indicating that ligase-deficient VACV can replicate in proliferating cell lines.
In order to investigate a role for a cellular DNA ligase in supporting replication of DNA ligase deficient VACV, we used siRNAs to Lig1, 3 and 4 mRNAs that had been previously shown to knock-down their expression in human cells (Muylaert and Elias, 2007). Transfection of each siRNA knocked down the corresponding mRNA by 45–70% (data not shown), which resulted in a concomitant target-specific reduction in the ligase protein (Fig. 1A). Next, we determined the effect of knocking-down cellular DNA ligases on replication of VACV. The yield of infectious vΔA50gfp was reduced approximately 1 log by siRNA to Lig1 but not by siRNA to Lig3 or 4 (Fig. 1B, upper left). None of the siRNAs strongly inhibited replication of vA50Rev (Fig. 1B, upper right), indicating that Lig1 was only required in the absence of the viral ligase. Control experiments demonstrated that the reduction in VACV replication caused by Lig1 siRNA could be partially alleviated by transfection of plasmids expressing Lig1 that was resistant to the siRNA because of silent mutations or VACV DNA ligase (Fig. S2).
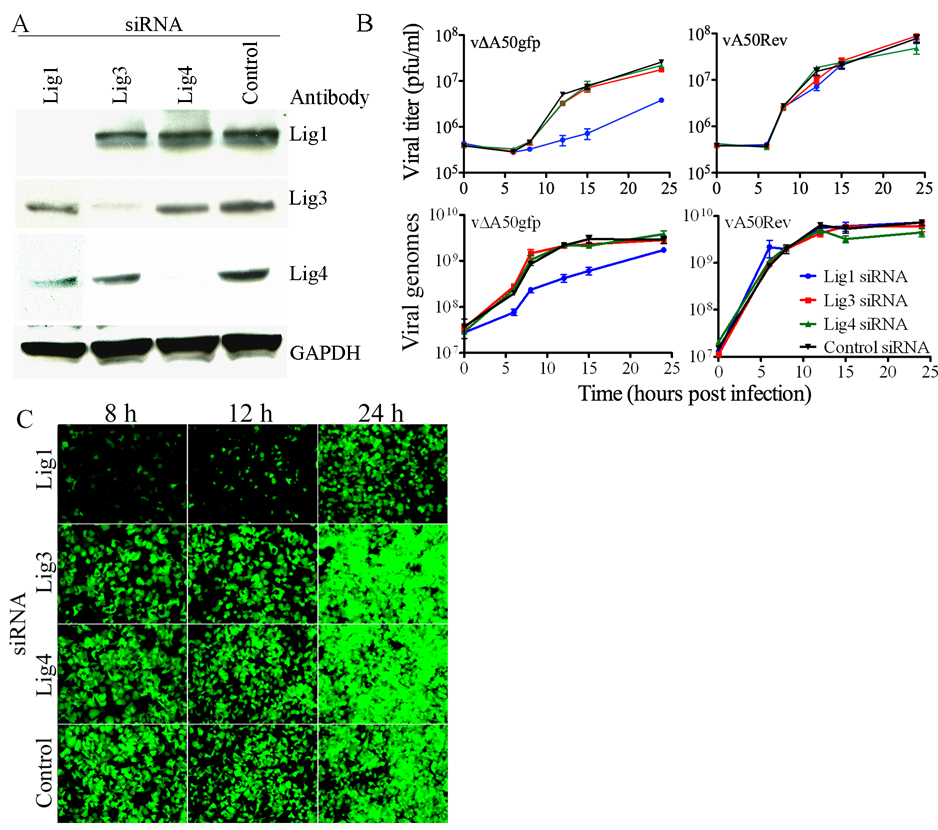
Figure 1. Knock-Down of Cellular Lig1 Inhibits Replication of VACV Ligase Deletion Mutant.
(A) Knock-down of cellular DNA ligases. HeLa cells were transfected with siRNAs to Lig1, 3 or 4 or a control non-targeting RNA. After 72 h, cell lysates were analyzed by Western blotting with antibodies to Lig1, 3, 4 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) used as a loading control. Antibody binding was detected by chemiluminescence. (B) Effects of siRNAs on replication of mutant and wild type VACV. HeLa cells were transfected with siRNAs to Lig1, 3, or 4 or with a control non-targeting siRNA. After 72 h, the cells were infected with 4 PFU per cell of vΔA50gfp or vA50Rev. At indicated times, virus titers were determined by plaque assay (upper panels) or viral DNA was quantified by real-time PCR (lower panels). Experiments were in triplicate and bars represent standard error of the mean. (C) Effect of Lig1 knock-down on late gene expression. HeLa cells were transfected with ligase-specific or control siRNA. After 72 h, the cells were infected with 4 PFU per cell of vΔA50gfp and visualized by fluorescence microscopy at indicated times.
