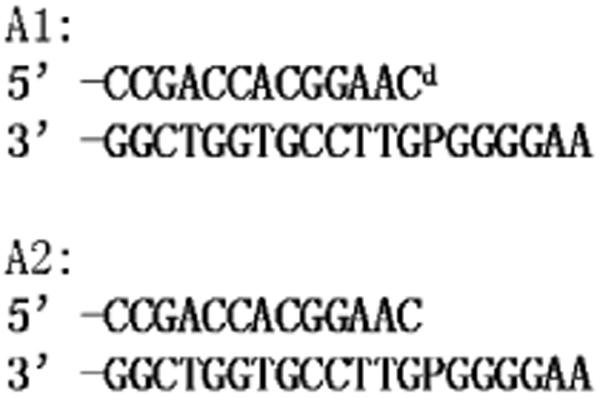Abstract
Rapid chemical quench assays, as well as equilibrium and stopped-flow fluorescence experiments, were performed with an RB69 DNA polymerase (RB69 pol)–primer-template (P/T) complex containing 2-aminopurine (dAP) and a metal exchange-inert Rh(III) derivative of a deoxynucleoside triphosphate (Rh • dTTP). The objective was to determine the effect of catalytic metal ion (A site) occupancy on the affinity of an incoming Rh • dTTP for the RB69 pol–P/T binary complex and on the rate of the conformational change induced by Rh • dTTP binding. With Ca2+ in the A site, the affinity of the incoming Rh • dTTP for the RB69 pol–P/T binary complex and the conformational change rate can be determined in the absence of chemistry. When Mg2+ was added to a ternary complex containing Rh • dTTP opposite dAP, the templating base, nucleotidyl transfer occurred, but the rate of product formation was only one-tenth of that found with Mg • dTTP, as determined by rapid chemical quench assays. Rates of conformational change subsequent to formation of a ternary complex, in the absence of chemistry, were estimated from the rate of change in dAP fluorescence with an increase in the Rh • dTTP concentration. We have shown that there is an initial rapid quenching of dAP fluorescence followed by a second phase of dAP quenching, which has nearly the same rate as that of dTMP incorporation, as estimated from rapid chemical quench experiments. We have also demonstrated that the affinity of Rh • dTTP for occupancy of the B metal ion site is dependent on the presence of Ca2+. However, a saturating Rh • dTTP concentration in the absence of Ca2+ results in full quenching of dAP fluorescence, whereas a saturating Ca2+ concentration in the absence of Rh • dTTP gives only partial quenching of dAP fluorescence. The implications of these results for the mechanism of Fingers closing, metal ion binding, and base selectivity are discussed.
A key feature of replicative DNA polymerases is the high level of dNTP1 discrimination exhibited during DNA synthesis. Reported error frequencies during replication are typically less than 10−6 due mainly to base selection during nucleotide incorporation (1–5). Several features of the enzymes and substrates contribute to base selectivity, including base-pair hydrogen bonding, base stacking, active site tightness, and base-pair geometry (6–10). These features manifest themselves kinetically as changes in the rate of prechemistry conformational changes and changes in the rate of chemistry (11–13). In addition, there is an editing function associated with many DNA polymerases that can excise incorrect nucleotide residues from the nascent primer strand, contributing further to fidelity (9, 14–18).
DNA polymerases catalyze a nucleotidyl transfer reaction that results in primer extension via a mechanism requiring two divalent metal ions (19). The metal ions occupy spatially distinct sites, A and B, although they share carboxylate ligands from two aspartyl side chains. The A site has been designated as the catalytic metal ion site because reactions occur only when this site is occupied by Mg2+ or Mn2+ ions. The presence of either one of these metal ions in the A site lowers the pKa of the 3′-terminal OH group of the primer, generating an oxyanion that can attack the α-phosphorus atom of the incoming dNTP (20). The metal ion occupying the B site enters with the incoming dNTP and is ligated to the negatively charged oxygen atoms in its triphosphate tail. The function of the B metal ion is to stabilize the developing negative charge in the pentavalent transition state and to assist in the departure of the pyrophosphate as the nascent phosphodiester is formed (20).
We have chosen RB69 pol as a model B family DNA polymerase for our investigation into the mechanisms used by DNA polymerases to achieve high-fidelity DNA synthesis for the following reasons. (i) Crystal structures of RB69 pol–P/T binary and ternary complexes have been determined at high resolution (21–25). (ii) Extensive kinetic studies have been carried out on RB69 pol and T4 pol, its close relative, as well as on many single-site mutants of these enzymes that have helped to delineate the functional role of various conserved residues (26–29). (iii) RB69 pol has extensive sequence similarities in the highly conserved regions that characterize replicative DNA polymerases belonging to the B family, suggesting that the results with RB69 pol could be applicable to other eukaryotic B family members, including human α and δ DNA polymerases (30–32). A diagram showing the location of the A and B metal ion sites as observed in the crystal structure of the RB69 pol ternary complex is shown in Figure 1. Although two divalent metal ions are essential for the nucleotidyl transfer reaction, it is not known whether one or both of these metal ions are required for the conformational change, nor is it clear how the occupancy of either the A or B site, alone, affects either the affinity of incoming dNTP for the E • D binary complex or the rate of the conformational change that precedes chemistry.
Figure 1.
RB69 pol active site with Ca2+ occupying the A metal site and Rh3+ occupying the B metal site. This figure was adapted from Figure 2A of ref (19).
The key to understanding the individual contributions of the two metal binding sites to isomerization is to separately analyze each site. Bahktina and Tsai (11) recently reported that rhodium(III) can be used as an exchange-inert metal ion to form a complex with a dNTP without largely compromising the catalytic efficiency of human polymerase β, effectively allowing the investigators to analyze the B site binding (Rh • dNTP) in a manner independent of A site binding (catalytic Mg2+). The conclusion was that B site binding of Rh • dNTP was sufficient for isomerization to a closed ternary complex (i.e., closing of the Fingers domain to form a preactive complex), but that addition of Mg2+ to occupy the A site was necessary for catalysis.
Their analysis however was incomplete, given the necessity to analyze the effect on the isomerization steps upon binding of Rh • dNTP with the catalytic A site already occupied. The use of Mg2+ to saturate the A site would be problematic since subsequent addition of Rh • dNTP leads to phosphodiester bond formation, complicating the interpretation of the kinetic data. Preventing catalysis has been achieved in several ways: the most frequently used approach has been the removal of the 3′-OH group of the primer, but the resulting space within the active site may not provide an accurate representation of the prechemistry steps due to the absence of this functionality that coordinates with the catalytic metal ion and water molecules that keep the reactive centers optimized for catalysis. Because so many crystal structures of DNA polymerase ternary complexes were obtained with dideoxy-terminated primers, we wanted to compare the results of kinetic studies using ddP/Ts versus dP/Ts to see whether the presence or absence of the primer's 3′-OH group affected the rate of the prechemistry conformational change.
Ca2+ has been shown to stabilize a ternary complex (i.e., by increasing the binding affinity of the incoming dNTP) without supporting catalysis and may be better suited as a mimic with the 3′-OH group present. However, a recent report by Egli and Guengerich (33) claimed that DNA pol IV from Sulfolobus solfataricus could utilize Ca2+ for phosphodiester bond formation, albeit at a much slower rate than Mg2+, but it was unclear if they had removed all of the Mg2+ from their assays.
In this report, we used catalytically inert ternary complexes (with Ca2+ as one of the metal ions, 3′-OH, a dideoxy-terminated primer, and Rh • dTTP) to analyze, in all combinations (i.e., through occupancy of the A site first, B site second, and vice versa), the contributions of each metal binding site to the formation of the final isomerized RB69 pol–P/T–Rh • dTTP complex using rapid chemical quench and stopped-flow fluorescence techniques. Our results showed that isomerization to a ternary complex (“closing” of the Fingers domain) occurred without occupancy of the A site. In the following paper (38), we show that reverse isomerization (Fingers “re-opening”) in the presence of Rh • dTTP is required before the catalytic metal ion can enter the empty A site. The importance of these results in understanding the kinetic mechanism of catalysis and base selectivity by this DNA polymerase is discussed.
