Summary
Genetic analyses previously implicated the matrix-localized protease ClpP in signaling the stress of protein misfolding in the mitochondrial matrix to activate nuclear encoded mitochondrial chaperone genes in C. elegans (UPRmt). Here we report that haf-1, a gene encoding a mitochondria-localized ATP-binding cassette protein, is required for signaling within the UPRmt and for coping with misfolded protein stress. Peptide efflux from isolated mitochondria was ATP-dependent and required HAF-1 and the protease ClpP. Defective UPRmt signaling in the haf-1 deleted worms was associated with failure of the bZIP protein, ZC376.7, to localize to nuclei in worms with perturbed mitochondrial protein folding, whereas zc376.7(RNAi) strongly inhibited the UPRmt. These observations suggest a simple model whereby perturbation of the protein-folding environment in the mitochondrial matrix promotes ClpP-mediated generation of peptides whose haf-1-dependent export from the matrix contributes to UPRmt signaling across the mitochondrial inner membrane.
Keywords: chaperones, unfolded proteins, heat shock, inter-organellar signaling, peptide transport
Introduction
Protein folding homeostasis in the various compartments of the eukaryotic cell is maintained by chaperones and enzymes that process and degrade proteins (Hartl and Hayer-Hartl, 2002; Bukau et al., 2006). Distinct signal transduction pathways, known as unfolded protein responses (UPRs), have evolved to couple the unfolded/misfolded protein load in the various compartments to the expression of the chaperones and enzymes that maintain protein folding homeostasis in that compartment. Major advances have been made in studies of the cytosolic and endoplasmic reticulum UPR (reviewed in: Kaufman, 2002; Barral et al., 2004), but we have only a rudimentary understanding of the mechanisms that maintain appropriate levels of chaperones in the mitochondrial matrix (Broadley and Hartl, 2008).
The mitochondrial matrix is burdened with unfolded proteins from two sources: local synthesis of mitochondrially-encoded proteins and import of nuclear-encoded precursors (Hartl and Neupert, 1990). A complement of nuclear-encoded mitochondrial chaperones, exemplified by mtHsp70/DnaK and Hsp60/GroE, assist in import, folding and multi-protein complex assembly in the matrix and on the matrix side of the inner-mitochondrial membrane. The balance between chaperones and clients is readily perturbed by over-expression of difficult-to-fold matrix proteins or by knock-down of components of the protein-handling machinery of the matrix (e.g. the protease SPG7), and these manipulations selectively activate the nuclear encoded mitochondrial chaperone genes via a mitochondrial UPR (UPRmt) (Zhao et al., 2002; Yoneda et al., 2004).
C. elegans with transcriptional reporter transgenes of mitochondrial Hsp60 (hsp-60pr∷gfp) and Hsp70 (hsp-6pr∷gfp) have been applied to the genome-wide analysis of the UPRmt. These studies have identified two genes, dve-1, encoding a DNA-binding protein, and ubl-5, encoding a small nuclear ubiquitin-like protein, whose loss interferes with signaling in the UPRmt (Benedetti et al., 2006; Haynes et al., 2007). In addition to these two genes, which likely affect downstream nuclear events, the screen also identified clpp-1, a gene encoding the mitochondrial matrix protease, ClpP, as playing an essential role in signaling the UPRmt.
Bacterial and mitochondrial ClpPs are chambered proteases that degrade soluble proteins in an ATP-dependent manner in conjunction with a partner AAA+ ATPase (Kang et al., 2002; Gottesman, 2003). The substrates of mitochondrial ClpP are not known, but the bacterial protease has a wide range of substrates whose degradation is enhanced under conditions that promote protein misfolding (Kruger et al., 2000; Flynn et al., 2003; Kock et al., 2004). Injection of worms with an inhibitor of ClpP's proteolytic activity acutely interfered with UPRmt signaling; that observation, and the localization of ClpP to the mitochondrial matrix, suggest a role for ClpP-mediated proteolysis in signaling a proximal step in the UPRmt (Haynes et al., 2007).
Bacterial ClpP degrades proteins to small peptides that are subsequently processed to amino acids via several cytoplasmic peptidases (Yu and Houry, 2007). However, Langer and colleagues discovered that peptides derived from protein degradation in the mitochondrial matrix of S. cerevisiae are actively extruded to the inter-membrane space and from there they diffuse to the cytosol (Young et al., 2001), and they went on to hypothesize that peptide efflux may be important for yet-to-be-determined signal transduction in yeast (Arnold et al., 2006). Mdl1p, the yeast mitochondrial peptide transporter is similar to the Transporters associated with Antigen Presentation (TAP transporters) that transfer peptides from the cytosol into the endoplasmic reticulum (ER) lumen for assembly on MHC-I complexes and antigen presentation on the plasma membrane of mammalian cells. Interestingly, mammals possess at least two TAP homologs, ABCB8 (ABC-me) and ABCB10 that are predicted to reside in the inner-mitochondrial membrane. Furthermore, the presentation of peptides derived from proteins that normally reside in the mitochondrial matrix on MHC-I complexes suggest that trafficking of peptides from the mitochondrial matrix to the cytosol (and from there to the ER) is a feature conserved from yeast to mammals (reviewed in Herget and Tampe, 2007). Therefore, we set out to test if ClpP-dependent proteolysis of matrix proteins, coupled with peptide efflux across the inner mitochondrial membrane, participates in UPRmt signaling.
Results
The ATP binding cassette transporter HAF-1 is required for UPRmt signaling
Both Mdl1p and TAP are ABC transporters with a transmembrane region and a single ATP-binding cassette (Sheps et al., 2004). The C. elegans genome encodes sixty predicted ABC transporters, nine of which bear similarity to S. cerevisiae Mdl1p and mammalian TAP (Supplementary Figure 1A). These nine genes were individually inactivated either by RNAi or homozygous deletion (when possible) and the impact on UPRmt induction was tested. Inactivation of only one gene (that was missed in the original RNAi screen), the most evolutionary similar to MDL1, haf-1, significantly affected the induction of the UPRmt reporter hsp-60pr∷gfp by mitochondrial stress. In two models of mitochondrial stress, a temperature-sensitive allele known to cause mitochondrial stress, zc32 (Benedetti et al., 2006), and feeding of spg-7(RNAi), GFP fluorescence increased over the course of post-embryonic development. The increase in fluorescence and GFP levels was attenuated in haf-1 (ok705) deletion animals (Figure 1A and 1B). Inhibition of the UPRmt reporter was mirrored by the effects of haf-1 (ok705) and haf-1 (tm843) deletions on the expression of the endogenous mitochondrial chaperone gene hsp-60, as determined by QRT-PCR (Figures 1C). Similar results were obtained for the mitochondrial HSP70 gene, hsp-6 (Supplementary figure 1C and 1D). Specificity for the UPRmt was revealed by the observation that induction of the UPRER was unaffected by haf-1 deletion (Figure 1D).
Figure 1. Impaired UPRmt in haf-1 mutant animals.
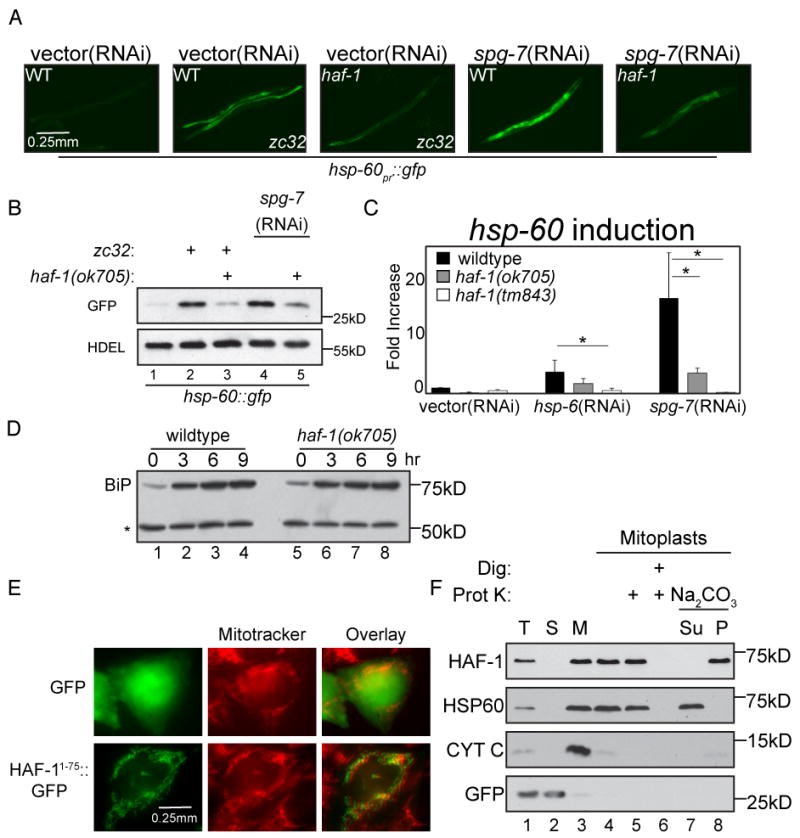
(A) Representative fluorescent photomicrographs of hsp-60pr∷gfp transgenic worms (reporting on the UPRmt) with a temperature sensitive mutation (zc32) that activates the UPRmt in an otherwise wildtype or haf-1(ok705) deleted background. Where indicated wildtype or haf-1(ok705) worms were raised on spg-7(RNAi) for 3 days to induce mitochondrial misfolded protein stress.
