Abstract
The use of microarray and comparative genomic technologies for the analysis of host–pathogen interactions has led to a greater understanding of the biological systems involved in infectious disease processes. Transcriptome analysis of intracellular pathogens at single or multiple time points during infection offers insight into the pathogen intracellular lifecycle. Host–pathogen transcriptome analysis in vivo, over time, enables characterization of both the pathogen and the host during the dynamic, multicellular host response. Comparative genomics using hybridization microarray-based comparative whole-genome resequencing or de novo whole-genome sequencing can identify the genetic factors responsible for pathogen evolutionary divergence, emergence, reemergence or the genetic basis for different pathogenic phenotypes. Together, microarray and comparative genomic technologies will continue to advance our understanding of pathogen evolution and assist in combating human infectious disease.
Keywords: genomics, host, microarray, microbe
Overview: intracellular pathogen transcriptome challenges & comparative genomics
Many different approaches can be used to study the complexity of host–pathogen interactions, including animal infection models, in vitro assays, confocal and electron microscopy, to name a few. Microarrays are a relatively new technology that have been employed to perform genome-wide host and/or pathogen transcriptome analyses. However, microarray analysis of intracellular pathogen gene expression in a host cell model can be challenging owing to the low amounts of pathogen RNA relative to the host RNA background. Enrichment is a new technology that involves the selective removal of host ribosomal transcripts and polyadenylated eukaryotic mRNAs. While some pathogen RNA loss occurs during enrichment, the ultimate goal of the process is to increase the ratio of pathogen RNA relative to that of the host. Amplification of the post-enriched RNA increases the amount of pathogen RNA such that it reaches a threshold concentration that is optimal for microarray performance (1 μg for Affymetrix platforms; Affymetrix, Inc., CA, USA). While host RNA is also amplified, microarray probe specificity and microarray design can mitigate cross hybridization effects. For pathogens characterized by low infectious doses, early time points can be particularly challenging because of the greater amounts of host RNA relative to pathogen RNA. Increasing the ratio of infectious organisms per host cell, also known as multiplicity of infection (MOI), is one solution to ensure enough pathogen RNA is isolated for microarray-based methods. Pretesting of higher MOIs to demonstrate that neither pathogenesis nor the timing of cellular events is significantly altered compared with lower MOIs is advised for this approach. Alternatively, higher levels of pathogen RNA can be isolated by sampling at later time points, by which time replicating bacteria have significantly increased in number. The use of quantitative PCR (Q-PCR) before, during and after enrichment and amplification is an essential tool for monitoring pathogen RNA levels during these sample preparation steps. La et al. review some of the technical challenges of host–pathogen transcriptome analysis and they discuss similar and alternative strategies to surmount those obstacles [1]. In addition to these strategies, experimental design and randomization are critical components of successful microarray experiments.
Our review focuses on the technical challenges and successful methods used in single or multiple time point transcriptome analyses of host–pathogen infection models. We highlight the discoveries from those efforts, with the goal of deciphering the complexity of the biological systems involved. We also review recent studies involving several comparative genomic technologies, such as comparative genome hybridization, microarray-based comparative whole-genome resequencing, and Sanger and next-generation comparative whole-genome sequencing. Finally, we will elaborate on how the discoveries and advances from these studies have led to a greater understanding of pathogen strain evolution, divergence and host pathoadaptation.
Intracellular transcriptome & phenotype microarray analysis
Microarray expression analysis coupled with metabolite typing or phenotype microarrays can coordinately define the genomic and biochemical range of pathogen survival in controlled environments. One such study involving these technologies focused on the obligate intracellular pathogen Coxiella burnetii, with the goal of developing an axenic (host cell-free) medium for in vitro cultivation.
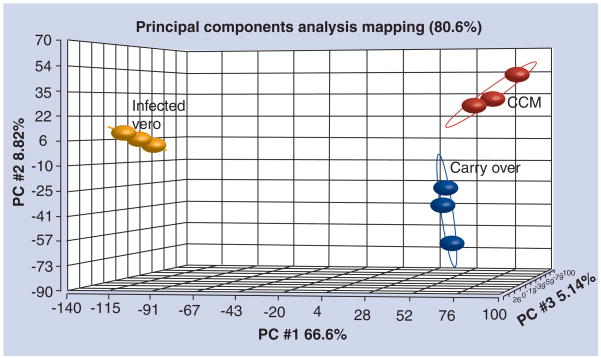
Coxiella burnetii is a prototypical obligate intracellular bacterial pathogen and the causative agent of human Q fever. Transmission of the infectious agent typically occurs through inhalation of contaminated aerosols. While the host range can be broad, in humans the infection usually presents as a self-limiting flu-like illness. The lack of axenic culture techniques for this organism has limited the physiological analysis of C. burnetii and development of a genetic system. These limitations become pronounced obstacles when attempting to identify bacterial virulence determinants. For example, to date, lipopolysaccharide remains the only identified virulence factor of the pathogen [2]. A medium named complex Coxiella medium (CCM) was developed recently and supports prolonged metabolic activity but not replication [3]. In an effort to determine the factors that prevent replication, microarray analysis, genome analysis and metabolite typing were performed with C. burnetii under specific growth conditions. C. burnetii RNAs were isolated from replicates of Vero cells 5 days postinfection and CCM 24 h postinoculation. Both sample sets were randomized and processed from RNA extraction through to microarray hybridization with enrichment and amplification performed to meet optimal levels of pathogen microarray performance. Principal components analysis (PCA) is used frequently for visualizing the three largest sources of variation in the array data relative to replicates and conditions or treatments. Figure 1 is an example of a new PCA graph of a previously published expression microarray dataset [4], with an updated interpretation or presentation provided here for the purposes of this review. The PCA represents the transcriptome of C. burnetii following 5 days of intracellular replication in Vero cells (infected Vero), 24 h incubation in CCM or obtained from the C. burnetii infectious inoculum, which is made by purifying C. burnetii from Vero cells infected for 7 days (carry-over). This latter RNA profile represents the RNA profile of C. burnetii (carried-over), prior to the pathogen’s exposure to new Vero cells (5 days incubation) or CCM-alone incubation (24 h). Each colored sphere is all the data from a single, biological sample and microarray. The tight grouping of biological replicates relative to the treatments indicates low variation between replicates. The distance between groups of replicates indicates large numbers of differentially expressed genes between the different conditions. Inspection of the microarray data identified substantially reduced expression of ribosomal genes of C. burnetii in CCM relative to Vero-cell cultivated organisms. Suspecting amino acid deficiency as the cause of low ribosomal gene expression, casamino acids and L-cysteine were added to CCM, creating acidified citrate cysteine medium (ACCM) [4]. The investigators discovered a 13-fold increase in C. burnetii protein synthesis as a result of growth in ACCM. Subsequent analysis of the C. burnetii genome revealed the presence of genes encoding different terminal cytochrome oxidases, suggesting that C. burnetii has the capability to respond to alterations in oxygen tension. A phenotype microarray system (BioLog, Inc., CA, USA) was then used to analyze the effect of different levels of oxygen tension on C. burnetii’s ability to metabolize a wide variety of substrates. The number of substrates oxidized by C. burnetii increased with decreasing amounts of oxygen [4]. Cultures were then analyzed for replication by genome equivalent analysis using Q-PCR at 24 h intervals over 6 days of incubation in ACCM under different oxygen tensions. Substantial C. burnetii replication occurred under microaerobic conditions (2.5% oxygen). ACCM cultivated organisms were highly infectious for Vero cells and they exhibited developmental forms comparable to in vivo-cultured organisms [4]. In addition, this study, through the use of expression microarrays, Q-PCR and phenotype microarrays, provided the framework for a systematic approach that could be applicable to the development of media that supports growth of other important, obligate intracellular pathogens. Subsequent microarray analysis of the C. burnetii transcriptome in human monocyte-derived macrophages compared with growth in ACCM has since revealed several genes that are dramatically upregulated during infection. These newly identified genes may encode virulence factors that contribute to the pathobiology of Q fever [Omsland A, Heinzen R, Unpublished Data]. Host cell-free growth of C. burnetii is a major scientific breakthrough for our ability to perform physiological and genetic analysis of this refractory pathogen.