MATERIALS AND METHODS
Escherichia coli strain BL21(DE3) was obtained from Stratagene Corp. DH5α cells and dNTPs were purchased from Invitrogen. Sequenase was from United States Biochemical Company. [γ-32P]ATP was from Perkin-Elmer Life Science Inc. Ni-NTA resin was obtained from Qiagen. Rhodium chloride hydrate was purchased from Sigma-Aldrich, Inc. AG1-X2-100 ion exchange resin was from ACROS Organics. Biogel P2 was ordered from Bio-Rad Laboratories. Other chemicals were analytical grade obtained from Sigma-Aldrich, Inc. RB69 DNA pol cDNA was a gift from J. Karam (Tulane University, New Orleans, LA).
Oligonucleotides were purchased from the W. M. Keck Foundation Biotechnology Resource Laboratory (Yale University). The sequences of the primer-templates (P/T) are shown in Figure 2.
Figure 2.
Sequences of the primer-templates used in this paper. Cd represents dideoxycytosine; P represents 2-aminopurine as the templating base.
Expression and Purification of Exo− RB69 pol
Site-directed mutagenesis, as previously reported, was used to substitute alanine for aspartic acid at positions 222 and 327 to create the exonuclease-deficient RB69 DNA pol that was used in all of our experiments, and that we term RB69 pol throughout the text (34). We subcloned the cDNA for the exo− RB69 pol into the pET21 vector (Invitrogen) so that the expressed protein would have six histidine residues appended to its C-terminus to facilitate purification which was carried out as previously described (29).
Preparation, Characterization, and Stability of the Rh•dTTP Complex
Rh• dTTP was prepared according to the procedure described for the synthesis of Rh • dCTP (11). To separate Rh • dTTP from the starting material and side products, the reaction mixture was loaded onto a 1.6 cm × 20 cm column of AG1-X2 in the chloride form. Prior to loading, the resin was washed with distilled, deionized (nanopure) water ([Mg2+] < 0.01 nM) that had been adjusted to a pH of 3 with HCl. After loading, elution was carried out by first passing nanopure water (pH 3) through the column until the eluate had an A260 of <0.05 optical density unit at 260 nm. The column was then eluted with 100 mM HCl. Two well-separated components with A260 absorption were obtained. The first component was a rhodium nucleotide complex that was inert as a substrate in primer extension reactions and was not characterized further. It was probably the dTTP equivalent of the dCTP derivative described by Dunlap and Tsai (35). The second component contained the desired Rh• dTTP. This material was concentrated by rotary evaporation, adjusted to pH 5 with NaOH, then loaded on a Biogel P2 gel filtration column, and eluted with a 2 mM sodium acetate solution (pH 5) to remove NaCl.
Rh• dTTP obtained using this procedure was characterized by 31P NMR using a NOVA-B400b NMR spectrometer. Spectra were recorded using a Rh • dTTP concentration of 20 mM in 2 mM sodium acetate buffer (pH 5) at 60 °C. The spectra showed the doublets expected for α- and γ-phosphates at −8.2 and −10.1 ppm with 20 Hz splitting for each peak (Figure 3A). The low-intensity peak at −7.8 ppm probably represents the β-phosphate. The peaks at 0.94 and 1.5 ppm are likely due to the complexation of dTTP with Rh3+. The spectra of Na • dTTP were recorded under the same conditions and concentrations as those of Rh• dTTP (Figure 3B). The spectrum has a triplet at −21.5 ppm representing the β-phosphate which is absent in the Rh•dTTP sample. There are also two doublets at −6 and −11 ppm with a 20 Hz split. These doublets represent the α- and γ-phosphates. The NMR results show that our Rh • dTTP sample was essentially free of Na • dTTP and had the expected chemical shift for the α- and γ-phosphates. The stability of Rh • dTTP was a concern even though it has been reported that Rh • dNTPs are stable up to pH 7 (11), which we have confirmed by 31P NMR; we did not know how Rh • dTTP would behave when it was incubated with RB69 pol–P/T complexes under conditions that were more stringent (5 h at pH 7 in MOPS buffer at 25 °C) than those used for our kinetic studies. Testing Rh • dTTP under these conditions showed that there was no exchange of rhodium with other cations. However, after incubation of Rh • dTTP with EDTA for 2 h at 25 °C, peaks corresponding to dTTP appeared in the 31P NMR spectra, indicating that EDTA competes with dTTP to sequester and chelate the rhodium cation. To test whether Rh • dTTP was stable under the conditions used for equilibrium and stopped-flow fluorescence experiments, we incubated a 20-fold molar excess of Rh • dTTP with an RB69 pol–dP/T binary complex and 2 mM Ca2+ for 16 h at 20 °C and then examined the 31P NMR spectra. There was no indication that the Rh • dTTP had been converted to Ca • dTTP.
Figure 3.
31P NMR spectra of Rh • dTTP (blue) and dTTP (black). (A) 31P NMR spectra of 20 mM Rh • dTTP in 2 mM acetate buffer (pH 5) at 60 °C. (B) 31P NMR spectra of 20 mM dTTP in 2 mM acetate buffer (pH 5) at 60 °C.
As a further test of purity, we subjected our Rh • dTTP preparation to thin layer chromatography. The Rh • dTTP sample was spotted on a thin layer, fluorescein-treated, polyethyleneimine plate which was then developed using a solution containing 0.5 M LiCl in 2 M acetic acid. This solution had been shown previously to separate mono-, di-, and trideoxynucleoside triphosphates (36). Only a single spot, which quenched the fluorescein fluorescence, appeared with a mobility that was somewhat greater than the dTTP control.
Incubation of this Rh • dTTP preparation with the RB69 pol–dP/T complex (dA as the templating base) for 30 min at 25 °C showed no evidence of dTMP incorporation. However, upon addition of 10 mM MgCl2, the Rh • dTTP was utilized and the primer was extended by one nucleotide residue, showing that the Rh • dTTP preparation was free of Mg2+ and that it could serve as a substrate when Mg2+ was present.
Preparation of Synthetic Oligonucleotide Substrates
The primer oligonucleotide used in the rapid chemical quench experiments was 5′-labeled with 32P with T4 polynucleotide kinase as per the manufacturer's instructions (New England Biolabs). Termination of the reaction was accomplished by heating the mixture to 95 °C for 5 min. We carried out primer-template annealing by mixing equimolar ratios of 13mer primer and 20mer template, heating the mixture to 95 °C, and allowing it to cool slowly to room temperature. The 20mer oligonucleotide templates used in the stopped-flow experiments, containing 2-aminopurine (dAP) at the templating position, were annealed to unlabeled 13mer primers as described above. The sequences of the primers and templates used are listed in Figure 2.
Chemical Quench Experiments
All rapid chemical quench reactions were preformed using a KinTek RQF-3 quench-flow instrument (KinTek Corp., Austin, TX). Single-turnover experiments were conducted at 24 °C by mixing equal volumes of a solution of 50 mM MOPS (pH 7), 10 mM MgCl2, 180 nM 5′-32P-labeled dP/T (dA or dAP at the templating position) (Figure 2), and 200 nM RB69 pol with a solution containing varying dTTP or Rh • dTTP concentrations in 50 mM MOPS buffer (pH 7) and 10 mM MgCl2. The reaction was quenched with 0.5 M EDTA at different times. Separation of the 32P-labeled substrate and product was achieved by gel electrophoresis (20% polyacrylamide/50% urea). The gel was visualized on a Storm 840 phosphoimager, and then substrate and product band intensities were quantified using Image QuaNT (Molecular Dynamics, Sunnyvale, CA).
Equilibrium Fluorescence Titrations
The emission spectra of deoxy- or dideoxy-P/Ts containing dAP were recorded at 24 °C with a Photon Technology International Alphascan scanning spectrofluorometer. The emission spectra of RB69 pol–P/T complexes were obtained in the same way. The reaction mixture contained 200 nM P/T (13/20mer) with dAP as the templating base (Figure 2), 1 μM RB69 pol, and either 10 mM MgCl2, 2 mM CaCl2, or no divalent cations. It also contained 50 mM MOPS buffer (pH 7.0) and varying dTTP concentrations. Samples were excited at 313 nm so that inner filter effects from nucleoside bases were negligible (37). Fluorescence emission spectra were collected from 330 to 460 nm. The intensities were corrected for the intrinsic fluorescence of RB69 pol and Rh • dTTP. No photobleaching could be detected after 10 min. Peak emission intensities at 365 nm were plotted as a function of dTTP or Rh • dTTP concentration and fit to hyperbolic or quadratic equations (see Data Analysis) to obtain the overall dissociation constants (Kdgapp) for dNTP binding.