(B) A corresponding immunoblot of GFP expressed by hsp-60pr∷gfp transgenic worms in whom mitochondrial unfolded protein stress was induced (as in “A”). The endogenous ∼55kDa ER protein, detected with an anti-HDEL monoclonal antibody (lower panel) serves as a loading control.
(C) Quantitative analysis (by QRT-PCR) of endogenous hsp-60 mRNA in wildtype or two different haf-1 deleted strains (ok705 and tm843) subjected to mitochondrial unfolded protein stress by hsp-6(RNAi) or spg-7(RNAi). Displayed is the mean +/- SEM, n=3, *p<0.05.
(D) Immunoblot of proteins reactive with an anti-HDEL monoclonal antibody in extracts of wildtype and haf-1 mutant worms in which ER stress was induced by exposure to elevated temperature (30°C) for the indicated time. The upper band, BiP (HSP-4), is a UPRER target gene whereas the lower invariant ∼55Kd band (*) serves as a loading control.
(E) Fluorescent photomicrographs of Chinese Hamster Ovary (CHO) cells expressing GFP (upper panels) or GFP fused to amino acids 1-75 of HAF-1 (HAF-11-75∷GFP, lower panels). The cells were co-stained with the vital dye Mitotracker, which stains mitochondria (middle panels).
(F) Immunoblot of extracts from HEK293T cells expressing GFP (as a cytosolic marker) and C-terminally-tagged HAF-1∷FLAG following cellular fractionation into total lysate (T), post-mitochondrial supernatant (S) and mitochondrial pellet (M). Lanes 4-8 are from mitochondria treated with hypotonic buffer to generate mitoplasts and further treated with digitonin and proteinase K where indicated. Lanes 7-8 are mitoplasts incubated in Na2CO3 followed by centrifugation at 150,000 * g and separated into the pellet (P) and supernatant (Su).
Members of the HAF family of ABC transporters (HAF-2 and HAF-6) are important in spreading of RNAi effects in C. elegans (Sundaram et al., 2006). Therefore, to exclude the trivial possibility that HAF-1 was altering the penetrance of the RNAi procedure, we tested the effect of the haf-1 mutation on the phenotype imposed by a weak RNAi. Whereas the positive control, eri-1 (mg366), markedly sensitized worms to dpy-13(RNAi), haf-1 deletion had no effect on the sensitivity of worms to the RNAi feeding procedure (Supplementary Figure 1E).
HAF-1 is a 677 amino acid protein with a putative N-terminal mitochondrial import signal, a transmembrane domain, predicted to span the membrane 4 times and a single ATP binding cassette. The haf-1(ok705) allele bears a 1273 base pair deletion encompassing exons 6-9 removing the majority of the ATP binding cassette while the haf-1(tm843) allele bears a 739 base pair deletion of exons 4-6, removing most of the transmembrane domains of HAF-1 (Supplementary Figure 1B). Thus deletion of either functional domain of HAF-1 results in a similar impairment of the UPRmt.
To function as an ABC transporter, HAF-1 is predicted to dimerize, either with itself or a homologous ABC transporter (Sheps et al., 2004). However, inactivation of haf-1's closest homologue haf-3 (Supplementary Figure 1A), did not affect UPRmt signaling. Furthermore, haf-3 deletion did not further compromise UPRmt signaling in haf-1 mutants or cause any detectable synthetic phenotypes (data not shown). The inability to implicate other ABC transporters in UPRmt signaling suggests that HAF-1 forms homodimers that possess some non-redundant function in UPRmt signaling.
HAF-1's N-terminal 66 amino acids are predicted to encode a mitochondrial targeting sequence (0.99 probability of mitochondrial localization, calculated by MITOPROT, Claros and Vincens, 1996). As we were unable to establish transgenic C. elegans lines expressing tagged HAF-1, to examine the predicted sub-cellular localization of the protein we fused the N-terminal 75 amino acids of C. elegans HAF-1 to GFP and expressed it in cultured Chinese Hamster Ovary cells. GFP lacking the HAF-1 targeting sequence distributed diffusely throughout the cytoplasm and nucleus as expected, however HAF-11-75∷GFP localized in serpentine cytoplasmic structures that were co-stained by the fluorescent dye Mitotracker (Figure 1E). The localization of HAF-1 to mitochondria was further supported by tracking the distribution of HAF-1 tagged at the C-terminus with a FLAG epitope (HAF-1∷FLAG) in cellular sub-fractionation experiments. HAF-1∷FLAG remained associated with the mitochondrial pellet fraction following mitoplast formation (which leads to loss of cytochrome C, an inter-membrane space marker) and was found in the membrane pellet following extraction of soluble proteins (e.g. HSP60) with Na2CO3. In mitoplasts HAF-1∷FLAG was substantially protected from proteinase K digestion, consistent with a topology whereby the FLAG epitope, and thus the ATP binding cassette, resides on the matrix side of the inner membrane (Figure 1F). These observations suggest that HAF-1's role in the UPRmt is played out at the inner-membrane of the stressed organelle and involves transport of cargo from the matrix.
Loss of haf-1 sensitizes worms to conditions that perturb mitochondrial protein folding
Apart from a slightly impaired growth rate, haf-1 mutants are indistinguishable from wildtype worms under normal laboratory conditions. However, development of haf-1 mutants was delayed when mitochondrial protein folding was perturbed either by spg-7(RNAi) or by hsp-60(RNAi), whereas conditions that perturb ER protein folding had no deleterious effects on development of haf-1 mutants (Figure 2A). Decreased expression of the mitochondrial chaperone HSP-6, in the adult worm, has been shown to decrease lifespan (Kimura et al., 2007) as well as activate the UPRmt (Yoneda et al., 2004; Benedetti et al., 2006). Compared with the wildtype, worms lacking haf-1 experienced further decrease in lifespan when exposed to hsp-6(RNAi) (Figure 2B). These observations are consistent with a role for haf-1 in defending protein folding homeostasis in mitochondria.
Figure 2. haf-1 is required for development and survival during mitochondrial stress.
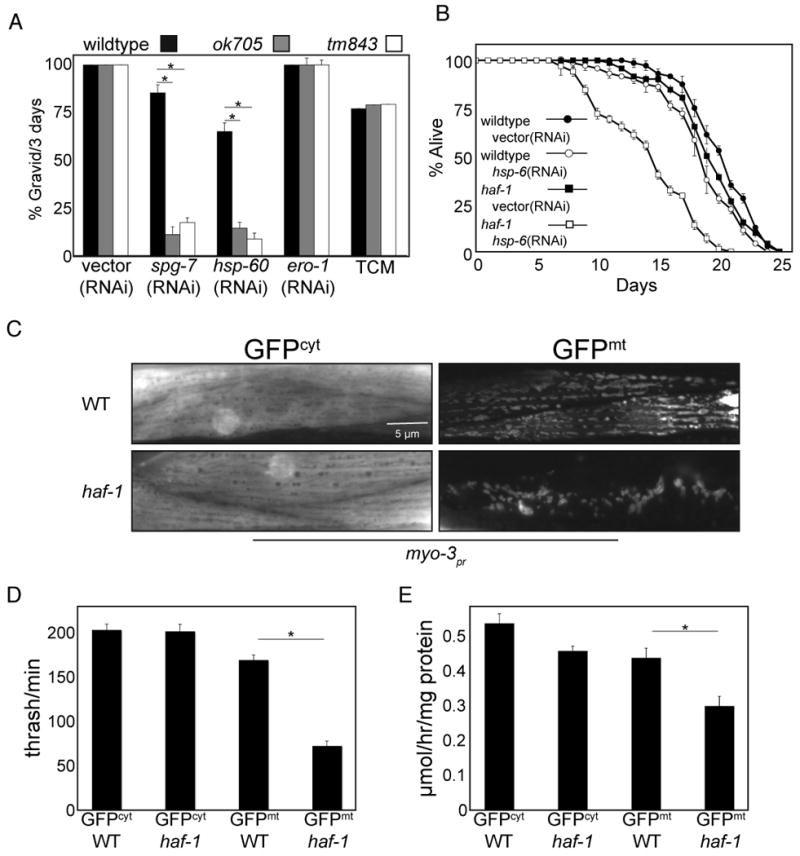
(A) Percent of animals that developed to gravid adults within 72 hours of hatching from wildtype, haf-1(ok705) and haf-1(tm843) eggs. The larva were exposed to bacteria expressing spg-7(RNAi) or hsp-60(RNAi) to induce mitochondrial stress or ero-1(RNAi) or tunicamycin (2 μg/ml) to induce ER stress. Shown is the mean ± SEM, n=3, *p<0.05.