Figure 1. Principal components analysis plot representing the whole-genome, differential expression profile of Coxiella burnetii under different growth conditions.
Each sphere in the figure is an individual biological replicate and represents all the data from one microarray chip. The orange spheres labeled ‘infected vero’ represent mRNA isolated from C. burnetii infected Vero cells following 5 days of intracellular infection. The red spheres labeled ‘CCM’ represent mRNA isolated from C. burnetii incubated in CCM for 24 h. The blue spheres labeled ‘carry over’ represent mRNA extracted from the original inoculum of C. burnetii cells that were made by purifying the pathogen from 7-day-infected Vero cells. Therefore, ‘carry over’ represents the mRNA content carried over initially into the new 5-day Vero cell-infected environment and the 24 h CCM environment. Principal components analysis plots allow rapid data visualization of groupings of biological replicates within their particular treatment, relative to the separation or distance between the different treatment groups. The tighter the groupings and the greater the distance between groups, the greater the list of genes demonstrating significant differential expression changes between treatments. CCM: Complex Coxiella medium; PC: Principal component.
Intracellular transcriptional analysis of different target cells
Elucidation of the pathogen transcriptome during invasion of target cells known to be involved in host disease or infection can provide a better understanding of infection in vivo. Lists of common or unique genes expressed during infection of different cell types can delineate portions of the genome tasked with promoting infection of a broad range of host cells or facilitating pathogen survival in unique cellular environments. If a genetic system exists for knocking out pathogen genes, then these different intracellular models can be tested with mutants and ‘cause and effect’ hypotheses generated.
Salmonella and its attendant species are one of the leading causes of gastrointestinal disease worldwide. Different forms of the disease may result and range from diarrhea to systemic infection. Salmonella enterica serotype Typhimurium (S. Typhimurium) is a useful model organism for the study of Salmonella infections because it can be modified by genetic methods and it produces a systemic infection in the mouse model that mimics typhoid fever in humans. Different subsets of genes in S. enterica are known to be integral to attachment to mucosal cells, invasion of epithelium cells and subsequent invasion and replication in monocytic cells [5]. One of these subsets of genes encodes the bacterial type III secretion system, which has the ability to translocate virulence proteins into the host cell, effectively altering normal host cell functions. A recent study analyzed the transcriptome of S. Typhimurium inside epithelial cells and compared these data with a previous study analyzing Salmonella expression in macrophages [5]. Microarray data of epithelial cells demonstrate that at 2 h postinfection the bacteria reside in a host cell vacuole without replicating, and at 4 and 6 h postinfection there is intracellular growth of the bacteria. When these data were compared with those from Luria–Bertani-cultured Salmonella and infected macrophages, genes belonging to similar functional categories had the same intracellular expression profiles, albeit at different levels of expression in the two different cell types. These data suggest that common cues for altering gene expression exist in both cellular environments. Several clusters of genes showed epithelial-specific expression, while other clusters of genes showed macrophage-specific patterns of expression. In addition, the data demonstrated that flagellin is expressed inside epithelial cells, a novel finding confirmed by other methods in the study. In conclusion, this time course comparative microarray study proposed a two-stage expression response of Salmonella in epithelial cells. The first stage involves cytotoxicity via adaptation to the Salmonella-containing vacuole. The second stage includes an increase in expression of genes that promote iron uptake, flagellin gene synthesis and production of the type III secretion system, suggesting that the pathogen senses imminent death of the epithelial cell, thereby preparing for invasion of neighboring uninfected cells [5].
Intracellular lifecycle transcriptome analysis
Intracellular microarray studies that focus on understanding global gene expression from the first minutes, hours or days of infection to the point of cellular lysis offer insight into the systems biology of the pathogen intracellular lifecycle. For obligate intracellular pathogens, for which no mutational genetic systems exist, important inferences into gene function can be gained from these lifecycle transcriptome studies. For organisms whose genome can be readily manipulated using molecular biology approaches, a gene whose function and expression correlates with a known cellular event can be mutagenized and studied in the context of a dynamic, closed system. Results can be as dramatic as the failure of a mutant to proceed to a subsequent step in its cellular lifecycle, or as subtle as a differential response that is environment or cell-model dependent. Both scenarios produce novel, hypothesis-generating outcomes and lead to a better understanding of the gene or gene pathways that are essential to pathogenesis.
Chlamydia trachomatis is an obligate intracellular pathogen, for which no genetic approaches are available for altering the genome of the pathogen. C. trachomatis infections are a leading cause of preventable blindness and bacterial sexually transmitted disease in humans [6]. Following phagocytosis, C. trachomatis is known to modify the phagosome such that entry into the lysosomal pathway is prevented. After multiple rounds of replication, the noninfectious reticulate body differentiates into the infectious elementary body (EB). At 40–60 h postinfection, the host cell lyses, thereby releasing EBs capable of infecting new cells. In a landmark study, Belland et al. analyzed the C. trachomatis developmental lifecycle using MOIs of 100 for early time points (1 and 3 h postinfection), and MOIs of 1 for the 8, 16, 24 and 40 h time points. This study identified specific genes that potentially play important roles in host–pathogen interactions and subsets of genes that control ‘immediate early’ and ‘late’ differentiation stages of the intracellular pathogen lifecycle [6]. A similar developmental lifecycle study involving a related organism, Chlamydophila (Chlamydia) pneumoniae (Cpn), a pathogen known to cause respiratory tract infections and chronic inflammatory diseases, such as asthma and atherosclerosis, expanded upon the work by Belland et al. [6] and was recently published [7]. The infection of Cpn in HEp-2 cells, was measured at 6, 12, 18, 24, 36, 60 and 72 h, where an MOI of 40 was used for time points 6 and 12 h, and an MOI of 15 for all other time points. The microarray data generated from these experiments were compared with:
The mRNA profile of EBs
Published EB proteasome data
mRNA expression under iron-depleted culture conditions [7]
This study assigned time-specific functions of genes to known events in the developmental cycle, enabling associations and potential ‘cause and effect’ hypotheses to be generated. The data demonstrated that categories of expressed genes could be broken down into early, mid, late and tardy profile categories [7]. Of interest were those genes identified in the late category that were associated with genes coding for EB proteins, while tardy genes were associated with EB mRNAs. The hypothesis generated was that EB proteins are made before initiation of redifferentiation and that redifferentiation itself is initiated by a different subset of genes identified in the tardy category [7].
A recent study of Francisella infection of macrophages accomplished lifecycle transcriptome analysis and, in the process, generated mutants interrupted in specific stages of their intracellular lifecycle [8]. Francisella tularensis is a facultative intracellular bacterial pathogen that infects a wide variety of hosts and cell types, including, but not limited to, endothelial cells and mononuclear phagocytes [8]. F. tularensis causes tularemia in humans and the pathogen is highly infectious, such that as few as ten bacteria can cause human disease. Left untreated, the pneumonic form of the disease can lead to 25% mortality. Owing to this pulmonary involvement and potential clinical outcome, intracellular macrophage infection and replication are believed to play an important role in this disease. Therefore, a detailed time course study involving intracellular transcriptional analysis of F. tularensis in mouse bone marrow-derived macrophage (BMM) cells was performed recently. The intracellular stages of Francisella infection of macrophages have been described and begin with phagocytic uptake, where interactions with early and late endosomal compartments occur [9]. Degradation of the phagosomal membrane follows with escape of the bacteria (within 1 to >4 h post-entry) into the cytoplasm, where extensive cytosolic replication occurs, after which some bacteria may re-enter the endocytic compartment to reside in large autophagic vacuoles [9]. Eventually, programmed cell death of the macrophage occurs, followed by bacterial release. At the time of this work, little was known about the intracellular biology of this bacterium, including the genes and encoded proteins that have increased expression and drive Francisella’s developmental cycle inside macrophages. In order to remove this hurdle, Wehrly et al. characterized the Francisella–macrophage intracellular lifecycle at defined time points by using confocal laser-scanning and transmission-electron microscopy, assessment of CFUs after macrophage infection and phagosomal disruption analysis [8]. Specific phases of the cycle of Francisella tularensis strain Schu S4 in BMMs were identified, such that sample collection for array analysis was focused on time points encompassing all intracellular stages of the Francisella cycle. The time zero sample, which involved bacteria added directly to BMMs and immediately harvested, was used to obtain initial bacterial expression profiles as a baseline for comparing genes upregulated within macrophages. Recovering enough bacterial RNA from the early time points for microarray analysis was a challenge. Therefore, a pilot study indicated that two different MOIs could be used with little-to-no change observed in the timing of events in the Francisella intracellular cycle. MOIs of either 200 (0, 1, 2 and 4 h time points), 50 (8 and 12 h time points) or 25 (16 and 24 h time points) were employed. The coordination of laboratory personnel and the timing for collection of all samples, with the goal of treating all samples in the same manner throughout the study, were concerns. Therefore, only four biological replicates at each time point were collected. Even with this biological replicate reduction, the ability to concurrently collect and process the samples was still a concern. Therefore, infections were performed in separate batches, where sample conditions were randomly assigned to the batches [8]. During sample processing, samples were randomized in 96-well plates in order to avoid any confounding factors owing to well location, time point, or biological replicate batch effects and to minimize error owing to plate edge effects and fluid transfers. The need for randomization during sample preparation as a means to avoid, or reduce, the likelihood of confounding biological factors with processing or procedural effects has been described for microarray-based platforms [10–13]. The tight groupings of replicates and the clear separation or distance between groups of replicates, indicative of low noise in the data, was demonstrated by the published PCA plot [8] and is a testament to the solid experimental design.