Real-time PCR was used to determine the effect of the siRNAs on viral DNA replication. Inhibition was specific for siRNA to Lig1 and occurred in cells infected with vΔA50gfp (Fig. 1B, lower left) but not with vA50Rev (Fig. 1B, lower right). The inhibition by Lig1 siRNA in cells infected with vΔA50gfp was more severe at early than at late times as viral DNA synthesis continued for 24 h under these conditions but reached a maximum by 12 h and plateaued in the control infected cells. At 13 h after infection, genome size vΔA50gfp DNA was almost undetectable by pulse-field gel electrophoresis and Southern blotting following knockdown of DNA Lig1 but not the other cellular ligases, further substantiating the above results (data not shown).
We took advantage of the presence of GFP regulated by the VACV late p11 promoter in vΔA50gfp to determine the effects of the siRNA on viral late protein synthesis, which is dependent on viral DNA replication. Cells transfected with control siRNA or siRNA to Lig3 or 4 and infected with vΔA50gfp exhibited green fluorescence at 8 h, which increased greatly with time (Fig. 1C). In contrast, few cells that had been transfected with siRNA to Lig1 showed green fluorescence at 8 to 12 h and the fluorescence was markedly less than the control even at 24 h (Fig. 1C). The latter result was consistent with the effect of siRNA to Lig1 on viral DNA replication in cells infected with the A50 deletion mutant.
Inhibitor of Lig1 reduces vΔA50gfp replication
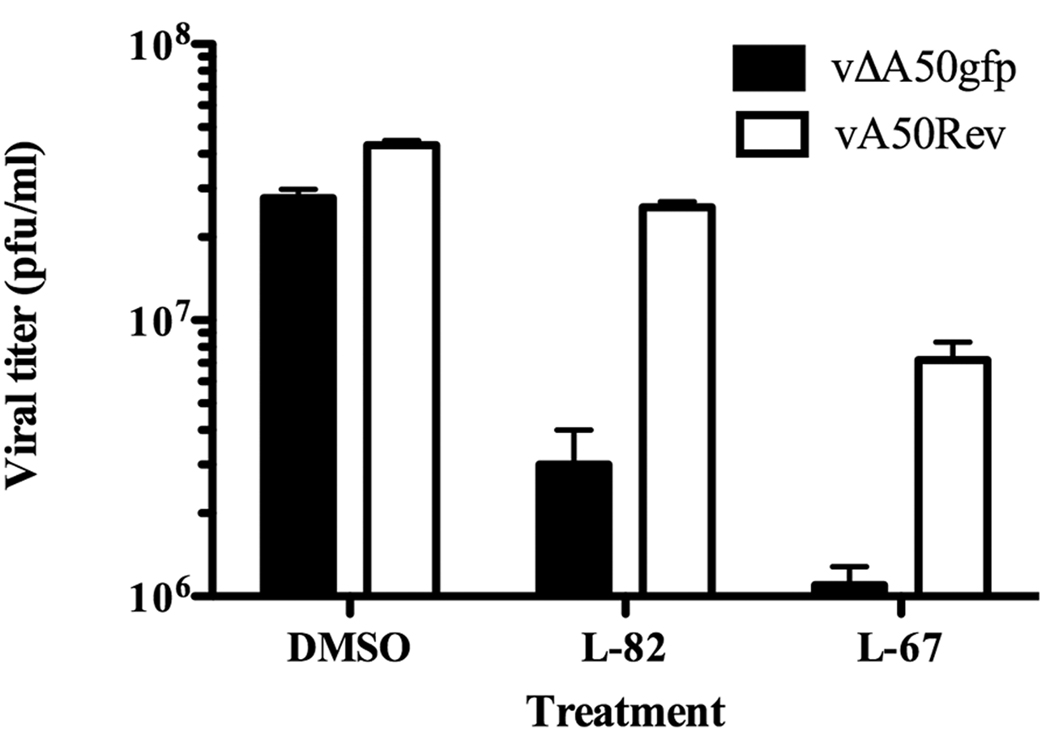
We tested the effects of the small molecule inhibitor L82, specific for Lig1, and L67, which inhibits Lig1 and Lig3 (Chen et al., 2008). Inhibition of Lig1 resulted in about 90% inhibition of replication of vΔA50gfp but had almost no effect on the replication of the revertant virus (Fig. 2). The less specific ligase inhibitor L67, however, reduced the replication of vΔA50gfp and the revertant virus (Fig. 2). Thus, use of the small molecule inhibitor L82 supported a role of Lig1 in compensating loss of the viral ligase.
Figure 2. Effects of Ligase Inhibitors.
HeLa cells were treated with 50 µM L82 or L67 or with dimethylsulfoxide (DMSO) carrier for 48 h and then infected with 4 PFU per cell of vΔA50gfp or vA50Rev. At 24 h after infection, the cells were harvested and the virus titers determined by plaque assay. Bars represent standard errors of the mean.
