Abstract
Phosphorylation is a universal mechanism for regulating cell behavior in eukaryotes. Although protein kinases are known to target short linear sequence motifs on their substrates, the rules for kinase substrate recognition are not completely understood. We used a rapid peptide screening approach to determine consensus phosphorylation site motifs targeted by 61 of the 122 kinases in Saccharomyces cerevisae. Correlation of these motifs with kinase primary sequence has uncovered previously unappreciated rules for determining specificity within the kinase family, including a residue determining P−3 Arg specificity among members of the CMGC group of kinases. Furthermore, computational scanning of the yeast proteome enabled the prediction of thousands of new kinase-substrate relationships. We experimentally verified several candidate substrates of the Prk1 family of kinases in vitro and in vivo, and we identified a protein substrate of the kinase Vhs1. Together, these results elucidate how kinase catalytic domains recognize their phosphorylation targets and suggest general avenues for the identification of new kinase substrates across eukaryotes.
INTRODUCTION
As one of the most widespread posttranslational modifications, protein phosphorylation is involved in virtually every basic cellular process, including DNA replication, gene transcription, protein translation, cell growth and metabolism, differentiation, and intercellular communication. With the advent of whole genome sequencing, the entire complement of kinases, or “kinome”, for multiple organisms have been cataloged, revealing that most eukaryotes devote ~2% of their protein coding capacity to these enzymes (1). Unraveling the function of each member of such a large family remains a challenge. Advances in phosphoproteomic methodologies, such as large-scale mass spectrometry (MS)-based phosphorylation site discovery, targeted siRNA screens, the use of analog-sensitive kinase alleles that are engineered to accept specific inhibitors and ATP analogs, and protein microarray analyses, have shed considerable light on the scope and complexity of phosphorylation-based signal transduction pathways in eukaryotes (2–5).
However, one aspect of protein kinase biology that remains poorly understood is how kinases achieve specificity for their target substrates. Understanding rules for substrate recognition by kinases has important applications in the mapping of phosphorylation sites in protein substrates, discovery of new substrates, and production of model substrates for small molecule inhibitor screening (6). In addition, a detailed understanding of how kinases interact with their substrates enables both deciphering and genetic re-wiring of kinase specificity, thereby uncovering fundamental ways in which signaling pathways are organized and propagated (7, 8).
In a typical eukaryotic cell, there are hundreds of thousands of Ser, Thr, and Tyr residues among the thousands of proteins. To ensure signaling fidelity, kinases must somehow discriminate among these vast numbers of potential phosphorylation sites. Mechanisms that influence substrate selection by a protein kinase include subcellular localization, substrate docking interactions, and binding to scaffold proteins (9). An important aspect of substrate recognition, however, is that the phosphorylation site on the substrate falls within a consensus amino acid sequence that is complementary to the active site of the kinase.
Consensus phosphorylation site motifs for protein kinases have been previously established on an individual basis through either the inspection of known phosphorylation sites, systematic mutagenesis of protein and peptide substrates, or screening of peptide libraries (10, 11). Although these studies have provided valuable insight into substrate recognition, such data is only available for a subset of known protein kinases. NetPhorest, which is the most comprehensive repository for kinase phosphorylation site motifs reported to date, includes motifs for only 35% of all human kinases (12). The incompleteness of available data and heterogeneity by which it was collected limits its application to elucidating cellular signaling pathways and modeling larger phosphorylation networks. For example, using motif scanning approaches to link specific kinases to the thousands of in vivo phosphorylation sites discovered through MS-based phosphoproteomics has proven difficult in targeted kinase studies because multiple kinases can potentially target the same or similar motifs.
We thus set out to catalog consensus phosphorylation site motifs for the kinome of the model organism Saccharomyces cerevisiae. We adapted a peptide library screening approach (13) to a miniaturized format that would enable rapid analysis of large numbers of kinases. With this method, we determined consensus phosphorylation motifs targeted by 61 of the 122 yeast kinases. This large collection of phosphorylation site motifs provided new insight into the structural basis for substrate recognition by protein kinases as a family in a manner not possible through analyses of individual kinases. Furthermore, we used our motif collection to predict new kinase-substrate relationships through database scanning and integration with other yeast proteomic and genomic datasets.
RESULTS
A rapid peptide-based approach for the high-throughput determination of kinase consensus phosphorylation site motifs
To determine phosphorylation motifs for yeast protein kinases, we developed a high-throughput approach using our previously reported positional scanning peptide library (13). This library consisted of 200 distinct peptide mixtures in which each 16-mer peptide contained a central fixed phosphorylation acceptor (phosphoacceptor) site (an equimolar mixture of Ser and Thr) flanked by degenerate positions consisting of equimolar mixtures of the 20 amino acids excluding Ser, Thr, and Cys, and a carboxy-terminal biotin tag (Fig. 1A). For each of the nine positions surrounding the phosphoacceptor site, there were 22 peptide mixtures in which each of the 20 unmodified amino acids, as well as phosphothreonine (pT) and phosphotyrosine (pY), were fixed. In addition to these 198 (9 × 22) peptide mixtures, two control peptide mixtures bearing either Ser or Thr alone as the fixed phosphoacceptor residue in the context of a fully degenerate sequence were also included. These control mixtures served as indicators of any preference the kinase had for either Ser or Thr residues at the phosphoacceptor site. Peptides were incubated with the kinase of interest in the presence of radiolabeled ATP. At the end of the incubation period, aliquots of each reaction were spotted simultaneously using a capillary pin-based liquid transfer device onto a streptavidin-coated membrane that captured the peptide substrates through their carboxy-terminal biotin tags. After extensive washing, the membrane was dried and exposed to a phosphor screen, allowing the extent of radiolabel incorporation for each peptide to be visualized and quantified. To enable high-throughput analysis, all steps were performed in a 1536-well format, thereby reducing the amount of kinase and peptide required and enabling simultaneous analysis of four kinases.
Fig. 1.
Miniaturized peptide array approach enables high-throughput analysis of kinase consensus phosphorylation motifs. (A) Scheme for kinase peptide screening. Capillary pin-based liquid transfer devices were used to add components to reactions (2 μl per well) and spot 0.2 μl aliquots onto the streptavidin-coated membrane following incubation. The 1536-well format allows four kinases to be analyzed simultaneously. (B) Representative peptide screening results for Atg1, Gin4, Mps1, and Prk1. (C) Phosphorylation of consensus peptide substrates by Atg1, Gin4, Mps1, and Prk1. The sequence of each peptide is as follows: ATGtide, YANWLAASIYLDGKKK; GINtide, YALRRSRSMWNLGKKK; MPStide, YADHDDDTMHFRGKKK; and PRKtide, YALKPQYTGPRGKKK. Peptide phosphorylation was assayed at 10 μM concentration by radiolabel kinase assay. Incorporation of radiolabeled phosphate into peptides was determined by phosphocellulose filter binding assay. Maximal rates for each kinase in these assays were: Atg1, 250 nM/min, Gin4, 510 nM/min, Mps1, 130 nM/min, Prk1, 330 nM/min. (D) Rates of Atg1 phosphorylation of ATGtide variants with individual point substitutions. Peptide phosphorylation was assayed as for panel C.
Three yeast kinases (Tpk1, Tpk2, and Ste20) were assayed with both the miniaturized and large volume formats, and we performed multiple replicates with one of these kinases, Tpk1. Identical results were observed with the two formats and in replicate assays with the 1536-well format (data for Tpk1 is shown in Fig. S1). These kinases also recapitulated preferences of their mammalian orthologs for basic residues upstream of the phosphorylation site (13, 14). These results confirm that the miniaturized peptide library screening system is reproducible and provides data that is quantitatively equivalent to lower throughput approaches.
Screening yeast kinases for their consensus phosphorylation site motifs
With our peptide array method, we screened 111 of the 122 yeast kinases. Kinases were initially purified from yeast strains that harbor galactose-inducible expression plasmids bearing either a C-terminal tandem affinity purification tag or an N-terminal glutathione S-transferase (GST) tag (15, 16). In a number of instances, it was necessary to perform the assay in the presence of known activating subunits [(for example, cyclins for cyclin-dependent kinases (CDKs)], phosphorylate the kinase in vitro or co-express it with an activating kinase, or purify the kinase from yeast grown under activating conditions. For kinases with which poor yields were obtained from yeast, we employed alternative bacterial and mammalian cell expression systems. Each kinase was assayed on the peptide substrates in duplicate on separate days. In total, we generated reproducible phosphorylation motifs for 61 of the 111 yeast kinases screened (Fig. 1B and table S1). Three distinct motifs were generated for the cyclin-dependent kinase Pho85 by analyzing separately in complex with different cyclin subunits (Pho80, Pcl1 and Pcl2). The remaining kinases were not sufficiently active to phosphorylate the peptides above background levels. These kinases may be highly specific for particular protein substrates and thus do not phosphorylate peptides efficiently. For example, in keeping with previous observations for their mammalian orthologs (17), we did not observe activity on our peptide substrates for the eight kinases in the mitogen-activated protein kinase kinase (MAPKK) and mitogen-activated protein kinase kinase kinase (MAPKKK) families. Other kinases were likely simply inactive under exponential growth conditions or when assayed in the absence of obligate binding partners and may be suitable for analysis once their activation mechanisms are more completely understood.
