Abstract
Nutrient starvation induces autophagy in eukaryotic cells through inhibition of TOR (target of rapamycin), an evolutionarily-conserved protein kinase. TOR, as a central regulator of cell growth, plays a key role at the interface of the pathways that coordinately regulate the balance between cell growth and autophagy in response to nutritional status, growth factor and stress signals. Although TOR has been known as a key regulator of autophagy for more than a decade, the underlying regulatory mechanisms have not been clearly understood. This review discusses the recent advances in understanding of the mechanism by which TOR regulates autophagy with focus on mammalian TOR (mTOR) and its regulation of the autophagy machinery.
Keywords: mTOR, Atg1, ULK1, ULK2, Atg13
1. Introduction
Nutrient starvation, stress, or reduced availability of growth factors alarms eukaryotic cells to adjust metabolism to survive. An early response of the cellular metabolic adjustments involves inhibition of growth and induction of macroautophagy (referred to as autophagy unless otherwise specified) to optimize the usage of limited energy supplies. Autophagy, as a cellular process mobilizing intracellular nutrient resource, plays an important role in contributing to survival during these growth unfavorable conditions. Eukaryotic cells have developed a mechanism through which autophagy induction is tightly coupled to the regulation of cell growth. Among the numerous components involved in the regulation of autophagy and growth, TOR is a key component that coordinately regulates the balance between growth and autophagy in response to cellular physiological conditions and environmental stress. Despite the significant progress in the autophagy study field, the mechanism by which TOR regulates autophagy remains not clearly understood. Here, we review recent findings that have made important progress in our understanding of the molecular link between TOR and autophagy in yeast and mammalian cells. We briefly describe the signaling pathways upstream of mTOR involving nutrient sensing pathways, growth factor signalling, and stress responses in mammalian cells. We then discuss the mechanisms that link TOR to autophagy in yeast and the recent advance in understanding the molecular linkage between mTOR and the autophagy machinery in mammalian cells.
2. mTOR as a sensor of cellular nutritional status
TOR is a serine/threonine protein kinase with a large molecular size near 300 kDa that belongs to the phosphatidylinositol kinase-related kinase (PIKK) family. The activity of TOR is inhibited under nutrient starvation, which has been known as a crucial step for autophagy induction in eukaryotes [1,2]. TOR was firstly described in 1991 as a target protein of the anti-fungal and immunosuppressant agent rapamycin [3]. Subsequent studies revealed many important functions of TOR in yeast and higher eukaryotes, making the protein kinase to gain its central position in the signaling network that regulates cell growth. The function of TOR in yeast and higher eukaryotes encompasses regulation of translation, metabolism, and transcription in response to nutrients and growth factors. The broad cellular functions of TOR have made the protein kinase to be an important subject for study of cancer, metabolism, longevity, and neurodegenerative diseases.
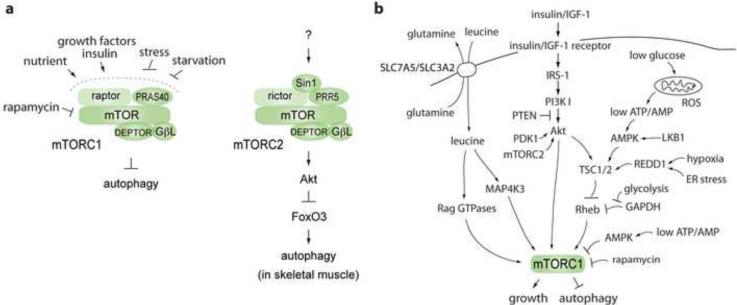
In yeast, TOR activity is regulated by the availability of nutrients such as nitrogen and carbon [4]. Similarly, mTOR activity is regulated by amino acid and glucose levels in mammalian cells [5,6]. The knowledge on the regulation of TOR has greatly increased since the discoveries of TOR binding partners. Saccharamyces cerevisiae contains two TOR proteins, TOR1 and TOR2. Both TOR proteins bind KOG1, LST8, and TCO89, whereas TOR2 also binds LST8, Avo1, Avo2, Avo3, and BIT61 to form a separate protein complex [4,7]. Similar to yeast TOR, mTOR binds several proteins to form two distinct protein complexes (Figure 1a). mTORC1 (mTOR complex 1) contains raptor (KOG1 ortholog), GβL/mLst8, PRAS40 and DEPTOR, whereas mTORC2 (mTOR complex 2) contains rictor (Avo3 ortholog), GβL/mLst8, Sin1 (Avo1 ortholog), PRR5/protor and DEPTOR [8–15].
Figure 1.
Regulation of the mTOR pathway by nutrients (amino acids, glucose), stress and insulin/IGF-1. (a) Two mTOR complexes, mTORC1 and mTORC2, and their components. mTORC1 is the protein complex responsible for autophagy induction in response to nutrient starvation, stress and reduced growth factor signaling. mTORC2 regulates autophagy via Akt-FoxO3 in skeletal muscle cells in response to a fasting condition [19,20]. (b) The signaling pathways upstream of mTORC1 that regulate cell growth and autophagy in response to nutrient levels, growth factors, and stress.
The yeast TOR complex TORC1, which contains KOG1, and the mammalian mTORC1 containing raptor are key regulators of translation and ribosome biogenesis in yeast and mammalian cells respectively, and they are responsible for autophagy induction in response to starvation. On the other hand, mTORC2 (mTOR-rictor complex), originally known as rapamycin insensitive but likely also targeted by rapamycin, is involved in the regulation of phosphorylation and activation of Akt/PKB, protein kinse C, serum- and glucocorticoid-induced protein kinase 1 [16–18]. Because Akt positively regulates mTORC1, it would be reasonable to speculate that mTORC2 acts as a negative regulator of autophagy. Indeed, mTORC2 inhibition induced autophagy and atrophy in skeletal muscle cells under a fasting condition [19,20]. However, the autophagy induction by mTORC2 inhibition is mediated mainly by FoxO3, a transcription factor downstream of Akt, that is involved in autophagy gene expression (Figure 1a). Although mTORC2 is involved in autophagy regulation as such, this review is mainly focused on mTORC1-dependent autophagy regulation that is responsive to nutrient starvation.
Availability of cellular amino acids, especially branched chain amino acids such as leucine, regulates mTOR activity (Figure 1b). Amino acid signaling is mediated by the RagA family small GTPases in mammalian cells and Drosophila [21,22]. Similarly, in budding yeast the EGO complex containing ras-related GTPases Gtr1 and Gtr2, which belong to the RagA family, and at least two other proteins Ego1 and Ego3 plays important roles in TOR regulation in response to amino acid starvation [23]. These molecules were initially known to function on the membrane of vacuoles in yeast to regulate growth and microautophagy [23], implying that the interface between growth and autophagy may occur on the vacoular membrane in yeast [23]. Another important link of nutrient signalling to mTORC1 involves MAP4K3 (mitogen-activated protein kinase kinase kinase kinase 3) in mammalian cells that is evolutionarily conserved and belongs to the Ste20 family protein kinase [24]. The kinase activity of MAP4K3 is regulated by amino acid (leucine) levels, indicating that the kinase might act upstream of mTORC1.