The amount of F. tularensis cRNA within each mixed RNA sample was estimated by Q-PCR of the FTT0243 locus, using a standard curve method (Applied Biosystems, CA, USA). The early time points contained insufficient amounts of bacterial RNA to allow for direct hybridization to the microarrays, a preferred approach for reducing noise in the data. Therefore, an RNA amplification step was performed. During initial data analysis, a time-dependent correlation between global gene expression profiles and specific intracellular stages was discovered. Microarray data from each time point were then normalized to those collected at time zero. A relatively constant number of genes were downregulated at 1 or 12 h postinoculation, while those significantly upregulated increased over time (from 2 h postinoculation to 24 h postinoculation). Over the entire time course, 658 genes had significant changes in expression, among which 298 were upregulated and 360 were downregulated. The study identified genes significantly upregulated either at all time points during phagosomal and late vacuolar stages, or during the cytosolic replication stages. These genes and their annotated functions were compared with temporal patterns of expression and associations with cellular events. Ten genes with hypothetical functions, which associated significantly with increased expression during the developmental lifecycle, were deleted in strain Schu S4. Seven of the ten isogenic mutant strains showed no defect in intracellular growth compared with the wild-type parental strain, while strains with deletion of FTT0383, FTT0369c and FTT1676 genes (ΔFTT0383, ΔFTT0369c and ΔFTT1676) demonstrated obvious intracellular growth defects. The authors hypothesized that increased intracellular killing of the mutant strains caused the observed large reductions in bacterial numbers [8]. Confocal laser-scanning microscopy analysis demonstrated that the ΔFFTT0383 mutant strain was trapped in a lysosomal LAMP-1-positive compartment, consistent with the mechanism of killing within macrophages, while ΔFFT0369c showed phagosomal escape suggesting that deletion of FFT0369c appears to mostly affect cytosolic growth [8]. ΔFFT1676 appears to be required for phagosomal escape and, perhaps more importantly, cytosolic replication. Trans-complementation of ΔFTT0383 and ΔFTT1676 restored wild-type-like phenotypes while trans-complementation of ΔFFT0369c with its native promoter also returned the mutant to a wild-type-like phenotype. Studies of mice infected with ΔFTT0383, ΔFTT0369c or ΔFFT1676 mutant Schu4 strains revealed that deletion of these genes abolished virulence in mice. In conclusion, this study is the first to characterize the intracellular transcriptome of a highly virulent strain of F. tularensis during its infection lifecycle within BMMs. Moreover, the study revealed key F. tularensis-specific, hypothetical genes that play integral roles in the pathogen’s adaptation to particular intracellular locations during its lifecycle.
In vivo, host–pathogen dual transcriptome analysis
Very few experiments published to date have involved simultaneous transcriptional analysis of the host and pathogen during an infection lifecycle analysis. Two murine studies in which defined end points were chosen for simultaneous analysis of host–pathogen transcriptomes were previously published [14,15]. While the challenges of an in vivo transcriptome experiment using repeated measurements over time can be enormous, the benefit is the potential to understand site-specific host–pathogen interactions in the context of a dynamic, multicellular host response. For the purpose of this review, we will focus on a microarray study that used 20 non-human primates that were their own within-model controls, repeatedly measured over time, and we will describe the systems-level biological discoveries that resulted [16].
Group A streptococcal (GAS) infections are endemic in developed and underdeveloped countries worldwide [16]. In 2000, acute pharyngitis was responsible for 11 million doctor’s office visits in the USA alone. While comparatively rare, invasive GAS infections can occur at a rate of approximately 3.5 per 100,000, with roughly 1500 of those resulting in death. To date, a vaccine remains elusive and new scientific approaches are needed to understand the infectious cycle of this important human pathogen. To better understand the molecular interactions involved in the natural course of acute pharyngitis, efforts were made to develop a large, nonhuman primate study designed to analyze all phases and stages of the infection from onset to host resolution. A pilot study analyzed post-streptococcal pharyngitis in three cynomolgus macaques to identify sources of error or variation that could be controlled for in a larger study [17]. The most significant source of variation was primate-to-primate variation, most likely due to these animals not being inbred. Therefore, 20 nonhuman primate study subjects were chosen for a subsequent study to accommodate subject-to-subject variation and maximize statistically significant gene expression changes (p < 0.05) [16]. A within-group experimental design was implemented involving both mock and infection treatments such that mock treatments were 5 weeks in length, separated by a 4-week, quiescent period of recovery, followed by GAS infection. This within-group framework allowed for each nonhuman primate to be its own control relative to the infection, thereby reducing the influence of individual immune and genetic backgrounds. During the study, 75% of the primates demonstrated persistent infection on day 32, allowing extension of the experiment and data collection for an additional 8 weeks. Critical to the success of the study was a rigorous experimental design and randomization procedure that minimized extraneous variables over time [16].
Five throat swabs were taken from each primate on 13 different study days (days 0, 1, 2, 4, 7, 9, 16, 23, 32, 45, 58, 72 and 86). One throat swab was used for GAS culture plating, two were combined for custom pathogen GeneChip® (Affymetrix, Inc.) target preparation, one was collected for use in host expression microarray analysis and one was used for TaqMan® (Applied Biosystems, Inc.) confirmation. Swabs were used consistently and in this specific order. At the time of this study, established RNA enrichment and amplification strategies were not available. Therefore, a complex statistical design strategy was used to optimize the GeneChip detection of low abundance pathogen transcripts (1000 less than in previous microarray studies).
In an effort to condense and interpret the large volume of significant differential host and pathogen expression data, pathogen gene category and host category analyses were performed [16]. Data were grouped by pairs of adjacent time points and analyzed to determine significant changes in gene expression from day to day. Analysis was performed for each of the eight consecutive pairs of time points [16]. A total of 13 out of 32 measured clinical parameters demonstrated a Bonferroni-level of statistical differential significance between mock- and GAS-infected animals. The combined clinical data identified three significant and distinct phases of disease:
Colonization
Acute
Asymptomatic disease [16]
Correlation analysis of gene expression patterns with the significant clinical data sets, number of CFUs and phases of disease were performed. Carbohydrate metabolism in the pathogen was a key contributor to initial pathogen growth, and a relationship was found to exist between prophage gene expression and high pathogen densities and inflammation. Three pyrogenic toxin superantigens were sequentially expressed in the posterior pharynx to assist colonization and significant decreased expression of these superantigens during the tertiary phase of disease contributed to asymptomatic clinical presentation. It was also discovered by Q-PCR that the increase in DNA copy numbers of streptococcal prophages during infection led to increased expression of their encoded virulence genes [16]. This finding led to the hypothesis that prophages contribute significantly to the pathogen lifecycle in vivo by altering the host immune response in the posterior pharynx. In addition, a gene encoding the response regulator of a novel two-component gene regulatory system, named sptR, was found to have a biphasic relationship with the known virulence gene regulator covR. The authors hypothesized that covR and sptR are key regulators of gene expression that act in concert to assist GAS in establishing or prolonging GAS–host interactions during different phases of disease [16].