(B) Survival of wildtype and haf-1(ok705) L4 worms on vector(RNAi) or hsp-6(RNAi) plates. The worms were raised to young adults on vector(RNAi) prior to the transfer to the indicated plate after 3 days. Shown is the mean ± S.E.M of fraction of survivors assessed daily (n=3).
(C) Representative fluorescent photomicrographs of body wall muscle of wildtype or haf-1(ok705) worms expressing either cytosolic (GFPcyt) or mitochondrial GFP (GFPmt), driven by the myo-3 promoter.
(D) Plot of the number of body strokes per minute (thrashing assay) of wildtype or haf-1(ok705) animals expressing either cytosolic (GFPcyt) or mitochondrial GFP (GFPmt) driven by the myo-3 promoter. Shown is the mean ± S.E.M obtained by counting strokes per minute of 3 day-old animals (n=5, *p<0.05).
(E) Oxygen consumption of wildtype or haf-1(ok705) animals expressing either cytosolic (GFPcyt) or mitochondrial GFP (GFPmt) driven by the myo-3 promoter. Shown is the mean ± S.E.M oxygen consumption normalized to protein content (n=3, *p<0.05).
Elevated temperature perturbs the protein-folding environment in the mitochondria and activates the UPRmt (Haynes et al., 2007). Compared with wildtype, haf-1 deleted worms displayed markedly impaired survival at the elevated temperature of 30°C (Supplementary Figure 2A). By contrast haf-1 deleted worms were indistinguishable from wildtype in their survival under reductive stress imposed by dithiothreitol (DTT), which promotes ER stress and activates the UPRER (Supplementary Figure 2B). To determine if the hypersensitivity of haf-1 deleted worms to elevated temperature correlates with a mitochondrial defect, we compared the rate of oxygen consumption by wildtype and mutant worms at physiological and elevated temperature. At normal temperature (20°C) haf-1 deleted worms consumed slightly (10%) less oxygen than wildtype worms, however following 24 hours of exposure to the elevated temperature of 30°C (a time point preceding the onset of mortality in the mutant) the oxygen consumption by the mutant worms fell to 45% of that of the wildtype. By contrast, when exposed to the ER stress-inducing agent, DTT, only minor difference in oxygen consumption were noted between wildtype and haf-1 mutant worms (Supplementary Figure 2C).
In addition to causing mitochondrial misfolded protein stress, elevated temperature, spg-7(RNAi), hsp-60(RNAi) and hsp-6(RNAi) broadly perturb worm physiology. To more directly assess mitochondrial protein homeostasis in haf-1 mutant worms we introduced GFP (a protein known to require chaperones for efficient folding, Wang et al., 2002), into the mitochondrial matrix or, as a control, into the cytoplasm of muscle cells. GFP with a mitochondrial targeting sequence was expressed from the muscle-specific myo-3 promoter, which is active from embryogenesis through adulthood. In wildtype worms, muscle mitochondria containing GFPmt formed an ordered array, neatly aligned with the muscle fibers, as described (Labrousse et al., 1999). By contrast, the mitochondria of myo-3∷GFPmt animals lacking haf-1 were severely deformed (Figure 2C).
To characterize the physiological consequences of GFPmt expression in the haf-1 deletion background, we measured the rates of thrashing and oxygen consumption, readouts for muscle activity (Miller et al., 1996). Thrashing rates were indistinguishable in wildtype and haf-1(ok705) mutant worms expressing GFPcyt, and were only modestly reduced by the expression of GFPmt in wildtype worms. However, compounding the expression of (the difficult to fold) GFPmt with the haf-1(ok705) mutation reduced thrashing rates by 50% (Figure 2D). A similar trend was also observed for oxygen consumption (Figure 2E).
HAF-1 -dependent peptide efflux from mitochondria
The observations described above are consistent with a role for HAF-1 in promoting protein folding homeostasis in mitochondria. To determine if they correlate with peptide export from mitochondria, we sought to measure HAF-1's role in that process. An in vitro system that reports on proteolysis in rat mitochondria (Desautels and Goldberg, 1982) was adapted to C. elegans. Mitochondria were isolated from wildtype and haf-1 mutant worms by differential sedimentation (Jonassen et al., 2002) and further purified by centrifugation through an Optiprep cushion to remove the lighter membrane fractions that include the ER (Figure 3A), and trypsin digestion to further remove ER fragments associated with mitochondria (Forner et al., 2006).
Figure 3. ATP-dependent degradation of proteins in mitochondria isolated from wildtype and haf-1 mutant worms.
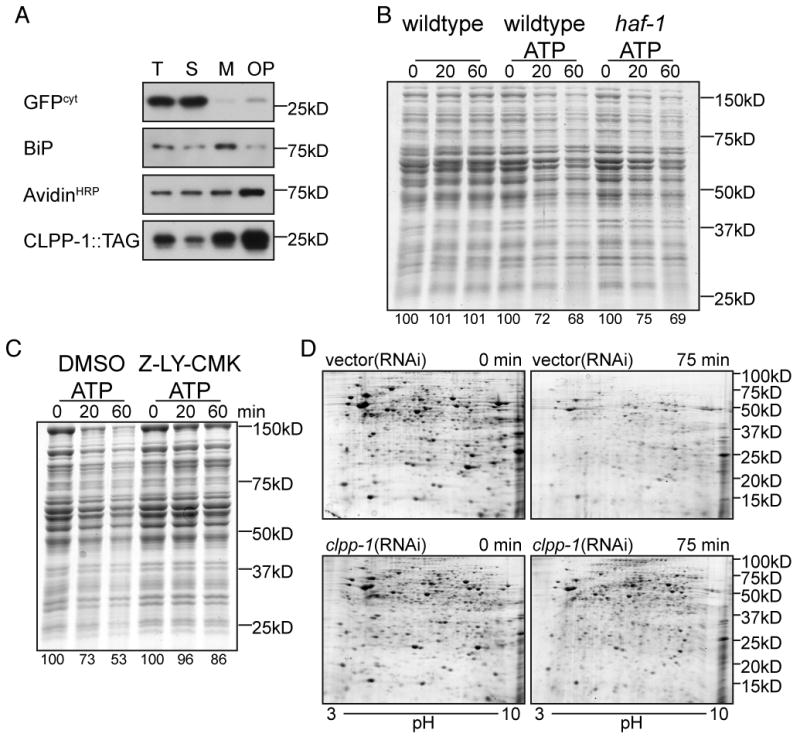
(A) Immunoblot of SDS-soluble proteins following fractionation of disrupted worms into total lysate (T), post-mitochondrial supernatant (S), mitochondrial pellet (M) and mitochondrial pellet further purified by centrifugation through 13% Optiprep (OP). The anti-GFP immunoblot of myo-3pr∷GFPcyt serves as a cytosolic marker (GFPcyt) whereas CLPP-1∷TAG (detected by a C-terminal Myc tag) and the avidinHRP ligand blot (that detects PCCa) serve as mitochondrial markers while the anti-HDEL immunoblot that detects worm BiP serves as an ER marker.
(B) Coomassie stained gel of SDS-solubilized proteins extracted from mitochondria purified from wildtype or haf-1(ok705) mutant animals following incubation in vitro for the indicated time at 30°C without or with ATP (3mM). The intensity of the protein stain, integrated across the entire lane is expressed as a percentage of the intensity at t=0.