Stopped-Flow Fluorescence Experiments
Experiments were performed using an Applied Photophysics (Leatherhead, U.K.) SX18MV-R stopped-flow apparatus thermostated at 24 °C. The excitation wavelength for dAP was 313 nm. A 345 nm long pass glass filter was used in monitoring of the dAP fluorescence emission. The final reaction mixtures consisted of 50 mM MOPS (pH 7.0), 400 nM P/T, 1 μM RB69 pol, varying dTTP or Rh • dTTP concentrations, and either 10 mM MgCl2, 2 mM CaCl2, or no divalent cation.
Data Analysis
Data from rapid chemical quench or stopped-flow experiments were fitted to single-exponential or double-exponential equations (Origin). The dNTP concentration dependence of kobs was fit to the hyperbolic equation kobs = kmax[dNTP]/(Kd + [dNTP]) + kr, where kobs is the observed rate of the reaction at different dNTP concentrations and kr is the extrapolated intercept at the Y axis when the dNTP concentration is zero. In experiments where chemistry is prevented, kr approximates the reverse isomerization rate. Data from the equilibrium fluorescence experiments were fit to either a hyperbolic or a quadratic equation as shown below:
where ΔQ is the fluorescence signal change, Qm is the maximum value for the fluorescence signal change at saturating substrate concentrations, E0 is the total enzyme concentration, and S0 is the total dNTP concentration. Kd is the ground-state dissociation constant for dissociation of dNTP from the RB69 pol–dP/T binary complex. The solution of these equations provided an estimate of the apparent dissociation constants of dNTP using Origin.
RESULTS
One of the goals of this study was to determine how occupancy of the A and/or B metal ion sites affected the affinity of Rh • dTTP for the RB69 pol–P/T binary complex and whether filling the A metal ion site influences the rate of Fingers closing after addition of a deoxynucleoside triphosphate. In addition, we wanted to determine whether the binary complex consisting of a dP/T or a ddP/T with RB69 pol would have comparable affinities for Rh • dTTP, and if both types of P/Ts would produce the same rates of conformational change in the presence or absence of sufficient Ca2+ to fill the A site. To monitor conformational changes subsequent to metal ion or nucleotide binding, we used 2AP as the templating base (dAP). For primer-templates (P/Ts), we used 13/20mers (Figure 2) that bound to RB69 pol with Kdgapp values in the range of 20 nM, approximately the same as those found previously for the T4 pol–13/20mer binary complexes (26).
Incorporation Efficiency of Rh • dTTP versus Templating dA or dAP
Prior to initiating equilibrium and stopped-flow fluorescence studies, we wanted to know how effective Rh• dTTP would be as a substrate for the nucleotidyl transfer reaction. For this purpose, we carried out rapid chemical quench experiments with four combinations of substrates and P/Ts that included Mg• dTTP or Rh • dTTP, as incoming dNTPs and dA or dAP as the templating base embedded in 13/20mer P/Ts. The kpol and Kdqapp values for this set of experiments are listed in Table 1. As shown in Figure 4, while Rh • dTTP can serve as a substrate, its kpol value (7 s−1), when paired opposite templating dAP, was much lower than the kpol value of 80 s−1 when Mg • dTTP was paired opposite dAP. Also, the Kdqapp for Rh • dTTP incorporation (150 μM) was almost 3 times higher than the Kdqapp for Mg• dTTP (45 μM) when it was paired opposite dAP. The observation that the kpol for Rh•dTTP was considerably lower than that for Mg• dTTP prompted us to examine the kinetic parameters for insertion of Mg• dTTP opposite dA and dAP. As shown in Table 1, the kpol value for Mg• dTTP incorporation, when paired opposite dAP, was only reduced by ~40% relative to the kpol for Mg• dTTP paired opposite dA under these conditions. These data provide a basis for interpreting the fluorescence results when Rh • dTTP was paired opposite dAP.
Table 1.
Re-Steady-State Kinetic Parameters for Incorporation of Different Metal Ion-dTTP Complexes Paired Opposite dA or dAP as the Templating Base
| Me•dNTP:templating base | kpol(s−1) | Kdqapp (μM) |
|---|---|---|
| Mg•dTTP:dA | 131 ± 8 | 27 ± 4 |
| Mg•dTTP:dAP | 80 ± 7 | 45 ± 16 |
| Rh•dTTP:dA | 76 ± 5 | 124 ± 40 |
| Rh•dTTP:dAP | 7 ± 0.6 | 150 ± 40 |
Figure 4.
Time course for the incorporation of Rh • dTTP opposite dAP catalyzed by RB69 pol. (A) The rate of product formation was measured at various Rh • dTTP concentrations, from 10 μM (black) to 1000 μM (dark yellow) from bottom to top. (B) The observed rate was plotted against Rh • dTTP concentration. The curve fit best to a hyperbolic equation with a kpol of 7.1 ± 0.6 s−1 and a Kdqapp of 150 ± 40 μM.
Equilibrium Fluorescence Experiments Using Rh • dTTP Paired Opposite a Templating dAP
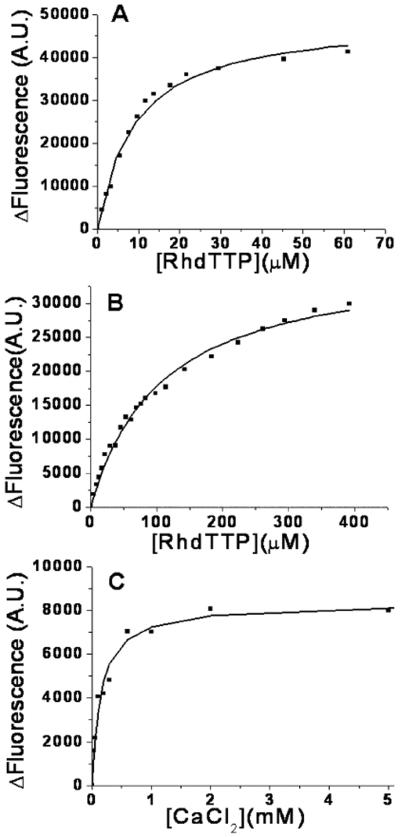
When Rh • dTTP was added to the binary E• dP/T complex in the presence of a divalent cation (2 mM CaCl2) (Figure 5) and the extent of dAP fluorescence quenching determined as a function of Rh • dTTP concentration, a Kdgapp for Rh • dTTP of 9.1 μM was obtained (Figure 6A). This experiment was repeated in the absence of divalent cations, and the Kdgapp for Rh • dTTP under these conditions was estimated to be 108 μM (Figure 6B). The reduction in Kdgapp for Rh • dTTP was attributed to Ca2+ occupancy of the A metal site which, we presume, stabilizes the closed ternary complex. In this state, the templating base was stacked in the dP/T duplex in the presence of the correct dNTP, as observed in the crystal structure of the RB69 pol–ddP/T–dNTP complex (21). The stacking of the templating base occurred even though the nascent incoming base was not covalently linked to the 3′ end of the primer (21). In the absence of Rh • dTTP, addition of Ca2+ results in partial quenching of dAP fluorescence in the binary RB69 pol–dP/T complex. The extent of dAP quenching depended on Ca2+ concentration, yielding a Kdgapp for Ca2+ of 152 μM (Figure 6C). The amplitude of the fluorescence change was only 8400 AU compared to a decrease of 37000 AU when a saturating concentration of Rh • dTTP was added to the binary complex in the absence of divalent cations. Whether this Kdgapp for Ca2+ actually represents the Ca2+ concentration required for half-occupancy of the A metal site only remains to be determined.
Figure 5.
Equilibrium fluorescence titration of the RB69 DNA pol–dP/T complex with an increasing Rh • dTTP concentration. Fluorescence emission spectra of the RB69 pol–dP/T–Rh • dTTP complex are shown as solid lines in the presence of 2 mM CaCl2. The concentration of dP/T was 200 nM and that of RB69 pol 1 μM. The Rh • dTTP concentrations (from top to bottom) were 0, 2.2, 5.5, 9.7, and 17.8 μM. The dotted line is the fluorescence spectrum of dP/T alone.
Figure 6.
Equilibrium fluorescence titrations of 1 μM RB69 pol complexed with 200 nM dP/T with an increasing Rh•dTTP or CaCl2 concentration. The change in fluorescence signal vs Rh • dTTP or CaCl2 concentration fit a hyperbolic equation. For the dP/T sequence, see Figure 2A2. (A) Titration of Rh • dTTP with the A site filled with Ca2+ (2 mM) gave a Kd2,gapp of 9.1 ± 0.9 μM. (B) With the A site empty, titration of Rh • dTTP gave a Kd1,gapp of 108 ± 6.6 μM. (C) Titration of Ca2+ with the B site empty gave a Kd3,gapp of 152 ± 19 μM. The fluorescence change (ΔF) with an increasing Rh•dTTP concentration indicates the extent of fluorescence quenching for the ternary RB69 pol–dP/T–Rh•dTTP complex compared with that of the binary RB69 pol–dP/T complex.