Since the publication of this work, analysis of the nonhuman primate host transcriptome has been performed with correlation analysis to clinical disease data, CFUs and phases of infection. To search for parallel and inverse expression relationships between the two datasets (host and pathogen), Spearman rank correlation analysis of all pair-wise combinations of individual pathogen and host gene expressions over time has been performed. The goal of this interactome-like analysis is to determine co-regulated host and pathogen genes, or ‘cause and effect’ relationships at the transcriptional level between the host and pathogen. These data, when compared with disease phases, clinical states of disease and CFU counts over time, have led to novel discoveries and testable hypotheses [O’Shea P & Musser J, Unpublished Data].
Comparative genomic hybridization analysis of pathogen isolates
Microarray-based comparative genomic hybridization (CGH) offers a number of advantages over conventional typing methods, such as restriction fragment length polymorphism, 16S ribosome sequencing, multilocus sequence typing or DNA–DNA hybridization. The advantages of microarray technology are greater throughput, less labor, genome-level scanning capabilities and parallel interpretation of the data. While several microarray-based technologies and approaches have been described [18], for the purposes of this review, our CGH discussion will focus on high-density, probe-set microarrays, specific to nearly all open reading frames (ORFs) from complete genomes. One drawback to microarray-based typing is that genes or DNA that are unique in tested isolates relative to the genome content on the array cannot be analyzed. That said, CGH is still a labor- and cost-efficient method for gaining valuable genome-wide data compared with known reference genomes. An additional advantage to the whole-genome array-based method of typing is that, when correlated to host origin or pathogenicity data, insight into the genetic factors responsible for phenotype, host adaptation or geographical location can be gained.
Infections with C. burnetii can occur nearly worldwide, indicating a near pan-global distribution of the pathogen. Isolates collected from a variety of locations reveal considerable genetic heterogeneity, as assessed by established typing methods. However, genome-wide variations responsible for genetic diversity or strain evolution are unknown. Therefore, Beare et al. used a DNA microarray containing the 2024 ORFs of the Nine Mile (NM) isolate (RSA493) genome to perform CGH analysis [19]. A total of 24 isolates of C. burnetii from diverse geographic and environmental origins were hybridized against the microarray, and the data were compared with restriction fragment length polymorphism-generated data for the same isolates. Interestingly, the CGH data showed that two isolates represented new genomic groups. ORF deletions, partial deletions, point mutations and insertions were discovered across the isolates during this analysis, which contributed to a better understanding of C. burnetii diversity and virulence potential [19]. This study was the first comprehensive whole-genome analysis performed for C. burnetii and revealed that genome conservation exists across a wide range of biologically and geographically diverse isolates. In addition, the data suggested that C. burnetii has undergone reductive genome evolution, which may, in part, explain some of the unique phenotypes observed within the isolates [19].
Comparative genomic hybridization also has relevance for newly emergent pathogens. An example is recent CGH work performed on the emerging Gram-negative pathogen Granulibacter bethesdensis. Chronic granulomatous disease (CGD) is a rare human genetic disorder caused by a defect in the phagocyte NADPH oxidase. As a result, phagocytes from CGD patients are unable to produce superoxide anions and any secondarily derived reactive oxygen species. Individuals with CGD develop reoccurring infections with Staphylococcus aureus, Burkholderia, Nocardia and other catalase-positive organisms. G. bethesdensis, a novel genus/species from the family Acetobacteraceae, was discovered in a CGD patient and has subsequently been isolated from five more patients, from different geographical locations. While 16S sequencing confirmed the organism belonged in the Acetobacteraceae, little other information about this unique bacterium was available. Following the genome sequencing of the isolate from the first patient, an antisense expression array was created [20]. The acquisition of several new patient isolates during this time allowed rapid microarray typing by CGH. The results demonstrated that, among the four isolates obtained from the first patient over a period of 2 years, little to no variation existed by CGH. However, isolates collected from three other patients showed unique hybridization patterns relative to each other and the reference genome. For one isolate, as many as 175 ORFs were lacking individual probe or probe-set signal relative to the reference genome on the chip [20]. The genes that showed differences across the isolates encoded DNA uptake proteins, transcriptional regulators, lipopolysaccharide synthesis proteins, hypothetically secreted proteins and transposases. New isolates from new patients are being collected at this time, and early CGH results suggest that there is a range of genetic diversity for this novel pathogen [Greenberg D et al., Unpublished Data]. The use of CGH in the rapid typing of isolates relative to a reference genome offers powerful information at a genome scale, providing interpretable discoveries into the genomic regions responsible for genetic diversity and pathoadaptation.
Microarray-based comparative whole-genome resequencing
Microarray-based comparative whole-genome resequencing (CGR) (NimbleGen Systems, WI, USA) involves the use of an approximately 30-bp oligonucleotide every seven bases for both strands of a reference genome, where each probe can overlap adjacent probes by approximately 22 bases. Following hybridization of isolate genome sequences, probes showing differences in signal intensity are flagged for possible mutations. A second array is designed for single base pair resolution of those regions such that all four possible bases are interrogated on both strands for that region. These resequencing arrays are then hybridized with isolate genomic DNA again and the data interpreted as described above. Other sequence differences, such as insertions/deletions (InDels) or sequential single-nucleotide polymorphisms (SNPs), are more difficult to interpret with this technology and the utility of this approach is dependent on high isolate homology to the interrogated reference genome on the array. First performed on Helicobacter pylori in 2005 [21], the evaluation by Herring et al. demonstrated a false-positive rate for SNP calls of one per 244,193 bp of sequence [22]. This level of accuracy and its relevance of use is best demonstrated by the analysis of ten clinical isolates of community-associated methicillin-resistant Staphylococcus aureus [23].
Community-associated methicillin-resistant Staphylococcus aureus is epidemic in the USA and infections are caused primarily by a strain known as pulsed-field type USA300 (USA300). Questions immediately arose as to whether the epidemic was caused by clonal emergence of similar USA300 isolates or coordinate evolution of independent, but similar, organisms (convergent evolution). Given the possibility that SNPs could drive either scenario, a microarray-based CGR system was chosen to analyze ten USA300 clinical isolates from eight different states associated with different types of human infection.
The results demonstrated that eight of the isolates had very few SNPs and were therefore closely related, suggesting recent clonal expansion and diversification as opposed to evolutionary convergence [23]. The authors pointed out that small numbers of SNPs can alter pathogen–host interaction and strain virulence in Streptococcus [24,25] and that a similar mechanism may be playing a role in these highly related S. aureus isolates. Therefore, using a mouse sepsis/bacteremia model, the investigators found significant variation in virulence among these 11 isolates (one reference and ten test isolates) [23]. Two of the eleven isolates that demonstrated reduced virulence also produced remarkably different exoprotein (secreted protein) profiles relative to the other nine. In conclusion, the results from this study demonstrate the utility and importance of microarray-based CGR technology to rapidly determine pathogen genetic factors driving an emergent, or re-emergent, epidemic.
Trachoma is caused by C. trachomatis serotypes A, B, Ba and C, where the disease is characterized by multiple stages and variant disease severity across infected individuals. In an effort to determine the pathogen genetic factors responsible for the variation in clinical disease, microarray-based sequencing was performed for three reference strains, serotypes A, B and C, and a recent serotype A isolate [26]. Polymorphisms in a subset of genes (~2% of the genome) were shown to have a relationship with the different virulence properties of these strains in cell culture and nonhuman-primate ocular-infection models [26]. This study demonstrates again the utility of a microarray-based sequencing technology applied against pathogen strain genomes known to have high genome homology towards the determination of genetic factors responsible for different pathogenic phenotypes.