Approximately half of the phosphorylation site motifs that we determined for yeast kinases were identical to known motifs, as they corresponded to yeast homologs of mammalian kinases that have been previously characterized (11, 12). In contrast, the remaining kinases and their mammalian homologs have either not been previously characterized (table S2 lists mammalian homologs and indicates which kinases have previously known motifs) or in one instance (Tos3) yielded a different motif from that reported. Representative spot arrays produced by four kinases for which phosphorylation motifs were not previously known (Atg1, Gin4, Mps1, and Prk1) are shown in Fig. 1B. Spot intensities from the peptide arrays were quantified, background corrected, and normalized to provide the selectivity values shown in Table 1. We verified the consensus phosphorylation motifs for these kinases by performing kinase assays using optimized peptide substrates (named ATGtide, GINtide, MPStide, and PRKtide, respectively) consisting of those residues that were most highly selected at each position. As shown in Figure 1C, each kinase was highly specific for its corresponding peptide substrate, thus providing independent validation of our mixture based peptide library screening approach.
Table 1.
Quantified selectivity values for protein kinases discussed in the text. Peptide array data were quantified and normalized to an average value of 1 within a position. Positively selected residues with values greater than 1.5 are shown. Complete quantified PWMs for all kinases are provided as Dataset S1.
| Position |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Kinase | −5 | −4 | −3 | −2 | −1 | +1 | +2 | +3 | +4 |
| Atg1 | W (1.8) | L (2.8) | A (1.6) | V (2.4) | Y (2.8) | L (1.7) | D (1.8) | ||
| M (2.4) | I (2.3) | I (2.3) | |||||||
| F (1.9) | F (2.1) | ||||||||
| M (1.9) | M (1.8) | ||||||||
| W (1.6) | |||||||||
| V (1.6) | |||||||||
| Gin4 | L (2.0) | R (2.7) | R (9.9) | S (7.9) | M (2.9) | M (2.6) | W (2.4) | N (2.8) | L (3.9) |
| I (1.8) | K (2.4) | K (1.6) | T (1.8) | K (1.8) | Y (2.2) | D (1.8) | I (2.3) | ||
| M (1.7) | H (1.8) | R (1.6) | P (1.8) | F (1.6) | |||||
| R (1.6) | |||||||||
| K (1.6) | |||||||||
| Mps1 | D (1.9) | D (4.5) | D (1.6) | M (1.9) | H (1.6) | ||||
| R (1.9) | E (2.4) | V (1.7) | |||||||
| R (2.2) | I (1.7) | ||||||||
| Prk1 | L (4.5) | R (4.0) | Q (3.4) | Y (2.2) | G (6.8) | ||||
| I (2.7) | P (1.9) | H (2.0) | H (1.9) | ||||||
| M (1.6) | R (1.9) | ||||||||
| Snf1 | L (2.5) | R (2.8) | R (7.5) | R (3.0) | R (2.2) | M (1.9) | N (2.1) | L (4.5) | |
| I (2.0) | K (2.5) | K (3.0) | T (2.4) | K (1.8) | F (1.9) | F (1.7) | I (2.1) | ||
| H (1.7) | S (2.2) | H (1.7) | C (1.8) | M (1.7) | |||||
| C (1.7) | Y (1.7) | Y (1.6) | |||||||
| V (1.6) | |||||||||
| Kin1 | K (1.7) | H (2.0) | H (2.2) | N (4.0) | R (2.0) | Q (1.6) | D (2.2) | L (5.5) | |
| W (2.0) | W (2.0) | S (2.3) | R (1.6) | N (1.7) | I (2.4) | ||||
| F (1.9) | C (1.6) | V (1.9) | |||||||
| Y (1.6) | |||||||||
| Kss1 | H (1.8) | P (6.2) | D (2.0) | P (8.5) | P (1.9) | K (1.7) | |||
| A (1.9) | |||||||||
| R (1.9) | |||||||||
| Yak1 | R (1.7) | R (2.4) | R (8.0) | R (3.0) | R (2.5) | P (5.5) | C (1.8) | P (1.7) | |
| C (2.5) | K (2.0) | R (2.3) | |||||||
| P (2.1) | H (1.6) | ||||||||
| A (1.6) | |||||||||
| Vhs1 | M (6.4) | R (2.1) | R (11) | S (3.1) | K (1.8) | S (2.7) | F (1.6) | ||
| K (1.6) | T (2.7) | H (1.7) | T (2.0) | ||||||
| V (1.6) | R (2.0) | ||||||||
Notably, the autophagy-linked kinase Atg1 has an atypical motif exhibiting selections for hydrophobic residues at multiple positions. We verified this motif by making targeted substitutions to the ATGtide substrate. As anticipated, substituting a different favorable hydrophobic residue (Met) at the most selective position (P−3) had no significant effect on the rate of ATGtide phosphorylation. Moreover, substituting unfavorable charged residues at any of three most strongly selective positions dramatically reduced the reaction rate (Fig. 1D).
Overall features of kinase phosphorylation signatures
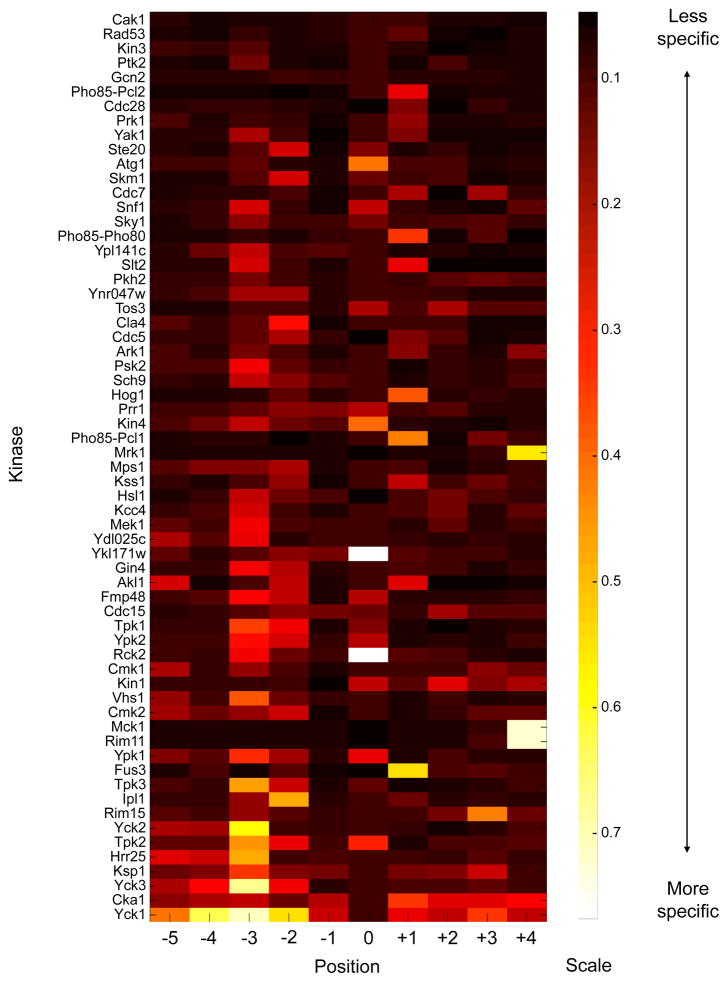
Normalized, background corrected phosphorylation signals for each kinase were assembled into position weight matrices (PWMs), which are quantitative representations of the phosphorylation motif. We scored each position for its total selectivity, and a specificity heat map of all kinases and positions revealed the wide range of selectivity exhibited by kinases (Fig. 2). At one extreme, Yck1 and Cka1 (yeast casein kinase 1 and casein kinase 2 homologs) were highly sequence specific, with requirements for particular amino acids at multiple positions. At the other extreme, Cak1 and Rad53 were the least selective in that, although the extent of substrate phosphorylation by these kinases is clearly dependent on peptide sequence, there were no residues that were absolutely required at any position surrounding the phosphoacceptor. Most kinases fell between these extremes, with a combination of required residues and more subtle propensities that influence the overall efficiency of phosphorylation. Furthermore, although each position surrounding the phosphorylation site was highly selective for by at least several kinases, kinases were most frequently selective at the P−3 position, followed by the P−2 and P+1 positions. By contrast, few kinases were selective at the P−1 position.
Fig. 2.
Heat map ranking kinases by their specificity quotients as calculated from their average PWMs. Kinases are ranked from least specific (top) to most specific (bottom). The specificity in each position is defined as the information content in each position, equivalent to the total height of the sequence logo (see table S1 for logos).