Although it has been well established that two divalent metal ions are essential for the nucleotidyl transfer reaction with DNA polymerases, it was not known whether occupancy of either the A or B metal ion site, or both, is absolutely necessary for the open to closed transition when a correct dNTP is introduced. To address this question, Rh • dTTP was selected as the incoming dNTP because of its metal exchange-inert properties and because it was complementary to dAP. This allowed us to determine the effect of B site occupancy, with the nucleotide binding pocket also filled, on the rate of the conformational change in the presence and absence of Ca2+ in the A site. It also allowed us to determine the affinity of Rh • dTTP for the binary complex when the A site was empty. To fill the A site, Ca2+ was chosen because it is presumably ligated by the same set of aspartate carboxyl groups as Mg2+ and because the crystal structure of the RB69 pol ternary complex was determined in the presence of Ca2+ (21). The amplitude of the quench with a saturating Rh • dTTP concentration was greater than the amplitude found when Ca2+ only was added to the binary complex. This implies that filling the B site and the nucleotide binding pocket with Rh • dTTP, the correct base, is sufficient to induce a complete transition to the closed ternary complex but also that Ca2+ alone may cause partial closure of the Fingers domain (Figure 7). A summary of the relationships between the affinities of Rh•dTTP for the binary RB69 pol–P/T complex in the presence and absence of Ca2+ and the affinity of Ca2+ for the RB69 pol–dP/T complex in the absence of Rh • dTTP is shown in Figure 7, and the Kd values are given in Table 2 for dP/T and ddP/T. The affinity of Ca2+ for the RB69 pol–dP/T–Rh • dTTP complex under conditions of saturating Rh • dTTP concentrations could not be determined because there was no further quenching of dAP when Ca2+ was added to the RB69 pol–dP/T–Rh• dTTP complex.
Figure 7.
Dependence of Ca2+ or Rh • dTTP binding to the RB69 pol–dP/T binary complex. The Kd for Ca2+ with the RB69 pol–dP/T complex was 152 μM. Kd1 and Kd2 values for Rh • dTTP binding to the RB69 pol–dP/T or RB69 pol–ddP/T complex are listed in Table 2.
Table 2.
Effect of A Metal Site Occupancy on the Binding Affinity of Rh•dTTP with a dP/T or ddP/T and Rates of Fingers Closing (k+3)
Since the amplitude of dAP quenching was nearly the same, with or without Ca2+, when a saturating concentration of RhTTP was added to the RB69 pol–dP/T complex, it implies that the fully closed ternary complex can form even when the A site is empty. The fluorescence observed with a saturating Rh • dTTP concentration was also consistent with the fluorescence of the RB69 pol–dP/T binary complex, as well as with that of the RB69 pol–ddP/T–Rh • dTTP ternary complex where the dAP stacks with its 3′ neighbor at saturating Rh • dTTP concentrations even though it does not become covalently linked to the nucleotide residue at the 3′ terminus of the primer.
Quench Amplitudes of 2AP Fluorescence Due to Rh • dTTP Binding to an RB69 pol–dP/T Binary Complex: Comparison of a dP/T with a ddP/T
The maximum amplitude for dAP quenching, when a ddP/T was a component of the binary complex, occurred with 2 mM Ca2+ and saturating Rh• dTTP or Ca • dTTP concentrations. The amplitude of the quench produced by saturating Rh • dTTP concentrations without Ca2+ was 26% of that observed when both Ca2+ and saturating Rh • dTTP concentrations were present and was 69% of that value when the mixture contained 10 mM Mg2+ instead of Ca2+. The higher affinity of Rh • dTTP for the E• D complex when Ca2+ rather than Mg2+ occupied the A site may be related to size differences and differences in coordination modes of the two metal ions. The quench amplitudes from the stopped-flow fluorescence scans, when a dP/T or a ddP/T was used, with Ca2+ in the A site, and either Rh • dTTP or Ca • dTTP in the B site, were dependent on the dTTP concentration.
Comparison of Rh • dTTP Affinities for RB69 pol–dP/T or RB69 pol–ddP/T Binary Complexes
While Rh • dTTP enabled us to use a dP/T, so that the contribution of the primer's 3′-OH group to the binding affinity of a deoxynucleoside triphosphate could be determined in the presence and absence of divalent cations, we still had to evaluate whether Rh • dTTP was able to mimic Mg• dTTP in its ability to induce the transition from the open to the closed state. In particular, we wanted to know (i) the rate at which Rh • dTTP induces the transition to the closed ternary complex and (ii) the Rh • dTTP concentration that would produce half-maximal quenching of dAP fluorescence. To do this, we compared the rates of dAP quenching induced by Rh • dTTP using both a dP/T and a ddP/T in the presence and absence of Ca2+. The effect on the affinity of Rh • dTTP for the binary E•D complex in the presence and absence of the primer's 3′-OH group could also be evaluated by determining the change in dAP fluorescence as a function of Rh • dTTP concentration, allowing us to estimate binding constants for Rh • dTTP in these two situations. As shown in Table 2, the Kdgapp for Rh • dTTP binding to the RB69 pol–ddP/T complex with the A site empty was 63 μM (Figure 1A of the Supporting Information) and 108 μM with a dP/T. When Ca2+ occupied the A site, the Kdgapp for Rh • dTTP was 9 μM with either a ddP/T (Figure 1B of the Supporting Information) or a dP/T, indicating that the presence or absence of an OH group at the 3′ terminus of the primer did not affect the affinity of Rh • dTTP for the E•D complex.
Rates of the Conformational Change When Rh • dTTP Binds to an E•D Complex: Comparison of a dP/T with a ddP/T on the Rate of Fingers Closing
Having determined the Kdgapp values for the different substrates and metal ion combinations, we then decided to compare the rates of conformational change when the A site was empty or filled with Ca2+ and when the B site was occupied by Rh3+ complexed with dTTP. At the same time, we wanted to determine what effect the 3′-OH group at the primer terminus would have on the rate of the conformational change. For this purpose, we compared a ddP/T with a dP/T. The first experiments were carried out with Rh • dTTP, dP/T, and 2 mM Ca2+. As can be seen in Figure 8A,B, the kmax was ~812 s−1, a value nearly equal to what we reported previously with Mg• dTTP and a ddP/T (37). In all these experiments, there was no change in start points and the quench rates followed a single-exponential equation. These results suggest that a single step was involved and that the presence or absence of the 3′-OH group at the primer terminus had only a minor effect on the rate of dAP quenching. The similarity between the results obtained with the RB69 pol–ddP/T–Mg• dTTP combination and the RB69 pol–dP/T–Mg• dTTP complex in the presence of Ca2+ suggests that the same process (Fingers closing) was being observed in both situations. Furthermore, the Y axis intercept was 6.7 s−1 (Figure 8B), a value consistent with the relatively slow rate of Fingers opening (~ 5s−1) that was derived from pulse chase chemical quench experiments with Mg2+ as described in the following paper (38). However, the apparent Kdrapp was 100 μM (Figure 8B), a value much higher than the Kdgapp for binding of Rh • dTTP to a dP/T in the presence of 2 mM Ca2+, determined from equilibrium fluorescence measurements (Figure 6A). The final equilibrium amplitudes of the dAP quenching, derived from the fluorescence end points at 0.1 s (Figure 8A), when plotted as a function of Rh • dTTP concentration, fit a hyperbolic equation giving a Kdaapp of 19.7 μM. The observation that the changes in amplitudes increased as a function of Rh • dTTP concentration (Figure 8C) is consistent with the reversibility of the EDN ⇌ FDN isomerization and the fact that chemistry did not occur. As in the case of Mg• dTTP with ddP/T, there was no change in start points, so that we were able to estimate the rate of the conformational change, which followed a single-exponential equation.
Figure 8.