(C) Where indicated, the ClpP inhibitor, Z-LY-CMK (10μM), or the carrier DMSO (0.3%, a control) were added to the suspension of wildtype mitochondria during the incubation at 30°C with ATP (3mM) (as in “B”, above).
(D) Coomassie blue stained 2D gel (pH: 3-10) gel of urea/SDS-solubilized proteins extracted from mitochondria from wildtype worms raised on vector(RNAi) or clpp-1(RNAi) following incubation in vitro for the indicated time at 30°C with ATP. The samples for the two-dimensional PAGE were prepared as described (Venkatraman et al., 2004).
Mitochondria isolated by this procedure were incubated at 30°C, to perturb protein folding. SDS-PAGE analysis of mitochondrial proteins showed little change when ATP was omitted from the incubation buffer, as expected (Desautels and Goldberg, 1982). However, in mitochondria procured from either wildtype or haf-1 mutant worms, addition of ATP lead to time-dependent degradation of most mitochondrial proteins (Figure 3B), which was blocked by LY-CMK, a known inhibitor of ClpP (Figure 3C). Furthermore, mitochondria isolated from animals raised on clpp-1(RNAi), have less protein degradation than wildtype mitochondria, suggesting that ClpP has a substantial role in proteolysis (Figure 3D).
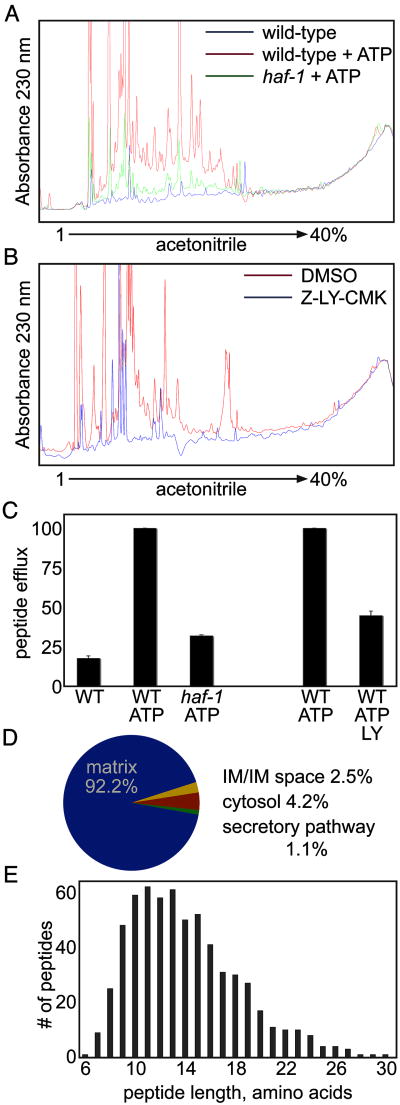
In yeast, the degradation of mitochondrial proteins is associated with efflux of peptides from the organelle (Young et al., 2001). To determine if this were also the case in C. elegans we analyzed the peptide content of the mitochondrial supernatant under conditions of protein degradation in isolated mitochondria. Peptides from the supernatant of purified mitochondria were resolved by reverse-phase HPLC on a C12 column and the column eluate was monitored by the absorption of the peptide backbone at 230 nm. The supernatant of mitochondria incubated with ATP revealed significantly more absorbance at 230 nm than that recovered from mitochondria incubated without ATP (Figure 4A) and recovery of material absorbing at 230 nm was inhibited by LY-CMK (Figure 4B and 4C). These observations established a correlation between degradation of mitochondrial proteins and recovery of peptides in the supernatant of wildtype mitochondria. Remarkably, the signal recovered in the supernatant of mitochondria isolated from haf-1 mutant worms was 3-fold lower than that of the wildtype (Figure 4A and 4C); this despite the fact that protein degradation was unimpaired by the mutation (Figure 3B).
Figure 4. haf-1-dependent peptide efflux from mitochondria.
(A) UV trace of peptides recovered from the supernatant of mitochondria isolated from wildtype or haf-1(ok705) mutant worms and resolved on HPLC. The isolated mitochondria were incubated at 30°C for 60 minutes with or without ATP (3mM, as described in figure 3B and 3C).
(B) Where indicated, the ClpP inhibitor, Z-LY-CMK (10μM), or the carrier DMSO (0.3%, a control) were added to the suspension of wildtype mitochondria during the incubation at 30°C with ATP (3mM) (as in (A), above).
(C) Quantification of the area-under-the-curve (AUC) of the HPLC traces from figures 3A and 3B. The AUC from wildtype mitochondria incubated with ATP was normalized to 100. Shown is the mean AUC ± S.E.M.
(D) A pie-diagram showing the distribution by origin of the identifiable peptides recovered in the supernatant of wildtype mitochondria (see table 1 for details).
(E) A graphic representation of the distribution of the length of peptides recovered in the supernatant of wildtype mitochondria.
The recovered peptides were separated and sequenced by electrospray mass spectrometry (LC-MS/MS). 646 unique peptides, derived from 51 proteins were identified (Table 1). Only peptides from proteins that appeared in two separate experiments are listed. 92.2% of the peptides came from proteins localized within the mitochondrial matrix, while only 2.5% came from proteins located within the inner membrane or inter-membrane space (Figure 4D). 4.2% and 1.1% of the peptides came from proteins residing in the cytosol or secretory pathway, indicating that the contamination of the mitochondrial fraction with cytosol and ER (noted in figure 3A) is of minor significance.
Table 1.
Identity of peptides recovered in the supernatant of wildtype mitochondria incubated at 30°C for 60 minutes with ATP.
| Protein | Number of unique peptides | Sequence Coverage % | Localization | Function |
|---|---|---|---|---|
| MDH-1 | 76 | 54 | matrix | malate dehydrogenase |
| ACO-2 | 51 | 37 | matrix | aconitase |
| GEI-7 | 41 | 24 | matrix | isocitrate lyase/malate synthase |
| ATP-2 | 37 | 32 | matrix | ATP synthase-beta |
| ECH-6 | 36 | 63 | matrix | enoyl-CoA hydratase |
| H28O16.1 | 35 | 39 | matrix | ATP synthase-alpha |
| CTS-1 | 29 | 31 | matrix | citrate synthase |
| SDHA-1 | 28 | 20 | matrix | complex II, subunit A |
| FUM-1 | 25 | 24 | matrix | fumarate hydratase |
| F27D4.1 | 24 | 49 | matrix | electron transfer flavoprotein, alpha |
| GTA-1 | 16 | 20 | matrix | 4-aminobutyrate aminotransferase |
| F43G9.1 | 15 | 21 | matrix | isocitrate dehydrogenase, subunit alpha |
| PYC-1 | 15 | 7 | matrix | pyruvate carboxylase 1 |
| MEL-32 | 14 | 19 | matrix | glycine/serine hydroxymethyltransferase |
| T05H10.6 | 13 | 15 | matrix | pyruvate dehydrogenase alpha 1 |
| F47B10.1 | 12 | 20 | matrix | succinyl-CoA synthetase, beta |
| F58F12.1 | 11 | 26 | matrix | ATP synthase-delta |
| UCR-1 | 9 | 15 | matrix | processing peptidase, beta |
| C05G5.4 | 9 | 22 | matrix | succinyl-CoA synthetase, alpha |
| PCCA-1 | 8 | 7 | matrix | propionyl-CoA carboxylase alpha |
| C05C10.3 | 8 | 10 | matrix | succinyl-CoA:alpha-ketoic acid-CoA transferase |
| CYN-1 | 8 | 26 | matrix | cyclophilin |
| MMCM-1 | 8 | 7 | matrix | methylmalonyl-CoA mutase |
| SDHB-1 | 8 | 22 | matrix | complex II, subunit b |
| ALH-8 | 6 | 11 | matrix | methylmalonate semialdehyde dehydrogenase |
| B0303.3 | 6 | 10 | matrix | 3-ketoacyl-CoA thiolase, beta |
| C04C3.3 | 5 | 11 | matrix | pyruvate dehydrogenase beta 1 |
| T08G2.3 | 5 | 6 | matrix | medium chain acyl-CoA dehydrogenase |
| ZC262.5 | 4 | 22 | matrix | ATP synthase-epsilon/R05D3.6 |
| SOD-2 | 4 | 4 | matrix | superoxide dismutase/SOD-3 |
| Y94H6A.8 | 4 | 18 | matrix | complex I component |
| T22B11.5 | 4 | 3 | matrix | alpha-ketoglutarate dehydrogenase |
| F40A3.3 | 4 | 9 | matrix | phosphatidylethanolamine binding protein |
| F22D6.4 | 4 | 7 | matrix | complex I component |
| MAI-2 | 3 | 23 | matrix | ATPase inhibitor |
| C34C12.8 | 3 | 15 | matrix | GrpE family |
| T22H6.2 | 3 | 3 | matrix | 1-pyrroline-5-carboxylate synthetase |
| BCAT-1 | 2 | 4 | matrix | branched chain amino acid aminotransferase |
| C16C10.11 | 2 | 14 | matrix | uncharacterized |
| PHB-2 | 1 | 4 | matrix | prohibitin |
| F54D8.2 | 8 | 27 | IM | complex IV, subunit VIa |
| F44G4.2 | 3 | 22 | IM | complex I component |
| MEV-1 | 2 | 6 | IM | complex II, subunit C |
| CCHL-1 | 2 | 7 | IMS | cytochrome c heme-lyase |
| DNJ-21 | 1 | 8 | IM | Tim14 orthologue |
| VIT-1 | 8 | 4 | secreted | vitellogenin |
| VIT-2 | 7 | 3 | secreted | vitellogenin |
| SPL-1 | 4 | 6 | ER | sphingosine-1-phosphate lyase |
| VIT-3 | 4 | 1 | secreted | vitellogenin |
| VIT-6 | 4 | 2 | secreted | vitellogenin |
| EFT-4 | 7 | 9 | cytoplasm | elongation factor |
The identified peptides were derived from highly expressed proteins involved in various metabolic pathways within the mitochondrial matrix. For example, 249 peptides came from 9 of the proteins in the TCA cycle and 87 peptides came from 4 of the 5 matrix-localized subunits of the ATP-synthase. The peptides ranged in length from 6 to 30 amino acids with a median of 14 amino acids (Figure 4E). This is within the size range previously reported for peptides effluxed by yeast Mdl1p (Augustin et al., 2005) and at the upper end of the size of artificial ClpP degradation products in vitro (Choi and Licht, 2005).