Replication of VACV in Resting Cells Requires Viral DNA Ligase
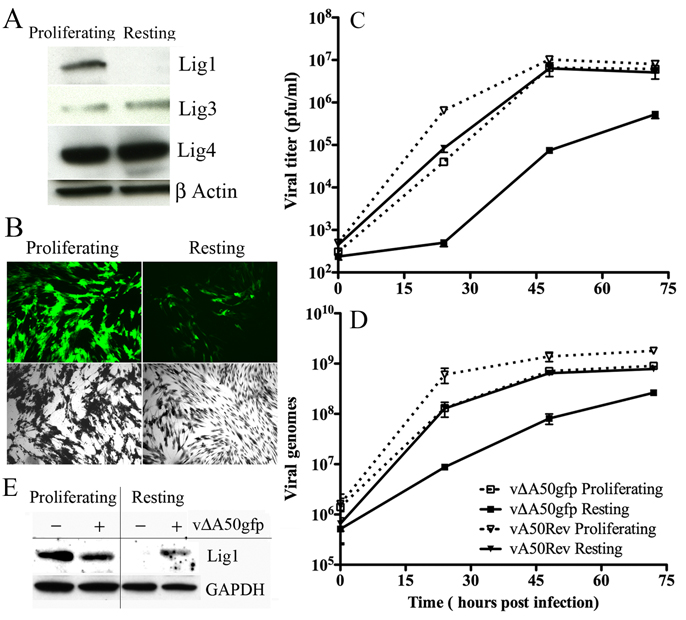
Unlike the situation in the laboratory, where cell lines are actively replicating, cells in vivo are mostly in a resting state. Moreover, Lig1 is the predominant DNA ligase in proliferating cells, whereas the other ligases are predominant in resting cells (Lindahl and Barnes, 1992; Montecucco et al., 1992). Comparison of the level of DNA ligases in proliferating and resting primary human foreskin fibroblasts (HFF) with ligase-specific antibodies confirmed that the level of Lig1 was much lower under the latter conditions, whereas the amounts of Lig3 and 4 were similar (Fig. 3A). Moreover, proliferating HFF supported the replication and spread of vΔA50gfp more highly than resting HFF as indicated by GFP expression (Fig. 3B).
Figure 3. Analysis of Cellular Ligases and Replication of vΔA50gfp Ligase Deletion Mutant in Resting and Proliferating HFF.
(A) Cellular ligases in resting and proliferating cells. HFF were maintained in medium containing 0.2% FBS for four days to induce the resting state or were passaged in medium containing 10% FBS to allow proliferation. The cells were lysed and the proteins analyzed by Western blotting with antibodies to Lig1, 3, or 4 or to β-actin and detected by chemiluminescence. (B) VACV late gene expression. Resting and proliferating HFF were infected with 0.02 PFU per cell of vΔA50gfp. After 48 h, the cells were visualized by fluorescence microscopy (green) and after 72 h by light microscopy after crystal violet staining (black and white). (C, D) Virus replication and DNA synthesis. Resting and proliferating HFF were infected with 0.02 PFU per cell of vΔA50gfp or vA50Rev. At indicated times, virus titers were determined by plaque assay or viral DNA was quantified by real-time PCR. Experiments were in duplicate and bars indicate standard error. (E) Induction of Lig1 in resting cells by VACV. Lig1 and GAPDH were analyzed by Western blotting of extracts from resting and proliferating HFF that were uninfected or infected for 48 h.
A time course analysis further indicated that replication and spread of vΔA50gfp was greatly reduced and delayed in resting HFF: at 24 and 48 h, the virus yields were about 2 logs lower than in proliferating cells, but the difference narrowed to about 1 log at 72 h (Fig. 3C). Furthermore, the yield of vA50Rev was 2 logs higher than vΔA50gfp at 24 h, indicating that the low replication of vΔA50gfp was largely due to the deletion of the viral ligase (Fig. 3C). A similar pattern was seen when viral DNA was measured. DNA synthesis with vΔA50gfp was higher in proliferating cells than in resting cells and lagged behind that of vA50Rev in the latter (Fig. 3D). In addition, the rise in viral DNA synthesis in resting cells infected with vΔA50gfp preceded that of infectious virus (Fig. 3D).
VACV Induces Lig1 in Resting Cells
We considered that the delayed rise in vΔA50gfp replication in resting HFF might be due to induction of Lig1. To check this possibility, resting and proliferating HFF were infected with vΔA50gfp or left uninfected and Lig1 levels were determined after 48 h by Western blotting. Indeed, Lig1 was detected in resting as well as proliferating cells following infection with vΔA50gfp, whereas only the uninfected proliferating cells had detectable Lig1 (Fig. 3E).