What upstream elements regulate Rag GTPases and MAP4K3 and how they functionally interact in mammalian cells remain unclear. Amino acid signaling upstream of these components may start at the surface of cells where the initial contact between amino acids and cells takes place. Amino acid uptake at the plasma membrane is performed by a group of membrane transport proteins classified into a family of solute-linked carrier (SLC) proteins. A recent study revealed that cellular uptake of L-glutamine by SLC1A5 and its subsequent rapid efflux by SLC7A5/SLC3A2, a bidirectional transporter that regulates the simultaneous efflux of L-glutamine and transport of leucine, are important for mTORC1 activation [25] (Figure 1b). The study showed that loss of SLC1A5 function inhibits cell growth and activates autophagy presumably due to inhibition of tranport of leucine into cells. This transport machinery provides an insight into its potential functional link to the EGO complex in yeast given the role of the EGO complex in the regulation of the general amino acid transporter GAP1 and TORC1 in response to glutamine on the vacuolar surface [23,26].
Another important element in the nutrient-signalling pathway upstream of mTORC1 involves hVps34 (human ortholog of yeast vacuolar protein sorting 34, Vps34, or named PI3KIII (phosphoinositide 3-kinase class III), a lipid kinase conserved throughout eukaryotes. In budding yeast, Vps34 was firstly identified as a component involved in vacuolar sorting [27]. Later, it was found to regulate autophagy in response to nutrients by producing and accumulating phosphatidylinositol 3-phosphate, PI(3)P, at specific locations during early steps of autophagy induction [28]. Deficiency of the mammalian hVps34 suppressed leucine-responsive activation of mTORC1, suggesting that hVps34 may act upstream of mTORC1 signaling [29,30]. In line with this finding, calcium and calmodulin dependent signaling was shown to regulate mTORC1 via hVps34 in response to leucine in HeLa cells [31]. However, hVps34 and mTORC1 seem to be in a complicated relationship since Vps34 is not required for TORC1 signalling in Drosophila [32]. Perhaps, the complex relation might be due to the difference between the mammalian and Drosophila systems. Alternatively, it could be due to the involvement of hVps34 into multiple different complexes [33]. It is possible that some of the hVps34 complexes function upstream and others do downstream of mTORC1. Further investigation is necessary to clarify the relation between mTORC1 and hVps34 complexes and their crosstalk during the whole autophagy processes.
While amino acids or leucine is regarded as a signalling molecule that coordinates cell growth with availability of cellular building blocks, glucose may be engaged in the pathway that coordinates growth not only with the availability of the building blocks but also the cellular energy state. It is noteworthy that the signalling pathway regulated by glucose is different from the one regulated by leucine while both pathways are integrated through mTORC1. Glucose starvation reduces the ratio of ATP and AMP in eukaryotic cells and thereby activates AMPK, the 5'-AMP-activated protein kinase, which is potentiated by LKB1, a protein kinase phosphorylating AMPK [34,35] (Figure 1b). As a consequence, the activated AMPK inhibits mTORC1 through phosphorylation and activation of tuberous sclerosis complex 2 (TSC2), a negative regulator of mTORC1 [6]. Consistent with the negative function of AMPK in mTORC1 signaling, AMPK positively regulates autophagy in mammalian cells [36].
A recent study revealed that AMPK can inhibit mTORC1 independently of TSC2 by phosphorylating raptor at Ser863 [37]. Thus, there are two separate pathways that transmit AMPK signalling to mTORC1 (Figure 1b). Another glucose-sensing pathway has been proposed to involve glyceraldehyde-3-phosphate dehydrogenase (GAPDH) that negatively regulates Rheb-mTORC1 signaling independently of TSC1/2 under low glucose influx [38]. This GAPDH-dependent pathway has a distinct feature relative to the above two AMPK-dependent pathways given that glycolytic influx, rather than energy state, is a signal that regulates mTORC1 (Figure 1b). Taken together, these studies suggest that glucose could regulate mTORC1 through at least three different pathways. Perhaps, the AMPK-mTORC1 pathways might be responsible for monitoring cellular energy status and stress conditions, whereas the GAPDH-mTORC1 pathway might be responsible for the cellular glucose metabolic status. It is important to note that mTORC1 is a converging point for these three pathways.
3. mTOR as a senor of cellular stress and growth factor signals
Autophagy is induced not only by nutrient starvation but also by reduced growth factor signaling. mTORC1 is also a key mediator of growth factor signalling to autophagy. The growth factor signaling that regulates mTORC1 mainly involves the insulin/insulin-like growth factor (IGF-1)-PI3K (phosphoinositide 3-kinase class I)-Akt pathway, which negatively regulates autophagy induction (Figure 1b). The insulin/IGF-1 pathway involves PDK1 and Rheb, the positive regulators of mTORC1 signaling, and PTEN and TSC2, the negative regulators of mTORC1 signaling. TSC2, in a complex with TSC1, acts as a GTPase activating protein (GAP) for the small GTPase Rheb and the GAP activity is regulated by Akt-mediated phosphorylation of TSC2 [39,40]. PRAS40, a component of mTORC1, also plays an important role in mediating insulin signaling to mTORC1 by serving as a substrate of Akt [12,41].
Autophagy is also induced by cellular stresses in a manner dependent upon mTORC1. Hypoxia regulates mTORC1 via REDD1 (regulated in development and DNA damage 1) and its related protein REDD2 in mammals and in Drosophila (Sofer et al., 2005) (Figure 1b). REDD1 negatively regulates mTORC1 via the TSC1-TSC2 complex through unknown mechanisms or a mechanism involving 14-3-3, a cytosolic anchor protein [42]. REDD1 is also involved in response of mTORC1 and autophagy to ER (endoplasmic reticulum) stress, which is associated with unfolded protein response and inflammatory signaling [43] (Figure 1b). Given the tight link between mitochondrial function and mTOR [9,44], reactive oxygen species (ROS) and their inhibitory effects on the mitochondrial function might somehow trigger the mitochondrial autophagy or mitophagy in a manner dependent upon mTORC1. The finding that mTORC1 is found in proximity of mitochondria and inhibited by oxidative stress and mitochondrial dysfunction implies a possibility that mTORC1 is involved in the mechanism through which damaged mitochondria induce autophagy [9,44]. Although it is possible that mitochondrial signal could directly target mTORC1, at least in mammalian cells the LKB1-AMPK-TSC1/2 branch seems to play a role in mTORC1 inhibition under oxidative stress [35] (Figure 1b).
