Abstract
Efficient delivery of siRNA to specific cell populations in vivo remains a formidable challenge to its successful therapeutic application. We describe a novel siRNA-based approach – synthetically linking siRNA to an oligonucleotide TLR9 agonist – that targets and silences genes in TLR9+ myeloid cells and B cells, both of which are key components of the tumor microenvironment. Because Stat3 in tumor-associated immune cells suppresses antitumor immune responses and hinders TLR9-induced immune stimulation, we tested CpG-Stat3siRNA conjugates for anti-tumor effects. When injected locally at the tumor site or systemically through an intravenous route, the CpG-Stat3siRNA conjugates access tumor-associated dendritic cells, macrophages and B cells, inhibit Stat3 expression, leading to activation of tumor-associated immune cells, and ultimately potent anti-tumor immune responses. Our findings demonstrate the potential of TLR agonist-siRNA conjugates for targeted gene silencing coupled with TLR stimulation and immune activation in the tumor microenvironment.
RNA interference provides compelling opportunities to control gene expression in cells and siRNAs therefore represent a family of new drugs with broad potential for the treatment of diverse human diseases. Several recent studies have demonstrated the feasibility of in vivo siRNA delivery, leading to therapeutic effects in mouse models1,2,3,4 and also in non-human-primates5,6. Nevertheless, efficient in vivo targeted delivery of siRNA into specific cell types, especially those of immune origin, which are important constituents of the tumor microenvironment and active players in promoting tumor progression, remains to be fully explored. One promising approach for targeted delivery of siRNA is the use of aptamers, which are oligonucleotide-based ligands that bind to specific receptors, such as those on tumor cells2.
The immune system can serve as extrinsic tumor suppressor7,8,9. However, the tumor microenvironment is characterized by lack of tumor-specific CD8+ T cells and an excess of regulatory T cells and myeloid-derived suppressor cells (MDSC) that promote tumor immune evasion10,11. Myeloid cells and other immune cells in the tumor microenvironment also produce growth factors and angiogenic/metastatic factors critical for tumor progression12. The orchestration of these processes in the tumor microenvironment appears to be highly dependent on the oncogenic transcription factor, Stat313,14,15,16,17. In particular, we and others have recently demonstrated a role of Stat3 in mediating tumor immune evasion18,10,17. Activated Stat3 in myeloid cells inhibits expression of a large number of immunostimulatory molecules related to Th1-type responses, while promoting production of several key immunosuppressive factors17,19,20 as well as angiogenic factors12. In addition, by mediating signaling of certain cytokines and growth factors, notably IL-6, Stat3 activation in myeloid cells activates Stat3 in tumor cells, enhancing tumor cell proliferation and survival21–24. Because Stat3 also restrains TLR-mediated Th1 immune responses10,25,17, we reasoned that simultaneously silencing Stat3 by siRNA and triggering TLRs by their agonists could effectively shift the tumor microenvironment from pro-oncogenic to anti-tumor. A recent study using polymer-mediated in vivo transfection of 5′-triphophate-Bcl2 siRNA has demonstrated the benefits of simultaneously inducing antitumor immunity and silencing a pro-oncogenic gene4.
In this study, we explored a strategy of linking siRNAs to synthetic oligonucleotide agonists for endosomal TLRs, which include TLR3, TLR7, TLR8 and TLR926,27,28, for targeted delivery of siRNA into immune cells, together with TLR-dependent activation of antitumor immune responses. The endosomal location of the oligonucleotide-binding TLRs might be advantageous in facilitating the siRNA component to reach the cytosol for efficient gene silencing in cells selectively expressing the cognate TLR. In order to model this concept, we chose TLR9-specific oligodeoxynucleotides containing an unmethylated CpG-motif (CpG ODN), because they are already in clinical testing29. Additionally, CpG ODNs are efficiently internalized by various antigen-presenting cells, such as dendritic cells (DCs), macrophages and B cells, and their binding to TLR9 can initiate a cascade of innate and adaptive immune responses30,28,29. Myeloid cells and B cells are also critical components of the tumor microenvironment that actively promote oncogenesis12,17,19,21,22. By linking the single-stranded CpG ODN with double-stranded siRNA, we have created a single synthetic molecule capable of delivering siRNA into myeloid and B cells, silencing an immune checkpoint and/or oncogenic gene, and activating TLR, leading to therapeutic antitumor immune responses.