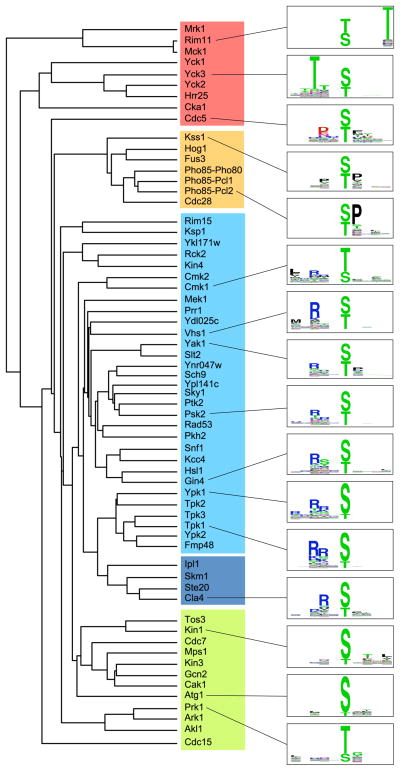
The 61 yeast kinases were clustered into groups on the basis of phosphorylation site selectivity (Fig. 3). 35 kinases were observed to target basophilic motifs. 31 of these showed a classic “basophilic” signature (10), with a strong selectivity primarily for an Arg residue at the P−3 position. This was the single most common feature found among all motifs (Fig. 3, table S1). Four other basophilic kinases, Ipl1, Skm1, Ste20, and Cla4, were selective for Arg at the P−2 position, but did not show strong selectivity for Arg at the P−3 position (Fig. 3 and table S1). The basophilic kinases however diverged with respect to the residues selected at other positions. For example, basophilic kinases are often reported to be selective primarily for either Leu or Arg at the P−5 position, as well as selective for Arg at P−3 (13, 18–20). Among the various kinases that selected Arg at the P−3 position, we observed a spectrum of residues selected at the P−5 position, including Leu (Cmk1 and Cmk2) and Arg (Ypk1), but also Met (Vhs1), Val or Ile (Prr1), and His (Psk2) (Fig. 3 and table S1). The seven proline-directed kinases, which primarily selected for Pro at the P+1 position, were also distinguishable on the basis of selectivity at other positions. For example, Kss1, Hog1, and Fus3 all showed a secondary selectivity for proline at the P−2 position that was not observed by Pho85 or Cdc28. Other motifs were less common, and include multiple distinct “acidophilic” motifs in which the strongest selectivity was for Asp, Glu, or pThr. Such acidophilic motifs have been previously seen for various mammalian kinases, including GSK3 (selectivity for acidic amino acids at the P+4 position), CK1 (P−5 through P−3), PLK (P−2), and CK2 (P+1 through P+3) (21–23). All yeast orthologs of these kinases recapitulated the motif found in their mammalian orthologs (table S2), but we also found additional yeast acidophilic kinases that were not anticipated (Mps1, Gcn2, and Cdc7). In addition, three kinases, Atg1, Kin1, and Kin3, exhibited their strongest selectivities for hydrophobic residues. The remaining kinases exhibited multiple strong selectivities and could not easily be categorized.
Fig. 3.
Dendrogram of yeast kinases clustered by specificity. Specificity categories are indicated by shading: red, acidophilic; orange, Pro-directed; cyan, P−3 Arg selecting; blue, P−2 Arg selecting; green, other. Because there were multiple distinct acidophilic motifs in which selectivity is varied by position, some kinases selecting primarily acidic residues clustered in the “other” category. Sequence logos (74) are shown for selected kinases from each group.
Connecting phosphorylation site motifs to kinase specificity determining residues
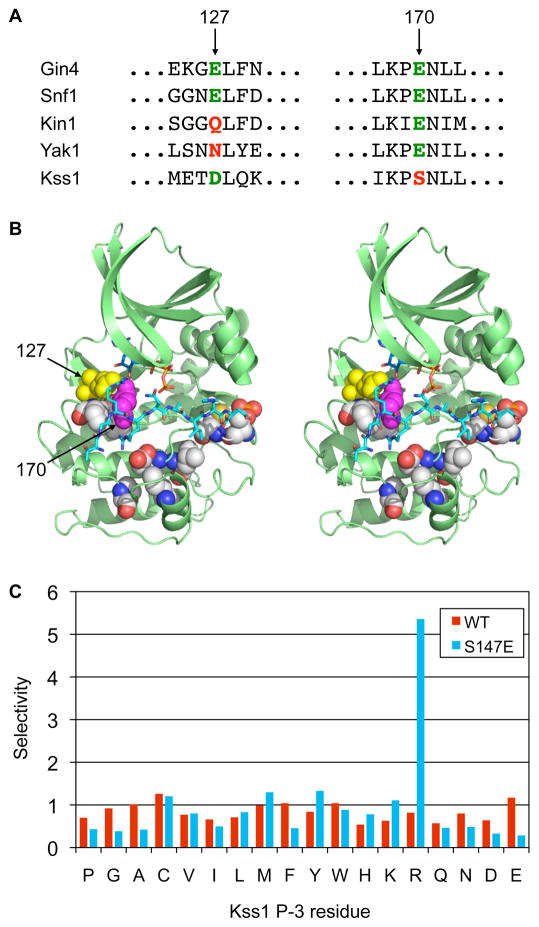
Yeast kinases have been classified into five groups on the basis of sequence homology: AGC (PKA/PKG/PKC), CAMK (calcium/calmodulin regulated and structurally similar kinases), CMGC (CDKs, MAPK, GSK, and CDK-like kinases), STE11/STE20, and STE7/MEK (MAPKK) (24). These groups have then been classified further into families that share a high degree of sequence similarity within their catalytic domains. Although related kinases generally recognized similar phosphorylation motifs, kinases within the same family occasionally exhibited differences, both subtle and striking. One family that illustrates striking differences is the Snf1 kinase family, which belongs to the CAMK group. In yeast, the Snf1 [also known as the AMPK (AMP-activated protein kinase)] family has six family members — Gin4, Hsl1, Kcc4, Kin1, Kin2, and Snf1. We identified consensus phosphorylation site motifs for each of these kinases with the exception of Kin2 (Table 1 and table S1). All five kinases had common features in their motifs, which are also shared with mammalian AMPKs (25, 26). For example, each one had preferences for a Ser residue as the phosphoacceptor site, a Ser residue at the P−2 position, an Asn residue at the P+3 position, and hydrophobic residues at the P+4 position (Gin4, Snf1, and Kin1 are summarized in Table 1; see Dataset S1 for quantitative data for Hsl1 and Kcc4). Strikingly, however, only four of the five Snf1 family kinases exhibited the hallmark basophilic P−3 Arg selectivity of the CAMK group, with Kin1 lacking this conserved feature. Instead, Kin1 had an additional preference for an Asn residue at the P−2 position. This difference correlated with a single amino acid substitution within the kinase catalytic domain (Fig. 4A). Gin4, Hsl1, Kcc4, and Snf1 each have a conserved Glu residue (corresponding to Glu127 in PKA, Fig. 4B). Crystal structures of multiple basophilic kinases in complex with peptide substrates have shown that this residue forms a salt bridge with the guanidino group of the P−3 Arg residue of the bound substrate (27–30). Unlike the other family members, Kin1 has a Gln residue in place of this conserved Glu. These observations are thus consistent with a role for Glu127 as the critical specificity-determining residue for Arg at the P−3 position in substrates, at least within the Snf1 family.
Fig. 4.
Comparison of kinase consensus phosphorylation site motifs to primary sequence reveals specificity-determining residues. (A) Sequence alignment of the regions surrounding residues 127 and 170 (human PKA numbering) in the catalytic domain of representative Snf1 family kinases (Gin4, Snf1, Kin1), and the CMGC kinases Yak1 and Kss1. The presence of an acidic residue at position 127 correlates with Arg selectivity at the P−3 position for the Snf1 family, but not the CMGC group. Conversely, a Glu residue at position 170 correlates with Arg selectivity for CMGC group kinases, but not for the Snf1 family. (B) Stereo view of the crystal structure of PKA with bound pseudosubstrate peptide (shown in cyan in stick representation; for clarity only the portion falling within the active site cleft is shown) highlighting predicted specificity determining residues (in sphere representation). Residues 127 and 170 are shown in yellow and magenta, respectively. The figure was generated using Pymol from the coordinates in PDB code 1ATP. (C) Kss1 mutagenesis. Mutation Kss1 Ser147 to Glu confers selectivity for Arg at P−3. The bar graph shows normalized spot intensities for the P−3 position taken from screens of the full peptide library (shown in Fig. S2).
However, crystallographic insight into specificity determinants in protein kinases is limited to a handful of cases where structures have been solved of kinase-peptide complexes. Although computational approaches have offered additional insight into structural features that control specificity (31, 32), the existence of alternative binding modes, even between kinases with similar specificity (30), makes it difficult to make general conclusions regarding the relationship of kinase sequence to specificity. Indeed, multiple sequence alignment of the yeast kinome and comparison with our experimentally determined motifs indicated that the presence of an acidic residue at position 127 is neither necessary nor sufficient to direct selectivity for Arg at the P−3 position in substrates. For example, within the CMGC group, members of the MAPK and CDK families (Fus3, Kss1, Hog1, Cdc28, and Pho85), which are proline-directed kinases, have an Asp residue at that position, despite a lack of selectivity for Arg at the P−3 position. Conversely, Yak1 within the same group is basophilic, yet lacks an acidic residue at that position (Table 1 and Fig. 4A). Presumably, other residues within the catalytic domain are responsible for dictating a basophilic signature within this group of kinases.
With our large collection of kinase motifs, we identified previously unknown specificity-determining residues, including, but not restricted to, residues that might confer P−3 Arg selectivity for kinases that are not part of the Snf1 family. We used an approach based on the idea of co-variation (33). We identified residues whose variation in the primary sequence of the catalytic domain significantly correlated with the variation in phosphorylation site specificity across kinases. To measure sequence variation, we used a simple pairwise similarity matrix, and to compare specificities, we calculated the Frobenius norm of the differences in PWMs (Table 2 and Fig. 4B). This approach reproduced several specificity-determining residues previously known from both structural and mutagenesis studies, including Glu127. In addition, we uncovered many previously unknown candidate specificity-determining residues, seven of which were predicted to be within ten angstroms to a bound protein substrate. Among these, an acidic Glu residue at position 170 (PKA numbering) correlated with P−3 Arg selectivity among CMGC kinases. This result contrasts with a previous prediction based on modeling of DYRK1A, the human homolog of Yak1 (34). To test our predictions, we examined the role of residue 170 in substrate selection. Indeed, a Ser to Glu mutation at the analogous position in the MAPK Kss1 (residue 147) conferred a basophilic signature (Fig. 4C and Fig. S2). This result validates our ability to predict new specificity-determining residues on the basis of our large motif dataset.