4. TOR regulation of Atg1 complex in yeast
The sections above summarized the components and pathways upstream of mTORC1. In the following sections, we discuss the mechanisms downstream of mTORC1 by which mTORC1 regulates the autophagy machinery. We first describe the finding in yeast that has tied TOR as a key molecule of autophagy regulation. According to the current knowledge, the first signalling component downstream of TOR in the autophagy pathway is Atg1, an evolutionarily-conserved serine/threonine kinase. Atg1 plays a key role perhaps at the most upstream step in the initial stages of autophagy induction such as the nucleation (the early event when membrane structures are initiated) and formation of the preautophagosome structures (PAS) [1,45,46]. In S. cerevisiae, Atg1 null or kinase-dead mutant strains have shown defects in autophagy even under nutrient starvation or when TOR is inhibited, supporting that Atg1 plays downstream of TOR [46,47]. Supporting this, starvation or rapamycin enhanced the kinase activity of Atg1 [47]. The significance of Atg1 and its homologues in autophagy induction has been demonstrated in several other eukaryotes including Dictyostelium discoideum, Drosophila melanogaster, and Caenorhabditis elegans [2,48–50].
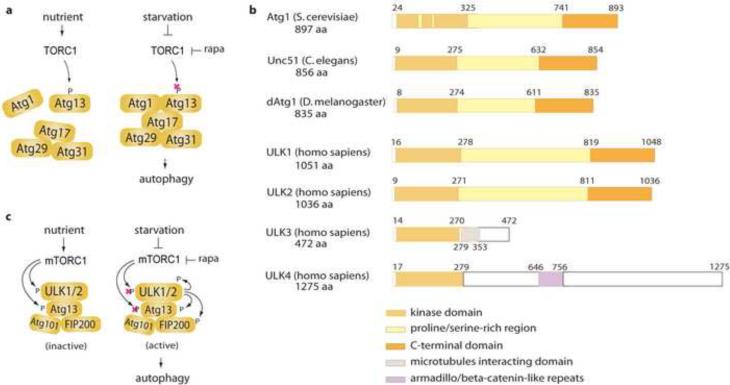
Since TOR was identified as an upstream kinase of Atg1, the mechanism by which TOR regulates Atg1 has been an important question regarding the molecular cascade of autophagy induction. In yeast, Atg1 interacts with several autophagy proteins Atg13, Atg17, Atg29 and Atg31 in a manner dependent upon nutrient status and TOR activity [47,51–54] (Figure 2a). Atg13 is required for Atg1 activity, autophagy induction and the cytoplasm-to-vacuole targeting (Cvt) that utilizes a small double-membrane vesicle (Cvt vesicle) similar to autophagosome [47]. Atg1 can localize to the PAS even under nutrient rich conditions [52,55], which is probably due to the Cvt pathway. Atg17 is an essential factor for the PAS localization of Atg1 and several other autophagy components [56,57]. It interacts with Atg1, Atg13, Atg29 and Atg31 under nutrient starvation and the interaction between Atg13 and Atg17 is required for Atg1 kinase activity and autophagosome formation [54,58] (Figure 2a). Atg17 also functions to recruit Atg9 to the PAS in a manner dependent upon Atg1 [57].
Figure 2.
Comparison of the Atg1/ULK machinery between yeast and higher eukaryotes. (a) Current model for the mechanism by which TOR regulates the Atg1 complex in S. cerevisiae. TOR phosphorylates Atg13 at multiple residues resulting in disruption of the complex and inhibition of autophagy. (b) Comparison of the domain structures of S. cerevisiae Atg1, C. elegans UNC51, D. melanogaster Atg1, and Homo sapiens ULK1, ULK2, ULK3 and ULK4. (c) Phosphorylational regulation of ULK1/2 complexes by mTORC1 in response to nutrient levels. mTORC1 phosphorylates ULK1/2 and Atg13 and inhibits the kinase activity of ULK1/2 under high nutrient conditions. Under starvation, mTORC1 phosphorylation of ULK complex is suppressed releasing ULK from mTORC1 inhibition, which subsequently induces ULK to phosphorylate Atg13, FIP200 and itself.
TOR phosphorylates Atg13 at multiple residues causing a reduced affinity between Atg1 and its binding proteins, resulting in repression of autophagy [47,52,54]. Under starvation when TOR is inactive, Atg13 phosphorylation is suppressed and the hypophosphorylated form of Atg13 acts to induce localization of Atg1, Atg17 and other essential autophagy factors to the PAS [59]. The interaction of Atg13 and Atg17 to Atg1 induced under rapamycin or nutrient starvation enhances the kinase activity of Atg1 essential for autophagosome formation. In sum, these studies suggested that the Atg1 complex constitutes an important node of the signaling pathway through which TOR triggers the events of autophagy in yeast.
5. Mammalian Atg1 complex
The counterpart of Atg1 in mammalian cells is known as ULK (UNC-51 like kinase). ULK1, a first member of the ULK family proteins, was identified as a homologue of C. elegans UNC-51, the gene named following a mutational phenotype “UNCoordinated locomotion” associated with defects in the mutant warm [60,61]. Subsequently, ULK2 was identified in mouse as a second member and cloned [62]. The domain structure showed unique features having the N-terminal kinase domain, the middle proline/serine-rich domain, and the C-terminal domain involved in protein-protein interaction (Figure 2b). ULK1 shows sequence similarity of 41 and 29% to C. elegans UNC-51 and S. cerevisiae Atg1, whereas ULK2 shows 33% amino acid identity to UNC-51 [54] (Supplemental Fig. 1). ULK1 and ULK2 share an overall 52% amino acid identity [62]. The human genome has at least two additional mammalian homologues of Atg1, which are ULK3 and ULK4. Both ULK3 and ULK4 are missing in the middle proline/serine-rich region and the C-terminal region that are present in ULK1 and ULK2. ULK3 contains a middle region predicted to interact with microtubules, whereas ULK4 contains a protein-protein interaction motif of armadillo/beta-catenin-like repeats in the middle region (Figure 2b). The C-terminal regions of ULK3 and ULK4 are largely divergent from ULK1 and ULK2. On the other hand, the kinase domains of ULK3 and ULK4 show high sequence similarity, 52% and 41%, to that of ULK1 respectively (Supplemental Fig. 1). Hereafter in this review, ULK will be referred to as both ULK1 and ULK2.
Several recent studies revealed that ULK1, ULK2 and ULK3 are involved in autophagy pathways [63–67]. ULK1 was identified as a kinase regulating autophagy through siRNA screens [63]. Overexpression of the kinase-dead mutants of ULK1 or ULK2 led to inhibition of autophagy in the NIH3T3 mouse embryonic fibroblasts [64]. Autophagy activity was suppressed in ULK1- or ULK2-silenced HeLa and HEK293 cells and ULK1-null mouse embryonic fibroblasts [66]. As for ULK3, its overexpression induced autophagy in a human fibroblast cell line IMR-90 [67]. ULK3 is co-localized with Atg12 (the ubiquitin-like protein involved in the formation of the autophagic isolation membrane) in punctate structures under amino acid and serum depletion [67]. On the other hand, any study has been reported yet about the function of ULK4.