RESULTS
Construction of the CpG-Stat3 siRNA conjugate molecule
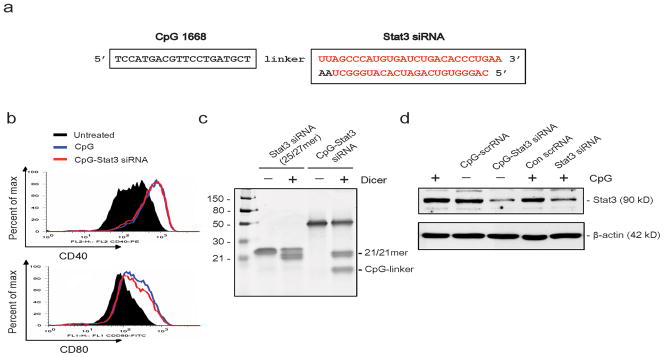
Synthesis of the antisense strand of the siRNA (27mer) was followed by CpG1668 ODN synthesis31,32, producing a single stranded oligonucleotide connected through a carbon chain linker (Fig. 1a). A 25/27mer siRNA was chosen over the conventional 21mer duplex to allow uncoupling of the siRNA from the CpG sequence by the Dicer enzyme once inside the cell. The asymmetric 25/27mer siRNA was optimized for specific processing by Dicer and was more potent in target gene silencing33,34. Adding either CpG1688 alone or CpG-Stat3 siRNA conjugate to cultured DC2.4 dendritic cells resulted in a similar increase in expression of co-stimulatory CD40 and CD80 molecules, suggesting that CpG-Stat3 siRNA conjugate retains its capacity to activate TLR9 (Fig. 1b). In addition, the immunostimulatory properties of CpG-siRNA conjugates do not differ from the effect of CpG alone as measured by production of inflammatory cytokines in primary cells and NF-κB/AP1 activation in a stable macrophage cell line (Supplementary Fig. 1). To assess whether linking siRNA with CpG moiety would still allow Dicer processing, we compared in vitro Dicer activity on CpG-Stat3 siRNA substrate versus 25/27mer Stat3 siRNA. Both the CpG-Stat3 siRNA and Stat3 siRNA were processed to 21mer siRNA by recombinant Dicer (Fig. 1c). Finally, to determine whether the CpG-Stat3 siRNA retains gene silencing function, the chimeric molecule was transfected into cells using Lipofectamine. Results from this experiment indicated that linking CpG ODN to siRNA did not interfere with Stat3 gene silencing (55% and 49% at protein level, respectively, as measured by densitometry) (Fig. 1d).
Figure 1.
Structure and function of the CpG-Stat3 siRNA conjugate. (a) Sequence of the CpG-linked mouse Stat3 siRNA conjugate (CpG1668-Stat3 siRNA): CpG1668 sequence (deoxyribonucleotides shown in black) were phosphothioated and connected through a carbon linker (6 of C3 units) to the antisense strand of Stat3 siRNA (ribonucleotides shown in red). (b) CpG-siRNA has similar immunostimulatory activity compared to uncoupled CpG ODN, as indicated by increased expression of costimulatory molecules, CD40 and CD80, on primary splenic DCs after 24 h incubation with or without ODNs; splenocytes were pooled from 2–3 mice and the experiment was done twice with similar results. (c) Linked CpG-Stat3 siRNA is processed to active 21mer siRNA by recombinant Dicer in vitro. Processing of conjugated CpG-Stat3 siRNA molecules and Stat3 siRNA were visualized on polyacrylamide gel through SYBRGold staining; position of the 21/21mer and the remaining part of the molecule (CpG plus carbon linker) are indicated. Full-length gel is presented in Supplementary Figure 13a. (d) Stat3 siRNA linked to CpG ODN retains the ability to mediate RNA interference. B16 cells were transfected, using Lipofectamine, with CpG-linked dsRNAs or unconjugated dsRNAs plus15 nM CpG ODN. Stat3 gene silencing effects were evaluated by Western blot analysis. Full-length blots are presented in Supplementary Figure 13b.