Lig1 Is Recruited to the Viral Factory
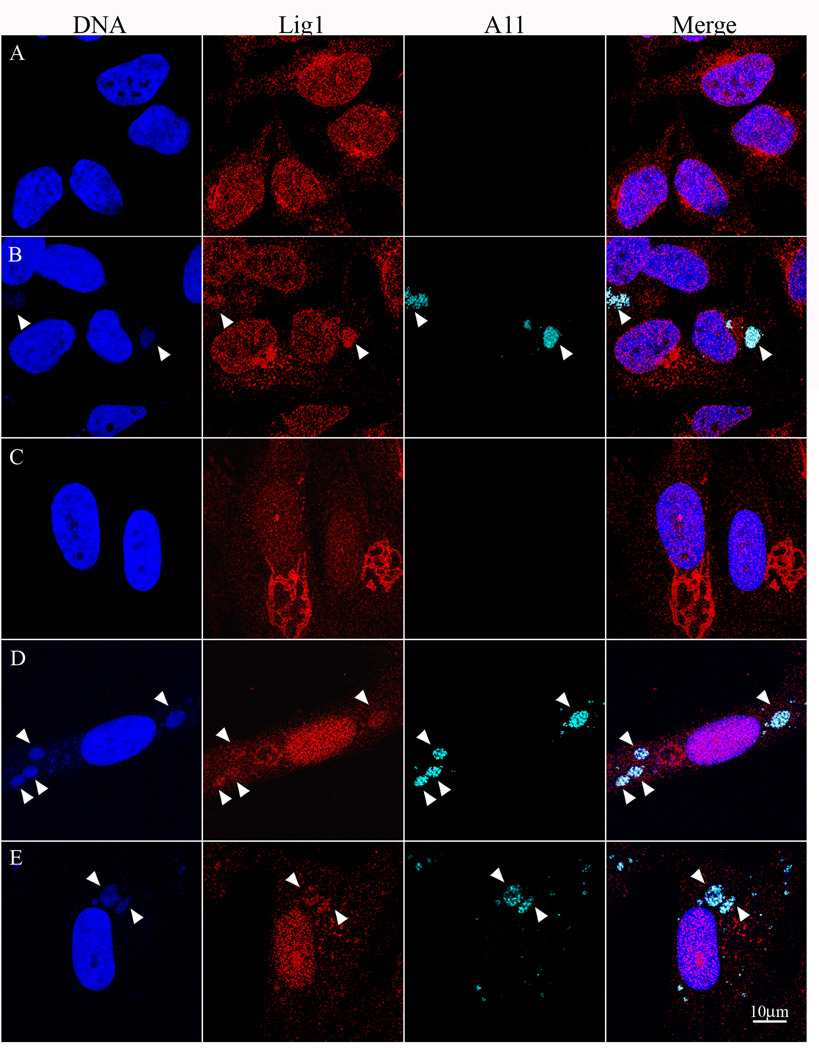
To determine the intracellular location of Lig1, we used the same MAb shown to be specific for Lig1 by Western blotting (Fig. 1A, Fig. 3A) and used previously for nuclear staining of replicating MCF-7 cells (Johansson et al., 2008). In uninfected HeLa cells, the Lig1 MAb showed predominant nuclear staining with some juxtanuclear staining that could represent aggresomes (Fig. 4A). Following low multiplicity infection of HeLa cells with VACV, cytoplasmic factories in infected cells were visualized by staining with DAPI and with rabbit polyclonal antibody to the VACV A11 protein. In addition to nuclear staining, the MAb to Lig1 stained factories in cells infected with vΔA50gfp (Fig. 4B) and vA50Rev (not shown).
Figure 4. Detection of Lig1 in Cytoplasmic VACV Factories.
Uninfected HeLa cells (A) or HeLa cells infected with vΔA50gfp for 12 h (B) were stained with mouse MAb to Lig1 and rabbit polyclonal antibody to the VACV A11 protein followed by secondary fluorescent antibodies and DAPI. Resting HFF were uninfected (C) or infected with vΔA50gfp for 48 h (D) or vA50Rev for 24 h (E) and stained as for panels A and B. Arrowheads point to cytoplasmic factories.
Similar experiments were carried out with HFF. In uninfected proliferating HFF, Lig1 was predominantly nuclear as was seen with HeLa cells (not shown). In the resting state, the stain was more diffusely located over the entire cell and cytoplasmic ribbon-like structures that could represent aggresomes were frequently more prominent than in proliferating HFF (Fig. 4C). At 48 h after infection with vΔA50gfp, Lig1 was induced and associated with cytoplasmic factories as well as the nucleus (Fig. 4D). The aggresome-like staining was less pronounced following virus infection but contained faint DAPI staining that could represent sequestered viral DNA. Similar results were obtained at 24 h, though the factories were smaller and less abundant and the staining of both Lig1 and the A11 were weaker (not shown). Association of Lig1 with factories could also be seen in resting HFF that were infected with vA50Rev (Fig. 4E), suggesting that induction of Lig1 may be important for wild type VACV infections in resting cells.
DISCUSSION
The role of the VACV DNA ligase has remained ambiguous since its discovery 20 years ago (Colinas et al., 1990; Kerr and Smith, 1989; Smith et al., 1989). In most cell lines, DNA ligase deletion mutants replicate as well or nearly as well as wild type virus and the only consistent defect is enhanced sensitivity to DNA damaging agents (Kerr et al., 1991; Parks et al., 1998). If DNA ligation were truly not required, as could be inferred from the above results, it would rule out poxvirus DNA replication mechanisms involving lagging strand synthesis. Here we showed that the apparent dispensability of DNA ligation is due to the ability of a cellular ligase to compensate for loss of the viral enzyme. Functional utilization of a cellular ligase was demonstrated with siRNA, a small molecule inhibitor, and resting cells; association of cellular ligase with viral factories was shown by confocal microscopy.
