Abstract
High-throughput genome-wide RNA interference (RNAi) screening is emerging as an essential tool to assist biologists in understanding complex cellular processes. The large number of images produced in each study make manual analysis intractable; hence, automatic cellular image analysis becomes an urgent need, where segmentation is the first and one of the most important steps. In this paper, a fully automatic method for segmentation of cells from genome-wide RNAi screening images is proposed. Nuclei are first extracted from the DNA channel by using a modified watershed algorithm. Cells are then extracted by modeling the interaction between them as well as combining both gradient and region information in the Actin and Rac channels. A new energy functional is formulated based on a novel interaction model for segmenting tightly clustered cells with significant intensity variance and specific phenotypes. The energy functional is minimized by using a multiphase level set method, which leads to a highly effective cell segmentation method. Promising experimental results demonstrate that automatic segmentation of high-throughput genome-wide multichannel screening can be achieved by using the proposed method, which may also be extended to other multichannel image segmentation problems.
Keywords: Fluorescent microscopy, high throughput, image segmentation, interaction model, level set, multichannel
I. Introduction
High-throughput screening using automated fluorescent microscopy is becoming an essential tool to assist biologists in understanding complex cellular processes and genetic functions [1]. By using the RNA interference (RNAi) process, the function of a gene can be determined by inspecting changes in a biological process caused by the addition of gene-specific double-stranded RNA (dsRNA) [2], [3]. The development of Drosophila RNAi technology to systematically disrupt gene expression enables screening of the entire genome for specific cellular functions. In a small-scale study using manual analysis of genome-wide screening [4], biologists were able to observe a wide range of phenotypes with affected cytoskeletal organization and cell shape. However, without the aid of computerized image analysis, it is almost intractable to quantitatively characterize morphological phenotypes and identify genes in high-throughput screening. For instance, in a typical study conducted by our biologists [1], approximately 21 000 dsRNAs specific to the predicted Drosophila genes are robotically arrayed in 384-well plates. Drosophila cells are plated, and take up dsRNA from culture media. After incubation with the dsRNA, cells are fixed, stained, and imaged by automated microscopy. Each screen generates more than 400 000 images, or even millions if replicas are included. Clearly, there is a growing need for automated image analysis as high-throughput technologies are being extended to visual screens.
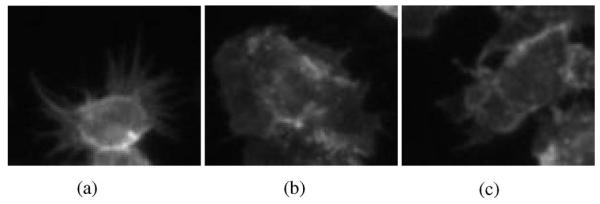
The key problem here is to automatically segment cells from cell-based assays in a cost-effective manner, since fast screening can generate hundreds of thousands of images in each study with rather poor image quality. Three main challenges [5], [6] related to this task are as follows. Firstly, cells with specific phenotypes need to be accurately segmented for further analysis. Samples of the three major phenotypes, S-spikey, R-ruffling, and A-actin acceleration at edge, to be identified in this study, are shown in Fig. 1. They are used for the gene clustering and scoring, and are of particular interest to our biologists. These shapes are not always convex, and violate the assumption of many algorithms [7], [8]. Secondly, significant intensity variations exist in each cell. Thus, areas inside each cell can no longer be assumed as homogeneous regions, which cause many intensity-based segmentation methods fail. Thirdly, cells are tightly clustered and will not likely show a strong border where they touch, which makes it difficult to separate the cells.
Fig. 1.
Patches of RNAi cell image in the Actin channel with specific phenotypes. (a) Spiky (S-type). (b) Ruffling (R-type). (c) Actin-acceleration-at-edge (A-type).
A novel cellular image segmentation scheme for RNAi fluorescent genome-wide screening is presented to address these challenges. In our biological study, cells are fixed and stained with different dyes to investigate the different components of cells. Each staining is captured by one image channel. The three channels, Actin, Rac, and DNA, are captured, as shown in Fig. 2. Only the nucleus of the cell is visible in the DNA channel, while the cytoplasm of the cell is only available in an Actin channel. The Rac channel contains some auxiliary information. In our scheme, nuclei are first extracted from the DNA channel and labeled, which are then used as the initialization for segmentation of cells in other channels. To separate the tightly clustered cells, a model describing the interaction between neighboring fronts is incorporated into the segmentation process. The segmentation scheme is then developed by using multiphase level sets. A new level set propagation scheme is obtained by minimizing the energy functional associated with the interaction model. The proposed algorithm is a general image segmentation method, which may also be applied to other multichannel imaging problems.
Fig. 2.
Sample RNAi fluorescent images. (a) DNA channels. (b) Actin channels. (c) Rac channels. Original image of each channel contains 1280 × 1024 pixels.
The rest of the paper is organized as follows. Section II provides a brief review of the related work. The proposed cell segmentation scheme is presented in Section III. Section IV provides the segmentation results and discussion, and finally, Section V concludes the work.
II. Related Work
A number of methods for nuclei and cell segmentation have been reported in recent years. According to the techniques used, existing methods can be broadly divided into two main categories, low-level and high-level information-based methods.