In vitro uptake and gene silencing effects of the CpG-siRNA conjugate molecules
To determine the specificity and efficiency of CpG-siRNA uptake, freshly prepared mouse splenocytes were incubated with CpG-linked Stat3 siRNA or an unconjugated Stat3 siRNA, in the absence of any transfection reagent(s). Both the CpG-Stat3 siRNA and unconjugated Stat3 siRNA were labeled with fluorescein isothiocyanate (FITC). Fluorescein-positive DCs, macrophages, B cells, granulocytes and T cells were assessed by flow cytometric analysis, which indicated that the chimeric CpG-Stat3 siRNA was efficiently taken up by both plasmacytoid (CD11c+B220+) and conventional (CD11c+B220−) splenic DCs, macrophages (F4/80+Gr1−) and B cells (B220+CD11c−), but only minimally by splenic granulocytes (Gr1+F4/80−) or T cells (CD3+) (Fig. 2a, Supplementary Fig. 2 and Supplementary Table 1). This uptake pattern reflects the known distribution of TLR9 expression in murine leukocyte subsets35,26. Intracellular staining of TLR9 in fixed splenocytes by flow cytometry CD11c+ DCs confirmed TLR9 expression (Fig. 2a, bottom right). Unconjugated Stat3 siRNA was not efficiently incorporated into DCs even after 24 h incubation, demonstrating that linkage to the TLR9 ligand facilitates siRNA uptake (Fig. 2a, bottom left).
Figure 2.
Uptake and gene silencing by CpG-Stat3 siRNA in vitro. (a) Targeted delivery: splenocytes were incubated for 3 h with CpG-Stat3 siRNA or for 24 h with unconjugated Stat3 siRNA labeled with fluorescein (bottom left panel), without any transfection reagents. Percentage of fluorescein-positive CD11c+B220− non-plasmacytoid (mDCs) and CD11c+B220+ plasmacytoid (pDCs) DCs, F4/80+Gr1− macrophages (MACs), B220+CD11c− B cells, Gr1+F4/80− granulocytes and CD3+ T cells was assessed by flow cytometric analysis (see also Supplementary Table 1). Splenic CD11c+ DCs express high levels of TLR9. Intracellular staining of TLR9 as shown in fixed splenic DCs by flow cytometry (bottom right panel). Similar results were obtained in two independent experiments. (b) Kinetics of CpG-siRNA internalization: CpG-Stat3 siRNA-FITC is quickly internalized by DCs without transfection reagents. The uptake by DC2.4 cells is analyzed by flow cytometry (top row) and confocal microscopy (two lower rows) after incubation with CpG-Stat3 siRNA-FITC (500 nM) for indicated times (two upper rows) or after 1 h incubation at indicated concentrations (bottom row); results are representative of 3 independent experiments; scale bar = 10 μm. (c) Internalized CpG-Stat3 siRNA colocalizes with TLR9 (two upper rows) and transiently interacts with Dicer (two lower rows). DC2.4 cells were incubated with 500 nM of CpG-Stat3 siRNA for indicated times. Shown are confocal microscopy images; green – CpG-Stat3 siRNA-FITC, red – immunofluorescent detection of endogenous TLR9 or Dicer, blue – nuclear staining with DAPI; scale bar = 10 μm. All confocal imaging studies were performed at least 3 times with similar results and the images acquired were characteristic for the majority of analyzed cells (Supplementary Fig. 4). (d) Dose-dependent gene silencing effects of CpG-Stat3 siRNA, comparing to GpC-Stat3 siRNA at the highest dose, as determined by quantitative real-time PCR in DC2.4 cells. Shown are the results of real-time PCR for Stat3, normalized to GAPDH expression levels. The level of Stat3 expression in CpG-scrambled RNA sample is set as 100%. Shown are means ± SEM from 3 independent experiments analyzed in duplicates. (e) Stat3 silencing is impaired in TLR9-deficient primary myeloid cells (top panel) and DCs (bottom panel). Shown are results of two independent experiments, analyzed in triplicates by real-time PCR; means ± SEM. (f) Stat3 DNA-binding is reduced following 48 h incubation of CpG-Stat3 siRNA, relative to CpG-scrambled RNA, in DC2.4 cells. Shown are results of electrophoretic mobility gel-shift assay using radiolabeled probe specifically bound by Stat3 and Stat1, data representing three independent experiments. Positions of Stat dimers are indicated. Full-length gel is presented in Supplementary Figure 13c.