Table 2.
Computationally predicted kinase specificity determining residues. Correlation values and peptide-kinase distance measurements are defined in the Materials and Methods section.
| Residue (PKA Numbering) | Correlation | Distance from peptide (Å) | Representative Kinase | Amino acid | Specificity | Reference |
|---|---|---|---|---|---|---|
| 170 | 0.195 | 5.3 | Tpk1 | Glu | −2 Arg | |
| Sch9 | Glu | −5 Arg | (27, 29, 70) | |||
| Yak1 | Glu | −3 Arg | This study | |||
| 169 | 0.186 | 7.2 | Psk2 | Asp | −5 basic | |
| Cmk1 | Pro | −5 aliphatic | (18, 30) | |||
| 197 | 0.176 | 9.5 | Cdc28 | (p)Thr | +3 basic | (71) |
| 129 | 0.172 | 6.4 | Tpk1 | Phe | −3 Arg | (27) |
| Atg1 | Thr | −3 aliphatic | This study | |||
| 127 | 0.157 | 7.4 | Tpk1 | Glu | −3 Arg | (27) |
| 199 | 0.145 | 5.7 | Tpk1 | Cys | 0 Ser | This study |
| 246 | 0.134 | 8.4 | Gcn2 | Arg | −2 acidic | This study |
| 205 | 0.122 | 8.4 | Kss1 | Arg | +1 Pro | (54) |
| Ipl1 | Leu | +1 hydrophobic | (27, 29, 72) | |||
| 237 | 0.120 | 9.9 | Yak1 | Leu | −2 Pro | This study |
| 203 | 0.108 | 6.1 | Yck1 | Arg | −3 pSer/pThr | (73) |
| Mps1 | Asn | −2 acidic | This study | |||
| Gin4 | His | −2 Ser | This study |
Connecting kinases to substrates on the basis of phosphorylation site motifs
Because in vivo phosphorylation sites on protein substrates tend to fall within the context of the phosphorylation site motif for a particular kinase, database scanning has been used to predict new substrates and to pinpoint sites of phosphorylation (14, 26, 35–39). However, simple sequence matching approaches are prone to false positives, because predicted sites may not be accessible for phosphorylation, and kinases can also depend on docking or scaffolding interactions for substrate recruitment. In addition, false negatives are frequent for kinases with low sequence specificity because their motifs occur in many proteins and are, thus present with high frequency in databases (14, 18). To increase the accuracy of such predictions, we generated and used a motif analysis pipeline, MOTIPS (http://motips.gersteinlab.org/). MOTIPS scans sequence databases for sites that most closely match the PWM for a particular kinase using a modified algorithm based on the program Scansite (40). Predicted sites are then scored on the basis of a panel of features (evolutionary conservation, predicted surface accessibility, and disordered structure) that are characteristic of known phosphorylation sites (41–43).
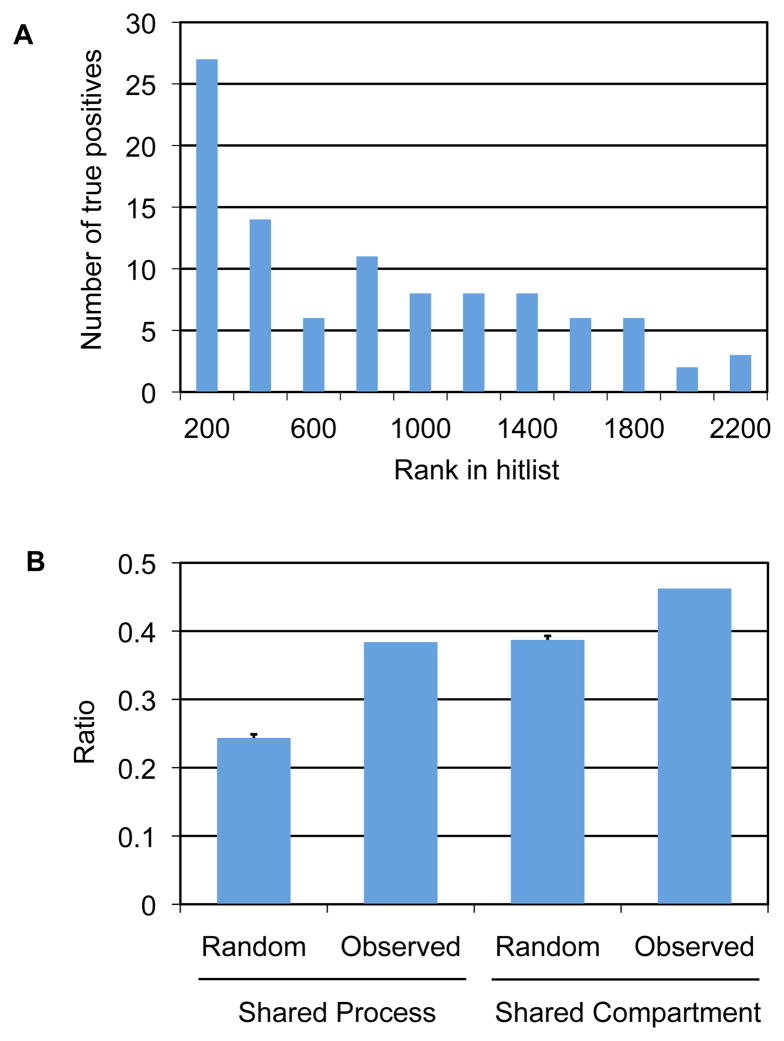
We first analyzed established kinase substrates for the presence of their respective phosphorylation site motifs with MOTIPS. From a sampling of 174 in vivo kinase-substrate relationships curated from the literature, 99 of the substrates ranked among the top 0.5% of predicted sites for their respective kinase, with 27 substrates falling within the top 200 sites (Fig. 5A). We next analyzed predicted substrates for each of the 61 yeast kinases for their associated biological processes and respective localization according to Gene Ontology (GO) assignments in the Saccharomyces Genome Database (44) (Fig. 5B; the full list of predicted substrates for each kinase with associated GO terms and MOTIPS features is provided as Dataset S2). We found that predicted substrates were more likely to be associated with the same biological process and to localize to the same subcellular compartment as their respective kinases than a randomly chosen set of proteins. Taken together, these observations suggest that motif scanning using our set of phosphorylation site motifs enriches for authentic kinase-substrate pairs.
Fig. 5.
MOTIPS ranking of known and predicted kinase-substrate pairs. (A) Bar graph showing the number of protein substrates reported in the literature (true positives) that have at least one phosphorylation site falling within the indicated rank value of predicted substrates for its respective kinase. Shown are the 99 sites of 174 known kinase-substrate pairs analyzed that fall within the top 0.5% predicted sites for that kinase among all Ser or Thr residues in the yeast proteome. (B) GO analysis of predicted kinase-substrate relationships that fall within the top 100 predicted substrates for all 61 kinases analyzed. The graph shows the ratio of predicted kinase-substrate pairs sharing either an annotated biological process (left bars) or subcellular compartment (right bars) in comparison to pairs of proteins chosen at random. For both pairs, the probability that the observed value falls within the random distribution is extremely low (p < 10−35) based on the calculated area under the Gaussian curve corresponding to the random distribution.
To establish directly that our bioinformatics analysis had uncovered authentic substrates, we examined more closely the predicted substrates of the protein kinase Prk1. Prk1 is a member of a small family of kinases conserved throughout eukaryotes that mediates reorganization of the actin cytoskeleton during endocytosis (45). Our peptide array analysis revealed an unusual phosphorylation site motif that included strong preferences for aliphatic residues at the P−5 position, Gly at the P+1 position, and Thr as the phosphoacceptor (Fig. 1B, Table 1). We selected 107 Prk1 candidate substrates identified by MOTIPS for further analysis. These substrates contained sites of high, middle, and low rank among the top 2,000 scoring sites. Because all five known Prk1 substrates undergo multisite phosphorylation (45–47), candidates were also chosen for having at least three predicted Prk1 phosphorylation sites. Of the 107 candidate substrates, we observed phosphorylation of 19 candidates in vitro with wild-type Prk1 but not with a Prk1 inactive mutant (Fig. S3). To identify additional candidates, we used these 19 candidates as positive data points in a training set to educate MOTIPS by machine learning. Negative data points in the training set included 81 of the original Prk1 candidates that were unambiguously not substrates in vitro, as well as about 400 proteins identified in the yeast protein database as localizing solely to non-cytosolic compartments (48).
This set of positive and negative data points was used to re-train the Bayesian algorithm in MOTIPS to integrate the motif matching, conservation, surface accessibility, and disorder scores for each site, along with an additional score based on the number of predicted sites. The five known in vivo substrates of Prk1, which were excluded from the training set, all fell within the top seven targets (Fig. 6A). Five additional candidates taken from the top 15 putative substrates in the new Prk1 hit list were tested by an in vitro kinase assay that used the purified candidates as substrates. These in vitro assays revealed three additional new substrates for Prk1— Gon7, a protein component of the EKC/KEOPS (Endopeptidase-like Kinase Chromatin-associated/Kinase, putative Endopeptidase and Other Proteins of Small size) complex involved in telomere regulation, Gph1, a protein involved in the mobilization of glycogen, and the key endocytic protein Las17. One of the five additional candidates tested was Ypl150w, which is a putative kinase that autophosphorylated in our assay and thus could not be confirmed or excluded as a substrate of Prk1. This second round of in vitro assays provides additional evidence that retraining our algorithm increased our success rate in predicting authentic kinase substrates. Furthermore, among the 22 in vitro confirmed Prk1 substrates, seven proteins (Bem2, Ede1, Las17, Sac3, Sla2, Syp1, and Yap1801) are reported to have roles in endocytosis or the regulation of the actin cytoskeleton, suggesting that they may be subject to regulation by Prk1 (Table 3).