Methods in the first category are based on techniques exploiting low-level image features, such as pixel intensity and image gradient. Since the watershed method has shown to produce good results in separating cells that are connected, it is the most commonly used technique for cell image segmentation in this category. A major problem associated with these methods is the over-segmentation of the image, although some algorithms have been proposed to deal with this problem. For example, Wählby et al. [9] used a rule-based approach for merging over-segmented regions. Zhou et al. [10] proposed Voronoi diagram to correct the overlapped regions produced by marker-controlled watershed. However, it is difficult to devise reliable universal rules to merge the over-segmented regions in different cases. The situation becomes even worse when the intensities vary within a large range.
The other class of segmentation methods includes those exploiting constraints derived from the image data together with high-level a priori knowledge of the objects. In this category, deformable-model-based methods have been quite successful, where contours driven by internal and external forces evolve in the image until they converge to the boundaries of nuclei or cytoplasm [11], [12]. Since higher level knowledge is incorporated in these methods, a more robust segmentation can be obtained. However, overlapping areas may exist between these evolving contours. To solve this problem, Zimmer et al. [7] introduced repulsive force between parametric active contours by modifying the edge map of each contour. However, their method may have difficulty in segmenting dense clustered cells, where the edge maps are difficult to distinguish. Furthermore, it will become very complex to use parametric active contours to segment a large number of cells. Ortiz de Solórzano et al. [13] employed a level set scheme [14], [15] for the segmentation of nuclei and cells. Each cell is approximated by a propagation front embedded in a level set map, and the crossing of fronts is prevented explicitly by considering the positions of other fronts. However, the gradient curvature flow is not powerful enough to deal with intensity variation inside each cell and to divide the cells at their boundaries. The multiphase level set scheme [16]-[19] was used by Dufour et al. [8] for segmenting and tracking cells in 3-D microscopy images. However, due to the strong constraints applied in their work for optimization, success of the algorithm has only been demonstrated on segmentation and tracking of sphere-like convex cells.
In this paper, a new interaction model is proposed for segmenting RNAi fluorescent cellular images. Large number of tightly clustered cells are successfully segmented by combining the shape, contrast, intensity, and gradient information, by the introduction of the competition and repulsion in the new interaction model using the multiphase level set [16], [20].
III. RNAi Image Segmentation
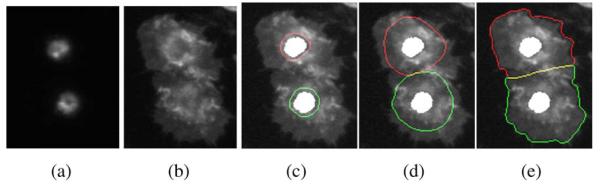
In this section, we present the proposed automatic scheme for RNAi fluorescent genome-wide screening segmentation. Since the number and locations of cells are unknown, it is difficult to directly segment cells from Actin and Rac channels, as shown in the sample images of Fig. 2. However, nuclei in the DNA channel are rather clearly imaged, and hence, are relatively easier to segment, which can then be used to count and position cells in other channels. Thus, the proposed segmentation scheme consists of two modules. In the first module, segmentation of nuclei is done by using a modified watershed method. The extracted nuclei are then used as initialization for the segmentation of cells in the second module. Since the Rac channel does not contain much useful information, it is summed into the Actin channel for cell segmentation to enhance the image contrast for segmentation. The challenges include weak boundaries between touching cells and intensity variations in each cell, which may cause many segmentation methods to fail, as shown by an example in Fig. 3. In order to deal with this problem, a novel interaction model characterizing the relationship between neighboring contours is proposed. The multiphase level set method is employed for minimizing the associated energy functional, which results in a robust cell segmentation method. The details of the scheme are given as follows.
Fig. 3.
(a) Initialization of the segmentation with nuclei. (b) Segmentation results using independent level sets. Due to the intensity similarity and the weak boundaries between cells, it is difficult to extract cells without considering the interaction between them.
A. Nuclei Segmentation
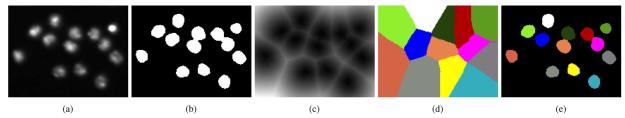
The first module of our RNAi image segmentation scheme is for extracting nuclei from the DNA channel. Since the nuclei appear much brighter than the background in the DNA channel, as shown in Fig. 4(a), Otsu's thresholding method is first applied to extract all the nuclei. A binary image with nuclei as foreground is obtained, as shown in Fig. 4(b). However, a global threshold value may not be optimal for the whole image. Note that some nuclei close to each other do not get separated due to the variance of pixel intensity [see Fig. 4(b)]. A higher threshold may help to separate the nuclei, but it will cause some other nuclei to be missed or split.
Fig. 4.
Segmenting of nuclei in the DNA channel. (a) Patch taken from an image of the DNA channel. (b) Binary thresholding result of (a). (c) Distance transform of (b). (d) Result of watershed on (c). (e) Labeling nuclei by combining (b) and (d).