We further evaluated CpG-Stat3 siRNA-FITC uptake by DC 2.4 mouse DCs. Flow cytometry and fluorescent microscopy indicated that without transfection reagents, the CpG-Stat3 siRNA-FITC was internalized by DC 2.4 cells, with kinetics similar to that CpG-ODN alone (Fig. 2b - two top rows and Supplementary Fig. 3) and reported previously36. By 60 min, greater than 80% DC 2.4 cells were positive for uptake of the conjugate, which was confirmed by confocal microscopic analysis. The uptake of the CpG-Stat3 siRNA-FITC was dose dependent (Fig. 2b, bottom row).
Confocal microscopic analyses further showed that at one hour after incubation, the CpG-Stat3 siRNA colocalized with TLR9 within perinuclear endocytic vesicles (Fig. 2c, two top rows; and Supplementary Fig. 4). This colocalization diminished at 2 and 4 h after CpG-siRNA treatment (Fig. 2c, two top rows. Previous studies have demonstrated that binding of the Dicer nuclease to the siRNA oligonucleotide is required for further siRNA processing to shorter 21mer fragments that mediate RISC complex-dependent mRNA degradation37. We observed transient colocalization of the CpG-Stat3 siRNA with Dicer within 2 h after adding the oligonucleotide to cultured DCs (Fig. 2c, two bottom rows and Supplementary Fig. 4). The colocalization of CpG-Stat3-siRNA and Dicer became weaker by 4 h (Fig. 2c) and undetectable after 24 h (data not shown).
Gene silencing effects of the CpG-Stat3 siRNA in DC2.4 cells were determined by quantitative real-time PCR analysis of the Stat3 mRNA (Fig. 2d). Maximum effect on Stat3 silencing was observed at relatively high concentration of CpG-Stat3 siRNA (1 μM) in serum-containing cell culture medium. GpC-conjugated Stat3 siRNA, which binds but does not activate TLR936, failed to silence Stat3, suggesting a possible requirement of TLR9 activation for the CpG-siRNA to be processed. Further experiments demonstrated that in TLR9−/− myeloid cells and DCs, while cellular uptake of CpG-Stat3 siRNA was normal (Supplementary Fig. 5), the gene silencing effect of CpG-siRNA was impaired (Fig 2e). We also confirmed the gene silencing effects using electrophoretic mobility shift assays (EMSA) to detect Stat3 DNA-binding activity in DC2.4 cells, which was induced by IL-10 stimulation (Fig. 2f). Note that, as indicated above, stimulation using CpG itself also resulted in Stat3 activation25,38 (Fig. 2f), thereby complicating the EMSA analysis for detection of Stat3 silencing. None-the-less, at higher concentrations, CpG-Stat3 siRNA diminished Stat3 DNA-binding activity relative to the conjugate scrambled RNA controls. To demonstrate the generality of this approach to gene silencing, we used TLR9-positive mouse A20 B cell lymphoma cells to test CpG-siRNA internalization (Supplementary Fig. 6a), and gene silencing, for which we tested a chimeric conjugate linking CpG1668 ODN with a Dicer substrate siRNA specific for firefly luciferase (Luc) in inhibiting luciferase activity in A20-Luc cells (Supplementary Fig. 6 b).
In vivo characterization of the CpG-Stat3siRNA conjugate molecule
Biodistribution experiments in naive tumor-free mice confirmed that CpG-Stat3 siRNA is specifically internalized by resident macrophages in different tissues as well as DCs and B cells in lymph nodes (Supplementary Fig. 7). We also assessed the uptake of the CpG-Stat3 siRNA by macrophages and DCs in tumor-bearing mice. C57BL/6 mice with B16 tumors were injected peritumorally with CpG-Stat3 siRNA (FITC) at 0.78 nmol (20 μg)/injection. Immunofluorescent staining showed myeloid cells positive for CpG-Stat3 siRNA accumulated at the injection site (Fig. 3a). Flow cytometry also indicated the presence of CpG-Stat3 siRNA in tumor-associated myeloid cells (Supplementary Fig. 8a). Furthermore, in vivo intravital two-photon microscopy showed the presence of FITC-positive cells in tumor draining lymph node (TDLN), as early as 1 h after injection of the labeled construct (Fig. 3b, Supplementary Fig. 8b and Supplementary Fig. 9), but not in the contralateral lymph nodes (Supplementary Fig. 9). Additionally, high resolution imaging by intravital two-photon microscopy revealed an increased number of FITC-positive cells in TDLNs, as well as an accumulation of FITC-labeled CpG-Stat3 siRNAs in perinuclear endocytic vesicles (Fig. 3b, bottom right), which was also observed in cultured DCs (Fig. 2c).