A role for ClpX in UPRmt signaling
The observations above, point to an important role for ClpP mediated proteolysis and peptide transport in UPRmt signaling. Studies in bacteria indicate that ClpP's proteolytic activity requires a partner AAA+ ATPase (Bewley et al., 2009) and proteolysis in our samples was highly ATP-dependent (Figure 3B). ClpX is the only ClpP-interacting partner known to exist in eukaryotes and C. elegans have two genes encoding proteins homologous to bacterial ClpX with predicted mitochondrial targeting sequences (d2030.2 and k07a3.3). Previous RNAi feeding experiments failed to uncover a role for these ClpX homologues in the UPRmt (Haynes et al., 2007). However, in light of the inherent weakness of RNAi as a tool for comprehensive analysis of gene function, we revisited ClpX's role in UPRmt signaling by exploiting a deletion strain of one of the two ClpX homologues, d2030.2(tm2183), and the powerful activation of the UPRmt in the zc32 mutant strain at the non-permissive temperature. d2030.2(tm2183) was crossed into the hsp-60pr∷gfp reporter strain in both a wildtype and zc32 background. Consistent with the previous RNAi experiments, neither the deletion d2030.2(tm2183) nor k07a3.3(RNAi) had a measurable impact on hsp-60pr∷gfp expression individually, however when d2030.2(tm2183) animals were exposed to k07a3.3(RNAi), stress-induced expression of hsp-60pr∷gfp was reproducibly attenuated (Figure 5A and 5B).
Figure 5. Impaired UPRmt signaling in animals with reduced ClpX activity.
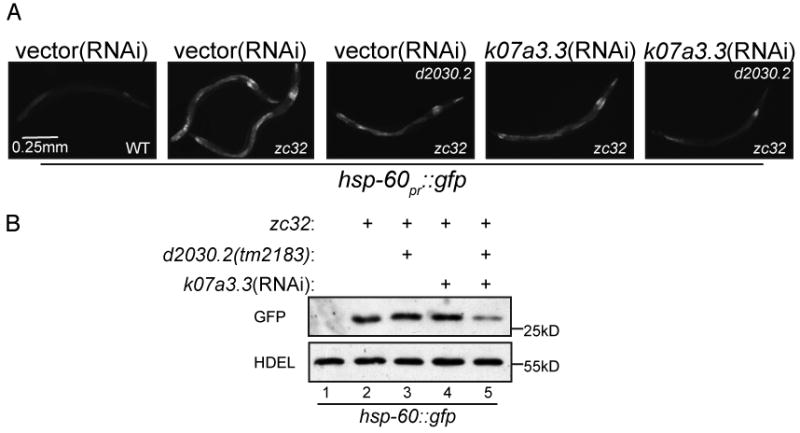
(A) Representative fluorescent photomicrographs of hsp-60pr∷gfp transgenic worms with a temperature sensitive mutation (zc32) that activates the UPRmt in an otherwise wildtype or d2030.2(tm2183) background. Where indicated wildtype or d2030.2(tm2183) worms were raised on k07a3.3(RNAi) for 3 days.
(B) A corresponding immunoblot of GFP expressed by hsp-60pr∷gfp transgenic worms in whom mitochondrial unfolded protein stress was induced. The worms were treated as indicated in (A).
These observations point to a role for a ClpXP holo-protease in UPRmt signaling and suggest that the proteolytic machine implicated in the C. elegans UPRmt is functionally related to the well-studied bacterial and mammalian systems.
Signaling downstream of HAF-1
Mitochondrial unfolded protein stress results in the redistribution of the transcription factor DVE-1 in the nucleus and the transcriptional induction of UBL-5, a small ubiquitin-like protein, both of which are required for hsp-60 induction (Haynes et al., 2007). To place HAF-1 in this presumptive pathway, we compared the localization of DVE-1∷GFP and expression of ubl-5pr∷gfp in stressed wildtype and haf-1 mutant worms. HAF-1 was required for ubl-5pr∷gfp expression in response to stress (Supplementary Figure 3A and 3B), however the haf-1 deletion did not affect the nuclear distribution of DVE-1∷GFP (Figure 6A), indicating that DVE-1 is not downstream of HAF-1 in UPRmt signaling and suggesting the existence of other UPRmt transcription factor(s) that had been missed in the original RNAi screen for genes that activate the UPRmt.
Figure 6. The bZIP protein, ZC376.7, is required for UPRmt signaling and functions downstream of both HAF-1 and ClpP.
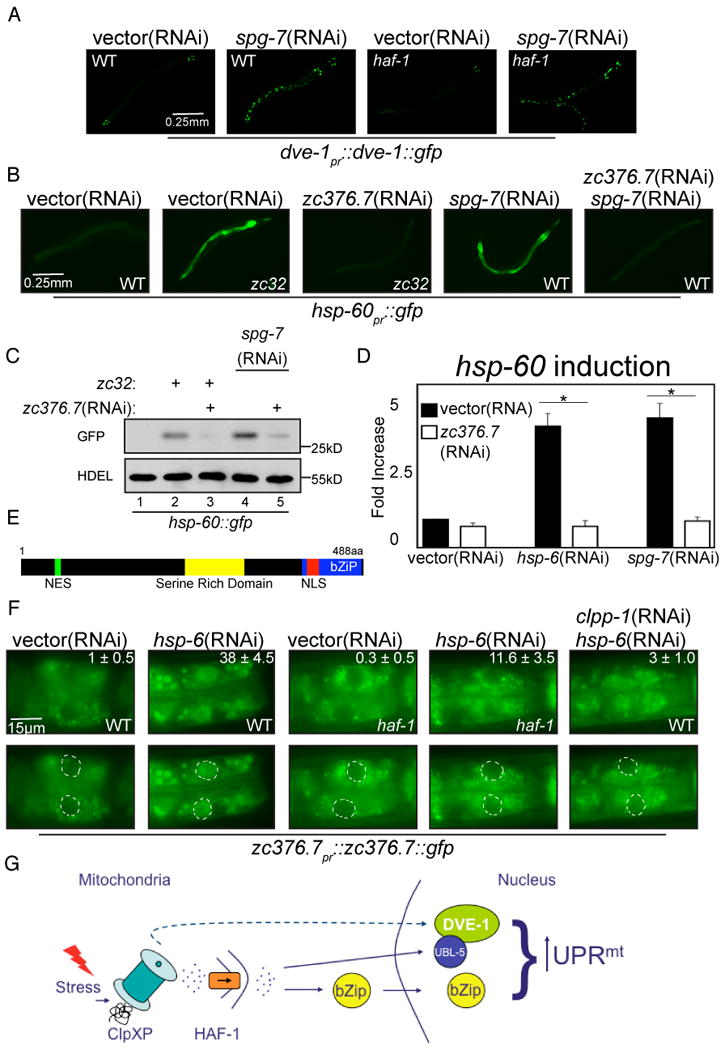
(A) Representative fluorescent photomicrographs of dve-1pr∷dve-1∷gfp transgenic worms in a wildtype or haf-1(ok705) background raised on vector(RNAi) or spg-7(RNAi).