In order to separate the connected nuclei after thresholding, a distance transform is applied to the binary image, as shown in Fig. 4(c). It can be seen that “basins” (dark areas) appear inside the nuclei, and “dams” (bright lines) are formed between them. The watershed algorithm [21] is then naturally employed to label the nuclei on the distance transform map. However, the simple watershed algorithm works well only when the shape of nuclei is exactly circular [22], [23]. Otherwise, multiple watershed regions may be produced for each single nucleus.
To handle this problem, an enhanced watershed algorithm is developed. In the first step, an initial classification of all points into catchment basin regions is done by tracing points down to their local minimum, following the path of steepest descent. Each segment consists of all the points whose paths of gradient descent terminate at the same minimum. The number of segments is equal to the number of local minima in the image. Obviously, too many regions are produced, which results in over-segmentation. To alleviate this, flood level thresholds are established. Flood level is a value that reflects the amount of prestored water in the catchment basins. In other words, the flood level threshold is the maximum depth of water that a region could hold without flowing into any of its neighbors. With the flood level established, a watershed segmentation algorithm can sequentially combine watersheds whose depths fall below the flood level. The minimum value of the flood level is zero, and its maximum value is the difference between the highest and the lowest values of the input image. In practice, we set the flood level as a scaled value between 0 and 1. Thus, the second step of the algorithm is to analyze neighboring basins and to compute the heights of their common boundaries for flood level thresholds.
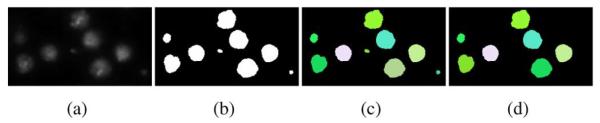
The segmentation results of our watershed method are shown in Fig. 4(d). By combining the results obtained by simple thresholding and the watersheding, touching nuclei can be separated, such that each nucleus has a unique label. The labeling results are shown in Fig. 4(e). After the initial labeling, there may be some small regions caused by noise, which may also be labeled as nuclei. To eliminate these unwanted regions, the average size range of nucleus is used as a criterion. Since the resolution of image acquisition equipment is known, the range can be estimated easily. If the size of a region is much smaller than the average nucleus size, the region will be considered as one produced by the noise. It will then be removed or merged into an adjacent region. Fig. 5 shows an example of the final segmentation results of our nuclei labeling algorithm.
Fig. 5.
Removing noise from the DNA channel. (a) Patch taken from an image of DNA channel. (b) Binary thresholding result of (a). (c) Initial labeling of nuclei using watershed. (d) Final labeling of nuclei after removing small regions.
B. Cell Segmentation
Once nuclei are properly identified, the segmentation of cells by using the proposed interaction model can proceed. The interaction involves mainly two types of mechanisms: repulsion and competition. The repulsion term is for separating the clustered cells and the competition is for defining the cell boundaries. These mechanisms are formulated as a new energy functional, which is then minimized by using the multiphase level set method to get cell segmentation.
1) Interaction Model
Consider an image I that has M clustered touching cells Si(i = 1, …, M). Let Ci(i = 1, …, M) denote the contours that evolve toward the boundaries of the cells. Instead of examining each contour independently, we integrate the interaction between neighboring contours into the segmentation process. The repulsion and competition are defined as follows.
- 1) Repulsion: The repulsive force prevents the contours from overlapping. Since the overlapping of the cell contours is not allowed, the repulsion needs to be emphasized to assure that the contours do not cross each other. This is to say that the union of all the regions {Ai|i = 1, 2, …, M} enclosed by contours Ci(i = 1, …, M) is empty
(1) - 2) Competition: Consider a pixel p of the image I among several cells. Which cell does p belong to? The answer lies in the competition between the evolving contours. In this paper, the membership of p is determined by computing the competition
where D is the difference measure metric and {mi|i = 1, 2, …,M,M + 1} denotes the membership. The background is indicated as the (M + 1)th region. The pixel p is assigned to the cell that produces the smallest difference.(2)
2) Energy Functional
To realize the interaction model, repulsion and competition are formulated by using an energy functional. Cells are segmented simultaneously by minimizing the joint energy functional. In our scheme, the repulsion can be naturally formulated as
| (3) |
where the positive parameter ω controls the repulsive force between the contours during the segmentation process.
When formulating the competition energy, the level set representation of the well-known Mumford–Shah model is employed [24], [25]. This model seeks to assign labels to pixels by minimizing the piecewise constant energy functional. Thus, the label of a pixel is decided by the competition of the surrounding contours and the background. The functional contains two data attachment terms that penalize the variance of the intensity inside and outside each object. Thus, it can be used to segment cells stained with fluorescent dyes, which appear as regions of average intensity larger than that of the background. However, due to the large intensity variation inside the cells, this model alone is not suitable to segment the cells from the background. To exploit the gradient information between the cells and the background, we add a geodesic length measure [26] in order to snap the contours to image edges. The membership of a pixel is then assigned by jointly considering the region-based and the gradient-based competition. Then, we have
| (4) |
where Ωb denotes the background, which is composed of the region outside of all the objects, i.e., out(C1) ∩ out(C2) ∩ … ∩ out(CM). The operators in() and out() give the regions inside and outside of an object, respectively. Equation (4) assumes that the image consists of M +1 homogeneous regions, M cells, and the background. The intensity means of the objects and the background are denoted by ci and cb, respectively, in (4). Parameters λo, λb, and μ are fixed weighting parameters. Function g : [0, +∞) → R+ is a strictly decreasing function, which is chosen to be a sigmoid function in our implementation
| (5) |
where α decides the slope of the output curve and β defines the point around which the output window is centered.