Sanger & next-generation genome sequencing & comparative analysis
Sanger-based DNA sequencing has been the gold standard for pathogen whole-genome sequencing since the Haemophilus influenzae genome was first completed in 1995. While improvements have been made and the quality is still unsurpassed, the technology remains labor intensive, time consuming and costly. As read lengths increase with next-generation sequencing technologies, so too does their promise of similar quality levels at comparatively reduced cost, labor and time. Because of the long read lengths, 800–900 bp per read, Sanger sequences can be assembled by de novo (i.e., no reference genome is needed for alignment) methods or finished to completion for novel pathogen genomes. Once assembled, these genomes can be compared with unique, conserved or convergent genetic elements, within or across pathovars or pangenomes. We have been involved in numerous Sanger-based, whole-genome-sequencing projects where the downstream comparative analyses resulted in numerous discoveries for pathogens such as Rickettsia [27], GAS [24,25,28–31], Chlamydia [32] and Streptococcus zooepidemicus [33]. For the purposes of this review, we will discuss a multi-genome comparative Sanger sequencing project performed for C. burnetii, diagnostic development for Chlamydia, and finish with a brief description of new, next-generation comparative pathogen genome sequencing efforts.
C. burnetii has isolate-specific, altered phenotypes and infectivity. In addition, this intracellular pathogen demonstrates a wide range, both geographically and in terms of animal reservoirs, and the presence of a 37–55-kb autonomously replicating plasmid or chromosomally integrated plasmid sequences. Therefore, in an effort to discover unique genetic factors responsible for isolate diversity, a whole-genome sequencing approach was warranted. The previously sequenced and characterized NM isolate is considered representative of a human clinical acute disease isolate [34]. Human endocarditis isolates, which cause less inflammation in mice, or inefficiently infect mouse fibroblasts, and a rodent isolate attenuated in virulence for guinea pigs, also exist. Therefore, representative genomes of the prototype IV and V human endocarditis isolates, K and G, were sequenced to completion along with the attenuated Dugway rodent isolate, with the goal of comparing these genomes to NM [35]. The study revealed high genome synteny, with recombination between abundant insertion elements a driving force in chromosomal rearrangement and DNA insertions and deletions between these four genomes [35]. Significant plasmid differences between the four isolates were discovered along with differences in genome size, ORF coding capacity, and numbers of encoded transposases. The attenuated Dugway strain genome proved to be interesting because, when compared with the other three genomes, the Dugway strain had the largest genome, plasmid and ORF coding capacity, but the fewest number of pseudogenes. These data suggest that the Dugway isolate may represent a more primitive lineage, or an earlier stage of pathoadaptation than the NM, K or G isolates. The four-way genome comparison performed in this study was the first of its kind for Coxiella. The results provide important insight into the genetic diversity and genome architecture of Coxiella that may be responsible for the different phenotypes and variations in Q-fever clinical disease.
Comparative genome sequencing of new or distinct pathogenic isolates can also have immediate application to diagnostic tests. Multiple serovariants with specific organotropism for the eye or urogenital tract exist for C. trachomatis. The C. trachomatis genome is approximately 1 Mbp in size and contains a cryptic 7.5 kb plasmid of unknown function. An oculotropic trachoma isolate (A/HAR-13) genome was sequenced to completion and the data was compared with the publically available genome of a genitotropic isolate (D/UW-3) [36]. The genomic comparison led to the discovery of a novel PCR diagnostic marker that can discriminate between ocular and genital strains, a profound development of immediate importance in nations where chlamydial sexually transmitted diseases and trachoma are endemic [36]. A new variant of genital C. trachomatis emerged in Sweden owing, in part, to a deletion event in the C. trachomatis plasmid, leading to negative PCR diagnosis. The sequence of the new isolate genome along with plasmids from six isolates, confirmed a 377 bp deletion in the emergent isolate plasmid that removed the diagnostic PCR target site from detection. Sequence analysis of the chlamydial plasmids demonstrated that they are not freely exchanged between isolates, but instead remain closely linked to their host chromosome [37]. This result is supportive of Carlson et al. who, through transcriptional analysis of two closely related plasmid-containing and plasmid-lacking strains, discovered that the plasmid is a virulence factor and a transcriptional regulator of chromosomal genes [38].
Next-generation sequencing
Next-generation DNA sequencing includes three platforms currently marketed: SOLiD (Applied Biosystems, Inc.), Solexa Genome Analyzer (Illumina, Inc., CA, USA) and 454 GSFLX Titanium (Roche Molecular Systems, CA, USA). All three platforms have the capability for great sequencing depth with considerable differences in read lengths as well as amount of reads – 454: 400–600 million bases/500 bp read lengths; Solexa: 10–20 Gigabases/35 bp, 2 × 106 bp read lengths; and SOLiD: 10–20 Gigabases/25 bp, 2 × 50 bp read lengths. While the Solexa platform and SOLiD are fairly comparable in overall performance, Solexa appears to have an edge on its inherent error rate, while SOLiD – due to its color space encoding scheme – exhibits a theoretical advantage in SNP detection accuracy. A good review regarding advanced sequencing technologies and their applications has been published [39].
Recent efforts to combine the higher volume and lower cost of 454 sequencing with the longer read lengths and quality of Sanger sequencing reads have led to successful closure and publication of the Mycoplasma conjunctivae genome [40], the Brucella microti genome [41] and the Helicobacter pylori strain G27 genome [42]. The use of a combination of Solexa sequencing depth and 454 read lengths, again in an attempt to lower costs, labor and time spent for de novo genome assembly, is a relatively new undertaking. However, a recent study highlighting the bioinformatics pipeline and success of this approach has been published for the rice pathogen Pseudomonas syringae pv. Oryzae genome [43]. The authors demonstrate that the combined approach produced 130 contigs by de novo assembly for this 5.6 Mb genome. Importantly, 87% of the genome was represented across 14 of the contigs, which is an impressive breakthrough and lends credence towards a dual technology approach for rapid closure of a large complex genome [43].
An alternative use of 454 and Solexa technology during sequencing of pathogen genomes does not focus on complete closure but, instead, on high volume coverage for SNP, InDel and unique sequence determination. The goals of these efforts are genome-level phylogenetics, assessment of plasticity zones, discovery of novel sequences or regions of difference and identification of genomic duplication events. Examples of this approach have recently been published, where researchers used deep 454 sequencing to analyze five unpublished Brucella species. Efforts in this study focused on a total or full genome orthologous SNP analysis among these five new genomes and eight previously sequenced Brucella isolate genomes [44]. In a study that used both Solexa and 454 sequencing technology, comparative SNP and phylogenetic analysis was performed across 19 Salmonella enterica serovar Typhimurium genomes [45]. This study demonstrated little variation or recombination between isolates, but suggested that evolution, through the loss of gene function, was occurring [45].
Conclusion
Transcriptome studies have the capability to facilitate a systems biology level of analysis of host–pathogen interactions. When applied to a time course or developmental cycle study, a systems-level view of the entire pathogenic process can be produced. When the microarray data are compared against coordinately collected clinical or empirically derived data, such as proteomic data, inferences or correlations with gene expression patterns can be discovered. In the latter example, a comprehensive systems biology network can be constructed where spatial gene expression changes can be interrogated in the context of protein expression and function, thereby enabling new insight into the molecular basis of host–pathogen interactions. In the end, the determination of molecular candidates for mutational analysis within the pathogenic lifecycle or animal model is possible, which can further the discovery of novel diagnostic, therapeutic or vaccine candidates.
Pathogen comparative genomics through the use of CGH, or microarray-based CGR, offers rapid genome-level typing capabilities towards the determination of the genetic basis for pathogen evolution, distribution or phenotype. Sanger genome sequencing technologies may eventually be replaced by newer, longer read length next-generation sequencing technologies where lower reagent costs and greater throughput is possible. Next-generation sequencing technology will undoubtedly expand in its repertoire of feasible applications and further facilitate the study of newly emergent or re-emergent pathogens, pathogen genome evolution and host niche adaptation.