Stopped-flow fluorescence scans of the RB69 pol–dP/T complex with increasing Rh • dTTP concentrations. For the dP/T sequence, see Figure 2A2. (A) One syringe contained 800 nM DNA and 4 μM RB69 pol in 50 mM MOPS buffer (pH 7) at 24 °C. The other syringe contained varying Rh • dTTP concentrations (0, 5, 10, 20, 40, 60, 75, 100, 125, 150, 200, and 250 μM from top to bottom) in MOPS buffer. Both syringes contained 2 mM CaCl2. The fluorescence signal vs time fit a single-exponential equation. The scans are shown in different colors for different Rh • dTTP concentrations, and the fitting curves are colored black. (B) Rates from the single-exponential vs Rh • dTTP concentration fit a hyperbolic equation with a kmax of 812 ± 38.9 s−1, a Kdrapp of 100 ± 18.3 μM, and a Y axis intercept of 7 ± 22 s−1. (C) The amplitude change for the fluorescence quench vs Rh• dTTP concentration also fit a hyperbolic equation with a Kdaapp of 19.7 ± 3.4 μM.
The same experiment was then performed under the same conditions except that a ddP/T was used instead of a dP/T so we could see how the primer's 3′-OH group affected the rate of dAP quenching. The fluorescence change fit best to a double exponential (Figure 2A of the Supporting Information). The faster rate of the first phase, obtained from the double exponential, when plotted against Rh • dTTP concentration, fit a hyperbolic equation. The average rate for the second phase was 21.5 ± 6.4 s−1 and was independent of Rh•dTTP concentration. This second phase was not observed in experiments with dP/T carried out under the same conditions, suggesting an off-pathway step or a local conformational change of dAP at the active site due to lack of the 3′-OH group at the primer terminus. As seen in Figure 8 and Figure 2B of the Supporting Information, the estimated kmax was faster with the dP/T (812 s−1) than with the ddP/T (511 s−1) for the first phase which we assumed was the rate of the Fingers domain closing. The Kdrapp was also larger with the dP/T than with the ddP/T. We note that the Y axis intercept for the experiments with ddP/T (8.4 s−1) (Figure 2 of the Supporting Information) was nearly the same as that found with the dP/T (6.7 s−1) (Figure 8), indicating that the rate (k−3) for the reversal of the isomerized FDN complex to the collision complex, EDN (Scheme 1), was not influenced by the nature of the 3′ terminus of the primer and appeared to be independent of the nature of the cation that occupies the B metal ion site.
Scheme 1.
Expanded Kinetic Scheme for Primer Extension
Effect of Ca • dTTP versus Rh • dTTP on the Rate of Conformational Change
From stopped-flow fluorescence experiments, which gave an estimate of the Rh • dTTP concentration required to provide 50% of the maximum quench rate, a Kdrapp of 100 μM was obtained with dP/T (Figure 8B). This value was 10 times greater than the Kdgapp for Rh • dTTP binding to a dP/T as estimated by equilibrium fluorescence titration (Figure 6A). We note that there was no change in start points (Figure 8A). This result resembled the situation that we observed previously when Mg• dTTP was added to the RB69 pol–ddP/T binary complex (37). Next we wanted to determine whether Ca • dTTP would behave in a fashion similar to that of Rh • dTTP so we repeated the stopped-flow fluorescence experiment using Ca • dTTP in place of Rh • dTTP. The fluorescence scans (Figure 3A of the Supporting Information) with increasing Ca • dTTP concentrations showed a time-dependent decrease in dAP fluorescence that followed a single exponential with no change in start points (Figure 2A of the Supporting Information). The plot of kobs versus Ca • dTTP concentration fit best to a hyperbolic equation giving a kmax of 1200 s−1 (Figure 3B of the Supporting Information). The Kdrapp for Ca • dTTP was estimated to be 6.8 μM (Figure 3B of the Supporting Information), a value 125 times that of the Kdgapp (53 nM) determined from equilibrium fluorescence experiments (Figure 4 of the Supporting Information). A plot of the final amplitudes from Figure 3A of the Supporting Information versus dTTP concentration fit a hyperbolic function with a Kdaapp of 1.9 ± 0.3 μM, consistent with the reversibility of the isomerization (Figure 3C of the Supporting Information).
Rates of Conformational Change and Chemistry When Mg2+ and an RB69 pol–dP/T Binary Complex Are Incubated with Increasing Rh • dTTP Concentrations
In all of the experiments described above, we investigated rates of the conformational change under conditions that did not allow chemistry. We now wanted to determine what would happen to the conformational change rate if we permitted chemistry to occur by using an RB69 pol–dP/T complex and adding increasing Rh • dTTP concentrations in the presence of 10 mM Mg2+. The results, depicted in Figure 9, show that the start points decrease as the Rh • dTTP concentration is increased, indicative of a quench rate of >1000 s−1, and that the second phase of the fluorescence quench has a rate approximately equal to the kpol obtained from the rapid chemical quench. These data are consistent with the kinetic scheme (Scheme 1), and the agreement between the fluorescent scans and those generated by KinTekSim using the parameters in Table 3 is quite good. The calculations supporting this assertion are given in the Supporting Information. These results should be compared with those of the same experiment that was carried out with Ca2+ instead of Mg2+. With Ca2+, there was a rapid quench phase where the decay of fluorescence followed single exponentials with different final amplitudes indicating the reversibility of the EDN ⇌ FDN isomerization. With Mg2+, the final amplitudes converged at the same value because all the Rh • dTTP was converted to product and where dAP was fully quenched because it was stacked against its 3′ neighbor in the P/T duplex. By reducing the rate of chemistry using Rh • dTTP instead of Mg• dTTP, we might have expected to see another fluorescent intermediate after Fingers closing, but none was found.
Figure 9.
Stopped-flow fluorescence for the incorporation of Rh • dTTP opposite dAP with a dP/T. One syringe contained 800 nM dP/T (Figure 2A2) and 4 μM RB69 pol in 50 mM MOPS buffer (pH 7) at 24 °C. The other syringe contained varying Rh • dTTP concentrations [0 (black), 20 (red), 60 (green), 200 (blue), and 400 μM (cyan)] in the same MOPS buffer. Both syringes contained 10 mM MgCl2. The fluorescence signal vs time fits the kinetic scheme shown in Scheme 1. The parameters for the kinetic scheme for Rh • dTTP incorporation, generated from the KinTek Simulator program, are listed in Table 3. The experimental scans and the fitting curves are shown in different colors for different Rh • dTTP concentrations.
Table 3.
Kinetic Parameters for Rh•dTMP Incorporation According to Scheme 1
| step | scheme | k+ | k − |
|---|---|---|---|
| 1 | 10 s−1 μM−1 | 0.01 s−1 | |
| 2 | 10 s−1 μM−1 | 4000000 s−1 | |
| 3 | 2000 s−1 | 4 s−1 | |
| 4 | 7 s−1 | 0 s−1 | |
| 5 | 0.01 s−1 | 10 s−1 μM−1 |
DISCUSSION
Effect of Metal Ions in the A and B Sites on the Affinity of Rh • dTTP for the E•D Binary Complex and on the Rate of dTMP Incorporation
The catalytic role for two divalent cations in the polymerase active site of DNA polymerases is well-established (39, 40). While Mg2+ is the physiologically relevant cation (41), Mn2+ can also efficiently catalyze the nucleotidyl transfer reaction; however, in its presence, the ability of replicative polymerases to discriminate against incorrect dNTPs is weakened, and thus, Mn2+ is considered to be a mutagenic agent (42–44). Attempts to crystallize RB69 pol–P/T–dNTP ternary complexes having a nonextendable primer with Mg2+ have failed, but the same types of complexes containing Ca2+ have provided high-quality crystals that have enabled RB69 pol structures to be determined at high resolution (21). This prompted us to use Ca2+ for our fluorescence studies since we could potentially relate our findings to the information available from X-ray crystallography, thus providing a structural context for interpreting our kinetic results.
Another advantage of using Ca2+ together with Rh • dTTP is that Ca2+ does not support catalysis. This made it possible to use a deoxy-terminated rather than a dideoxy-terminated primer to investigate the effect of filling the A or B metal ion sites individually on the rates of conformational change, and on the binding affinity of Rh • dTTP. It also allowed us to compare deoxy- and dideoxy-P/Ts to determine what role the primer's 3′-OH group has on the affinity of the incoming Rh • dNTP, and on the rate of Fingers closing. Nevertheless, we are aware that Ca2+ is an imperfect mimic for Mg2+ in a number of respects. Its ionic radius is larger than Mg2+, making it less effective in polarizing the α P–O bond of the incoming dNTP for nucleophilic attack by the 3′-OH group of the primer terminus (33). This is likely to be part of the reason that Ca2+ is unable to catalyze the nucleotidyl transfer reaction. In addition, the coordination mode of Ca2+ is more variable than that of Mg2+, which is usually octahedral. Ca2+ has been observed to be coordinated between four and eight ligands. There is also a difference in hydration enthalpy, −1577 kJ for Ca2+ versus −1921 kJ for Mg2+ (45). This results in inner-sphere coordination of Ca2+ to DNA (46). Ca2+ does not lower the pKa of a water molecule as much as Mg2+ [12.8 and 11.4, respectively (47, 48)], and this likely also contributes to the catalytically inert behavior of Ca2+ in phosphoryl transfer reactions. These properties may also account for the fact that Ca2+ in the A site increases the affinity of Rh • dTTP for the RB69 pol–P/T complex.