(B) Representative fluorescent photomicrographs of hsp-60pr∷gfp transgenic worms, wildtype (WT) or with a temperature sensitive mutation (zc32) that activates the UPRmt raised on vector, zc376.7, spg-7 or zc376.7 and spg-7(RNAi).
(C) A corresponding immunoblot of GFP expressed by hsp-60pr∷gfp transgenic worms in whom mitochondrial unfolded protein stress was induced. The genotypes and treatments are as in (B), above.
(D) Quantitative analysis (by QRT-PCR) of endogenous hsp-60 mRNA in vector or zc376.7(RNAi) fed N2 worms subjected to mitochondrial unfolded protein stress by hsp-6(RNAi) or spg-7(RNAi). Displayed is the mean +/- SEM, n=3, *p<0.05.
(E) Predicted features of ZC376.7. A basic-region leucine zipper domain extends from amino acid 420 to 483 and a serine rich domain exists at positions 261-312. Amino acids 75-83 are a predicted leucine-rich nuclear export signal (NES) and residues 425-441 encode a predicted bipartite nuclear localization signal (NLS).
(F) Representative fluorescent photomicrographs of the most proximal two intestinal cells in wildtype (WT) or haf-1(ok705) L3 stage animals expressing an extra-chromosomal array of zc376.7pr∷zc376.7∷gfp. Where indicated, the animals were exposed to vector, hsp-6 or hsp-6 and clpp-1(RNAi). The inset at the upper right hand corner reports on the mean percentage ± SEM number of worms in which ZC376.7∷GFP is apparent in intestinal nuclei in each condition. In the lower panels each nuclei has been outlined.
(G) Scheme of the hypothesized relationships of the gene products implicated in UPRmt signaling.
To identify previously overlooked genes (that might have been missing from the original library used, Kamath et al., 2003) we screened all 485 genes annotated as encoding worm transcription factors from a different RNAi library (Reece-Hoyes et al., 2005). A single RNAi clone that attenuated hsp-60pr∷gfp (Figure 6B and C) and endogenous hsp-60 and hsp-6 mRNA induction in response to mitochondrial stress (Figure 6D, Supplementary Figure 3C) was identified. The RNAi clone was directed to the coding sequence of a gene, zc376.7, which was missing in the library used in the original screen (conversely, the dve-1(RNAi) clone was absent from the new library).
zc376.7 encodes a 488 amino acid protein with a C-terminal basic-region leucine zipper domain that bears distant similarity to mammalian ATF5, a stress-induced transcription factor (Zhou et al., 2008). To examine expression patterns of ZC376.7, we generated transgenic lines expressing zc376.7pr∷zc376.7∷gfp in which the native promoter drives expression of a ZC376.7∷GFP fusion protein. Transgenic animals expressing high levels of ZC376.7∷GFP died as embryos, but stable extra-chromosomal arrays expressing low levels of ZC376.7∷GFP were recovered. GFP signal was present diffusely in the majority of cells and tissues, most prominently in the embryonic intestine and larval pharynx (data not shown).
ZC376.7 has both a predicted nuclear localization sequence as well as a nuclear export sequence (Figure 6E), suggesting that subcellular localization may be important for regulation of its transcriptional activity. In unstressed worms ZC376.7∷GFP exhibited a diffuse cytoplasmic localization, best observed in the intestinal cells. However, when raised on hsp-6(RNAi), ZC376.7∷GFP localized to the nuclei (Figure 6F). To explore the relationship between ZC376.7, ClpP and HAF-1, animals expressing the zc376.7pr∷zc376.7∷gfp extra-chromosomal array were crossed with haf-1(ok705) worms or raised on clpp-1(RNAi). Nuclear localization of ZC376.7∷GFP, observed in many intestinal cells of mitochondrially-stressed wildtype worms, was attenuated both by clpp-1(RNAi) and by haf-1 deletion thus placing ZC376.7 downstream of ClpP and HAF-1 in the UPRmt signaling pathway (Figure 6G).
Discussion
The experiments described here implicate the mitochondrially-localized peptide transporter, HAF-1, in signaling the stress of unfolded/misfolded proteins in the mitochondrial matrix to the nucleus and establish a correlation between mitochondrial unfolded protein stress and HAF-1-dependent efflux of peptides derived from proteolysis of matrix proteins. Importantly, HAF-1's contribution to peptide efflux from stressed mitochondria is readily integrated with the requirement for ClpP-mediated proteolysis in the UPRmt. Together these findings suggest a model whereby the efflux of peptides derived from stress-induced proteolysis is important to signaling the UPRmt.
The combination of impaired activation of mitochondrial chaperone genes and selectively enhanced sensitivity to mitochondrial stress observed in haf-1 deleted worms is precisely the phenotype expected of loss of a gene implicated in signaling the UPRmt. It is a rare phenotype, as there are many more genes whose loss-of-function causes mitochondrial stress and increased sensitivity to further stress; but these are associated with enhanced signaling in the UPRmt (Yoneda et al., 2004; Benedetti et al., 2006; Haynes et al., 2007).
HAF-1's involvement in peptide export from the mitochondrial matrix is consistent both with its homology to the yeast mitochondrial peptide transporter, Mdl1p, and with the defect in peptide export observed in vitro from mitochondria isolated from haf-1 mutant worms. The assay used to measure peptide export has several features that bear on the plausibility of a link between UPRmt signaling and peptide export: Peptide export is dependent on ATP, which is consistent with a role for ClpXP in peptide generation and HAF-1 (an ATPase) in their export. The minor differences in average peptide size we observed, compared with those reported for bacterial ClpAP in vitro (Choi and Licht, 2005) may reflect different enzymes (ClpXP in C. elegans mitochondria), different substrates (endogenous mitochondrial proteins in our experiments, versus GFP-SsrA in the in vitro experiments) or a bias introduced by the HPLC purification procedure.
Furthermore, peptide efflux is highly temperature dependent and therefore consistent with a role for protein unfolding in the generation of substrates for proteolysis and peptide export. This temperature dependence has also been observed in yeast, where there is an additional suggestion that labile, unfolded proteins are the preferred source of peptides for Mdl1-dependent export (Augustin et al., 2005). Thus, the rate of peptide export is suited to report on the protein-folding environment in the mitochondrial matrix and to serve a proximal role in signaling the UPRmt.
The significance of peptide export may be different in yeast and C. elegans. In the former, many of the peptides exported originate from proteins that have domains in the inter-membrane space (Augustin et al., 2005). And, the inter-membrane space protease, Yme1p, makes an important contribution to the generation of peptides which then diffuse through the semi-porous mitochondrial outer membrane to the cytosol (Young et al., 2001). By contrast, in C. elegans, matrix proteins appear to be the major contributors to the flux of peptides from mitochondria under conditions of thermal stress (Table 1). It is interesting to note that a loss-of-function mutation of ymel-1(tm1920), the only identifiable C. elegans homolog of the yeast inter-membrane space protease Yme1p, had no effect on UPRmt signaling and exhibited no synthetic interactions with the haf-1(ok705) mutation (data not shown). Thus, the discovery of HAF-1's role in peptide export and the UPRmt provides a plausible clarification for the previously-noted role for ClpP in the UPRmt, though other proteases that were missed in our screen may make important contributions to the process.
Several caveats apply: The phenotype of ClpP (clpp-1) deficiency is much more severe than that of haf-1 deficiency. In the context of the UPRmt, ClpP loss-of-function blocks the redistribution of the transcription factor DVE-1 in the nuclei of cells experiencing mitochondrial stress, whereas DVE-1 redistribution is not discernibly affected by haf-1 deletion. Together these suggest that dve-1 contributes to the UPRmt independently of haf-1 and that the latter may signal in the nucleus by alternative means. A more thorough survey of transcription factor encoding genes by RNAi feeding identified an additional gene required for UPRmt signaling. The encoded bZIP-containing protein, ZC376.7, is concentrated in the nuclei of stressed worms in a manner that is dependent on clpp-1 and haf-1. Further analysis of the mechanisms regulating ZC376.7 may provide clues to missing components of the signal transduction pathway downstream of HAF-1.