Until recently, the lack of knowledge on Atg13, Atg17, Atg29 and Atg31 homologues in higher eukaryotes has limited our understanding of the mechanism underlying autophagy induction. A putative mammalian homologue of Atg13 was recently predicted by a gapped-blast homologue search [68]. Several recent studies revealed that the predicted mammalian homologue of Atg13 interacts with ULK1 and ULK2 and plays an essential role in autophagosome formation [66,69–73] (Figure 2c). On the other hand, the mammalian counterpart of Atg17 has not been clearly defined. FIP200 (focal adhesion kinase family interacting protein of 200 kDa, also called as RB1CC1) was identified as a protein that interacts with ULK1 and ULK2 and plays an important for autophagosome formation [64] (Figure 2c). FIP200 was proposed as a counterpart of Atg17 in mammalian cells given its functional similarity to yeast Atg17, although they do not show high sequence similarity [64]. Given that FIP200 interacts with several proteins such as stathmin, Pyk2, FAK, p53, TSC1, ASK1, and TRAF2 involved in microtubule dynamics, stress response, and cell growth [74], it is possible that FIP200 mediates ULK functions in diverse biological processes not directly related to autophagy. Another novel protein named Atg101 was identified as an Atg13 binding protein [73,75] (Figure 2c). Atg101 was named as such given no apparent homologue present in S. cerevisiae, although its homologues are present in Mus musculus, D. melanogaster, C. elegans, and Schizosaccharomyces pombe [73,75]. Atg101 interacts with ULK1 in an Atg13-dependent manner and plays an important role for autophagosome formation [73,75]. Atg101 was also shown to play a role in protection of Atg13 from the proteasomal degradation [73].
Although Atg13 and FIP200 were shown to play important roles for ULK kinase activity, stability and localization to autophagosome [66,69,71,72], the exact mechanism underlying the ULK regulation by the binding proteins remains unclear. A likely mechanism involves the role of Atg13 as a bridge molecule to mediate the interaction between ULK and FIP200 [66] (Figure 2c). With this regard, it is noteworthy that ULK1 and ULK2 have different binding affinities toward Atg13 and FIP200. In contrast to the direct interaction between ULK1 and Atg13, the interaction between ULK2 and Atg13 was only detected in the presence of FIP200 in vitro. This indicates that Atg13 may bind ULK1 preferentially to ULK2 and the ULK2-Atg13 interaction may depend highly on FIP200. Possibly, ULK1 and ULK2 might have redundant functions so that ULK2 could compensate for the role of ULK1 when ULK1 is disrupted. Supporting this, mice lacking ULK1 were viable and did not display any apparent developmental defect, which is in contrast with the mouse models lacking other autophagy gene such as Atg5 or Atg7 [65]. Alternatively, ULK1 and ULK2 might have distinct functions in autophagy regulation. Indeed, ULK2 phosphorylation and membrane association upon starvation occurred more readily compared to those of ULK1 [69]. Given that only one Atg1 exists in yeast and Drosophila [2,47], the two ULK complexes imply a higher order of complexity for the Atg1 complex machinery in mammalian cells. Furthermore, human cells have more than three splicing variants of Atg13 and at least two of them are capable of binding ULK1 and ULK2 [66]. Therefore, the Atg1 complex machinery is likely highly divergent in the composition and perhaps also the functions in mammalian cells.
6. mTOR regulation of ULK complex phosphorylation
Following the discovery of ULK complexes, several groups showed that mTORC1, similar to yeast TOR, phosphorylates mammalian Atg13 [66,71,72]. Indeed, mTORC1 could phosphorylate not only Atg13 but also ULK1 and ULK2 in vitro [66,71,72] (Figure 2c). Inhibition of mTORC1 by rapamycin or starvation, the conditions that induce autophagy, led to dephosphorylation of ULK1, ULK2, and Atg13 in human cells [66,71,72]. These studies suggested that ULK1/2 and Atg13 would be direct effectors of mTORC1. On the other hand, the phosphorylation state of FIP200 inversely correlated with the activity of mTORC1 [66]. The inverse correlation implied that FIP200 phosphorylation is induced when ULK1 and ULK2 are active. Indeed, ULK1 and ULK2 could phosphorylate FIP200 in vitro [66]. Furthermore, the interaction between ULK1/2 and Atg13 was important for FIP200 phosphorylation by ULK1/2 in cells [66], suggesting that FIP200 binding to ULK via Atg13 is required for FIP200 phosphorylation by ULK.
Although the Atg1-Atg13 complex is conserved in eukaryotes, the protein complex seems to have distinct regulatory mechanisms between yeast and mammals. Unlike the interaction between the yeast proteins Atg1 and Atg13 that is regulated by nutrient starvation, the interaction between ULK1 and mammalian Atg13 was not affected by rapamycin or starvation [66,72] (Figure 2c). The ULK1 complex was indeed found to undergo stronger association with mTORC1 under nutrient-enriched conditions [72] (Figure 3). This is in line with the findings that mTORC1 induces phosphorylation of ULK1 under nutrient-enriched conditions [66,71,72]. The recruitment of ULK1 complex to mTORC1 under nutrient-enriched conditions may facilitate the phosphorylation of ULK1 complex by mTOR thereby inhibiting ULK1 function. A likely mechanism is that mTOR-mediated phosphorylation would not cause a drastic conformational change in ULK and Atg13 but rather induce a local perturbation of the protein-protein interaction that interferes with ULK recognition of its substrate FIP200 or other unidentified substrates.
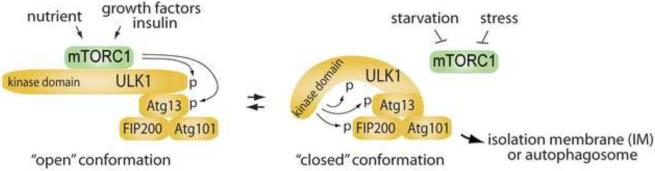
Figure 3.
(a) Two-state hypothesis for ULK1 conformation and activity. ULK1 interacts with Atg13, FIP200 and Atg101 through its C-terminal region [66,69,72,73,75]. Under high nutrient conditions, mTORC1 interacts with the ULK1 complex and phosphorylates ULK1 and Atg13 [66,71,72]. The phosphorylation may induce ULK1 to take an “open” conformation, an inactive form [69]. Under stress or starvation, mTORC1 is dissociated from the ULK1 complex. When mTORC1 could no longer phosphorylate ULK1 and Atg13 under the condition, ULK1 may take a “closed” conformation, an active form. In the closed-conformation state, the kinase domain of ULK1 may phosphorylate Atg13 and FIP200 and trigger the downstream events for autophagosome formation.
Despite the evidence that mTORC1 phosphorylates ULK1/2 and Atg13 in vitro and in living cells, the mTOR-mediated phosphorylation sites remain undetermined. Recently, more than ten phosphorylation sites of ULK1 were identified by high resolution, tandem mass spectrometry [76]. Serine-87, -195, and -224 were identified as sites of phosphorylation in the kinase domain. Serine-224 is predicted to lie near the end of a subdomain of the kinase domain where it functions to stabilize the catalytic loop. Phosphorylation of serine-224 is therefore presumed to disrupt the active site of ULK1 potentially. Serine-341 was identified as a site of phosphorylation, probably of autophosphorylation. Serine-341 lies within a region encompassing residues 279–427 required for the binding of ULK1 with γ-aminobutyric acid receptor associated protein (GABARAP) and Golgi-associated ATPase enhancer of 16 kDa (GATE-16), the mammalian homologues of Atg8 [77]. Thus, Ser341 phosphorylation might potentially be involved in the binding of these proteins to ULK1. Serine-867 and -913 were identified in the C-terminal region where Atg13, FIP200 and Atg101 bind [66,69,72,73], suggesting that these sites might regulate the association of ULK1 with the binding proteins. Two serine residues near the C-terminus, Ser1043 and Ser1047, were identified and the former is likely a PKA site and the latter a site for autophosphorylation. It is important to determine if these residues are indeed phosphorylated in vivo for endogenous protein and if they play crucial roles in ULK1 function. Moreover, which residues are mTOR and ULK1 sites is a remaining question.