Combining the repulsion and competition terms, we obtain a joint energy functional E for cell segmentation
| (6) |
The segmentation is achieved by minimizing the energy functional (6). Explicit parametric active contours can be used to minimize the energy aforementioned; however, since there are hundreds of cells in each image, it is extremely complex to deal with the interaction between them using parametric active contours. In this paper, we use an efficient multiphase level set scheme for energy minimization, which is detailed as follows.
3) Evolution of Multiphase Level Set
In the level set formulation, Ci, the contour of the ith object, is embedded as the zero level set of a level set function ψi, i.e., Ci = {(x, y)|ψi(x, y) = 0}. The level set function ψi is defined to be positive outside of Ci and negative inside of Ci. The value of the function ψi at each point is computed as the Euclidean distance to the nearest point on the contour Ci. Each of the M objects being segmented in the image has its own contour Ci and corresponding ψi.
In order to express the energy functional (6) using level sets, we first bring in the Heaviside function H and the Dirac function δ
| (7) |
The inside and outside operators can then be formulated as
| (8) |
We are then able to express the energy functional (6) using level sets as
| (9) |
where Ω denotes the image domain.
The energy functional (9) can be minimized iteratively. In each iteration, we first fix ci (i = 1, 2, …,M) and cb and minimize the energy functional E with respect to ψi(x, y) (i = 1, 2, …,M). We employ the gradient descent method for minimization. The evolution equation for each ψi(t, x, y) is then obtained by deducing the associated Euler–Lagrange equation as
| (10) |
After evolving the level sets, the means ci and cb of the regions can be updated.
The new evolution equation has several advantages that are particularly useful for RNAi fluorescent cellular image segmentation. First of all, the overlapping of the evolving contours is prevented by the repulsion term, which helps to isolate the touching cells in the dense clusters. Besides that, the information from both the image intensity and the gradient is exploited by the new scheme. This helps to separate the cells from the background and deal with the dramatical intensity variation between different cells. Furthermore, the fourth term of (10) keeps the curves smooth, and makes the algorithm more robust to noise.
IV. Results and Discussion
In this section, we present our experimental results on the segmentation of Drosophila RNAi fluorescent cellular images. Since each image contains all kinds of cells, fixed universal parameters are needed for automatic segmentation. Different parameter settings are tested, and the one with the best performance is then used for the segmentation of all the images. The values of the parameters used in our experiments are shown in Table I. It should be noted that our algorithm is not sensitive to the parameters. In our experiments, the performance of the algorithm varies not more than 5%, when the values of the parameters change by approximately 25%. Currently, the segmentation of each image with 1280 × 1024 pixels takes approximately 2–3 s on a PC with 3 GHz CPU. The computational time can be significantly shortened by using high-performance computer or parallel computing techniques.
TABLE I.
Parameters Used in the Experiments
| Parameter | λ o | λ b | μ | ν | ω | α | β |
|---|---|---|---|---|---|---|---|
| Value | 1.0 | 1.0 | 0.8 | 0.3 | 0.7 | 2.0 | 7.0 |
Selected patches of the segmentation results containing cells with specific phenotypes “S-type,” “R-type,” and “A-type” are shown in Figs. 6-8 to demonstrate the performance of our algorithm. In Fig. 7, spiky cells are successfully segmented (notice the spiky edges of the cells). In addition, the tightly clustered cells are separated, thanks to the interaction model. Furthermore, cells with large intensity variations are extracted, and the contours are able to stop at the weak boundaries between the cells and the background, since both image intensity and gradient information is used. Similar observations can be made in Fig. 8, where segmentation results of “R-type” and “A-type” cells are demonstrated. The quantitative performance evaluation was performed by comparing our results to the ground truth. For comparison purposes, the segmentation results of the widely used open source software CellProfiler [27] on our data sets were also obtained. The performance of our method and that of CellProfiler were then evaluated and compared. In CellProfiler, thresholding was first applied to extract cells from the image. In order to divide the clustered cells, the propagation algorithm is employed [28]. This method is similar to the watershed method, but can be considered as an improvement in that it combines the distance to the nearest nucleus and intensity gradients to divide cells. However, the cells are divided at the places most likely with strong gradients.
Fig. 6.
Segmentation of ruffling cells. (a) Patch from the DNA channel shows the nuclei. (b) Two touching ruffling cells. (c) and (d) Intermediate segmentation results of the cells. (e) Final segmentation results of the ruffling cells.
Fig. 8.
Cell segmentation process. (a) Improved image of Actin channel with labeled nuclei as initialization. (b)–(d) Evolution process of level sets. (e) Final segmentation results with ruffling and actin cells successfully segmented.