The dissection of the effect of A and B site occupancy on the affinity of the incoming dNTP for the E •D complex and on the rate of conformational change can be investigated only when a nonexchangeable metal ion is complexed with an incoming dNTP. Despite the fact that the phosphate oxygen–rhodium bond distances are similar to those of Mg •dNTPs (11), it is not possible to conclude, a priori, that Rh • dNTPs are good substrates compared to dNTPs complexed with Mg2+. The fact that full quenching could be achieved with the A site empty suggests that A site occupancy is not a prerequisite for formation of the closed ternary complex. It would seem that dAP can be fully stacked in the duplex, even when the incoming Rh • dTTP is not covalently attached to the primer. The low Kdgapp for Rh • dTTP, when Ca2+ is present, suggests that filling the A site stabilizes the closed, ternary structure. Note that Ca2+ alone, in the absence of any dNTP, results in dAP quenching, but with only 26% of the full amplitude that is observed when the Rh • dTTP concentration is at saturating levels. However, we do not have any independent evidence to show that the dAP quenching is actually caused by Ca2+ occupancy of the A site only.
Rapid chemical quench experiments clearly show that Rh • dTTP was incorporated at less than one-tenth of the rate of Mg• dTTP, when matched opposite a dAP as the templating base, supporting the notion that Rh • dTTP did not undergo metal ion exchange under the conditions used for the chemical quench (Table 1). It is interesting to compare the kinetic parameters obtained from rapid chemical quench experiments with Rh• dTTP and Mg• dTTP when matched opposite a templating dA or dAP. As shown in Table 1, the largest difference in kpol (10-fold) was observed when Rh • dTTP was matched opposite dA or dAP, favoring dA. In contrast, when Mg• dTTP was matched against dA or dAP, these differences in kpol values were only 1.5-fold, in favor of dA. This difference is in line with what has been reported previously (37, 49). On the other hand, when the kpol for Rh • dTTP was compared with that of Mg• dTTP, positioned opposite dA, there was only a small difference (1.5-fold) in favor of Mg• dTTP, but the enzyme exhibits less base discrimination in the presence of dAP rather than dA when Rh • dTTP is used. This suggests that dAP is a good mimic of dA when opposite Mg• dTTP, but not when opposite Rh • dTTP. While Rh • dTTP does not behave exactly like Mg• dTTP, even when Mg2+ is in the A site, it still serves as a substrate and presumably as a ligand for inducing the critical conformational change when the A site is empty or occupied by a divalent cation that does not permit catalysis (11). It is likely that the differences in coordination between Rh3+ and Mg2+ for dTTP can account for changes in rates of dTMP incorporation but have little effect on the rates of the conformational change. Comparison of the Kdqapp values for Mg• dTTP and Rh • dTTP is instructive in that they show approximately the same 2:1 ratios of apparent binding affinities for dA relative to dAP. The single substitutions show 2.8- and 7.9-fold effects, compared with a 104-fold effect for the double substitution (2.8 × 7.9) = 22, which is 5-fold less than 104). These data and the molecular dynamics simulation suggest a 1 Å movement of dAP opposite Rh • dTTP under this condition compared to dA opposite Mg• dTTP, which is consistent with the observed cooperativity in binding and/or recognition of templating base and metal B. This confirms previous reports that when dAP is the templating base, the complementarity with dTTP is altered compared to that when the templating base is dA (49, 50). The decrease in kpol for the Rh • dTTP–dAP pair is probably due to imperfect alignment of the attacking 3′-oxygen at the terminus of the primer with the R-phosphorus atom of Rh • dTTP. These results suggest that the nature of the metal ion in the B site affects the catalytic rate in addition to facilitating dNTP binding.
Effect of Rh • dTTP on the Rate of the Conformational Change
It is interesting to compare the rates of conformational change induced when Mg• dTTP or Rh • dTTP is the incoming dNTP. We have previously shown that there were two phases for the rate of conformational change when Mg• dTTP was matched opposite dAP: a very rapid phase with a rate of >1000 s−1 which we assigned to Fingers closing and a slower phase (rate of 290 s−1) that was approximately the same as the rate of dTMP incorporation as determined by rapid chemical quench (37). With Rh • dTTP opposite dAP, there was also a rapid fluorescence quench (>1000 s−1) followed by a slower quench with approximately the same rate as dTMP incorporation. This slower phase of fluorescence quenching may be reporting a change in dAP stacking that occurs just prior to or simultaneously with the chemical step. Modeling the RB69 pol complex as it proceeds to the transition state shows that there must be an ~1 Å shift of the templating base with respect to its 3′ neighbor that could create additional stacking of dAP, causing an increase of ~20% in the amplitude of the quench if the 3′ neighboring base is a purine (J. Wang, personal communication). This was indeed the case with our template sequence, and the amplitude of the second phase was ~25% of the total quench amplitude. If there is a concerted movement of the substrates in their approach to the transition state, and in progressing through it to product, the rate of the additional stacking interaction cannot be separated from the rate of the chemical step. Note that this second phase is not observed when chemistry does not occur and that the first rate, which reflects Fingers closing, is reduced from >1000 to ~500 s−1 at a saturating Rh • dTTP concentration. In contrast to the situation with dP/Ts, where the quench amplitudes of stopped-flow fluorescence scans converge at a single value, the amplitudes of the scans with ddP/Ts are dependent on the Rh • dTTP concentration because the quenched state is reversible, as depicted in Scheme 1.
Among the reports that have estimated the rate of Fingers closing, the most recent have come from FRET experiments with Klentaq (51, 52) and with T7 DNA polymerase (T7 pol) (53) which serve to illustrate the scope and limitation of this approach. With Klentaq, the rate of Fingers closing was very rapid, exceeding the rate of dNMP incorporation, as monitored by chemical quench experiments, suggesting that chemistry, or a kinetically silent step before chemistry, limited the nucleotidyl transfer rate (51, 52). With T7 pol, labeling with a fluorescent dye at a single Cys residue in the Fingers domain provided a convenient probe for monitoring conformational changes. Upon addition of a correct dNTP, the fluorescence was partially quenched, whereas addition of an incorrect dNTP caused fluorescence enhancement. The rates of these fluorescent changes were measured and ascribed to conformation changes involving opening and closing of the Fingers domain (53). Other papers that have dealt with the rate of Fingers closing include studies with Klenow fragment (13), T4 pol (54), RB69 pol (37), and pol β (55).
Effect of Individual Metal Ions on the Rates of Conformational Change in pol β
Attempts have been made previously to dissect the potential roles of the two metal ions, known to be essential for catalysis, in facilitating the conformational change that occurs after dNTP binding. Experiments with this aim were carried out using polymerase β (pol β) and Cr(III) nucleotide complexes because of their exchange-inert properties (56). These authors suggested that binding of the Cr •dNTP to the pol β–-P/T complex facilitated Mg2+ binding to the catalytic metal ion site (site B in the nomanclature Zhong et al. but termed the A metal ion site in our paper). Our results with RB69 pol differed from those reported for pol β in that we observed closing of the Fingers domain with Rh • dTTP even when the A site was empty. Instead of Rh • dTTP facilitating Mg2+ binding in the A site, Mg2+ binding did not occur until the Fingers reopened, a step that occurred more slowly than Fingers closing (38). We also found that the rate of Fingers reopening limited the rate of dTMP incorporation under conditions where Mg2+ was added to a ternary RB69 pol–dP/T–Rh • dTTP complex that had been assembled in the absence of divalent cations (38).