A recent study in yeast revealed that TOR phosphorylates several serine residues in Atg13 [59]. The TOR-mediated phosphorylation of Atg13 inhibited Atg1 kinase activity, Atg13-Atg17 interaction and autophagy induction. It is not clear whether the phosphorylation sites in yeast Atg13 are conserved in higher eukaryotes given low amino acid similarity between yeast and higher eukaryotic Atg13 proteins [68]. Perhaps, Atg13 would be phosphorylated less extensively in mammalian cells than in yeast. This is because mammalian Atg13 contains 517 amino acids, which is about 200 amino acids shorter than yeast Atg13, and the missing region contains a few phosphorylation sites in yeast Atg13 [59]. Knowing that Atg13 phosphorylation is crucial for autophagy in yeast cells, we assume that identification of Atg13 phosphorylation sites in mammalian cells will be important for understanding the mechanism by which mTOR regulates autophagy.
7. mTOR regulation of ULK activity and localization
Given the lesson from the studies in yeast that TOR induces hyperphosphorylation of Atg13 and inhibits Atg1 kinase activity [47], we anticipate that a similar mechanism would occur with the mammalian counterparts. Indeed, mTORC1 regulated the kinase activity of ULK1 and ULK2 [64,66,71,72] (Figure 2c). Conditions that inhibit mTORC1, such as leucine deprivation or rapamycin, enhanced the kinase activity of ULK1/2 and triggered phosphorylations of Atg13 and FIP200 and autophosphorylation of ULK under starvation conditions [66,71,72]. On the other hand, Rheb overexpression, which enhances mTORC1 activity, reduced the kinase activity of ULK1 [66,71,72]. In line with this finding, ULK-dependent phosphorylation of FIP200 as well as ULK autophosphorylation became more apparent under the condition of mTORC1 inhibition. These results support that mTORC1 regulates ULK kinase activity and thereby it suppresses autophagy induction (Figure 2c).
Although ULK kinase activity seems to be important for autophagy in mammalian cells, a study in yeast revealed that yeast Atg1 could induce autophagy in a manner independent of the kinase activity [78]. Consistently, Atg1 kinase activity was not necessary for Atg17-dependent PAS localization of Atg9 [57]. These observations may need to be taken into account along with the finding that ULK kinase activity is not required for ULK1/2 association with membrane under starvation in mammalian cells [69]. A possible explanation is that ULK has a function independent of its kinase activity, whereas the kinase activity becomes important after it is recruited to autophagosomal membrane under starvation. The function of ULK kinase activity on the autophagosomal membrane might depend on its autophosphorylation that is potentially important for downstream events for the autophagosome formation [69,76]. Autophosphorylation of ULK could be important for the activity of ULK or interaction with ULK binding proteins. Like many tyrosine kinase family members, autophosphorylation could be involved in activation of ULK possibly following dimerization of ULK, although no evidence has been provided about this possibility yet.
With regard to the issue about ULK kinase activity, it is worthwhile to note that mTORC1 reduces the kinase activity of ULK1 and ULK2 to a moderate level in vitro [64,66,71,72]. A likely basis for this moderate change is that the kinase activity itself might not be altered drastically by mTORC1. Instead, mTORC1 might regulate ULK function through altering ULK cellular localization. Indeed, nutrient starvation, a condition that inhibits mTOR, induces localization of ULK1, ULK2 and Atg13 to the autophagic isolation membrane [50,63,69,72]. In an agreement with these findings, rapamycin induces accumulation of ULK1 in membrane fractions of HeLa cells [66]. It is possible that mTOR phosphorylates residues of ULK and Atg13 that are involved in translocation of ULK to autophagosomes. Alternatively, mTOR-mediated phosphorylation of ULK may inhibit ULK activity thereby suppressing phosphorylation of Atg13 and FIP200 or autophosphorylation of ULK at residues crucial for ULK localization. However, the latter possibility seems not to reconcile with the finding that starvation could still induce localization of the kinase-dead ULK to membrane [69].
It is also possible that Atg13 and FIP200 could bind currently unknown components involved in various downstream events including the formation of the autophagic isolation membrane [50,63,69,72]. Given that ULK1 binds multiple proteins that are localized to autophagosomes [77], it is a logical assumption that ULK1 regulates the autophagosomal localization of its binding proteins. The starvation-induced phosphorylations of ULK might alter the protein-protein interaction mediated by ULK for its binding proteins. Alternatively, the ULK1 binders may recruit ULK1 to autophagosomal membranes where ULK1 can phosphorylate proteins involved in the downstream events of autophagy. GATE-16 and GABARAP, the mammalian homologues of Atg8, are candidate proteins for such roles in the regulation of localization of ULK complexes to autophagosome given their implicated functions in vesicle transport and fusion [79,80]. It is also possible that proteins interacting with the ULK C-terminal domain (CTD), such as Atg13, FIP200, and Atg101, might regulate ULK localization as evidenced for Atg13 and FIP200 and discussed above [64,69,71–73,75]. Whether ULK2 behaves as similarly as ULK1 in binding these molecules remains unknown.
8. Two-state hypothesis for ULK conformation and activity
Further mechanical insight on ULK regulation came from a study on ULK CTD [69]. The C-terminal region was shown to be important for the autophagosomal localization of ULK1 [63,69]. ULK C-terminal residues regulate not only autophagosomal localization of ULK but also the kinase activity of ULK toward phosphorylation of its substrates [69]. Two possible functions are anticipated with regard to the C-terminal region: (1) mediation of ULK binding proteins; (2) regulation of ULK conformation. When the C-terminal region was deleted in ULK1, the inhibitory effects of the kinase-dead mutant ULK1 on autophagy were rescued partly [69]. This result is consistent with the possibility that ULK1 sequestrates essential ULK1-binding proteins, which may include Atg13, FIP200, Atg101 or unknown factors. In addition to the function of mediating ULK-binding proteins, the C-terminal region plays an important role in the conformational change of the protein [69]. A study using limited-proteolysis assay revealed that ULK1 C-terminal region appears to fold back with the N-terminal kinase domain to keep the protein in a “closed” conformation in a manner dependent upon autophosphorylation, which seems to be important for phosphorylation of ULK substrates [69] (Figure 3). In accordance with this model, kinase dead mutant of ULK1 was more accessible to proteases compared to wild type, a potential indication of an “open” conformation of kinase-dead mutant. A triggering signal for the autophosphorylation and subsequent conformational change of ULK might be inhibition of mTORC1-mediated phosphorylation of ULK1 or Atg13 or dissociation of mTORC1 from ULK1 complex (Figure 3).