Fig. 7.
Cell segmentation process. (a) Improved image of Actin channel. The nuclei obtained by segmenting the image of DNA channel are marked in different colors, which are used as the initialization of level sets. (b)–(e) Evolution process of level sets. (f) Final segmentation results with spiky cells successfully segmented.
Fig. 9 shows some randomly chosen image patches and the corresponding segmentation results of our method. Manual segmentation results from biologists are included as ground truth for validation. The segmentation results using CellProfiler with default parameter setting are also displayed. It can be seen that our segmentation method outperforms CellProfiler in two ways. One is that CellProfiler tends to oversegment the nuclei, where large nucleus may be divided into several small regions. Since the segmentation result of nuclei is used to determine the number of cells in the Actin channel, this directly results in the over-segmentation of the cells. More cells are produced than there actually are. The other problem of CellProfiler is that it tends to put circular shape constraints on segmentation of each cell. Thus, it is not able to get the specific cell shapes present in our data sets. For each cell, the segmented region from CellProfiler is normally round in shape with a smaller area than the ground truth. Thus, compared to CellProfiler, our method obtains better segmentation results.
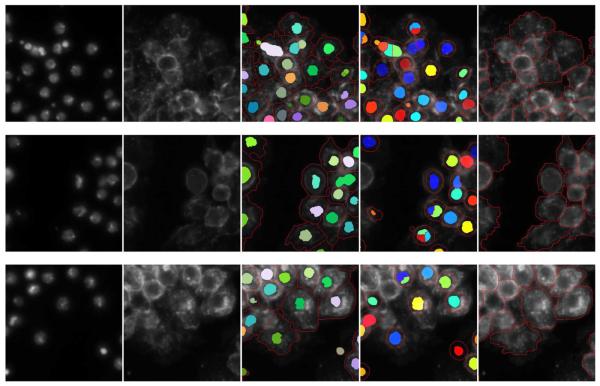
Fig. 9.
Three sets of segmentation results from the validation data sets. Each row shows the segmentation results of one set of images. First column: patches of DNA channel. Second column: patches of actin channel. Third column: our segmentation results. Fourth column: segmentation results of CellProfiler. Last column: manual segmentation results for comparison. Compared with the ground truth, one can see that CellProfiler tends to oversegment the nuclei, which leads to the oversegmentation of the cells. In addition, CellProfiler tends to put circular shape constraints on cell segmentation. Thus, it is not able to get the specific cell shapes present in our data sets. Evaluated against the ground truth, our method is able to get better segmentation results than CellProfiler.
In order to quantitatively analyze the performance of the proposed approach, an F-score is employed as our performance measure. The F-score is an evaluation measure computed from the precision and the recall values that are standard techniques used to evaluate the quality of the results of a system against the ground truth. Let X denote the segmentation result and Y be the ground truth. The precision p and recall r are given by
| (11) |
The F-score is then computed by taking the harmonic average of the precision and the recall [29]
| (12) |
where β is the weighting parameter. In order to give equal weight to precision and recall, a common approach is to set β = 1, such that F1 = 2pr/(p + r).
The F1-scores of our method and the CellProfiler are shown in Fig. 10. When computing the scores, 20 sets of images are randomly chosen from all the data sets. Since each image contains about 100–500 cells, thousands of cells are segmented and measured in our experiments. It can be seen that our method gets a higher score on all of the images. This can be explained by the results shown in Fig. 9. Evaluated against the ground truth, the CellProfiler may have higher precision values because it produces smaller cells so that most of its results are inside of the ground truth. However, it has very low recall values, because a large part of the cells are not obtained. The precision of the proposed method may not be as high as the CellProfiler,butthe recall values are generally much higher. Thus, our method has higher overall F1-scores.
Fig. 10.
F1-score of our method and the CellProfiler. Our method gets a higher score on all the validation data sets.
V. Conclusion
In this paper, we proposed an automatic scheme for the segmentation of high-throughput RNAi fluorescent cellular images. Since the precision of the following cell analysis largely depends on the performance of cell segmentation, it is of great importance to have a good segmentation scheme. In this paper, the proposed automated segmentation scheme starts extracting nuclei from the DNA channel and uses the results as the initialization for the segmentation of cells from other channels. A novel interaction model was formulated for segmenting tightly clustered cells with significant intensity variance and specific phenotypes. The energy functional was minimized by using a multiphase level set method, which leads to a highly effective cell segmentation method. Promising segmentation results on RNAi fluorescent cellular images are presented. In our future work, we will explore more image information for better cell segmentation. We believe that the incorporation of more image information, e.g., cell texture, into the interaction model will lead to a better segmentation performance. In addition, the proposed method can be extended to other multichannel image segmentation applications, but not limited to RNAi cellular image segmentation. We will explore the use of our method on solving other problems of multisource imaging systems.
Acknowledgment
The authors would like to express their appreciation of the rewarding collaborations with our biological colleagues; in particular, Dr. P. Bradley and Dr. N. Perrimon, Department of Genetics, Harvard Medical School. The raw image data described in this paper were obtained from their laboratory. The authors would also like to thank the research members, especially Mr. F. Li of the Life Science Imaging Group in the Center for Bioinformatics, Harvard Center for Neurodegeneration and Repair (HCNR), Harvard Medical School, and Functional and Molecular Imaging Center, Radiology, Brigham and Women's Hospital, Harvard Medical School.