Future perspective
Transcriptome technologies have contributed significantly to a greater understanding of the systems biology of host–pathogen interactions. Further refinement of microarrays through improved assay conditions, the development of new analysis algorithms, and by increasing data density through continued feature-size reduction will enable increased sensitivity and accuracy. Enrichment and amplification technologies will continue to improve the use of lower amounts of starting material and help expand the range of testable systems. Of great interest are the molecular interactions and initial events occurring at very early stages of infection, where pathogen numbers are low, transcriptional changes subtle and the promise of interventional therapeutics and vaccines greatest. While next-generation DNA sequencing technologies do offer the potential to survey these environments with unparalleled sensitivity, the requirement for larger input nucleic acid amounts and significant bioinformatics resources, relative to microarrays, still make microarray platforms an effective method of choice for surveying these environments. We believe that microarrays will continue to serve a vital purpose and role in infectious disease research, owing to their low cost, multiple-sample throughput, low requirements for initial starting material and their well-established bioinformatics infrastructure. This is particularly the case where experimental settings or institutional infrastructures do not allow for the costs and labor of high-throughout sample processing and rapid bioinformatic data analysis on next-generation sequencing platforms. Nevertheless, and while still relatively new on scene, next-generation DNA sequencing is poised to supersede current high-throughput analyses, as it is quantitative in nature, requires fewer replicates for analysis of variation and is less subject to experimental design artifacts. In addition, this technology is gaining momentum as it improves in read length, which, in itself, is critical for de novo assemblies and alignment to reference genomes. Next-generation DNA sequencing technologies will continue to further improve in providing even greater sequencing depth, and hopefully, requiring lower sample input amounts. Owing to the massive amount of data produced by next-generation sequencing, two imperative goals for wider-spread use and greater success of this technology are:
The parallelization of processing algorithms on high-RAM workstations
The ability to store and transfer hundreds of terabytes of data in an efficient manner
Lastly, it cannot be stressed enough that new, better and established bioinformatics tools are needed for next-generation DNA sequencing. Algorithms have to be far more time efficient, robust, accurate and scalable to allow for meaningful analysis in a timely manner. Better graphical interfaces and data outputs will speed interpretation and publication of meaningful results. As these pressing issues evolve, next-generation DNA sequencing has the potential to position itself as an established standard for systems biology-level analysis of host–pathogen interactions and comparative microbial genomics in many laboratories.
Executive summary
Intracellular pathogen transcriptome challenges
There are low amounts of pathogen RNA relative to host RNA.
There is a need for enrichment and amplification strategies for increasing the ratio of pathogen RNA to host RNA.
Studies may involve single or multiple (time point or treatment) sample collection at defined end points (terminal tissue or animal collection).
Time course studies involve collecting samples at multiple specific time points related to the biology or course of disease.
Repeated measures in vivo studies involve repeated sample collection, in vivo, over time, from single or multiple sites related to the biology or course of disease.
Separate controls are external and provide a normal or uninfected baseline to compare other infected samples to.
Within-subject controls are the same host or sample first mock-treated and then infection-treated. They offer a ‘within-subject’ level of pre- and postinfection analysis.
Experimental design and randomization are important for successful microarray data.
Cell-specific transcriptome studies
Intracellular models of infection are dynamic, closed systems useful for host and/or pathogen transcriptome profiling.
Predetermined or selected single time point analysis is cost and labor effective.
Multiple time point or time course analysis allows identification of upregulated genes responsible for cellular events or actions during the lifecycle.
Mutational inactivation of expressed genes can lead to interruption of the pathogen lifecycle.
Ultimate confirmation is mutant analysis in animal models.
In vivo,host–pathogen, dual transcriptome analysis
Site or tissue-specific in vivo host–pathogen interaction analysis in the context of a dynamic, open-system (multicellular) host response can be analyzed over time (during the natural course of the infection).
An increase in sample size is necessary in order to manage the influence of animal-to-animal variation in the data, particularly in the context of noninbred animals.
There is a need for experimental design and randomization relative to sample conditions and treatments.
The benefit of in vivo, host–pathogen dual transcriptome analysis is that co-regulated and expressed host and pathogen genes can be analyzed in the context of time, pathogenesis and clinical factors towards providing a systems level of understanding.
Comparative genomic hybridization
The advantages of comparative genomic hybridization (CGH) include high throughput, low labor, low cost, genome-wide level of analysis and parallel processing of many samples at one time.
CGH is dependent upon the relatedness of the reference genome on the microarray to the isolates/strains being tested.
CGH cannot determine unique or novel sequences in the strains or isolates being tested, relative to the reference genome on the microarray.
Correlation analysis of the CGH data against pathogenicity, geographical distribution of isolates and isolate phenotypes can be performed.
Microarray-based comparative whole-genome resequencing
Nimblegen is the platform of choice for comparative whole-genome resequencing (CGR).
CGR utilizes dual-step array process for the screening and interrogation of data.
The advantages of CGR include high data accuracy and great sequencing depth (full-tile genome array).
CGR is similar to CGH in that a reference genome is required and no unique sequence detection is possible in isolates relative to the reference genome on the microarray.
Sanger genome sequencing
This well-established technology is capable of long read lengths but is labor and cost intensive.
Owing to high accuracy and long read lengths, Sanger genome sequencing is capable of de novo assembly and analysis.
Next-generation comparative genome sequencing
Next-generation sequencing technologies that produce accurate, long read lengths hold the most promise for Sanger-like quality, de novo assembly and discovery of unique sequences.
Next-generation technologies show greater flexibility, less cost and less labor than Sanger-based methods.
Next-generation technologies are capable of interrogating at ultra-high sensitivity host–pathogen interaction environments and producing novel transcriptome or sequence discoveries.
A high input amount of nucleic acids is required.
Next-generation technologies are bioinformatically complex and intensive, but software and out-of-the-box capabilities are improving.
Improvements are also being made in terms of a lower sample input, greater read length and greater sequencing depth.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
Contributor Information
Daniel E Sturdevant, Email: dsturdevant@niaid.nih.gov, Genomics Unit, Research Technologies Section, Research Technologies Branch, Rocky Mountain Laboratories, NIH, 904 South 4th Street, Hamilton, MT 59840, USA, Tel.: +1 406 363 9248, Fax: +1 406 363 9427.
Kimmo Virtaneva, Email: kvirtaneva@niaid.nih.gov, Genomics Unit, Research Technologies Section, Research Technologies Branch, Rocky Mountain Laboratories, NIH, 904 South 4th Street, Hamilton, MT 59840, USA, Tel.: +1 406 363 9430, Fax: +1 406 363 9427.
Craig Martens, Email: cmartens@niaid.nih.gov, Genomics Unit, Research Technologies Section, Research Technologies Branch, Rocky Mountain Laboratories, NIH, 904 South 4th Street, Hamilton, MT 59840, USA, Tel.: +1 406 363 9430, Fax: +1 406 363 9415.
Daniel Bozinov, Email: bozinovd@niaid.nih.gov, Genomics Unit, Research Technologies Section, Research Technologies Branch, Rocky Mountain Laboratories, NIH, 904 South 4th Street, Hamilton, MT 59840, USA, Tel.: +1 406 375 7444, Fax: +1 406 363 9415.
Olajumoke M Ogundare, Email: ogundarem@niaid.nih.gov, Genomics Unit, Research Technologies Section, Research Technologies Branch, Rocky Mountain Laboratories, NIH, 904 South 4th Street, Hamilton, MT 59840, USA, Tel.: +1 406 375 9744, Fax: +1 406 363 9415.
Nina Castro, Email: castroni@niaid.nih.gov, Genomics Unit, Research Technologies Section, Research Technologies Branch, Rocky Mountain Laboratories, NIH, 904 South 4th Street, Hamilton, MT 59840, USA, Tel.: +1 406 375 9744, Fax: +1 406 363 9415.
Kishore Kanakabandi, Email: kanakabandik@niaid.nih.gov, Genomics Unit, Research Technologies Section, Research Technologies Branch, Rocky Mountain Laboratories, NIH, 904 South 4th Street, Hamilton, MT 59840, USA, Tel.: +1 406 375 9744, Fax: +1 406 363 9415.