It is noteworthy that association of ULK1 and ULK2 with membrane structures is dependent upon the ULK CTD domain but unlikely the kinase activity [69]. A mechanism that reconciles with this observation might be that ULK kinase activity becomes important for the molecular steps following ULK localization to autophagosomal membrane. With this regard, it is noteworthy that Ser1047 was identified as a putative autophosphorylation site near the C-terminus of ULK1 [76]. Perhaps, this phosphorylation would be important for ULK1 to bind its substrates. Alternatively, the phosphorylation could induce a conformational change in ULK1 that allows for recruitment of unknown factors to the complex. These unknown factors might not be Atg13 or FIP200 given that they could bind ULK1 in a manner independent of ULK1 kinase activity [66,72]. A deletion mapping analysis identified a 7-residue motif (IERRLSA) in the last 14-amino acids at C-terminus that is required for the dominant negative effect of kinase dead mutant of ULK1 and 2 on autophagy [69]. This motif is entirely conserved in ULK1 and ULK2 from mouse to human [69]. The 7-residue motif is not involved in the interaction with Atg13, implying that this motif might bind an unknown factor essential for autophagy [69].
9. Crosstalk between ULK complexes and other autophagy machinery
According to the current knowledge, ULK1 and ULK2 are localized to the isolation membrane and are involved in the early steps for nucleation of autophagosome formation [64]. The nucleation process involves the formation of PI(3)P-enriched membrane compartment that seems to be derived from ER membranes in response to amino acid starvation [81,82]. The hVps34 complex containing Atg14L and Beclin1 is likely responsible for the formation of PI(3)P at the early step that recruits downstream effectors to the site where the nucleation event occurs [83–86]. A missing gap in knowledge lies in understanding the crosstalk between ULK1/2 and hVps34 complexes at the early step of autophagy induction. Perhaps, the two protein complexes regulate each other so that they could coordinate the processes of autophagy induction. Alternatively, they may sense different cellular signals in parallel pathways to regulate the nucleation machinery. With this regard, it is noteworthy that Atg1 regulates localization of PI(3)P to autophagosomes upon starvation in Drosophila [32]. Therefore, it is possible that the ULK1 complex might function upstream of Atg14L-hVps34 complex to regulate activity of hVps34 and accumulation of PI(3)P on the membrane compartments thereby recruiting PI(3)P effectors involved in the nucleation events.
The function of the ULK1 complex involves regulation of mammalian Atg9 (mAtg9) trafficking [87]. The transmembrane protein mAtg9 localized on TGN and endosomes is redistributed to the isolation membrane or autophagosome in response to amino acid starvation [87]. In yeast, Atg9 is an essential component for autophagosome formation and it is recruited to the PAS by Atg17 in a manner dependent upon Atg1 [57]. It is a logical speculation that the mammalian homologue mAtg9 is essential for autophagy, which has been supported by knockdown experiments [87,88], and its redistribution may be regulated by ULK1 and ULK1 binding proteins.
How about the crosstalk between the ULK complexes and the Atg5-Atg12 and Atg4-Atg7-Atg3-Atg8 conjugation systems that are also involved in the early events of autophagosome formation? Whereas disruption of ULK complexes did not inhibit the phosphatidylethanolamine (PE) modification of LC3 (the mammalian homologue of yeast Atg8 localizing to autophagosome, which is a process mediated by the Atg4-Atg7-Atg3 conjugation system), the disruption indeed inhibited autophagic flux [64,66,71,72]. This result implies that the ULK complexes regulate a certain crucial step during autophagy flux without directly affecting the activity of the conjugation machinery. It is noteworthy that ULK1 and ULK2 colocalize with Atg16L and that ULK1 regulates autophagosomal localization of Atg16L [64]. Given that Atg16L is the protein interacting with the Atg5-Atg12 conjugation machinery on the isolation membrane and is tightly coupled with Atg4-Atg7-Atg3-Atg8 conjugation machinery [80,89–91], a likely mechanism might involve ULK1 and perhaps ULK2 in the formation of the autophagic isolation membrane through regulating the conjugation systems.
In line with the role of ULK in the regulation of the two conjugation systems, Vps34 complex was found to play an essential role in the recruitment of Atg8–PE and the Atg5–Atg12 conjugates to the preautophagosomal structure in yeast [92]. Taking into account that Atg1 appears to regulate Vps34 function in Drosophila [32], we could speculate that Vps34 and Atg1 complexes may act upstream of the Atg8 and Atg5-Atg12 conjugation systems. However, ULK1 and ULK2, unlike Atg1 in yeast, depend on Atg5 for their localization to the isolation membrane [64]. Therefore, in mammalian cells ULK complexes likely act both upstream and downstream of the Atg5-Atg12 conjugation machinery. With this regard, there are at least two different pathways that regulate macroautophagy, one dependent and the other independent upon Atg5/Atg7 [93]. The Atg5/Atg7-independent pathway requires ULK1 and Beclin1, a protein interacting with hVps34 [93], suggesting that ULK1 and hVps34 complexes seem to have broader roles in the regulation of autophagy in mammalian cells encompassing both Atg5/Atg7-dependent and independent autophagy pathways.
Concluding remarks
The studies described above have emphasized that the ULK complexes function as an important node through which nutrient and growth factor signalling and stress response pathways are transmitted to the autophagy machinery via mTORC1. Many important questions are imposed by the recent discovery of the ULK complexes. What are ULK1 and ULK2 substrates? Would ULK1 and ULK2 complexes be solely the mediators of mTORC1 signaling to autophagy? What are the exact sites and functions of ULK, Atg13 and FIP200 phosphorylations by mTORC1 and ULK? What are the functions of Atg13 and FIP200 in the regulation of ULK? Do ULK1 and ULK2 have their specific binding partners, substrates and functions? How do ULK complexes crosstalk with hVps34 complexes, Atg5 and Atg7 conjugation systems and mAtg9?
Although starvation signaling is not always mediated by mTORC1 and it is possible that ULK complexes are regulated by mTORC1-independent pathways [94–96], the current knowledge suggests that mTORC1 and Atg1/ULK complexes constitute the central axis of the pathways that coordinately regulates growth and autophagy in response to cellular physiological and nutritional conditions. With this regard, it is noteworthy that Atg1/ULK negatively regulates TORC1 and growth in Drosophila and several types of human cells [50,66,97]. It is unclear how the Atg1/ULK complexes negatively regulate TORC1 and growth and whether this negative function generally occurs in other cell types. A likely importance of this regulation is that the Atg1/ULK complexes might contribute further to autophagy by inhibiting the activity of TORC1 to ensure more favourable conditions for autophagy induction and cell growth inhibition. The negative regulation of TORC1 by Atg1/ULK stresses further the intimate crosstalk between the autophagy and cell growth pathways. Future studies may unveil more functions of the Atg1/ULK complexes involved in the regulation of cell growth and autophagy.