The work of X. Zhou was supported by the Harvard Center for Neurodegeneration and Repair (HCNR), Center for Bioinformatics Research, Harvard Medical School, Boston, MA. The work of S. T. C. Wong was supported by the HCNR Center for Bioinformatics Research, Harvard Medical School, Boston, MA, and also by the National Institutes of Health (NIH) under Grant R01 LM008696.
Biographies
 Pingkun Yan (S'04–M'06) received the B.Eng. degree in electronics engineering from the University of Science and Technology of China (USTC), Hefei, China, in 2001, and the Ph.D. degree in electrical and computer engineering from the National University of Singapore (NUS), Singapore, in 2006.
Pingkun Yan (S'04–M'06) received the B.Eng. degree in electronics engineering from the University of Science and Technology of China (USTC), Hefei, China, in 2001, and the Ph.D. degree in electrical and computer engineering from the National University of Singapore (NUS), Singapore, in 2006.
He is currently a Research Associate with the Computer Vision Laboratory, School of Electrical Engineering and Computer Science, University of Central Florida (UCF), Orlando. His current research interests include computer vision, pattern recognition, machine learning, statistical image processing, and information theory with their applications to biomedical image analysis.
Dr. Yan is the recipient of the Medical Image Computing and Computer Assisted Intervention (MICCAI) 2005 Student Award for the best presentation on image segmentation and analysis at the 8th International Conference on MICCAI, 2005. He is a member of the Sigma Xi.
 Xiaobo Zhou (S'98–A'02–M'03) received the B.S. degree in mathematics from Lanzhou University, Lanzhou, China, in 1988, and the M.S. and Ph.D degrees in mathematics from Peking University, Beijing, China, in 1995 and 1998, respectively.
Xiaobo Zhou (S'98–A'02–M'03) received the B.S. degree in mathematics from Lanzhou University, Lanzhou, China, in 1988, and the M.S. and Ph.D degrees in mathematics from Peking University, Beijing, China, in 1995 and 1998, respectively.
From 1988 to 1992, he was a Lecturer at the Training Center, 18th Building Company, Chongqing, China. From 1992 to 1998, he was a Research Assistant and a Teaching Assistant with the Department of Mathematics, Peking University. From 1998 to 1999, he was a Postdoctoral Research Fellow in the Department of Automation, Tsinghua University, Beijing. From 1999 to 2000, he was a Senior Technical Manager with the 3G Wireless Communication Department, Huawei Technologies Company, Ltd., Beijing. From February 2000 to December 2000, he was a Postdoctoral Research Fellow in the Department of Computer Science, University of Missouri-Columbia, Columbia. From 2001 to 2003, he was a Postdoctoral Research Fellow in the Department of Electrical Engineering, Texas A&M University, College Station. From 2003 to 2006, he was a Research Scientist and Instructor with the Harvard Center for Neurode-generation and Repair in Harvard Medical School and Radiology Department in Brigham and Women's Hospital, Boston, MA. In 2007, he was an Assistant Professor at Harvard University. Since 2007, he has been an Associate Professor at Cornell University and Methodist Hospital, Houston, TX. His current research interests include image and signal processing for high-content molecular and cellular imaging analysis, informatics for integrated multiscale and multimodality biomedical imaging analysis, molecular imaging informatics, neuroinformatics, and bioinformatics for genomics and proteomics.
 Mubarak Shah (M'81–F'03) received the B.E. degree in electronics from Dawood College of Engineering and Technology, Karachi, Pakistan, in 1979. He completed the E.D.E. diploma at Philips International Institute of Technology, Eindhoven, The Netherlands, in 1980, and received the M.S. and Ph.D. degrees in computer engineering from Wayne State University, Detroit, MI, in 1982 and 1986, respectively.
Mubarak Shah (M'81–F'03) received the B.E. degree in electronics from Dawood College of Engineering and Technology, Karachi, Pakistan, in 1979. He completed the E.D.E. diploma at Philips International Institute of Technology, Eindhoven, The Netherlands, in 1980, and received the M.S. and Ph.D. degrees in computer engineering from Wayne State University, Detroit, MI, in 1982 and 1986, respectively.
He is the Chair Professor of Computer Science and the founding Director of the Computer Visions Laboratory, University of Central Florida (UCF), Orlando. He is a Researcher in a number of computer vision areas. He is a Guest Editor of the special issue of International Journal of Computer Vision on Video Computing. He is an Editor of the international book series on Video Computing, the Editor-in-Chief of the Machine Vision and Applications Journal, and an Associate Editor of the Association for Computer Machinery (ACM) Computing Surveys Journal. He was an IEEE Distinguished Visitor Speaker from 1997 to 2000.