How might peptide efflux be sensed to produce downstream signaling and influence ZC376.7 localization? In the cytosol peptides are rather short lived, but in mammalian cells some mitochondrial-derived peptides survive long enough to be transported into the ER and are presented on the cell surface in MHC-I complexes (Herget and Tampe, 2007). Thus, a cytosolic/nuclear peptide receptor with some specificity for mitochondrial-derived peptides could signal downstream of the ClpP and HAF-1 steps suggested by this study (Figure 6G). Alternatively, signaling might proceed through a sensor, not of the peptides themselves but of the rate at which they flux through HAF-1. HAF-1 and its yeast homolog Mdl1p appear to have broad substrate specificity, but this last hypothetical mechanism side-steps the specificity problem by its indifference to the identity of the peptides transported. It is important to keep in mind that HAF-1 may also transport other substrates, in addition to peptides. For example, it has been suggested that the related mammalian ABC-me transports heme across the mitochondrial inner membrane (Shirihai et al., 2000). In one fanciful scenario, HAF-1 could transport a ligand (e.g. heme) that is released from a matrix carrier protein by ClpP digestion under stress conditions and it is such a ligand that activates signaling in the UPRmt. Either way, the aforementioned hypotheses predict the existence of downstream components of the UPRmt that respond to a peptide ligand, peptide efflux or a non peptide ligand transported by HAF-1.
Experimental Procedures
Transgenic and mutant C. elegans strains
Strains containing the UPRmt reporters hsp-6pr∷gfp(zcIs13) V, hsp-60pr∷gfp(zcIs9) V and ubl-5pr∷gfp(zcIs42) X, dve-1pr∷dve-1∷gfp(zcIs39) II, the UPRER reporter, hsp-4pr∷gfp(zcIs4) V, the expression of cytosolic and mitochondrial GFP myo-3pr∷gfpcyt(zcIs21) and myo-3pr∷gfpmt(zcIs14) and hsp-16pr∷clpp-1WT∷TAG(zcIs21) V have all been previously described (Yoneda et al., 2004; Benedetti et al., 2006; Haynes et al., 2007). The haf-1(ok705) and haf-3(gk549) alleles were provided by the CGC and haf-1(tm843), d2030.2(tm2183) and ymel-1(tm1920) were provided by the National Bioresource Project for C. elegans. haf-1 mutant alleles ok705 and tm843 were backcrossed into the N2 background (4 backcrosses) creating strains SJ4201 and SJ4202. The zc376.7pr∷zc376.7∷gfp expressing strain was generated by co-injecting the plasmid zc376.zc376.GFP (2.5 μg/ml) and the lin-15 rescuing plasmid, pSK1 (25 μg/ml) into a lin-15(n765ts) strain.
Plasmids
The mammalian HAF-11-75∷GFP expression plasmid (pEGFP-haf-1ss) was constructed by ligating a PCR generated cDNA fragment encoding amino acids 1-75 of HAF-1 N-terminal of GFP in the mammalian expression vector pEGFP-N1 (Clontech). The mammalian HAF-1∷FLAG expression plasmid (phaf-1-flag) was constructed by ligating a PCR generated cDNA fragment encoding the full-length C. elegans HAF-1 coding sequence in frame with a C-terminal FLAG peptide in the plasmid cFLAG.pCEFLneo. The C. elegans zc376.zc376.GFP expression plasmid was constructed by PCR amplifying a 5.4 kb fragment containing the promoter and entire coding sequence of zc376.7 from C. elegans genomic DNA in frame with the GFP-encoding gene of pPD95.75.
Development and survival assays
N2 or haf-1 mutant eggs were placed on RNAi or tunicamycin-containing plates and allowed to develop. After 3 days, the percentage of gravid animals was determined. For each point, 3 plates containing roughly 100 worms each were counted. For lifespan studies, animals were synchronized and allowed to develop for 3 days on vector(RNAi) and then, where indicated, the animals were transferred to plates containing hsp-6(RNAi). For the survival assays, 24 L4 animals were placed onto the described plates. Every other day the animals were transferred to a new plate containing fresh chemical.
Oxygen consumption
Oxygen consumption was measured using a Clark type electrode (Hansatech) as described (Braeckman et al., 2002). Eggs were synchronized by bleaching and the hatchlings grown for three days at 20°C. Prior to measurements, animals were treated as indicated and washed three times to remove bacteria. Worms were re-suspended in 1 ml M9 buffer and the rate of oxygen consumption was measured for at least 10 minutes at 20°C. Animals were recovered and total protein was determined using the Bradford assay.
Mitochondrial purification and degradation assays
5g of 3-day old synchronized worms were harvested from liquid culture via sucrose flotation. The worms were resuspended in 8mls of mitochondria buffer (70mM sucrose, 1mM EGTA, 210mM sorbitol, 10mM MOPS ph 7.4) and crude mitochondria were purified as described (Jonassen et al., 2002). The pellet was re-suspended in mitochondria buffer, layered over 1ml of 13% Optiprep and centrifuged at 12,000*g for 10 minutes. The supernatant was discarded and the pellet was again run through 13% Optiprep. The pellet was washed once in mitochondria buffer and resuspended in mitochondria buffer with 1mg/ml trypsin and incubated on ice for 30 minutes. The mitochondria were washed three times with mitochondria buffer and resuspended in import buffer (250mM sorbitol, 150mM KCl, 10mM MgCl2, 2.5mM EDTA, 3mM ATP (where indicated) and 20mM HEPES pH 7.4). Mitochondria were incubated at 30°C for the indicated amount of time and pelleted at 9500*g. The pellet was resuspend in 7M urea, 50mM DTT, 4% ChAPs, 2M thiourea and incubated on ice for 30 min. Sample buffer was added and samples were boiled for 3 minutes prior to loading on a 12% acrylamide SDS-PAGE.
Disrupted CHO and HEK293T cells were fractionated in a similar manner to C. elegans, differing only in the lysis procedure. Transfected cells were pelleted, resuspended in mitochondria buffer, and passed through a 27.5 gauge needle 10 times to lyse. Generation of mitoplasts, proteinase K and Na2CO3 treatment were performed as described (Prasannan and Appling, 2009). 35μg of protein was loaded in lanes 1-4, while mitochondria-equivalent volumes were loaded in lanes 5-8 of figure 1F.
Peptide purification and mass spectrometry
Supernatants were collected following proteolysis, adjusted to 0.1% TFA (trifluoracetic acid) and loaded onto a HyperSep C18 column (Thermo). Peptides were eluted using 20% acetonitrile and dried. Peptides were resuspended in water and separated on a reverse-phase C12 Jupiter 10u Proteo column (Phenomenex) on an HPLC (Waters, 1-40% acetonitrile gradient in 0.1% TFA). Fractions were collected, dried and subjected to mass spectrometry. Identification of peptides was achieved using an LTQ-Orbitrap mass spectrometer (Thermo Scientific, San Jose, CA) in line with a 15 cm × 75 um inner diameter BEH C18 75 um/300 Å capillary column (Waters). A nanoAcquity ultra performance liquid chromatography (Waters) system was used to deliver a 120 minute gradient from 1% to 40% mobile phase B (mobile phase A = 0.1% formic acid in HPLC grade water; mobile phase B = 0.1% formic acid in acetonitrile) at a flow rate of 400nl/min. Tandem mass spectra from the raw data were extracted using BioWorks Browser (version 3.3, Thermo Scientific) and used to search the SwissProt (version 51.6) database using Mascot (version 2.2, Matrix Science, London, UK) search engine (taxonomy = C. elegans, cleavage enzyme = none, variable modification = oxidation (M), mass tolerances (MS) = 10ppm and (MS/MS) = 0.7Da). The Mascot score cut-off for 95% confidence in the peptide matches was 35. Only proteins with Mascot scores above this threshold are listed in Table 1.