Dynamic simulations were also performed with pol β by Yang et al. (56), leading to the conclusion that the dNTP–metal ion complex alone cannot induce pol β closing before chemistry (56). Although a comparable dynamic simulation study has not been performed with RB69 pol, our experimental results with RB69 pol differ from the pol β simulations, suggesting that either the two polymerases behave very differently with respect to Fingers closing. In a subsequent report (57), the authors revised their scheme indicating that closing of the Fingers could be induced by Cr • dNTP binding alone with the A site empty but that the slow step reflected Mg2+ binding when the polymerase was still in the closed state (57). This revised model is closer to what we have proposed for RB69 pol but still differs, in that we suggest that the Fingers have to reopen, at least partially, to allow for Mg2+ binding and that it is the rate of Fingers reopening that is slow, rather than the rate of Mg2+ binding to the A site in the closed ternary complex (38).
In a more recent report, Rh(III) rather than Cr(III) • dNTP complexes were employed with pol β because of reasons stated here and in the paper by Bakhtina et al. (11). In this version, the authors stated that the slow phase following Fingers closing occurs after Mg2+ binding and that the slow step reflected the rate of the chemical step. While this revised scheme seems to be the one that best fits with all of the pol β data, it still does not square completely with our RB69 pol results. Further experimentation along the lines suggested by Bakhtina et al. (11) will be required to determine whether pol β represents a special case or whether the conclusions drawn from the extensive studies on pol β can be applied generally to all DNA polymerases.
SUMMARY
By employing Rh • dTTP as the incoming nucleotide matched opposite dAP, we have been able to separate the effect of A and B metal ion site occupancy on the rates of Fingers closing and on the affinity of Rh • dTTP for the E • D binary complex. We show the dependence of RhdTTP binding as a function of Ca2+ concentration. We show that the presence or absence of the 3′-OH group on the primer does not appreciably affect the rate of Fingers closing with the A site empty or when it was filled with Ca2+ when Rh • dTTP is the substrate. In the following paper (38), we show that the rate of Fingers reopening is slow in the presence of correct dNTPs but very rapid when an incorrect dNTP is part of the ternary complex. The implications for mechanisms that control polymerase fidelity are discussed in that paper.
Supplementary Material
ACKNOWLEDGMENT
We are indebted to Dr. Enrique De La Cruz and Dr. Jeff Beckman for informative discussion and for critical reading of the manuscript.
This work was supported by National Institutes of Health Grant GM063276.
Footnotes
Abbreviations: T4 pol, T4 DNA polymerase; RB69 pol, RB69 DNA polymerase; dP/T, 3′-deoxy primer-template; ddP/T, 3′-dideoxy primer-template; dNTP, deoxynucleoside triphosphate; E, DNA polymerase; D, either dP/T or ddP/T; N, any dNTP; Rh • dTTP, rhodium(III) complexed with dTTP; 2AP, 2-aminopurine; dA, adenine as the templating base; dAP, 2-aminopurine as the templating base; KF, Klenow fragment; Kdqapp, dNTP concentration at the half-maximal rate of dNMP incorporation as determined by rapid chemical quench assays; Kdgapp, dNTP concentration at the midpoint of dAP quenching with an increasing dNTP concentration from equilibrium fluorescence titrations; Kdrapp, dNTP concentration at the half-maximal rate of dAP quenching from stopped-flow fluorescence experiments; Kdaapp, dNTP concentration at the half-maximal amplitude for dAP quenching from stopped-flow fluorescence experiments; kpol, maximum rate of nucleotide incorporation from rapid chemical quench assays; kmax, maximum rate of dAP quenching from stopped-flow fluorescence experiments; kobs, observed rate constant.
SUPPORTING INFORMATION AVAILABLE Calculation of kinetic parameters for Scheme 1 and Figures 1–4 for equilibrium fluorescence titration and stopped-flow fluorescence experiments. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Echols H, Goodman MF. Fidelity mechanisms in DNA replication. Annu. Rev. Biochem. 1991;60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- 2.Johnson KA. Conformational coupling in DNA polymerase fidelity. Annu. Rev. Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- 3.Joyce CM, Benkovic SJ. DNA polymerase fidelity: Kinetics, structure, and checkpoints. Biochemistry. 2004;43:14317–14324. doi: 10.1021/bi048422z. [DOI] [PubMed] [Google Scholar]
- 4.Kunkel TA, Bebenek K. Recent studies of the fidelity of DNA synthesis. Biochim. Biophys. Acta. 1988;951:1–15. doi: 10.1016/0167-4781(88)90020-6. [DOI] [PubMed] [Google Scholar]
- 5.Kunkel TA, Bebenek K. DNA replication fidelity. Annu. Rev. Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 6.Echols H. Mutation rate: Some biological and biochemical considerations. Biochimie. 1982;64:571–575. doi: 10.1016/s0300-9084(82)80089-8. [DOI] [PubMed] [Google Scholar]
- 7.Goodman MF. Hydrogen bonding revisited: Geometric selection as a principal determinant of DNA replication fidelity. Proc. Natl. Acad. Sci. U.S.A. 1997;94:10493–10495. doi: 10.1073/pnas.94.20.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kool ET. Active site tightness and substrate fit in DNA replication. Annu. Rev. Biochem. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]
- 9.Kunkel TA. DNA replication fidelity. J. Biol. Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 10.Sloane DL, Goodman MF, Echols H. The fidelity of base selection by the polymerase subunit of DNA polymerase III holoenzyme. Nucleic Acids Res. 1988;16:6465–6475. doi: 10.1093/nar/16.14.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bakhtina M, Lee S, Wang Y, Dunlap C, Lamarche B, Tsai MD. Use of viscogens, dNTPRS, and rhodium(III) as probes in stopped-flow experiments to obtain new evidence for the mechanism of catalysis by DNA polymerase β. Biochemistry. 2005;44:5177–5187. doi: 10.1021/bi047664w. [DOI] [PubMed] [Google Scholar]
- 12.Hariharan C, Bloom LB, Helquist SA, Kool ET, Reha-Krantz LJ. Dynamics of nucleotide incorporation: Snapshots revealed by 2-aminopurine fluorescence studies. Biochemistry. 2006;45:2836–2844. doi: 10.1021/bi051644s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Purohit V, Grindley ND, Joyce CM. Use of 2-aminopurine fluorescence to examine conformational changes during nucleotide incorporation by DNA polymerase I (Klenow fragment) Biochemistry. 2003;42:10200–10211. doi: 10.1021/bi0341206. [DOI] [PubMed] [Google Scholar]
- 14.Drake JW. Comparative rates of spontaneous mutation. Nature. 1969;221:1132. doi: 10.1038/2211132a0. [DOI] [PubMed] [Google Scholar]
- 15.Goodman MF. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 16.Kunkel TA, Loeb LA, Goodman MF. On the fidelity of DNA replication. The accuracy of T4 DNA polymerases in copying ϕX174 DNA in vitro. J. Biol. Chem. 1984;259:1539–1545. [PubMed] [Google Scholar]
- 17.Muzyczka N, Poland RL, Bessman MJ. Studies on the biochemical basis of spontaneous mutation. I. A comparison of the deoxyribonucleic acid polymerases of mutator, antimutator, and wild type strains of bacteriophage T4. J. Biol. Chem. 1972;247:7116–7122. [PubMed] [Google Scholar]
- 18.Showalter AK, Tsai MD. A reexamination of the nucleotide incorporation fidelity of DNA polymerases. Biochemistry. 2002;41:10571–10576. doi: 10.1021/bi026021i. [DOI] [PubMed] [Google Scholar]