It had been noted previously that poxvirus ligases are more closely related in sequence to Lig3 than to Lig1 or Lig4 (Wang et al., 1994). However, we found that only knock-down of Lig1 inhibited the replication of a VACV ligase deletion mutant, and that this occurred at the level of viral DNA synthesis. In addition, the specific Lig1 inhibitor L82 (Chen et al., 2008) severely reduced replication of vΔA50gfp. A second inhibitor L-67, which has less specificity (Chen et al., 2008) reduced replication of both ligase deletion mutant and wild type VACV. However, it is not known whether L-67 inhibits the VACV DNA ligase. From a functional point of view it makes sense that Lig1 can substitute for the VACV ligase since Lig1 is mainly involved in DNA replication (Ellenberger and Tomkinson, 2008). With the same set of siRNAs used here, however, Muylaert and Elias (Muylaert and Elias, 2007) found that knock-down of Lig4 but not Lig1 or Lig3 in human cells inhibited herpes simplex virus 1 DNA replication. They suggested that Lig4 is needed for joining to make herpes simplex virus circular DNA templates, leaving open the question of which ligase is involved in lagging strand DNA synthesis in that system.
We confirmed by confocal microscopy the predominant nuclear localization of Lig1 in replicating cells (Johansson et al., 2008; Lasko et al., 1990) and discovered that Lig1 localized in cytoplasmic viral factories in cells infected with a VACV ligase deletion mutant as well as with wild type virus. In uninfected replicating cells, Lig1 localizes to nuclear foci of DNA replication through interaction with proliferating cell nuclear antigen (PCNA) (Montecucco et al., 1998). Both Lig1 and PCNA are part of a large complex with other proteins involved in Okazaki fragment formation and processing (Applegren et al., 1995). Whether PCNA and other nuclear proteins accompany Lig1 to viral factories and whether Lig1 interacts with some viral protein(s) remain to be determined. VACV ligase has been shown to interact with cellular topoisomerase II and recruit the latter to viral factories (Lin et al., 2008). Coincidently, it has been suggested that topoisomerase II also interacts with PCNA (Niimi et al., 2001). A variety of additional cellular proteins including translation initiation and transcription factors have been shown to localize in virus factories (Broyles, 1991; Katsafanas and Moss, 2007; Walsh et al., 2008). Whether the latter proteins simply associate with viral DNA and RNA or have specific viral protein binding partners is not known.
Replication of the VACV ligase deletion mutant was not inhibited or only modestly inhibited in all proliferating cell lines tested. The better replication of ligase-deficient VACV in BS-C-1 cells compared to HeLa cells (Figure S1) correlated with a relatively higher level of Lig1 compared to Lig4 in the former (T.G.S. unpublished) but other contributing factors cannot be ruled out. If Lig1 can compensate for the viral ligase, why does VACV encode its own ligase? An answer came from studies with resting primary cells, which exhibited a specific deficiency in Lig1 but not Lig3 or Lig4. Replication of vΔA50gfp was severely impaired and delayed compared to a control VACV that expressed the viral ligase. Thus, the viral ligase provides an important advantage in resting cells. Interestingly, the delayed onset of replication of ligase-deficient virus corresponded to the induction of Lig1 shown by Western blotting and the localization of Lig1 in cytoplasmic virus factories. The mechanism of Lig1 induction by VACV remains to be determined.
The opposite question to the one posed above, is whether there is an advantage for wild type VACV to recruit the cellular ligase when it encodes its own. While additional ligase may be beneficial, another possibility is that Lig1 brings along an associated cellular protein. The plasticity of DNA ligase usage is underscored by the fact that, although an ATP-dependent ligase was likely encoded by the common ancestor of all eukaryotic nucleocytoplasmic large DNA viruses (Iyer et al., 2001), this gene was lost in several chordopoxviruses (molluscipoxvirus, parapoxviruses, and yatapoxviruses) and replaced by an NAD-dependent ligase in entomopoxviruses.
Our data can explain the previously described decreased pathogenicity of a VACV ligase deletion mutant (Kerr et al., 1991), since the majority of cells in an animal are in the quiescent state and consequently have low levels of Lig1 (Vitolo et al., 2005). The early expression of the viral ligase presumably provides VACV with a head start in DNA replication.
MATERIALS AND METHODS
Cells and Virus
HeLa cells were propagated in Dulbecco’s Modified Eagles medium (D-MEM, Quality Biologicals, Inc. Gaithersburg, MD) containing 10% fetal bovine serum (FBS). HFF were maintained as previously described (De Silva and Moss, 2008) and seeded into 6-well tissue culture plates in D-MEM supplemented with 10% FBS. When monolayers were confluent, culture medium was replaced with fresh medium supplemented with 0.2% FBS and maintained for 96 h to induce the resting state. HFF were infected with virus using 0.2% FBS-supplemented medium. Alternatively, cells were passaged with 10% FBS and as soon as confluent were infected with virus in 2.5% FBS-supplemented media.