Supplementary Material
Acknowledgments
We thank Jane Hubbard (NYU), Shoei Mitani (National Bioresource Project for C. elegans, Tokyo, Japan) and the C. elegans Genetics Center (Minneapolis, MN) for access to resources and reagents. Luke Wiseman for advice on HPLC analysis and Helene Cardasis for mass spectrometry advice. Supported by NIH grants DK47119 & ES08681 to DR, and NS050276 & RR017990 to TAN, and a NRSA to CMH (F32-NS050901).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arnold I, Wagner-Ecker M, Ansorge W, Langer T. Evidence for a novel mitochondria-to-nucleus signalling pathway in respiring cells lacking i-AAA protease and the ABC-transporter Mdl1. Gene. 2006;367:74–88. doi: 10.1016/j.gene.2005.09.044. [DOI] [PubMed] [Google Scholar]
- Augustin S, Nolden M, Muller S, Hardt O, Arnold I, Langer T. Characterization of peptides released from mitochondria: evidence for constant proteolysis and peptide efflux. J Biol Chem. 2005;280:2691–2699. doi: 10.1074/jbc.M410609200. [DOI] [PubMed] [Google Scholar]
- Barral JM, Broadley SA, Schaffar G, Hartl FU. Roles of molecular chaperones in protein misfolding diseases. Semin Cell Dev Biol. 2004;15:17–29. doi: 10.1016/j.semcdb.2003.12.010. [DOI] [PubMed] [Google Scholar]
- Benedetti C, Haynes CM, Yang Y, Harding HP, Ron D. Ubiquitin Like Protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics. 2006;174:2229–2239. doi: 10.1534/genetics.106.061580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley MC, Graziano V, Griffin K, Flanagan JM. Turned on for degradation: ATPase-independent degradation by ClpP. J Struct Biol. 2009;165:118–125. doi: 10.1016/j.jsb.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braeckman BP, Houthoofd K, De Vreese A, Vanfleteren JR. Assaying metabolic activity in ageing Caenorhabditis elegans. Mech Ageing Dev. 2002;123:105–119. doi: 10.1016/s0047-6374(01)00331-1. [DOI] [PubMed] [Google Scholar]
- Broadley SA, Hartl FU. Mitochondrial stress signaling: a pathway unfolds. Trends Cell Biol. 2008;18:1–4. doi: 10.1016/j.tcb.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Choi KH, Licht S. Control of peptide product sizes by the energy-dependent protease ClpAP. Biochemistry (Mosc) 2005;44:13921–13931. doi: 10.1021/bi0505060. [DOI] [PubMed] [Google Scholar]
- Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Desautels M, Goldberg AL. Demonstration of an ATP-dependent, vanadate-sensitive endoprotease in the matrix of rat liver mitochondria. J Biol Chem. 1982;257:11673–11679. [PubMed] [Google Scholar]
- Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol Cell. 2003;11:671–683. doi: 10.1016/s1097-2765(03)00060-1. [DOI] [PubMed] [Google Scholar]
- Forner F, Arriaga EA, Mann M. Mild protease treatment as a small-scale biochemical method for mitochondria purification and proteomic mapping of cytoplasm-exposed mitochondrial proteins. J Proteome Res. 2006;5:3277–3287. doi: 10.1021/pr060361z. [DOI] [PubMed] [Google Scholar]
- Gottesman S. Proteolysis in bacterial regulatory circuits. Annu Rev Cell Dev Biol. 2003;19:565–587. doi: 10.1146/annurev.cellbio.19.110701.153228. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Neupert W. Protein sorting to mitochondria: evolutionary conservations of folding and assembly. Science. 1990;247:930–938. doi: 10.1126/science.2406905. [DOI] [PubMed] [Google Scholar]
- Haynes CM, Petrova K, Benedetti C, Yang Y, Ron D. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev Cell. 2007;13:467–480. doi: 10.1016/j.devcel.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Herget M, Tampe R. Intracellular peptide transporters in human--compartmentalization of the “peptidome”. Pflugers Arch. 2007;453:591–600. doi: 10.1007/s00424-006-0083-4. [DOI] [PubMed] [Google Scholar]
- Jonassen T, Marbois BN, Faull KF, Clarke CF, Larsen PL. Development and fertility in Caenorhabditis elegans clk-1 mutants depend upon transport of dietary coenzyme Q8 to mitochondria. J Biol Chem. 2002;277:45020–45027. doi: 10.1074/jbc.M204758200. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kang SG, Ortega J, Singh SK, Wang N, Huang NN, Steven AC, Maurizi MR. Functional proteolytic complexes of the human mitochondrial ATP-dependent protease, hClpXP. J Biol Chem. 2002;277:21095–21102. doi: 10.1074/jbc.M201642200. [DOI] [PubMed] [Google Scholar]
- Kaufman RJ. Orchestrating the unfolded protein response in health and disease. J Clin Invest. 2002;110:1389–1398. doi: 10.1172/JCI16886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Tanaka N, Nakamura N, Takano S, Ohkuma S. Knockdown of mitochondrial heat shock protein 70 promotes progeria-like phenotypes in caenorhabditis elegans. J Biol Chem. 2007;282:5910–5918. doi: 10.1074/jbc.M609025200. [DOI] [PubMed] [Google Scholar]
- Kock H, Gerth U, Hecker M. The ClpP peptidase is the major determinant of bulk protein turnover in Bacillus subtilis. J Bacteriol. 2004;186:5856–5864. doi: 10.1128/JB.186.17.5856-5864.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger E, Witt E, Ohlmeier S, Hanschke R, Hecker M. The clp proteases of Bacillus subtilis are directly involved in degradation of misfolded proteins. J Bacteriol. 2000;182:3259–3265. doi: 10.1128/jb.182.11.3259-3265.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrousse AM, Zappaterra MD, Rube DA, van der Bliek AM. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol Cell. 1999;4:815–826. doi: 10.1016/s1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- Miller KG, Alfonso A, Nguyen M, Crowell JA, Johnson CD, Rand JB. A genetic selection for Caenorhabditis elegans synaptic transmission mutants. Proc Natl Acad Sci U S A. 1996;93:12593–12598. doi: 10.1073/pnas.93.22.12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasannan P, Appling DR. Human mitochondrial C1-tetrahydrofolate synthase: submitochondrial localization of the full-length enzyme and characterization of a short isoform. Arch Biochem Biophys. 2009;481:86–93. doi: 10.1016/j.abb.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Deplancke B, Shingles J, Grove CA, Hope IA, Walhout AJ. A compendium of Caenorhabditis elegans regulatory transcription factors: a resource for mapping transcription regulatory networks. Genome Biol. 2005;6:R110. doi: 10.1186/gb-2005-6-13-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheps JA, Ralph S, Zhao Z, Baillie DL, Ling V. The ABC transporter gene family of Caenorhabditis elegans has implications for the evolutionary dynamics of multidrug resistance in eukaryotes. Genome Biol. 2004;5:R15. doi: 10.1186/gb-2004-5-3-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirihai OS, Gregory T, Yu C, Orkin SH, Weiss MJ. ABC-me: a novel mitochondrial transporter induced by GATA-1 during erythroid differentiation. EMBO J. 2000;19:2492–2502. doi: 10.1093/emboj/19.11.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram P, Echalier B, Han W, Hull D, Timmons L. ATP-binding cassette transporters are required for efficient RNA interference in Caenorhabditis elegans. Mol Biol Cell. 2006;17:3678–3688. doi: 10.1091/mbc.E06-03-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatraman A, Landar A, Davis AJ, Chamlee L, Sanderson T, Kim H, Page G, Pompilius M, Ballinger S, Darley-Usmar V, Bailey SM. Modification of the mitochondrial proteome in response to the stress of ethanol-dependent hepatotoxicity. J Biol Chem. 2004;279:22092–22101. doi: 10.1074/jbc.M402245200. [DOI] [PubMed] [Google Scholar]
- Wang JD, Herman C, Tipton KA, Gross CA, Weissman JS. Directed evolution of substrate-optimized GroEL/S chaperonins. Cell. 2002;111:1027–1039. doi: 10.1016/s0092-8674(02)01198-4. [DOI] [PubMed] [Google Scholar]
- Yoneda T, Benedetti C, Urano F, Clark SG, Harding HP, Ron D. Compartment specific perturbation of protein folding activates genes encoding mitochondrial chaperones. J Cell Sci. 2004;117:4055–4066. doi: 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
- Young L, Leonhard K, Tatsuta T, Trowsdale J, Langer T. Role of the ABC transporter Mdl1 in peptide export from mitochondria. Science. 2001;291:2135–2138. doi: 10.1126/science.1056957. [DOI] [PubMed] [Google Scholar]
- Yu AY, Houry WA. ClpP: a distinctive family of cylindrical energy-dependent serine proteases. FEBS Lett. 2007;581:3749–3757. doi: 10.1016/j.febslet.2007.04.076. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. A mitochondrial specific stress response in mammalian cells. EMBO J. 2002;21:4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Pallam LR, Jiang L, Narasimhan J, Staschke KA, Wek RC. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J Biol Chem. 2008;283:7064–7073. doi: 10.1074/jbc.M708530200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.