Fig. 6.
Prediction and confirmation of kinase-substrate relationships. (A) Top 15 hits from the trained Prk1 MOTIPS output. The Prk1 hit list of candidate substrates was subjected to machine learning using a training set consisting of 19 true positives (experimentally derived) and ~480 true negatives (experimentally derived and supplemented with those proteins that are known to solely localize to non-cytosolic compartments). Known in vivo substrates of Prk1 are highlighted in yellow. (B) Electrophoretic mobility shift analyses of Bem2 and Ede1. TAP−tagged Bem2 and Ede1 were purified from WT or prk1 Δ ark1 Δ strains by immobilized IgG, and then incubated in the presence or absence of phosphatases followed by immunoblotting against the TAP tag. (C) Mobility shift confirms Sol2 as an in vivo substrate of Vhs1. Lysates from WT or vhs1 Δ strains expressing TAP−tagged Sol2 were fractionated on denaturing polyacrylamide gels impregnated with Phos-tag (57), which retards the mobility of phosphoproteins, followed by immunoblotting against the TAP tag.
Table 3.
Proteins phosphorylated by Prk1 in vitro. Proteins functionally associated with actin rearrangement or endocytosis are highlighted. Ub, ubiquitin; RhoGAP, Rho guanosine triphosphatase-activating protein; SCF, Skp1-Cullin-F-box; UPR, unfolded protein response; WASP, Wiscott-Aldrich Syndrome protein.
| ORF | Protein name | Function |
|---|---|---|
| YBL024W | Ncl1 | m5C-methyltransferase |
| YBL047C | Ede1 | Key endocytic protein; binds plasma membrane in a Ub-dependent manner |
| YCR030C | Syp1 | Overexpression suppresses a pfy1 (profilin) null mutation |
| YDL001W | Rmd1 | Cytoplasmic protein required for sporulation |
| YDR159W | Sac3 | Nuclear pore-associated protein; suppressor of actin mutations |
| YER155C | Bem2 | RhoGAP involved in the control of cytoskeleton and morphogenesis |
| YGR094W | Vas1 | Mitochondrial and cytoplasmic valyl-tRNA synthetase |
| YHL030W | Ecm29 | Major component of the proteasome |
| YHR161C | Yap1801 | Protein involved in clathrin cage assembly; binds clathrin and Pan1 |
| YHR098C | Sfb3 | Involved in sorting Pma1 into COPII vesicles |
| YJL090C | Dpb11 | Subunit of DNA polymerase II epsilon complex |
| YJL129C | Trk1 | Component of the Trk1-Trk2 potassium transport system |
| YJL184W | Gon7 | Component of the EKC/KEOPS complex |
| YJR137C | Ecm17 | Sulfite reductase beta subunit involved in amino acid biosynthesis |
| YML088W | Ufo1 | F-box protein subunit of the SCF E3 ubiquitin ligase complex |
| YML103C | Nup188 | Subunit of the nuclear pore complex |
| YNL243W | Sla2 | Transmembrane protein that links actin to clathrin and endocytosis |
| YNL287W | Sec21 | Gamma subunit of coatomer complex |
| YOR093C | Yor093c | Function unknown; deletion causes sensitivity to UPR-inducing agents |
| YOR181W | Las17 | Actin assembly factor; homolog of human WASP |
| YPL131W | Rpl5 | Component of the large 60S ribosomal subunit |
| YPR160W | Gph1 | Glycogen phosphorylase |
We next investigated whether our predicted Prk1 candidate substrates represented bona fide substrates. Because a closely related kinase, Ark1, has an overlapping biological function and shares a nearly identical phosphorylation site motif with Prk1, we examined the phosphorylation state of candidate substrates in yeast strains deleted for both PRK1 and ARK1. Changes in phosphorylation were monitored by electrophoretic mobility shifts in immunoblots of purified substrates, with phosphatase-treated samples serving as a control for the unphosphorylated species. We observed a change in mobility for two candidate substrates, Bem2 and Ede1, suggesting that they are in vivo targets of Prk1 or Ark1, or both (Fig. 6B). Although we did not observe gel shifts for other substrates, it is likely that some are authentic Prk1/Ark1 substrates as well but simply do not change mobility upon phosphorylation. Notably, previous mass spectrometry (MS) phosphoproteomic analysis identified three of the in vitro Prk1 substrates (Ede1, Syp1, and Rpl5) as phosphorylated at Prk1 consensus sites in vivo (49–54) (the MOTIPS output for all kinases, which is available as Dataset S2, indicates which candidate phosphorylation sites have been identified by MS).
We also validated kinase-substrate pairs through integration with other proteomic datasets. We found that the kinase Vhs1, for which limited functional information is known, exhibited selectivity for the phosphorylation site motif MXRXXS (table 1 and table S1. Fourteen in vitro substrates for the kinase Vhs1 (55) were previously identified by protein microarray analysis (4), and six of these, Mga1, Pfk26, Sef1, Sol1, Sol2, and Utr1, contain the Vhs1 consensus phosphorylation site motif. MS phosphoproteomic analysis (49) revealed that Sef1 was phosphorylated in vivo at a Vhs1 consensus phosphorylation site and in an immunoprecipitation-MS analysis Sef1 and Vhs1 physically interacted (56). In addition, MS phosphoproteomic analysis identified Sol1 as phosphorylated at a Vhs1 consensus phosphorylation site in vivo (50), and its homolog Sol2 was the most highly phosphorylated Vhs1 in vitro substrate identified by protein microarray analysis (4). Mobility shift analysis of VHS1 deletion strains using Phos-tag SDS-PAGE (57) was consistent with Sol2 as a substrate for Vhs1 in vivo (Fig. 6C). Though the presence of multiple Sol2 species in the presence and absence of Vhs1 indicates phosphorylation at multiple sites, likely by more than one kinase, the mobility shift indicates that in vhs1 mutant cells, Sol2 is phosphorylated at fewer sites. Sol2, which promotes nucleocytoplasmic tRNA transport (58), is the first reported in vivo substrate for Vhs1 and suggests a role for this kinase in regulating this process. These results illustrate how integration of data from multiple proteomic approaches can shed light on the biology of poorly characterized molecules.
DISCUSSION
The elucidation of the mechanisms underlying kinase specificity remains an integral part of understanding phosphorylation-based signal transduction pathways. Previous methods for determining consensus phosphorylation site motifs have not been suitable for large-scale screening of a eukaryotic kinome. Here, we have described an approach for the high-throughput identification of consensus phosphorylation site motifs in which multiple kinases, with no previously known substrates, can be analyzed simultaneously. We have used this approach to provide comprehensive analysis of kinase specificity in a single eukaryotic organism, the yeast Saccharomyces cerevisiae. Among other applications, this large dataset has provided much broader insight into the structural basis for kinase selectivity than has been possible through individual analyses of single kinases.
With our data, we linked protein kinases to previously unknown substrates, thus elucidating mechanisms of phosphorylation-dependent signaling. A limitation to our approach, however, is that the peptide arrays treat each position in the substrate independently, and thus the potential interdependence between multiple positions is ignored. This approach is nonetheless a valuable first pass screen for analyzing kinase specificity because it involves the systematic and exhaustive analysis of each amino acid residue at each position surrounding the phosphorylation site. Preferences observed with this approach can provide the basis for the design of kinase-specific peptide libraries to uncover positional interdepedence. Furthermore, the presence of a consensus phosphorylation sequence alone is insufficient to direct phosphorylation of a protein by a particular kinase, and accordingly identification of previously unknown substrates on the basis of motif scanning is difficult. However, integration with other proteomic datasets provides a means of increasing confidence in predicted kinase-substrate relationships. In addition, specific kinase-substrate pairs can be inferred through computational methods that make use of non-sequence-based “contextual” features, such as subcellular localization and molecular function (38). For example, predicting substrates targeted by relatively nonspecific kinases using phosphorylation site motifs alone is unlikely to be successful because these sequences occur frequently in proteomes. In such cases, selection of authentic substrates is driven by docking or scaffolding interactions, and consensus sequences for substrate recruitment can be used in combination with phosphorylation site motifs to identify new substrates (59, 60).
For previously characterized kinases, we observed a high degree of conservation of phosphorylation site motifs between yeast and mammalian orthologs. These similarities suggest that the many previously unknown consensus motifs reported here are also conserved. Therefore, this dataset will serve as a resource for studies of phosphorylation-dependent signaling in higher eukaryotes, as well as yeast.
MATERIALS AND METHODS
Details regarding yeast strain information, kinase preparation, characterization of purified kinases, in vitro kinase assays, and electrophoretic mobility shift analyses are available in the Supplementary Material.