Antibodies
Antibodies used were: mouse anti-Lig1 (clone 10H5) (Santa Cruz Biotechnology, Santa Cruz, CA), Mouse anti-Lig3 (Abcam, Cambridge, UK) and rabbit anti-lig4 (Proteintech Group Inc, Chicago, IL), mouse anti-glyceraldehyde-3-phosphate dehydrogenase (Abcam), mouse anti-alpha actin (Santa Cruz Biotechnologies), goat anti-Mouse IgG conjugated to horse radish peroxidase, goat anti-rabbit IgG conjugated to horse radish peroxidase (Santa-Cruz Biotechnologies), rabbit polyclonal anti-VACV A11 (Resch et al., 2005), Alexa Fluor 594 and Alexa Fluor 633 conjugated to anti-IgG of appropriate species (Molecular probes, Eugene OR).
siRNAs
Double-stranded siRNAs specific for Lig1, 3 and 4 [Lig1, 5’-AAGGGCAAGACAGCAGAGGCC; Lig3, 5’-AACUGCAACCCAGAUGAUAUG; Lig4, 5’-AAGCCAGACAAAAGAGGUGAA] with dT dinucleotides added at the 3’ends (Muylaert and Elias, 2007) and control non-targeting siRNA were purchased from Dharmacon (Lafayette, CO).
Recombinant Viruses
Recombinant viruses were constructed as described (Earl et al., 1998a; Earl et al., 1998b) except that DNA made by overlapping PCR (Senkevich et al., 2000) was used for homologous recombination. vΔA50gfp was constructed by replacing the A50R ORF with the GFP ORF controlled by the VACV P11 promoter. DNA containing part of the A49R ORF, the VACV P11 promoter and GFP ORF, and VACV A51R sequences were prepared and assembled into a single A49-p11GFP-A51 PCR product. BS-C-1 cells infected with VACV were transfected with the DNA and plaques containing recombinant viruses were identified by green fluorescence and clonally purified. vA50Rev was constructed by replacing the P11 promoter and GFP of vΔA50gfp with the natural A50R sequence. DNA containing the complete A50R ORF and flanking A49R and A51R sequences was assembled by PCR and transfected into BS-C-1 cells that were infected with vΔA50gfp. Plaques containing recombinant virus were identified by absence of green fluorescence. vA50Stop was made by replacing the P11 promoter and GFP of vΔA50gfp with the A50R ORF with a stop codon located after amino acid 75. Overlapping PCR products with nucleotides 226–228 of the A50R ORF changed from CTA to TGA were used to assemble DNA for transfection. Plaques containing recombinant virus were identified by absence of green fluorescence.
Virus Yield Experiments
HeLa cells at 90% confluency in 12-well plates were transfected with 170 pmol of siRNA using Lipofectamine 2000 (Invitrogen) in triplicate. After 72 h the cells were infected with 0.25 or 4 PFU of virus per cell and maintained at 4°C for 1 h. Inocula were removed, cells washed three times, and overlaid with 2 ml of E-MEM containing 2.5% FBS. The cells were maintained at 37°C, harvested at various times, disrupted by three cycles of freezing and thawing and two 30-s bursts of sonication. Virus yields were determined by titration on BS-C-1 cells.
Real-time PCR
Cells from one well of a 12-well plate were disrupted by three freeze-thaw cycles and two 30-sec sonications. DNA was isolated with the Blood mini kit (Qiagen, Hilden, Germany). Reactions were carried out using SYBR Green PCR master mix (Applied Biosystems), 10 µM of each primer, and 2 ml of DNA in a total volume of 50 µl with the RealPlex sequence detection system and software (Eppendorf, Westbury, NY). DNA was amplified with 40 cycles at 95°C for 15 s and 55° C for 60 s. For determination of genome equivalents, standard curves were made using DNA from virus of known titer assuming 50 particles per PFU.
Western Blot
Lysates from 106 cells were treated with micrococcal nuclease (Worthington, Lakewood, NJ) for 30 min on ice in buffer containing 10 mM NaCl, 10 mM Tris-HCl pH 7.4, 10 mM CaCl2, 0.2% NP40, followed by three 30-sec sonications. Proteins were separated on 4–12% SDS NuPage gel (Invitrogen), and transferred to a nitrocellulose filter. After binding primary and secondary antibodies, SuperSignal West Femto chemiluminescent substrate was used for detection (Pierce Biotechnology, Rockford, IL).
DNA Ligase Inhibitors
The chemical inhibitors L82 and L67 (Chen et al., 2008) were dissolved in dimethylsulfoxide at 50 µM and stored at −80° C. Confluent monolayers of HeLa cells were pre-treated for 48 h with 50 µM L82 or L67 or mock treated with dimethylsulfoxide in DMEM supplemented with 10% FBS. After 48 h the cells were infected with 4 PFU per cell of VACV in E-MEM supplemented with 2.5% FBS and 50 µM inhibitor. The cells were collected at 24 h after infection and the virus titers were determined by plaque assay after at a dilution of at least four logs to reduce the concentration of inhibitor.