Paul A Beare, Email: pbeare@niaid.nih.gov, Coxiella Pathogenesis Section, Laboratory of Intracellular Parasites, Rocky Mountain Laboratories, NIH, 904 South 4th Street, Hamilton, MT 59840, USA, Tel.: +1 406 375 9671, Fax: +1 406 375 9380.
Anders Omsland, Email: omslanda@niaid.nih.gov, Coxiella Pathogenesis Section, Laboratory of Intracellular Parasites, Rocky Mountain Laboratories, NIH, 904 South 4th Street, Hamilton, MT 59840, USA, Tel.: +1 406 375 9656, Fax: +1 406 375 9380.
John H Carlson, Email: jcarlson@niaid.nih.gov, Laboratory of Intracellular Parasites, Rocky Mountain Laboratories, NIH, 904 South 4th Street, Hamilton, MT 59840, USA, Tel.: +1 406 363 9296, Fax: +1 406 375 9380.
Adam D Kennedy, Email: kennedyadam@niaid.nih.gov, Laboratory of Human Bacterial Pathogenesis, Rocky Mountain Laboratories, NIH, 904 South 4th Street, Hamilton, MT 59840, USA, Tel.: +1 406 363 9439, Fax: +1 406 375 9394.
Robert A Heinzen, Email: rheinzen@niaid.nih.gov, Coxiella Pathogenesis Section, Laboratory of Intracellular Parasites, Rocky Mountain Laboratories, NIH, 904 South 4th Street, Hamilton, MT 59840, USA, Tel.: +1 406 375 9695, Fax: +1 406 375 9380.
Jean Celli, Email: jcelli@niaid.nih.gov, Tularemia Pathogenesis Section, Laboratory of Intracellular Parasites, Rocky Mountain Laboratories, NIH, 904 South 4th Street, Hamilton, MT 59840, USA, Tel.: +1 406 375 9713, Fax: +1 406 375 9640.
David E Greenberg, Email: degreenberg@niaid.nih.gov, Immunopathogenesis Section, Laboratory of Clinical Infectious Diseases, National Institute of Allergy and Infectious Diseases, NIH, 33 North Dr., Room 2W10A.3, Bethesda, MD 20892, USA, Tel.: +1 301 402 6923, Fax: +1 310 480 4506.
Frank R Deleo, Email: fdeleo@niaid.nih.gov, Laboratory of Human Bacterial Pathogenesis, Rocky Mountain Laboratories, NIH, 904 South 4th Street, Hamilton, MT 59840, USA, Tel.: +1 406 363 9448, Fax: +1 406 375 9394.
Stephen F Porcella, Email: sporcella@niaid.nih.gov, Genomics Unit, Research Technologies Section, Research Technologies Branch, Rocky Mountain Laboratories, NIH, 904 South 4th Street, Hamilton, MT 59840, USA, Tel.: +1 406 363 9271, Fax: +1 406 363 9415.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.La MV, Raoult D, Renesto P. Regulation of whole bacterial pathogen transcription within infected hosts. FEMS Microbiol Rev. 2008;32(3):440–460. doi: 10.1111/j.1574-6976.2008.00103.x. [DOI] [PubMed] [Google Scholar]
- 2.Voth DE, Heinzen RA. Coxiella type IV secretion and cellular microbiology. Curr Opin Microbiol. 2009;12 (1):74–80. doi: 10.1016/j.mib.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Omsland A, Cockrell DC, Fischer ER, Heinzen RA. Sustained axenic metabolic activity by the obligate intracellular bacterium Coxiella burnetii. J Bacteriol. 2008;190(9):3203–3212. doi: 10.1128/JB.01911-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4▪▪.Omsland A, Cockrell DC, Howe D, et al. Host cell-free growth of the Q fever bacterium Coxiella burnetii. Proc Natl Acad Sci USA. 2009;106(11):4430–4434. doi: 10.1073/pnas.0812074106. Presents a systematic approach to developing media that supports axenic growth of obligate intracellular pathogens. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5▪.Hautefort I, Thompson A, Eriksson-Ygberg S, et al. During infection of epithelial cells Salmonella enterica serovar Typhimurium undergoes a time-dependent transcriptional adaptation that results in simultaneous expression of three type 3 secretion systems. Cell Microbiol. 2008;10(4):958–984. doi: 10.1111/j.1462-5822.2007.01099.x. Salmonella transcriptome paper providing comparative analysis of pathogen transcriptomes from two different target cell infections. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6▪▪.Belland RJ, Zhong G, Crane DD. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc Natl Acad Sci USA. 2003;100(14):8478–8483. doi: 10.1073/pnas.1331135100. Early, landmark paper for transcriptional analysis of chlamydial intracellular lifecycle. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mäurer AP, Mehlitz A, Mollenkopf HJ, Meyer TF. Gene expression profiles of Chlamydophila pneumoniae during the developmental cycle and iron depletion-mediated persistence. PLoS Pathog. 2007;3(6):e83. doi: 10.1371/journal.ppat.0030083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8▪▪.Wehrly TD, Chong A, Virtaneva K, et al. Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages. Cell Microbiol. 2009;(7):1128–1150. doi: 10.1111/j.1462-5822.2009.01316.x. Well-designed, very comprehensive transcriptome profile and mutational analysis of Francisella intracellular lifecycle. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chong A, Wehrly TD, Nair V. The early phagosomal stage of Francisella tularensis determines optimal phagosomal escape and Francisella pathogenicity island expression. Infect Immun. 2008;76(12):5488–5499. doi: 10.1128/IAI.00682-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10▪.Yang H, Harrington CA, Vartanian K, Coldren CD, Hall R, Churchill GA. Randomization in laboratory procedure is key to obtaining reproducible microarray results. PLoS ONE. 2008;3(11):e3724. doi: 10.1371/journal.pone.0003724. Importance of randomization in Affymetrix chip processing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kerr MK. Design considerations for efficient and effective microarray studies. Biometrics. 2003;59(4):822–828. doi: 10.1111/j.0006-341x.2003.00096.x. [DOI] [PubMed] [Google Scholar]
- 12.Hsu JC, Chang J, Wang T, Steingrímsson E, Magnússon MK, Bergsteinsdottir K. Statistically designing microarrays and microarray experiments to enhance sensitivity and specificity. Brief Bioinform. 2007;8(1):22–31. doi: 10.1093/bib/bbl023. [DOI] [PubMed] [Google Scholar]
- 13.Verdugo RA, Deschepper CF, Muñoz G, Pomp D, Churchill GA. Importance of randomization in microarray experimental designs with Illumina platforms. Nucleic Acids Res. 2009;37(17):5610–5618. doi: 10.1093/nar/gkp573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Motley ST, Morrow BJ, Liu X, et al. Simultaneous analysis of host and pathogen interactions during an in vivo infection reveals local induction of host acute phase response proteins, a novel bacterial stress response, and evidence of a host-imposed metal ion limited environment. Cell Microbiol. 2004;6(9):849–865. doi: 10.1111/j.1462-5822.2004.00407.x. [DOI] [PubMed] [Google Scholar]
- 15.Lovegrove FE, Peña-Castillo L, Mohammad N, Liles WC, Hughes TR, Kain KC. Simultaneous host and parasite expression profiling identifies tissue-specific transcriptional programs associated with susceptibility or resistance to experimental cerebral malaria. BMC Genomics. 2006;7:295. doi: 10.1186/1471-2164-7-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16▪▪.Virtaneva K, Porcella SF, Graham MR, et al. Longitudinal analysis of the Group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc Natl Acad Sci USA. 2005;102(25):9014–9019. doi: 10.1073/pnas.0503671102. Well-designed, pathogen transcriptome profile of streptococcal infection, over time, in non-human primates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Virtaneva K, Graham MR, Porcella SF, et al. Group A Streptococccus gene expression in humans and cynomolgus macaques with acute pharyngitis. Infect Immun. 2003;71(4):2199–2207. doi: 10.1128/IAI.71.4.2199-2207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu L, Liu X, Fields MW, et al. Microarray-based whole-genome hybridization as a tool for determining procaryotic species relatedness. ISME J. 2008;2(6):642–655. doi: 10.1038/ismej.2008.23. [DOI] [PubMed] [Google Scholar]