Supplementary Material
Acknowledgements
We acknowledge the supports of the American Diabetes Association 7-07-CD-08, the National Institutes of Health DK072004, and Minnesota Obesity Center P30DK50456. We thank T. Neufeld, Y.-Y. Chang, and Kim lab members for helpful discussion and comments. We apologize that we could not include many original publications due to limited space.
Abbreviations
- mTOR
mammalian target of rapamycin
- mTORC1
mTOR complex 1
- mTORC2
mTOR complex 2
- raptor
regulatory associated protein of mTOR
- rictor
rapamycin insensitive companion of mTOR
- Atg
Autophagy-related gene
- TSC
tuberous sclerosis complex
- Rheb
Ras homolog enriched in brain
- PKB
protein kinase B
- GAP1
general amino acid permease 1
- AMPK
5'-AMP-activated protein kinase
- hVps34
human ortholog of yeast vacuolar protein sorting 34
- ULK1
UNC-51-like kinase 1
- ULK2
UNC-51-like kinase 2
- FIP200
focal adhesion kinase (FAK) family interacting protein of 200 kDa
- GATE-16
Golgi-associated ATPase enhancer of 16 kDa
- GABARAP
γ-aminobutyric acid (GABA) receptor associated protein
- TGN
Trans-Golgi Network
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCE
- [1].Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273:3963–6. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- [2].Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7:167–78. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- [3].Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–9. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- [4].Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–84. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- [5].Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–94. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- [6].Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–90. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- [7].Loewith R, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–68. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- [8].Hara K, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–89. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- [9].Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–75. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- [10].Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- [11].Jacinto E, et al. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–37. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- [12].Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9:316–23. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- [13].Pearce LR, et al. Identification of Protor as a novel Rictor-binding component of mTOR complex-2. Biochem J. 2007;405:513–22. doi: 10.1042/BJ20070540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Woo SY, et al. PRR5, a novel component of mTOR complex 2, regulates platelet-derived growth factor receptor beta expression and signaling. J Biol Chem. 2007;282:25604–12. doi: 10.1074/jbc.M704343200. [DOI] [PubMed] [Google Scholar]
- [15].Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–86. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- [17].Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–68. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- [18].Garcia-Martinez JM, Alessi DR. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1) Biochem J. 2008;416:375–85. doi: 10.1042/BJ20081668. [DOI] [PubMed] [Google Scholar]
- [19].Mammucari C, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–71. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- [20].Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–83. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- [21].Kim E, Guan KL. RAG GTPases in nutrient-mediated TOR signaling pathway. Cell Cycle. 2009;8:1014–8. doi: 10.4161/cc.8.7.8124. [DOI] [PubMed] [Google Scholar]
- [22].Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Dubouloz F, Deloche O, Wanke V, Cameroni E, De Virgilio C. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol Cell. 2005;19:15–26. doi: 10.1016/j.molcel.2005.05.020. [DOI] [PubMed] [Google Scholar]
- [24].Findlay GM, Yan L, Procter J, Mieulet V, Lamb RF. A MAP4 kinase related to Ste20 is a nutrient-sensitive regulator of mTOR signalling. Biochem J. 2007;403:13–20. doi: 10.1042/BJ20061881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Nicklin P, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–34. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Chen EJ, Kaiser CA. Amino acids regulate the intracellular trafficking of the general amino acid permease of Saccharomycescerevisiae. Proc Natl Acad Sci U S A. 2002;99:14837–42. doi: 10.1073/pnas.232591899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Schu PV, Takegawa K, Fry MJ, Stack JH, Waterfield MD, Emr SD. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- [28].Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3'-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275:992–8. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- [29].Nobukuni T, et al. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc Natl Acad Sci U S A. 2005;102:14238–43. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Byfield MP, Murray JT, Backer JM. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J Biol Chem. 2005;280:33076–82. doi: 10.1074/jbc.M507201200. [DOI] [PubMed] [Google Scholar]
- [31].Gulati P, et al. Amino acids activate mTOR complex 1 via Ca2+/CaM signaling to hVps34. Cell Metab. 2008;7:456–65. doi: 10.1016/j.cmet.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Juhasz G, Hill JH, Yan Y, Sass M, Baehrecke EH, Backer JM, Neufeld TP. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J Cell Biol. 2008;181:655–66. doi: 10.1083/jcb.200712051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Backer JM. The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem J. 2008;410:1–17. doi: 10.1042/BJ20071427. [DOI] [PubMed] [Google Scholar]
- [34].Corradetti MN, Inoki K, Bardeesy N, DePinho RA, Guan KL. Regulation of the TSC pathway by LKB1: evidence of a molecular link between tuberous sclerosis complex and Peutz-Jeghers syndrome. Genes Dev. 2004;18:1533–8. doi: 10.1101/gad.1199104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Shaw RJ, Bardeesy N, Manning BD, Lopez L, Kosmatka M, DePinho RA, Cantley LC. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell. 2004;6:91–9. doi: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- [36].Meley D, Bauvy C, Houben-Weerts JH, Dubbelhuis PF, Helmond MT, Codogno P, Meijer AJ. AMP-activated protein kinase and the regulation of autophagic proteolysis. J Biol Chem. 2006;281:34870–9. doi: 10.1074/jbc.M605488200. [DOI] [PubMed] [Google Scholar]
- [37].Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–26. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lee MN, et al. Glycolytic flux signals to mTOR through glyceraldehyde-3-phosphate dehydrogenase-mediated regulation of Rheb. Mol Cell Biol. 2009;29:3991–4001. doi: 10.1128/MCB.00165-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell. 2002;10:151–62. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- [40].Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–57. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- [41].Kovacina KS, Park GY, Bae SS, Guzzetta AW, Schaefer E, Birnbaum MJ, Roth RA. Identification of a proline-rich Akt substrate as a 14-3-3 binding partner. J Biol Chem. 2003;278:10189–94. doi: 10.1074/jbc.M210837200. [DOI] [PubMed] [Google Scholar]