Confocal Microscopy
HFF cells on glass coverslips were allowed to replicate or rest by serum starvation (0.2% FBS) for 96 h. HeLa cells were allowed to reach 70% confluence and then infected or transfected with siRNA for 60 h before infection. Cells were fixed with paraformaldehyde, permeabilized with triton X 100 and blocked with phosphate buffered saline (PBS) containing 1% bovine serum albumin for 1 h at room temperature. Primary antibodies were incubated overnight at 4° C; after washing secondary antibody was added for 45 min at room temperature. DAPI was added at a 5 mg/ml for 20 min at room temperature and the cells were washed and mounted with ProLong Gold antifade reagent (Invitrogen). Images were collected on a Leica SP2 AOBS confocal microscope (Leica Microsystems, Bannockburn, IL). Fluorochromes were excited using 405 nm for DAPI, 594 nm for Alexa Fluor 594 and 63 nm for Alexa Fluor 647. Detector slits were configured to minimize crosstalk between the channels.
Supplementary Material
ACKNOWLEDGEMENTS
We thank A. E. Tomkinson, University of Maryland, Baltimore for ligase inhibitors and the following members of NIAID: A. McBride for HFF cells, C. Cotter for cell-cultures and M. Gastinger, S. Becker and L. Koo for help with confocal microscopy imaging. Research was supported by the Division of Intramural Research, NIAID, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Applegren N, Hickey RJ, Kleinschmidt AM, Zhou Q, Coll J, Wills P, Swaby R, Wei Y, Quan JY, Lee MY, et al. Further characterization of the human cell multiprotein DNA replication complex. J. Cell. Biochem. 1995;59:91–107. doi: 10.1002/jcb.240590111. [DOI] [PubMed] [Google Scholar]
- Broyles SS. A role for ATP hydrolysis in vaccinia virus early gene transcription. J. Biol. Chem. 1991;266:15545–15548. [PubMed] [Google Scholar]
- Chen X, Zhong S, Zhu X, Dziegielewska B, Ellenberger T, Wilson GM, MacKerell AD, Jr, Tomkinson AE. Rational design of human DNA ligase inhibitors that target cellular DNA replication and repair. Cancer Res. 2008;68:3169–3177. doi: 10.1158/0008-5472.CAN-07-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colinas RJ, Goebel SJ, Davis SW, Johnson GP, Norton EK, Paoletti E. A DNA ligase gene in the Copenhagen strain of vaccinia virus is nonessential for viral replication and recombination. Virology. 1990;179:267–275. doi: 10.1016/0042-6822(90)90295-3. [DOI] [PubMed] [Google Scholar]
- De Silva FS, Lewis W, Berglund P, Koonin EV, Moss B. Poxvirus DNA primase. Proc. Natl. Acad. Sci. USA. 2007;104:18724–18729. doi: 10.1073/pnas.0709276104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva FS, Moss B. Effects of vaccinia virus uracil DNA glycosylase catalytic site and deoxyuridine triphosphatase deletion mutations individually and together on replication in active and quiescent cells and pathogenesis in mice. Virol. J. 2008;5:145. doi: 10.1186/1743-422X-5-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva FS, Paran N, Moss B. Products and substrate/template usage of vaccinia virus DNA primase. Virology. 2009;383:136–141. doi: 10.1016/j.virol.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl PL, Cooper N, Wyatt LS, Moss B, Carroll MW. Preparation of cell cultures and vaccinia virus stocks. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York: John Wiley and Sons; 1998a. pp. 16.16.11–16.16.13. [Google Scholar]
- Earl PL, Moss B, Wyatt LS, Carroll MW. Generation of recombinant vaccinia viruses. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York: Greene Publishing Associates & Wiley Interscience; 1998b. pp. 16.17.11–16.17.19. [Google Scholar]
- Ellenberger T, Tomkinson AE. Eukaryotic DNA ligases: structural and functional insights. Annu. Rev. Biochem. 2008;77:313–338. doi: 10.1146/annurev.biochem.77.061306.123941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban M, Holowczak JA. Replication of vaccinia DNA in mouse L cells. I. In vivo DNA synthesis. Virology. 1977;78:57–75. doi: 10.1016/0042-6822(77)90078-2. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Aravind L, Koonin EV. Common origin of four diverse families of large eukaryotic DNA viruses. J. Virol. 2001;75:11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson VM, Miniotis MF, Hegardt C, Jonsson G, Staaf J, Berntsson PS, Oredsson SM, Alm K. Effect of polyamine deficiency on proteins involved in Okazaki fragment maturation. Cell Biol. Int. 2008;32:1467–1477. doi: 10.1016/j.cellbi.2008.08.018. [DOI] [PubMed] [Google Scholar]