Dr. Shah is the recipient of the IEEE Outstanding Engineering Educator Award in 1997. He also received of the Harris Corporation's Engineering Achievement Award in 1999, the Transfer of Knowledge Through Expatriate Nationals (TOKTEN) Awards from the United Nations Development Programme (UNDP) in 1995, 1997, and 2000, the Teaching Incentive Program Awards in 1995 and 2003, the Research Incentive Award in 2003, Millionaires' Club Awards in 2005 and 2006, the University Distinguished Researcher Award in 2007, Honorable Mention for the International Conference on Computer Vision (ICCV) 2005, and was also nominated for the Best Paper Award in the ACM Multimedia Conference in 2005. In 2006, he was awarded the Pegasus Professor Award, the highest award at the UCF. He was an Associate Editor of the IEEE Transactions on Pattern Analysis and Machine Intelligence (PAMI).
 Stephen T. C. Wong received the Ph.D. degree in electrical engineering and computer science (EECS) from Lehigh University, Bethlehem, PA, in 1991.
Stephen T. C. Wong received the Ph.D. degree in electrical engineering and computer science (EECS) from Lehigh University, Bethlehem, PA, in 1991.
He was the Founding Director of the Havard Center for Neurodegeneration and Repair (HCNR) Center for Bioinformatics; the Founding Director of the Functional and Molecular Imaging Center, Department of Radiology, Brigham and Women's Hospital; and an Associate Professor of radiology, Harvard Medical School, Boston, MA. He is currently the Vice Chairman and the Chief of Medical Physics, Department of Radiology, Methodist Hospital, Houston, TX, and the Program Director of Bioinformatics, and Senior Member of the Methodist Hospital Research Institute, Weill Cornell Medical College of Cornell University, Ithaca, NY. He has over two decades of research and development experience with leading institutions in academia and industry, including Hewlett-Packard (HP), AT&T Bell Laboratories, Japanese Fifth Generation Computing Project, Royal Philips Electronics of California, San Francisco (UCSF), Charles Schwab, and Harvard, and has made original contributions in the fields of bioinformatics and biomedical image computing. He is the author or coauthor of more than 200 peer-reviewed articles, two books, and six patents. He has also been a member of the editorial boards of seven international journals. His current research interests include biomedical informatics, biomedical imaging, and molecular image-guided therapy.
Dr. Wang has been a PE (registered engineer since 1991).
Contributor Information
Pingkum Yan, School of Electrical Engineering and Computer Science, University of Central Florida, Orlando, FL 32816 USA.
Xiaobo Zhou, Center for Bioinformatics, Harvard Center for Neurodegeneration and Repair, Harvard Medical School and the Functional and Molecular Imaging Center, Department of Radiology, Brigham and Women's Hospital and Harvard Medical School, Boston, MA 02115 USA..
Mubarak Shah, School of Electrical Engineering and Computer Science, University of Central Florida, Orlando, FL 32816 USA.
Stephen T. C. Wong, Center for Bioinformatics, Harvard Center for Neurodegeneration and Repair, Harvard Medical School and the Functional and Molecular Imaging Center, Department of Radiology, Brigham and Women's Hospital and Harvard Medical School, Boston, MA 02115 USA.
REFERENCES
- 1.Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, Haas SA, Paro R, Perrimon N. Genome-wide RNAi analysis of growth and viability in drosophila cells. Science. 2004;303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- 2.Agaisse H, Burrack L, Philips J, Rubin E, Perrimon N, Higgins DE. Genome-wide RNAi screen for host factors required for intra-cellular bacterial infection. Science. 2005 Aug.309(5738):1248–1251. doi: 10.1126/science.1116008. [DOI] [PubMed] [Google Scholar]
- 3.Sonnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume A-M, Artelt J, Bettencourt P, Cassin E, Hewitson M, Holz C, Khan M, Lazik S, Martin C, Nitzsche B, Ruer M, Stamford J, Winzi M, Heinkel R, Röder M, Finell J, Häantsch H, Jones SJM, Jones M, Piano F, Gunsalus KC, Oegema K, Gönczy P, Coulson A, Hyman AA, Echeverri CJ. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- 4.Kiger A, Baum B, Jones S, Jones M, Coulson A, Echeverri C, Perrimon N. A functional genomic analysis of cell morphology using RNA interference. J. Biol. 2003;2:27. doi: 10.1186/1475-4924-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou X, Wong S. High content cellular imaging for drug development. IEEE Signal Process. Mag. 2006 Mar.23(2):170–174. [Google Scholar]
- 6.Zhou X, Wong S. Informatics challenges of high-throughput microscopy. IEEE Signal Process. Mag. 2006 May;23(3):63–72. [Google Scholar]