- [42].Sofer A, Lei K, Johannessen CM, Ellisen LW. Regulation of mTOR and cell growth in response to energy stress by REDD1. Mol Cell Biol. 2005;25:5834–45. doi: 10.1128/MCB.25.14.5834-5845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Whitney ML, Jefferson LS, Kimball SR. ATF4 is necessary and sufficient for ER stress-induced upregulation of REDD1 expression. Biochem Biophys Res Commun. 2009;379:451–5. doi: 10.1016/j.bbrc.2008.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Desai BN, Myers BR, Schreiber SL. FKBP12-rapamycin-associated protein associates with mitochondria and senses osmotic stress via mitochondrial dysfunction. Proc Natl Acad Sci U S A. 2002;99:4319–24. doi: 10.1073/pnas.261702698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Straub M, Bredschneider M, Thumm M. AUT3, a serine/threonine kinase gene, is essential for autophagocytosis in Saccharomyces cerevisiae. J Bacteriol. 1997;179:3875–83. doi: 10.1128/jb.179.12.3875-3883.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–50. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- [47].Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–13. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–91. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- [49].Otto GP, Wu MY, Kazgan N, Anderson OR, Kessin RH. Dictyostelium macroautophagy mutants vary in the severity of their developmental defects. J Biol Chem. 2004;279:15621–9. doi: 10.1074/jbc.M311139200. [DOI] [PubMed] [Google Scholar]
- [50].Scott RC, Juhasz G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Funakoshi T, Matsuura A, Noda T, Ohsumi Y. Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene. 1997;192:207–13. doi: 10.1016/s0378-1119(97)00031-0. [DOI] [PubMed] [Google Scholar]
- [52].Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell. 2008;19:2039–50. doi: 10.1091/mbc.E07-10-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kabeya Y, Noda NN, Fujioka Y, Suzuki K, Inagaki F, Ohsumi Y. Characterization of the Atg17-Atg29-Atg31 complex specifically required for starvation-induced autophagy in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2009;389:612–5. doi: 10.1016/j.bbrc.2009.09.034. [DOI] [PubMed] [Google Scholar]
- [54].Kabeya Y, Kamada Y, Baba M, Takikawa H, Sasaki M, Ohsumi Y. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell. 2005;16:2544–53. doi: 10.1091/mbc.E04-08-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Cheong H, Nair U, Geng J, Klionsky DJ. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:668–81. doi: 10.1091/mbc.E07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–18. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- [57].Sekito T, Kawamata T, Ichikawa R, Suzuki K, Ohsumi Y. Atg17 recruits Atg9 to organize the pre-autophagosomal structure. Genes Cells. 2009;14:525–38. doi: 10.1111/j.1365-2443.2009.01299.x. [DOI] [PubMed] [Google Scholar]
- [58].Cheong H, Yorimitsu T, Reggiori F, Legakis JE, Wang CW, Klionsky DJ. Atg17 regulates the magnitude of the autophagic response. Mol Biol Cell. 2005;16:3438–53. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Kamada Y, Yoshino KI, Kondo C, Kawamata T, Oshiro N, Yonezawa K, Ohsumi Y. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol. 2009 doi: 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Yan J, Kuroyanagi H, Kuroiwa A, Matsuda Y, Tokumitsu H, Tomoda T, Shirasawa T, Muramatsu M. Identification of mouse ULK1, a novel protein kinase structurally related to C. elegans UNC-51. Biochem Biophys Res Commun. 1998;246:222–7. doi: 10.1006/bbrc.1998.8546. [DOI] [PubMed] [Google Scholar]
- [61].Kuroyanagi H, Yan J, Seki N, Yamanouchi Y, Suzuki Y, Takano T, Muramatsu M, Shirasawa T. Human ULK1, a novel serine/threonine kinase related to UNC-51 kinase of Caenorhabditis elegans: cDNA cloning, expression, and chromosomal assignment. Genomics. 1998;51:76–85. doi: 10.1006/geno.1998.5340. [DOI] [PubMed] [Google Scholar]
- [62].Yan J, et al. Mouse ULK2, a novel member of the UNC-51-like protein kinases: unique features of functional domains. Oncogene. 1999;18:5850–9. doi: 10.1038/sj.onc.1202988. [DOI] [PubMed] [Google Scholar]
- [63].Chan EY, Kir S, Tooze SA. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem. 2007;282:25464–74. doi: 10.1074/jbc.M703663200. [DOI] [PubMed] [Google Scholar]
- [64].Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Kundu M, et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Young AR, et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Meijer WH, van der Klei IJ, Veenhuis M, Kiel JA. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy. 2007;3:106–16. doi: 10.4161/auto.3595. [DOI] [PubMed] [Google Scholar]
- [69].Chan EY, Longatti A, McKnight NC, Tooze SA. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol Cell Biol. 2009;29:157–71. doi: 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Chang YY, Neufeld TP. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol Biol Cell. 2009;20:2004–14. doi: 10.1091/mbc.E08-12-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Hosokawa N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–91. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Mercer CA, Kaliappan A, Dennis PB. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy. 2009;5:649–62. doi: 10.4161/auto.5.5.8249. [DOI] [PubMed] [Google Scholar]
- [74].Gan B, Guan JL. FIP200, a key signaling node to coordinately regulate various cellular processes. Cell Signal. 2008;20:787–94. doi: 10.1016/j.cellsig.2007.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Hosokawa N, Sasaki T, Iemura S, Natsume T, Hara T, Mizushima N. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy. 2009;5:973–9. doi: 10.4161/auto.5.7.9296. [DOI] [PubMed] [Google Scholar]
- [76].Dorsey FC, Rose KL, Coenen S, Prater SM, Cavett V, Cleveland JL, Caldwell-Busby J. Mapping the phosphorylation sites of Ulk1. J Proteome Res. 2009;8:5253–63. doi: 10.1021/pr900583m. [DOI] [PubMed] [Google Scholar]
- [77].Okazaki N, Yan J, Yuasa S, Ueno T, Kominami E, Masuho Y, Koga H, Muramatsu M. Interaction of the Unc-51-like kinase and microtubule-associated protein light chain 3 related proteins in the brain: possible role of vesicular transport in axonal elongation. Brain Res Mol Brain Res. 2000;85:1–12. doi: 10.1016/s0169-328x(00)00218-7. [DOI] [PubMed] [Google Scholar]
- [78].Abeliovich H, Zhang C, Dunn WA, Jr., Shokat KM, Klionsky DJ. Chemical genetic analysis of Apg1 reveals a non-kinase role in the induction of autophagy. Mol Biol Cell. 2003;14:477–90. doi: 10.1091/mbc.E02-07-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–12. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- [80].Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–78. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- [81].Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–7. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- [83].Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–72. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A. 2008;105:19211–6. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Matsunaga K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–96. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- [86].Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–76. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Young AR, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- [88].Yamada T, Carson AR, Caniggia I, Umebayashi K, Yoshimori T, Nakabayashi K, Scherer SW. Endothelial nitric-oxide synthase antisense (NOS3AS) gene encodes an autophagy-related protein (APG9-like2) highly expressed in trophoblast. J Biol Chem. 2005;280:18283–90. doi: 10.1074/jbc.M413957200. [DOI] [PubMed] [Google Scholar]
- [89].Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282:37298–302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- [90].Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Sou YS, et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell. 2008;19:4762–75. doi: 10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–81. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Nishida Y, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–8. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- [94].He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Stephan JS, Yeh YY, Ramachandran V, Deminoff SJ, Herman PK. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc Natl Acad Sci U S A. 2009;106:17049–54. doi: 10.1073/pnas.0903316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Sarkar S, Ravikumar B, Floto RA, Rubinsztein DC. Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differ. 2009;16:46–56. doi: 10.1038/cdd.2008.110. [DOI] [PubMed] [Google Scholar]
- [97].Lee SB, Kim S, Lee J, Park J, Lee G, Kim Y, Kim JM, Chung J. ATG1, an autophagy regulator, inhibits cell growth by negatively regulating S6 kinase. EMBO Rep. 2007;8:360–5. doi: 10.1038/sj.embor.7400917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.