- Katsafanas GC, Moss B. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host & Microbe. 2007;2:221–228. doi: 10.1016/j.chom.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr SM, Johnston LH, Odell M, Duncan SA, Law KM, Smith GL. Vaccinia DNA ligase complements Saccharomyces cerevisiae Cdc9, localizes in cytoplasmic factories and affects virulence and virus sensitivity to DNA damaging agents. EMBO J. 1991;10:4343–4350. doi: 10.1002/j.1460-2075.1991.tb05012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr SM, Smith GL. Vaccinia virus encodes a polypeptide with DNA ligase activity. Nucleic Acids Res. 1989;17:9039–9050. doi: 10.1093/nar/17.22.9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr SM, Smith GL. Vaccinia virus DNA ligase is nonessential for virus replication: recovery of plasmids from virus-infected cells. Virology. 1991;180:625–632. doi: 10.1016/0042-6822(91)90076-n. [DOI] [PubMed] [Google Scholar]
- Lasko DD, Tomkinson AE, Lindahl T. Mammalian DNA ligases. Biosynthesis and intracellular localization of DNA ligase I. J. Biol. Chem. 1990;265:12618–12622. [PubMed] [Google Scholar]
- Lin YC, Li J, Irwin CR, Jenkins H, DeLange L, Evans DH. Vaccinia virus DNA ligase recruits cellular topoisomerase II to sites of viral replication and assembly. J. Virol. 2008;82:5922–5932. doi: 10.1128/JVI.02723-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T, Barnes DE. Mammalian DNA ligases. Annu. Rev. Biochem. 1992;61:251–281. doi: 10.1146/annurev.bi.61.070192.001343. [DOI] [PubMed] [Google Scholar]
- Montecucco A, Biamonti G, Savini E, Focher F, Spadari S, Ciarrocchi G. DNA ligase I gene expression during differentiation and cell proliferation. Nucleic Acids Res. 1992;20:6209–6214. doi: 10.1093/nar/20.23.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecucco A, Rossi R, Levin DS, Gary R, Park MS, Motycka TA, Ciarrocchi G, Villa A, Biamonti G, Tomkinson AE. DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigen: identification of a common targeting mechanism for the assembly of replication factories. EMBO J. 1998;17:3786–3795. doi: 10.1093/emboj/17.13.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B. Poxviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2905–2946. [Google Scholar]
- Moss B, De Silva F. Poxvirus DNA replication and human disease. In: DePamphilis ML, editor. DNA Replication & Human Disease. Cold Spring Hrbor: Cold Spring Harbor Laboratory Press; 2006. pp. 707–727. [Google Scholar]
- Moyer RW, Graves RL. The mechanism of cytoplasmic orthopoxvirus DNA replication. Cell. 1981;27:391–401. doi: 10.1016/0092-8674(81)90422-0. [DOI] [PubMed] [Google Scholar]
- Muylaert I, Elias P. Knockdown of DNA ligase IV/XRCC4 by RNA interference inhibits herpes simplex virus type I DNA replication. J. Biol. Chem. 2007;282:10865–10872. doi: 10.1074/jbc.M611834200. [DOI] [PubMed] [Google Scholar]
- Niimi A, Suka N, Harata M, Kikuchi A, Mizuno S. Co-localization of chicken DNA topoisomerase IIalpha, but not beta, with sites of DNA replication and possible involvement of a C-terminal region of alpha through its binding to PCNA. Chromosoma. 2001;110:102–114. doi: 10.1007/s004120100140. [DOI] [PubMed] [Google Scholar]
- Olgiati DD, Pogo BG, Dales S. Evidence for RNA linked to nascent DNA in HeLa cells. J. Cell Biol. 1976;68:557–566. doi: 10.1083/jcb.68.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks RJ, Winchcombe-Forhan C, DeLange AM, Xing X, Evans DH. DNA ligase gene disruptions can depress viral growth and replication in poxvirus-infected cells. Virus Res. 1998;56:135–147. doi: 10.1016/s0168-1702(98)00055-0. [DOI] [PubMed] [Google Scholar]
- Sekiguchi J, Shuman s. Nick sensing by vaccinia virus DNA ligase requires a 5' phosphate at the nick and occupancy of the adenylate binding site on the enzyme. J. Virol. 21997;71:9679–9684. doi: 10.1128/jvi.71.12.9679-9684.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senkevich TG, Weisberg A, Moss B. Vaccinia virus E10R protein is associated with the membranes of intracellular mature virions and has a role in morphogenesis. Virology. 2000;278:244–252. doi: 10.1006/viro.2000.0656. [DOI] [PubMed] [Google Scholar]
- Smith GL, Chan YS, Kerr SM. Transcriptional mapping and nucleotide sequence of a vaccinia virus gene encoding a polypeptide with extensive homology to DNA ligases. Nucleic Acids Res. 1989;17:9051–9062. doi: 10.1093/nar/17.22.9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitolo B, Lidonnici MR, Montecucco C, Montecucco A. A new monoclonal antibody against DNA ligase I is a suitable marker of cell proliferation in cultured cell and tissue section samples. Eur. J. Histochem. 2005;49:349–354. doi: 10.4081/962. [DOI] [PubMed] [Google Scholar]
- Walsh D, Arias C, Perez C, Halladin D, Escandon M, Ueda T, Watanabe-Fukunaga R, Fukunaga R, Mohr I. Eukaryotic translation initiation factor 4F architectural alterations accompany translation initiation factor redistribution in poxvirus-infected cells. Mol. Cell. Biol. 2008;28:2648–2658. doi: 10.1128/MCB.01631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YC, Burkhart WA, Mackey ZB, Moyer MB, Ramos W, Husain I, Chen J, Besterman JM, Tomkinson AE. Mammalian DNA ligase II is highly homologous with vaccinia DNA ligase. Identification of the DNA ligase II active site for enzyme-adenylate formation. J. Biol. Chem. 1994;269:31923–31928. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.