- 7.Zimmer C, Labruyère E, Meas-Yedid V, Guillén N, Olivo-Marin J-C. Segmentation and tracking of migrating cells in videomicroscopy with parametric active contours: A tool for cell-based drug testing. IEEE Trans. Med. Imag. 2002 Oct.21(10):1212–1221. doi: 10.1109/TMI.2002.806292. [DOI] [PubMed] [Google Scholar]
- 8.Dufour A, Shinin V, Tajbakhsh S, Guillén-Aghion N, Olivo-Marin J-C, Zimmer C. Segmenting and tracking fluorescent cells in dynamic 3-D microscopy with coupled active surfaces. IEEE Trans. Image Process. 2005 Sep.14(9):1396–1410. doi: 10.1109/tip.2005.852790. [DOI] [PubMed] [Google Scholar]
- 9.Wählby C, Lindblad J, Vondrus M, Bengtsson E, Björkesten L. Algorithms for cytoplasm segmentation of fluorescence labelled cells. Anal. Cell. Pathol. 2002;24:101–111. doi: 10.1155/2002/821782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou X, Liu K-Y, Bradley p., Perrimon N, Wong ST. Towards automated cellular image segmentation for RNAi genome-wide screening. In: Duncan J, Gerig G, editors. Proc. Med. Image Comput. Comput.-Assisted Intervention. Palm Springs; CA: 2005. pp. 885–892. [DOI] [PubMed] [Google Scholar]
- 11.Bamford P, Lovell B. Unsupervised cell nucleus segmentation with active contours—An efficient algorithm based on immersion simulations. Signal Process. 1998;71:203–213. [Google Scholar]
- 12.Garrido A, Pérez de la Blanca N. Applying deformable templates for cell image segmentation. Pattern Recog. 2000;33(5):821–832. [Google Scholar]
- 13.Ortiz de Solórzano C, Malladi R, Leliévre SA, Lockett SJ. Segmentation of nuclei and cells using membrane related protein markers. J. Microsc. 2001;201:404–415. doi: 10.1046/j.1365-2818.2001.00854.x. [DOI] [PubMed] [Google Scholar]
- 14.Osher S, Sethian JA. Fronts propagating with curvature-dependent speed: Algorithms based on Hamilton–Jacobi formulations. J. Comput. Phys. 1988;79:12–49. [Google Scholar]
- 15.Malladi R, Sethian JA, Vermuri BC. Shape modeling with front propagation: A level set approach. IEEE Trans. Pattern Anal. Mach. Intell. 1995 Feb.17(2):158–174. [Google Scholar]
- 16.Zhao H-K, Chan TF, Merriman B, Osher S. A variational level set approach to multiphase motion. J. Comput. Phys. 1996;127:179–195. [Google Scholar]
- 17.Samson C, Blanc-Feraud L, Aubert G, Zerubia J. A level set model for image classification. Int. J. Comput. Vis. 2000;40(3):187–197. [Google Scholar]
- 18.Brox T, Weickert J. Pattern Recognition (LNCS 3175) Springer-Verlag; Berlin, Germany: 2004. Level set based image segmentation with multiple regions; pp. 415–423. [Google Scholar]
- 19.Vese LA, Chan TF. A multiphase level set framework for image segmentation using the mumford and shah model. Int. J. Comput. Vis. 2002;50(3):271–293. [Google Scholar]
- 20.Yan P, Shen W, Kassim AA, Shah M. Segmentation of neighboring organs in medical image with model competition. In: Duncan J, Gerig G, editors. Proc. Med. Image Comput. Comput.-Assisted Intervention. Vol. 1. Palm Springs; CA: 2005. pp. 270–277. [DOI] [PubMed] [Google Scholar]
- 21.Vincent L, Soille P. Watersheds in digital spaces: An efficient algorithm based on immersion simulations. IEEE Trans. Pattern Anal. Mach. Intell. 1991 Jun.13(6):583–598. [Google Scholar]
- 22.Yang Q, Parvin B. Harmonic cut and regularized centroid transform for localization of subcellular structures. IEEE Trans. Biomed. Eng. 2003 Apr.50(4):469–475. doi: 10.1109/TBME.2003.809493. [DOI] [PubMed] [Google Scholar]
- 23.Wählby C, Sintorn I-M, Erlandsson F, Borgefors G, Bengtsson E. Combining intensity, edge and shape information for 2D and 3D segmentation of cell nuclei in tissue sections. J. Microsc. 2004 Jul.215:67–76. doi: 10.1111/j.0022-2720.2004.01338.x. [DOI] [PubMed] [Google Scholar]
- 24.Mumford D, Shah J. Optimal approximation by piecewise smooth functions and associated variational problems. Commun. Pure Appl. Math. 1989;42:577–685. [Google Scholar]
- 25.Chan TF, Vese LA. Active contours without edges. IEEE Trans. Image Process. 2001 Feb.10(2):266–277. doi: 10.1109/83.902291. [DOI] [PubMed] [Google Scholar]
- 26.Caselles V, Kimmel R, Sapiro G. Geodesic active contours. Int. J. Comput. Vis. 1997;22(1):61–79. [Google Scholar]
- 27.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang I, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland p., Sabatini DM. Cellprofiler: Image analysis for high throughput microscopy. doi: 10.1186/gb-2006-7-10-r100. [Online]. Available: http://www.cellprofiler.org. [DOI] [PMC free article] [PubMed]
- 28.Jones T, Carpenter A, Golland P. Proc. ICCV Workshop Comput. Vis. Biomed. Image Appl. Beijing, China: 2005. Voronoi-based segmentation of cells on image manifolds; pp. 535–543. [Google Scholar]
- 29.van Rijsbergen C. Information Retrieval. 2nd ed. Butterworth; London, U.K.: 1979. [Google Scholar]