Peptide library screening
The peptide library (Anaspec, Inc.) has been previously reported (13). For this study, fresh stock solutions were made from 5 mg powder by dissolving peptides in DMSO, quantifying by absorbance at 280 nm, and adjusting to a stock concentration of 10 mM by adding the appropriate volume of DMSO. Stock solutions were stored at −20°C in microcentrifuge tubes. Working 0.6 mM aqueous stocks were prepared by diluting the DMSO stock in 20 mM HEPES, pH 7.4 and arrayed into 1536-well stock plates containing 5 μl aliquots in each well. Plates were sealed with adhesive foil and stored at −20°C.
Peptides (0.2 μl per well) were transferred to assay plates containing 2 μl of kinase reaction buffer (generally 20 mM HEPES, pH 7.4, 10 mM MgCl2, 1 mM DTT, 0.1% Tween 20) from stock plates manually using a 48 × 6 slot pin replicator (VP Scientific). Reactions were initiated by adding a solution (0.2 μl per well) containing purified kinase and γ-[33P ]-ATP (0.55 mM, 0.3–0.4 μCi/μl, Perkin Elmer) using a 48 × 1 slot pin replicator (VP Scientific). Plates were sealed and incubated for 1 to 8 hr at 30°C. The final concentrations of the reaction components in each well were 50 μM peptide and 50 μM ATP at a specific activity of 0.55–0.73 mCi/μmol. After incubation, 0.2 μl from each well was spotted onto streptavidin-coated membrane (SAM2 Biotin Capture Membrane, Promega) simultaneously using the 48 × 6 slot pin replicator. Membranes were washed three times with 10 mM Tris-HCl, pH 7.5 with 140 mM NaCl and 0.1% SDS, twice with 2 M NaCl, twice with 2 M NaCl with 1% H3PO4, and twice with water, then dried and exposed to a phosphor storage screen. Processing of final images of the spot arrays consisted of copying the 4 × 22 grid corresponding to the P+1, P+2, P+3, and P+4 peptide mixtures and pasting it below the 5 × 22 grid corresponding to the P−5, P−4, P−3, P−2, and P−1 peptide mixtures using Adobe Photoshop to provide the 9 × 22 spot grids shown in Figure 1 and table S1.
PWM generation
For each array, peptide phosphorylation signals were quantified using Genepix Pro 6.0 (Molecular Devices) by manually aligning a 48 × 8 grid of circles onto each scanned phosphorimage to calculate the median intensity for each spot. These median intensity values were then background corrected by subtracting the median intensity value corresponding to the negative control spot (reaction carried out in the absence of any peptide substrate). Signal scores for each amino acid at each position were then normalized by the following equation
where Zca stands for the normalized score of amino acid a at position c having a signal score Sca, and m stands for the total number of amino acids. Sci is the signal score of amino acid i at position c where i is defined in the summation of all the m amino acids.. The PWM is an N × 20 matrix of N positions with the normalized, background corrected value given as the weight for each amino acid at each position. To account for spurious phosphorylation of Ser and Thr residues at other positions, the PWM entries in all Ser and Thr positions were set to one (equivalent to neutral selection at that position) with subsequent renormalization of the PWM.
Proteome scanning
The entire yeast proteome was scanned to identify the best matches to each PWM. Our approach used a window-sliding method based on the normalized PWM similar to the method used in Scansite (40). Briefly, it extracted every possible 15-mer sequence from the yeast proteome and calculated the match score to the PWM, based on the formula:
where i stands for the position in the motif and ri stands for the residue that is present at position i in the peptide in question. Mia is the normalized PWM as described above. The resulting score was then normalized, such that zero stands for an optimal match to the motif and larger positive scores correspond to weaker matches. The top 10,000 potential phosphorylation sites for each kinase are reported in the Dataset S2. This algorithm was implemented in a modular form in Java. All sequences and features were loaded into a SQL database that is interactively queried by the Java search module.
Feature collection
A number of different genomic features were gathered to supplement the initial match score. To compute the conservation score, we collected all orthologs for 13 proteomes of related yeast species (Saccharomyces paradoxus as the closest and Schizosaccharomyces pombe as the farthest) using the comparative genomics algorithm implemented in INPARANOID (61). We then aligned these orthologs using the automated alignment method MUSCLE (62) (the full set of alignments is available as Dataset S3). For each PWM hit, we calculated the conservation score by estimating the entropy at each position based on the aligned orthologs with the AL2CO program. The disorder score was based on the prediction program DISOPRED (63). DISOPRED was run for each protein in the yeast proteome. We used the DISOPRED probability score, corresponding to the likelihood of the residue in question being in a disordered region, as the measure of disorder. Finally, the surface accessibility score was calculated using the prediction program SABLE for each protein in the yeast proteome (64). The simple numerical surface score was used as the measure of surface accessibility.
Feature integration
An integration algorithm based on the Naïve Bayes framework was used to integrate the four features. We used a number of experimentally determined gold-standard kinase substrate pairs, “positives,” to train the algorithm. For gold-standard negatives, we supplemented a set of experimentally determined negatives with a set of randomly chosen protein pairs. Each of these pairs is a pair of proteins that are annotated to always localize to two different compartments (for example, nucleus only and cytoplasm only). Thus, we biased the randomly chosen set of protein pairs further towards a set that was highly unlikely to contain any spurious positive interactions. The conditional probability was calculated from the four features according to the following formula:
where I denotes either interaction or non-interaction and D1 through D4 denote the four features. Data were thus integrated under the assumption that the four features are independent. To formally assess independence of the features, we calculated pairwise correlation coefficients. The results showed the pairwise correlation coefficients ranging from 0.01 to 0.57 (absolute values) have an average of 0.18, indicating the features are to a large extent independent (see table S3). Moreover, we performed Principle Component Analysis (PCA) using the statistical software R to transform the possibly correlated values of the five features (hits per protein, match score, disorder score, accessibility score, and conservation score) of the PRK1 targets into uncorrelated values. The first three vectors were chosen to build a Naïve Bayes model followed by a 10-fold stratified cross validation. The Area Under Curve (AUC; 75.9%) of the Receiver Operating Curve (ROC) resulting from the PCA validation was then compared to the AUC (78.6%) of PRK1 without the PCA transformation. The very close performance of the two further indicated a certain level of independency of the features. Bayesian integration was implemented using the Java machine learning package Weka (65). The entire methodology is available as the modularized software packages MOTIPS (URL: http://motips.gersteinlab.org/).
Covariation calculation to estimate specificity-determining residues
Sequences of the 61 yeast kinase catalytic domains (obtained from the kinase.com database) were initially aligned using ClustalW2 (66). A high quality sequence alignment was generated by manual editing of the initial alignment in Jalview (67) on the basis of multiple pairwise alignments with kinases of known 3D structure and conserved catalytic residues (table S4). In addition, 89 orthologous kinases from S. pombe, D. discoideum, and H. sapiens were added and manually aligned. For these orthologs, the PWM was inferred to be identical to its yeast counterpart. A correlation-based methodology was implemented to identify specificity determining residues:
For each pairs of sequence positions (n) and positions in the PWM (m), two -dimensional vectors were generated; k is the total number of kinases in the alignment and is equal to the number of PWMs. The first vector contained all pairwise similarities between the primary sequences of the kinases in that position, based on the McLachlan matrix (that is the similarity of the amino acid in position X in kinase A to the similarity of the amino acid in the same position in kinase B) (68). The McLachlan matrix was chosen because it scores for residue substitutions based on chemical similarity (i.e., physico-chemical properties). The second vector contained the pairwise similarity of all PWMs to each other, based on the Frobenius norm (69): .
Each position was then scored with the Pearson correlation coefficient of these two vectors (listed under “correlation” in Table 2). This method was implemented in the programming package MATLAB. Distances of the residue in question from bound peptide were estimated by mapping the residue onto the PKA-PKI structure (PDB ID: 1ATP) using the program VMD. The peptide-kinase distances were measured as the closest distances between the geometric centers of the residue on the kinase, as mapped to the PKA structure, to the bound peptide, as in the PKA structure.
Supplementary Material
Materials and Methods
Fig. S1. Assay reproducibility.
Fig. S2. Representative peptide array screening results for the Kss1 S147E mutant.
Fig. S3. Representative Prk1 in vitro assays.
Table S1. Representative peptide array screening results and sequence logos for each of the 61 kinases assayed.
Table S2. Protein kinases analyzed in this study.
Table S3. Pairwise correlation coefficients for each of four genomic features and the Scansite match score.
Table S4. Alignment of yeast kinases analyzed in this study.
References
Dataset S1. Average PWMs for each of the 61 kinases assayed (plain text files).
Dataset S2. MOTIPS output for each of the 61 kinases assayed (plain text files).
Dataset S3. MUSCLE alignment of all predicted S. cerevisiae ORFs with orthologs from 12 other yeast species (clustal alignment files).
Dataset S4. Alignment of yeast kinases analyzed in this study (clustal alignment file).
Acknowledgments
Supported by US National Institutes of Health grants to M.S., B.E.T. (GM079498), N.M.H. (GM50717), D.F.S. (CA82257) and by a Swiss National Science Foundation grant to C.D.
Footnotes
Editor’s Summary: Exploring Kinase Selectivity
Kinases are master regulators of cellular behavior. Because of the large number of kinases and even larger number of substrates, approaches that permit global analysis are valuable tools for investigating kinase biology. With a miniaturized peptide library screening approach, Mok et al. identified the phosphorylation site selectivity for 61 of the 122 kinases in Saccharomyces cerevisiae. By integrating this data with other datasets and structural information, they revealed information about the relationship between kinase catalytic residues and substrate selectivity. They also identified and experimentally verified substrates for kinases, including one in which limited functional information was previously available, demonstrating the potential for this type analysis as a launching point for the exploration of the biological functions of kinases.