- 19.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. U.S.A. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steitz TA. A mechanism for all polymerases. Nature. 1998;91:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 21.Franklin MC, Wang J, Steitz TA. Structure of the replicating complex of a pol α family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 22.Freisinger E, Grollman AP, Miller H, Kisker C. Lesion (in)tolerance reveals insights into DNA replication fidelity. EMBO J. 2004;23:1494–1505. doi: 10.1038/sj.emboj.7600158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hogg M, Wallace SS, Doublie S. Crystallographic snapshots of a replicative DNA polymerase encountering an abasic site. EMBO J. 2004;23:1483–1493. doi: 10.1038/sj.emboj.7600150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shamoo Y, Steitz TA. Building a replisome from interacting pieces: Sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell. 1999;99:155–166. doi: 10.1016/s0092-8674(00)81647-5. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Sattar AK, Wang CC, Karam JD, Konigsberg WH, Steitz TA. Crystal structure of a pol R family replication DNA polymerase from bacteriophage RB69. Cell. 1997;89:1087–1099. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- 26.Capson TL, Peliska JA, Kaboord BF, Frey MW, Lively C, Dahlberg M, Benkovic SJ. Kinetic characterization of the polymerase and exonuclease activities of the gene 43 protein of bacteriophage T4. Biochemistry. 1992;31:10984–10994. doi: 10.1021/bi00160a007. [DOI] [PubMed] [Google Scholar]
- 27.Reha-Krantz LJ, Stocki S, Nonay RL, Dimayuga E, Goodrich LD, Konigsberg WH, Spicer EK. DNA polymerization in the absence of exonucleolytic proofreading: in vivo and in vitro studies. Proc. Natl. Acad. Sci. U.S.A. 1991;88:2417–2421. doi: 10.1073/pnas.88.6.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang G, Franklin M, Li J, Lin TC, Konigsberg W. Correlation of the kinetics of finger domain mutants in RB69 DNA polymerase with its structure. Biochemistry. 2002;41:2526–2534. doi: 10.1021/bi0119924. [DOI] [PubMed] [Google Scholar]
- 29.Zakharova E, Wang J, Konigsberg W. The activity of selected RB69 DNA polymerase mutants can be restored by manganese ions: The existence of alternative metal ion ligands used during the polymerization cycle. Biochemistry. 2004;43:6587–6595. doi: 10.1021/bi049615p. [DOI] [PubMed] [Google Scholar]
- 30.Braithwaite DK, Ito J. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 1993;21:787–802. doi: 10.1093/nar/21.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cann IK, Ishino Y. Archaeal DNA replication: Identifying the pieces to solve a puzzle. Genetics. 1999;152:1249–1267. doi: 10.1093/genetics/152.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, Prakash L, Prakash S, Todo T, Walker GC, Wang Z, Woodgate R. The Y-family of DNA polymerases. Mol. Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 33.Irimia A, Zang H, Loukachevitch LV, Eoff RL, Guengerich FP, Egli M. Calcium is a cofactor of polymerization but inhibits pyrophosphorolysis by the Sulfolobus solfataricus DNA polymerase Dpo4. Biochemistry. 2006;45:5949–5956. doi: 10.1021/bi052511+. [DOI] [PubMed] [Google Scholar]
- 34.Yang G, Lin T, Karam J, Konigsberg WH. Steady-state kinetic characterization of RB69 DNA polymerase mutants that affect dNTP incorporation. Biochemistry. 1999;38:8094–8101. doi: 10.1021/bi990653w. [DOI] [PubMed] [Google Scholar]
- 35.Dunlap CA, Tsai MD. Use of 2-aminopurine and tryptophan fluorescence as probes in kinetic analyses of DNA polymerase β. Biochemistry. 2002;41:11226–11235. doi: 10.1021/bi025837g. [DOI] [PubMed] [Google Scholar]
- 36.Pedersen PL, Catterall WA. The use of thin-layer chromatography on poly(ethyleneimine) cellulose to facilitate assays of ATP-ADP exchange, ATP-Pi exchange, adenylate kinase, and nucleoside diphosphokinase activity. Methods Enzymol. 1979;55:283–289. doi: 10.1016/0076-6879(79)55032-0. [DOI] [PubMed] [Google Scholar]
- 37.Zhang H, Cao W, Zakharova E, Konigsberg W, De La Cruz EM. Fluorescence of 2-aminopurine reveals rapid conformational changes in the RB69 DNA polymerase-primer/template complexes upon binding and incorporation of matched deoxynucleoside triphosphates. Nucleic Acids Res. 2007;35:6052–6062. doi: 10.1093/nar/gkm587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee H, Wang M, Konigsberg W. The Reopening Rate of the Fingers Domain Is a Determinant of Base Selectivity for RB69 DNA Polymerase. Biochemistry. 2009;48:2087–2098. doi: 10.1021/bi8016284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joyce CM, Steitz TA. Function and structure relationships in DNA polymerases. Annu. Rev. Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- 40.Steitz TA. DNA polymerases: Structural diversity and common mechanisms. J. Biol. Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 41.Kornberg A, Baker TA. DNA Replication. 2nd ed. Freeman; San Francisco: 1992. [Google Scholar]
- 42.Copeland WC, Lam NK, Wang TS. Fidelity studies of the human DNA polymerase α. The most conserved region among α-like DNA polymerases is responsible for metal-induced infidelity in DNA synthesis. J. Biol. Chem. 1993;268:11041–11049. [PubMed] [Google Scholar]
- 43.Goodman MF, Keener S, Guidotti S, Branscomb EW. On the enzymatic basis for mutagenesis by manganese. J. Biol. Chem. 1983;258:3469–3475. [PubMed] [Google Scholar]
- 44.Vaisman A, Ling H, Woodgate R, Yang W. Fidelity of Dpo4: Effect of metal ions, nucleotide selection and pyrophosphorolysis. EMBO J. 2005;24:2957–2967. doi: 10.1038/sj.emboj.7600786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cotton FA, Wilkinson G, Murillo C, Bochmann M. Advanced Inorganic Chemistry. 6th ed. John Wiley & Sons; New York: 1999. [Google Scholar]
- 46.Minasov G, Tereshko V, Egli M. Atomic-resolution crystal structures of B-DNA reveal specific influences of divalent metal ions on conformation and packing. J. Mol. Biol. 1999;291:83–99. doi: 10.1006/jmbi.1999.2934. [DOI] [PubMed] [Google Scholar]
- 47.Kragten J. Atlas of Metal-Ligand Equilibria in Aqueous Solution. Halsted Press; Chichester, U.K.: 1978. [Google Scholar]
- 48.Westermann K, Naser K-H, Brandes G. Inorganic Chemistry. 12th ed. VEB Deutscher Verlag fur Grundstoffindustire; Leipzig, Germany: 1986. pp. 53–55. [Google Scholar]
- 49.Fidalgo da Silva E, Mandal SS, Reha-Krantz LJ. Using 2-aminopurine fluorescence to measure incorporation of incorrect nucleotides by wild type and mutant bacteriophage T4 DNA polymerases. J. Biol. Chem. 2002;277:40640–40649. doi: 10.1074/jbc.M203315200. [DOI] [PubMed] [Google Scholar]
- 50.Mandal SS, Fidalgo da Silva E, Reha-Krantz LJ. Using 2-aminopurine fluorescence to detect base unstacking in the template strand during nucleotide incorporation by the bacteriophage T4 DNA polymerase. Biochemistry. 2002;41:4399–4406. doi: 10.1021/bi015723p. [DOI] [PubMed] [Google Scholar]
- 51.Rothwell PJ, Mitaksov V, Waksman G. Motions of the fingers subdomain of klentaq1 are fast and not rate limiting: Implications for the molecular basis of fidelity in DNA polymerases. Mol. Cell. 2005;19:345–355. doi: 10.1016/j.molcel.2005.06.032. [DOI] [PubMed] [Google Scholar]
- 52.Rothwell PJ, Waksman G. A pre-equilibrium before nucleotide binding limits fingers subdomain closure by Klentaq1. J. Biol. Chem. 2007;282:28884–28892. doi: 10.1074/jbc.M704824200. [DOI] [PubMed] [Google Scholar]
- 53.Tsai YC, Johnson KA. A new paradigm for DNA polymerase specificity. Biochemistry. 2006;45:9675–9687. doi: 10.1021/bi060993z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hariharan C, Reha-Krantz LJ. Using 2-aminopurine fluorescence to detect bacteriophage T4 DNA polymerase-DNA complexes that are important for primer extension and proofreading reactions. Biochemistry. 2005;44:15674–15684. doi: 10.1021/bi051462y. [DOI] [PubMed] [Google Scholar]
- 55.Bakhtina M, Roettger MP, Kumar S, Tsai MD. A unified kinetic mechanism applicable to multiple DNA polymerases. Biochemistry. 2007;46:5463–5472. doi: 10.1021/bi700084w. [DOI] [PubMed] [Google Scholar]
- 56.Yang L, Arora K, Beard WA, Wilson SH, Schlick T. Critical role of magnesium ions in DNA polymerase β's closing and active site assembly. J. Am. Chem. Soc. 2004;126:8441–8453. doi: 10.1021/ja049412o. [DOI] [PubMed] [Google Scholar]
- 57.Arndt JW, Gong W, Zhong X, Showalter AK, Liu J, Dunlap CA, Lin Z, Paxson C, Tsai MD, Chan MK. Insight into the catalytic mechanism of DNA polymerase β: Structures of intermediate complexes. Biochemistry. 2001;40:5368–5375. doi: 10.1021/bi002176j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.