- 19▪.Beare PA, Samuel JE, Howe D, Virtaneva K, Porcella SF, Heinzen RA. Genetic diversity of the Q fever agent, Coxiella burnetii, assessed by microarray-based whole-genome comparisons. J Bacteriol. 2006;188(7):2309–2324. doi: 10.1128/JB.188.7.2309-2324.2006. Thorough introduction to comparative genome hybridization technology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20▪.Greenberg DE, Porcella SF, Zelazny AM, et al. Genome sequence analysis of the emerging human pathogenic acetic acid bacterium Granulibacter bethesdensis. J Bacteriol. 2007;189:8727–8736. doi: 10.1128/JB.00793-07. Of interest in terms of rapid genome analysis and genome-level typing of a novel, newly emergent pathogen. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21▪.Albert TJ, Dailidiene D, Dailide G, et al. Mutation discovery in bacterial genomes: metronidazole resistance in Helicobacter pylori. Nat Methods. 2005;2(12):951–953. doi: 10.1038/nmeth805. Landmark paper describing Nimblegen pathogen comparative genome sequencing. [DOI] [PubMed] [Google Scholar]
- 22▪.Herring CD, Palsson BØ. An evaluation of comparative genome sequencing (CGS) by comparing two previously-sequenced bacterial genomes. BMC Genomics. 2007;(8):274. doi: 10.1186/1471-2164-8-274. Important paper that describes the accuracy and errors in Nimblegen array sequencing as compared with Sanger-based genome sequencing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23▪▪.Kennedy AD, Otto M, Braughton KR, et al. Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification. Proc Natl Acad Sci USA. 2008;105(4):1327–1332. doi: 10.1073/pnas.0710217105. Important paper that describes the use of comparative genome sequencing technology and the important findings generated that relate to epidemic methicillin-resistant Staphylococcus aureus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sumby P, Porcella SF, Madrigal AG, et al. Evolutionary origin and emergence of a highly successful clone of serotype M1 Group A Streptococcus involved multiple horizontal gene transfer events. J Infect Dis. 2005;192(5):771–782. doi: 10.1086/432514. [DOI] [PubMed] [Google Scholar]
- 25.Beres SB, Richter EW, Nagiec MJ, et al. Molecular genetic anatomy of inter- and intraserotype variation in the human bacterial pathogen Group A Streptococcus. Proc Natl Acad Sci USA. 2006;103(18):7059–7064. doi: 10.1073/pnas.0510279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kari L, Whitmire WM, Carlson JH, et al. Pathogenic diversity among Chlamydia trachomatis ocular strains in nonhuman primates is affected by subtle genomic variations. J Infect Dis. 2008;197(3):449–456. doi: 10.1086/525285. [DOI] [PubMed] [Google Scholar]
- 27.Ellison DW, Clark TR, Sturdevant DE, Virtaneva K, Porcella SF, Hackstadt T. Genomic comparison of virulent Rickettsia rickettsii Sheila Smith and avirulent Rickettsia rickettsii Iowa. Infect Immun. 2007;6:542–550. doi: 10.1128/IAI.00952-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28▪.Smoot JC, Barbian KD, Van Gompel JJ, et al. Genome sequence and comparative microarray analysis of serotype M18 Group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc Natl Acad Sci USA. 2002;99(7):4668–4673. doi: 10.1073/pnas.062526099. Thorough, highly cited, streptococcal genome and comparative microarray paper. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beres SB, Sylva GL, Barbian KD, et al. Genome sequence of a serotype M3 strain of Group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc Natl Acad Sci USA. 2002;99(15):10078–10083. doi: 10.1073/pnas.152298499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banks DJ, Porcella SF, Barbian KD, et al. Progress toward characterization of the Group A Streptococcus metagenome: complete genome sequence of a macrolide-resistant serotype M6 strain. J Infect Dis. 2004;190(4):727–738. doi: 10.1086/422697. [DOI] [PubMed] [Google Scholar]
- 31.Green NM, Zhang S, Porcella SF, et al. Genome sequence of a serotype M28 strain of Group A Streptococcus: potential new insights into puerperal sepsis and bacterial disease specificity. J Infect Dis. 2005;192(5):760–770. doi: 10.1086/430618. [DOI] [PubMed] [Google Scholar]
- 32.Carlson JH, Porcella SF, McClarty G, Caldwell HD. Comparative genomic analysis of Chlamydia trachomatis oculotropic and genitotropic strains. Infect Immun. 2005;73(10):6407–6418. doi: 10.1128/IAI.73.10.6407-6418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beres SB, Sesso R, Wyton S, et al. Genome sequence of a Lancefield Group C Streptococcus zooepidemicus strain causing epidemic nephritis: new information about an old disease. PLoS ONE. 2008;3(8):e3026. doi: 10.1371/journal.pone.0003026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seshadri R, Paulsen IT, Eisen JA, et al. Complete genome sequences of the Q-fever pathogen Coxiella burnetii. Proc Natl Acad Sci USA. 2003;100(9):5455–5460. doi: 10.1073/pnas.0931379100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35▪.Beare PA, Unsworth N, Andoh M, et al. Comparative genomics reveal extensive transposon-mediated genomic plasticity and diversity among potential effector proteins within the genus. Coxiella Infect Immun. 2008;77(2):642–656. doi: 10.1128/IAI.01141-08. Detailed, Sanger-based, multigenome sequence analysis of an important intracellular pathogen. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carlson JH, Porcella SF, McClarty G, Caldwell HD. Comparative genomic analysis of Chlamydia trachomatis oculotropic and genitotropic strains. Infect Immun. 2005;73(10):6407–6418. doi: 10.1128/IAI.73.10.6407-6418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seth-Smith HM, Harris SR, Persson K, et al. Co-evolution of genomes and plasmids within Chlamydia trachomatis and the emergence in Sweden of a new variant strain. BMC Genomics. 2009;10:239. doi: 10.1186/1471-2164-10-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carlson JH, Whitmire WM, Crane DD, et al. The Chlamydia trachomatis plasmid is a transcriptional regulator of chromosomal genes and a virulence factor. Infect Immun. 2008;76(6):2273–2283. doi: 10.1128/IAI.00102-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hall N. Advanced sequencing technologies and their wider impact in microbiology. J Exp Biol. 2007;210(9):1518–1525. doi: 10.1242/jeb.001370. [DOI] [PubMed] [Google Scholar]
- 40.Calderon-Copete SP, Wigger G, Wunderlin C, et al. The Mycoplasma conjunctivae genome sequencing, annotation and analysis. BMC Bioinformatics. 2009;10(Suppl 6):S7. doi: 10.1186/1471-2105-10-S6-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Audic S, Lescot M, Claverie JM, Scholz HC. Brucella microti: the genome sequence of an emerging pathogen. BMC Genomics. 2009;10(1):352. doi: 10.1186/1471-2164-10-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baltrus DA, Amieva MR, Covacci A, et al. The complete genome sequence of Helicobacter pylori strain G27. J Bacteriol. 2009;191(1):447–448. doi: 10.1128/JB.01416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43▪.Reinhardt JA, Baltrus DA, Nishimura MT, Jeck WR, Jones CD, Dangl JL. De novo assembly using low-coverage short read sequence data from the rice pathogen Pseudomonas syringae pv. Oryzae Genome Res. 2009;19(2):294–305. doi: 10.1101/gr.083311.108. Important paper that describes a process/pipeline for Solexa and 454 sequencing and de novo assembly of a pathogen genome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Foster JT, Beckstrom-Sternberg SM, Pearson T, et al. Whole-genome-based phylogeny and divergence of the genus Brucella. J Bacteriol. 2009;191(8):2864–2870. doi: 10.1128/JB.01581-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45▪.Holt KE, Parkhill J, Mazzoni CJ, et al. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat Genet. 2008;40(8):987–993. doi: 10.1038/ng.195. Thorough paper describing the use of 454 technology for high-throughput sequencing of bacterial pathogen genomes. [DOI] [PMC free article] [PubMed] [Google Scholar]