Author contributions: J.M., S.P., G.R.J., D.L.S., S.A.P., V.D. and B.E.T. performed experiments; P.M.K., H.Y.K.L. and M.B.G. performed computational work; J.M., X.Z., G.R.J., S.A.P., V.D., M.J., E.C., H.N., M.G., A.R., J.N.M., Y.S., H.E.S., R.S., C.S.M.C., C.D., N.M.H., W.A.L., D.F.S., B.S., and B.J.A. prepared and characterized protein kinases and expression constructs; J.M., P.M.K., H.Y.K.L., M.B.G., M.S. and B.E.T. designed experiments, analyzed data and wrote the paper.
Competing interests: M.S. consults for Affomix, which has an interest in proteomics, including phosphoproteomics.
REFERENCES AND NOTES
- 1.Manning G, Plowman GD, Hunter T, Sudarsanam S. Evolution of protein kinase signaling from yeast to man. Trends Biochem Sci. 2002;27:514–520. doi: 10.1016/s0968-0004(02)02179-5. [DOI] [PubMed] [Google Scholar]
- 2.Moffat J, Sabatini DM. Building mammalian signalling pathways with RNAi screens. Nat Rev Mol Cell Biol. 2006;7:177–187. doi: 10.1038/nrm1860. [DOI] [PubMed] [Google Scholar]
- 3.Schmelzle K, White FM. Phosphoproteomic approaches to elucidate cellular signaling networks. Curr Opin Biotechnol. 2006;17:406–414. doi: 10.1016/j.copbio.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, McCartney RR, Schmidt MC, Rachidi N, Lee SJ, Mah AS, Meng L, Stark MJ, Stern DF, De Virgilio C, Tyers M, Andrews B, Gerstein M, Schweitzer B, Predki PF, Snyder M. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- 5.Elphick LM, Lee SE, Gouverneur V, Mann DJ. Using chemical genetics and ATP analogues to dissect protein kinase function. ACS Chem Biol. 2007;2:299–314. doi: 10.1021/cb700027u. [DOI] [PubMed] [Google Scholar]
- 6.Turk BE. Understanding and exploiting substrate recognition by protein kinases. Curr Opin Chem Biol. 2008;12:4–10. doi: 10.1016/j.cbpa.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park SH, Zarrinpar A, Lim WA. Rewiring MAP kinase pathways using alternative scaffold assembly mechanisms. Science. 2003;299:1061–1064. doi: 10.1126/science.1076979. [DOI] [PubMed] [Google Scholar]
- 8.Skerker JM, Perchuk BS, Siryaporn A, Lubin EA, Ashenberg O, Goulian M, Laub MT. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133:1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ubersax JA, Ferrell JE., Jr Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol. 2007;8:530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- 10.Pinna LA, Ruzzene M. How do protein kinases recognize their substrates? Biochim Biophys Acta. 1996;1314:191–225. doi: 10.1016/s0167-4889(96)00083-3. [DOI] [PubMed] [Google Scholar]
- 11.Amanchy R, Periaswamy B, Mathivanan S, Reddy R, Tattikota SG, Pandey A. A curated compendium of phosphorylation motifs. Nat Biotechnol. 2007;25:285–286. doi: 10.1038/nbt0307-285. [DOI] [PubMed] [Google Scholar]
- 12.Miller ML, Jensen LJ, Diella F, Jorgensen C, Tinti M, Li L, Hsiung M, Parker SA, Bordeaux J, Sicheritz-Ponten T, Olhovsky M, Pasculescu A, Alexander J, Knapp S, Blom N, Bork P, Li S, Cesareni G, Pawson T, Turk BE, Yaffe MB, Brunak S, Linding R. Linear motif atlas for phosphorylation-dependent signaling. Sci Signal. 2008;1:ra2. doi: 10.1126/scisignal.1159433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutti JE, Jarrell ET, Chang JD, Abbott DW, Storz P, Toker A, Cantley LC, Turk BE. A rapid method for determining protein kinase phosphorylation specificity. Nat Methods. 2004;1:27–29. doi: 10.1038/nmeth708. [DOI] [PubMed] [Google Scholar]
- 14.Rennefahrt UE, Deacon SW, Parker SA, Devarajan K, Beeser A, Chernoff J, Knapp S, Turk BE, Peterson JR. Specificity profiling of Pak kinases allows identification of novel phosphorylation sites. J Biol Chem. 2007;282:15667–15678. doi: 10.1074/jbc.M700253200. [DOI] [PubMed] [Google Scholar]
- 15.Gelperin DM, White MA, Wilkinson ML, Kon Y, Kung LA, Wise KJ, Lopez-Hoyo N, Jiang L, Piccirillo S, Yu H, Gerstein M, Dumont ME, Phizicky EM, Snyder M, Grayhack EJ. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 2005;19:2816–2826. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, Lan N, Jansen R, Bidlingmaier S, Houfek T, Mitchell T, Miller P, Dean RA, Gerstein M, Snyder M. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 17.Dent P, Chow YH, Wu J, Morrison DK, Jove R, Sturgill TW. Expression, purification and characterization of recombinant mitogen-activated protein kinase kinases. Biochem J. 1994;303:105–112. doi: 10.1042/bj3030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manke IA, Nguyen A, Lim D, Stewart MQ, Elia AE, Yaffe MB. MAPKAP kinase-2 is a cell cycle checkpoint kinase that regulates the G2/M transition and S phase progression in response to UV irradiation. Mol Cell. 2005;17:37–48. doi: 10.1016/j.molcel.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 19.Obata T, Yaffe MB, Leparc GG, Piro ET, Maegawa H, Kashiwagi A, Kikkawa R, Cantley LC. Peptide and protein library screening defines optimal substrate motifs for AKT/PKB. J Biol Chem. 2000;275:36108–36115. doi: 10.1074/jbc.M005497200. [DOI] [PubMed] [Google Scholar]
- 20.Alessi DR, Caudwell FB, Andjelkovic M, Hemmings BA, Cohen P. Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 kinase. FEBS Lett. 1996;399:333–338. doi: 10.1016/s0014-5793(96)01370-1. [DOI] [PubMed] [Google Scholar]
- 21.Fiol CJ, Haseman JH, Wang YH, Roach PJ, Roeske RW, Kowalczuk M, DePaoli-Roach AA. Phosphoserine as a recognition determinant for glycogen synthase kinase-3: phosphorylation of a synthetic peptide based on the G-component of protein phosphatase-1. Arch Biochem Biophys. 1988;267:797–802. doi: 10.1016/0003-9861(88)90089-6. [DOI] [PubMed] [Google Scholar]
- 22.Songyang Z, Lu KP, Kwon YT, Tsai LH, Filhol O, Cochet C, Brickey DA, Soderling TR, Bartleson C, Graves DJ, DeMaggio AJ, Hoekstra MF, Blenis J, Hunter T, Cantley LC. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol Cell Biol. 1996;16:6486–6493. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson EF, Stewart KD, Woods KW, Giranda VL, Luo Y. Pharmacological and functional comparison of the polo-like kinase family: insight into inhibitor and substrate specificity. Biochemistry. 2007;46:9551–9563. doi: 10.1021/bi7008745. [DOI] [PubMed] [Google Scholar]
- 24.Hunter T, Plowman GD. The protein kinases of budding yeast: six score and more. Trends Biochem Sci. 1997;22:18–22. doi: 10.1016/s0968-0004(96)10068-2. [DOI] [PubMed] [Google Scholar]
- 25.Dale S, Wilson WA, Edelman AM, Hardie DG. Similar substrate recognition motifs for mammalian AMP-activated protein kinase, higher plant HMG-CoA reductase kinase-A, yeast SNF1, and mammalian calmodulin-dependent protein kinase I. FEBS Lett. 1995;361:191–195. doi: 10.1016/0014-5793(95)00172-6. [DOI] [PubMed] [Google Scholar]
- 26.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knighton DR, Zheng JH, Ten Eyck LF, Xuong NH, Taylor SS, Sowadski JM. Structure of a peptide inhibitor bound to the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:414–420. doi: 10.1126/science.1862343. [DOI] [PubMed] [Google Scholar]
- 28.Lowe ED, Noble ME, Skamnaki VT, Oikonomakos NG, Owen DJ, Johnson LN. The crystal structure of a phosphorylase kinase peptide substrate complex: kinase substrate recognition. EMBO J. 1997;16:6646–6658. doi: 10.1093/emboj/16.22.6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J, Cron P, Good VM, Thompson V, Hemmings BA, Barford D. Crystal structure of an activated Akt/protein kinase B ternary complex with GSK3-peptide and AMP-PNP. Nat Struct Biol. 2002;9:940–944. doi: 10.1038/nsb870. [DOI] [PubMed] [Google Scholar]
- 30.Bullock AN, Debreczeni J, Amos AL, Knapp S, Turk BE. Structure and substrate specificity of the Pim-1 kinase. J Biol Chem. 2005;280:41675–41682. doi: 10.1074/jbc.M510711200. [DOI] [PubMed] [Google Scholar]
- 31.Brinkworth RI, Breinl RA, Kobe B. Structural basis and prediction of substrate specificity in protein serine/threonine kinases. Proc Natl Acad Sci USA. 2003;100:74–79. doi: 10.1073/pnas.0134224100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brinkworth RI, Munn AL, Kobe B. Protein kinases associated with the yeast phosphoproteome. BMC Bioinformatics. 2006;7:47. doi: 10.1186/1471-2105-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li L, Shakhnovich EI, Mirny LA. Amino acids determining enzyme-substrate specificity in prokaryotic and eukaryotic protein kinases. Proc Natl Acad Sci USA. 2003;100:4463–4468. doi: 10.1073/pnas.0737647100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Himpel S, Tegge W, Frank R, Leder S, Joost HG, Becker W. Specificity determinants of substrate recognition by the protein kinase DYRK1A. J Biol Chem. 2000;275:2431–2438. doi: 10.1074/jbc.275.4.2431. [DOI] [PubMed] [Google Scholar]
- 35.Yaffe MB, Leparc GG, Lai J, Obata T, Volinia S, Cantley LC. A motif-based profile scanning approach for genome-wide prediction of signaling pathways. Nat Biotechnol. 2001;19:348–353. doi: 10.1038/86737. [DOI] [PubMed] [Google Scholar]
- 36.Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/Akt pathway. Mol Cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- 37.Holt LJ, Hutti JE, Cantley LC, Morgan DO. Evolution of Ime2 phosphorylation sites on Cdk1 substrates provides a mechanism to limit the effects of the phosphatase Cdc14 in meiosis. Mol Cell. 2007;25:689–702. doi: 10.1016/j.molcel.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Linding R, Jensen LJ, Ostheimer GJ, van Vugt MA, Jorgensen C, Miron IM, Diella F, Colwill K, Taylor L, Elder K, Metalnikov P, Nguyen V, Pasculescu A, Jin J, Park JG, Samson LD, Woodgett JR, Russell RB, Bork P, Yaffe MB, Pawson T. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129:1415–1426. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hutti JE, Shen RR, Abbott DW, Zhou AY, Sprott KM, Asara JM, Hahn WC, Cantley LC. Phosphorylation of the tumor suppressor CYLD by the breast cancer oncogene IKKe promotes cell transformation. Mol Cell. 2009;34:461–472. doi: 10.1016/j.molcel.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Budovskaya YV, Stephan JS, Deminoff SJ, Herman PK. An evolutionary proteomics approach identifies substrates of the cAMP-dependent protein kinase. Proc Natl Acad Sci USA. 2005;102:13933–13938. doi: 10.1073/pnas.0501046102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iakoucheva LM, Radivojac P, Brown CJ, O’Connor TR, Sikes JG, Obradovic Z, Dunker AK. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- 44.Hong EL, Balakrishnan R, Dong Q, Christie KR, Park J, Binkley G, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hitz BC, Krieger CJ, Livstone MS, Miyasato SR, Nash RS, Oughtred R, Skrzypek MS, Weng S, Wong ED, Zhu KK, Dolinski K, Botstein D, Cherry JM. Gene Ontology annotations at SGD: new data sources and annotation methods. Nucleic Acids Res. 2008;36:D577–581. doi: 10.1093/nar/gkm909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toshima J, Toshima JY, Martin AC, Drubin DG. Phosphoregulation of Arp2/3-dependent actin assembly during receptor-mediated endocytosis. Nat Cell Biol. 2005;7:246–254. doi: 10.1038/ncb1229. [DOI] [PubMed] [Google Scholar]
- 46.Huang B, Zeng G, Ng AY, Cai M. Identification of novel recognition motifs and regulatory targets for the yeast actin-regulating kinase Prk1p. Mol Biol Cell. 2003;14:4871–4884. doi: 10.1091/mbc.E03-06-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watson HA, Cope MJ, Groen AC, Drubin DG, Wendland B. In vivo role for actin-regulating kinases in endocytosis and yeast epsin phosphorylation. Mol Biol Cell. 2001;12:3668–3679. doi: 10.1091/mbc.12.11.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 49.Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, Zhou H. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics. 2008;7:1389–1396. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 51.Gruhler A, Olsen JV, Mohammed S, Mortensen P, Faergeman NJ, Mann M, Jensen ON. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol Cell Proteomics. 2005;4:310–327. doi: 10.1074/mcp.M400219-MCP200. [DOI] [PubMed] [Google Scholar]
- 52.Li X, Gerber SA, Rudner AD, Beausoleil SA, Haas W, Villen J, Elias JE, Gygi SP. Large-scale phosphorylation analysis of a-factor-arrested Saccharomyces cerevisiae. J Proteome Res. 2007;6:1190–1197. doi: 10.1021/pr060559j. [DOI] [PubMed] [Google Scholar]
- 53.Chi A, Huttenhower C, Geer LY, Coon JJ, Syka JE, Bai DL, Shabanowitz J, Burke DJ, Troyanskaya OG, Hunt DF. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc Natl Acad Sci USA. 2007;104:2193–2198. doi: 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smolka MB, Albuquerque CP, Chen SH, Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci USA. 2007;104:10364–10369. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Munoz I, Simon E, Casals N, Clotet J, Arino J. Identification of multicopy suppressors of cell cycle arrest at the G1-S transition in Saccharomyces cerevisiae. Yeast. 2003;20:157–169. doi: 10.1002/yea.938. [DOI] [PubMed] [Google Scholar]
- 56.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 57.Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics. 2006;5:749–757. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- 58.Stanford DR, Whitney ML, Hurto RL, Eisaman DM, Shen WC, Hopper AK. Division of labor among the yeast Sol proteins implicated in tRNA nuclear export and carbohydrate metabolism. Genetics. 2004;168:117–127. doi: 10.1534/genetics.104.030452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Snead JL, Sullivan M, Lowery DM, Cohen MS, Zhang C, Randle DH, Taunton J, Yaffe MB, Morgan DO, Shokat KM. A coupled chemical-genetic and bioinformatic approach to Polo-like kinase pathway exploration. Chem Biol. 2007;14:1261–1272. doi: 10.1016/j.chembiol.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sheridan DL, Kong Y, Parker SA, Dalby KN, Turk BE. Substrate discrimination among mitogen-activated protein kinases through distinct docking sequence motifs. J Biol Chem. 2008;283:19511–19520. doi: 10.1074/jbc.M801074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Remm M, Storm CE, Sonnhammer EL. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- 62.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ward JJ, McGuffin LJ, Bryson K, Buxton BF, Jones DT. The DISOPRED server for the prediction of protein disorder. Bioinformatics. 2004;20:2138–2139. doi: 10.1093/bioinformatics/bth195. [DOI] [PubMed] [Google Scholar]
- 64.Adamczak R, Porollo A, Meller J. Accurate prediction of solvent accessibility using neural networks-based regression. Proteins. 2004;56:753–767. doi: 10.1002/prot.20176. [DOI] [PubMed] [Google Scholar]
- 65.Frank E, Hall M, Trigg L, Holmes G, Witten IH. Data mining in bioinformatics using Weka. Bioinformatics. 2004;20:2479–2481. doi: 10.1093/bioinformatics/bth261. [DOI] [PubMed] [Google Scholar]
- 66.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 67.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 68.McLachlan AD. Tests for comparing related amino-acid sequences. Cytochrome c and cytochrome c 551. J Mol Biol. 1971;61:409–424. doi: 10.1016/0022-2836(71)90390-1. [DOI] [PubMed] [Google Scholar]
- 69.Golub GH, Van Loan CF. Matrix computations. 3. Johns Hopkins University Press; Baltimore: 1996. [Google Scholar]
- 70.Zhu G, Fujii K, Liu Y, Codrea V, Herrero J, Shaw S. A single pair of acidic residues in the kinase major groove mediates strong substrate preference for P−2 or P−5 arginine in the AGC, CAMK, and STE kinase families. J Biol Chem. 2005;280:36372–36379. doi: 10.1074/jbc.M505031200. [DOI] [PubMed] [Google Scholar]
- 71.Holmes JK, Solomon MJ. The role of Thr160 phosphorylation of Cdk2 in substrate recognition. Eur J Biochem. 2001;268:4647–4652. doi: 10.1046/j.1432-1327.2001.02392.x. [DOI] [PubMed] [Google Scholar]
- 72.Zhu G, Fujii K, Belkina N, Liu Y, James M, Herrero J, Shaw S. Exceptional disfavor for proline at the P + 1 position among AGC and CAMK kinases establishes reciprocal specificity between them and the proline-directed kinases. J Biol Chem. 2005;280:10743–10748. doi: 10.1074/jbc.M413159200. [DOI] [PubMed] [Google Scholar]
- 73.Xu RM, Carmel G, Sweet RM, Kuret J, Cheng X. Crystal structure of casein kinase-1, a phosphate-directed protein kinase. EMBO J. 1995;14:1015–1023. doi: 10.1002/j.1460-2075.1995.tb07082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and Methods
Fig. S1. Assay reproducibility.
Fig. S2. Representative peptide array screening results for the Kss1 S147E mutant.
Fig. S3. Representative Prk1 in vitro assays.
Table S1. Representative peptide array screening results and sequence logos for each of the 61 kinases assayed.
Table S2. Protein kinases analyzed in this study.
Table S3. Pairwise correlation coefficients for each of four genomic features and the Scansite match score.
Table S4. Alignment of yeast kinases analyzed in this study.
References
Dataset S1. Average PWMs for each of the 61 kinases assayed (plain text files).
Dataset S2. MOTIPS output for each of the 61 kinases assayed (plain text files).
Dataset S3. MUSCLE alignment of all predicted S. cerevisiae ORFs with orthologs from 12 other yeast species (clustal alignment files).
Dataset S4. Alignment of yeast kinases analyzed in this study (clustal alignment file).