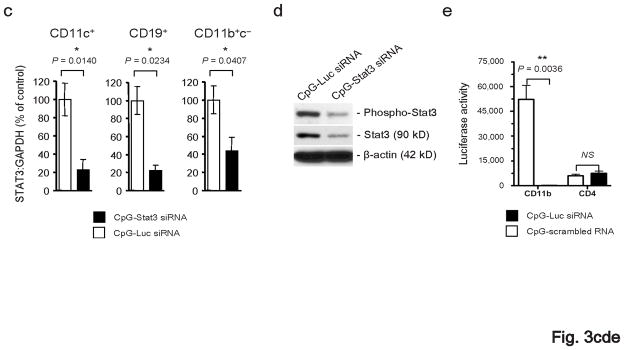
Figure 3.
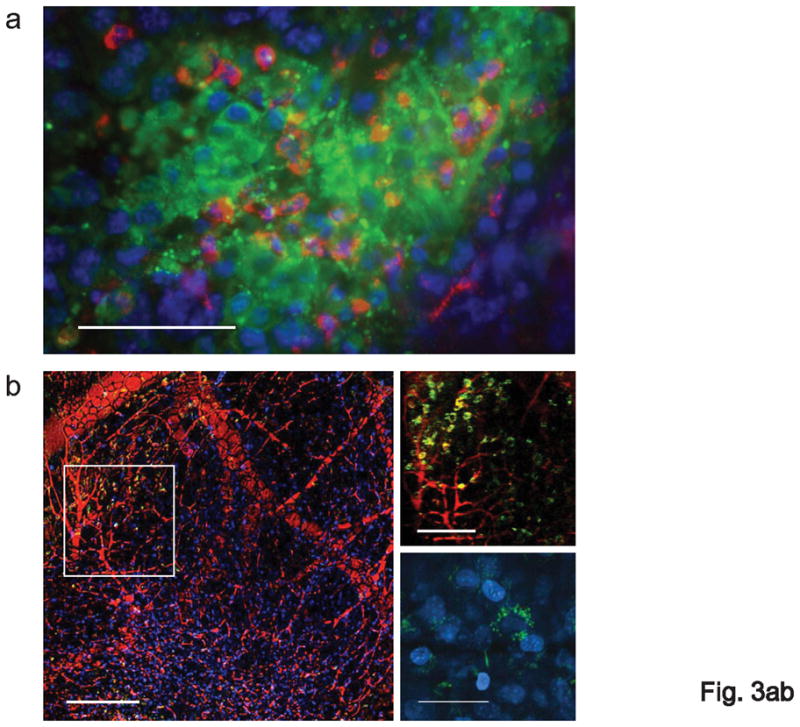
Treatment with CpG-Stat3 siRNA leads to cell-specific gene silencing in vivo. (a) In vivo uptake of intratumorally injected CpG-Stat3 siRNA by myeloid cells. Shown is immunofluorescent staining of frozen tumor tissue section 6 h after injection of CpG-siRNA conjugate. Green: FITC-labeled CpG-Stat3 siRNA; red: myeloid cells stained with anti-CD11b antibodies: blue: nuclear staining with Hoechst; scale bar = 50 μm. (b) Left panel - intravital two-photon microscopy imaging on TDLN at 1 h after intratumoral injection of FITC-labeled CpG-Stat3 siRNA (green); blood vessels stained with dextran-rhodamine (red); Hoechst-stained nuclei (blue); scale bar = 200 μm. Top right panel: close-up of the lymph node tissue to visualize FITC-positive cells entering the lymph node; scale bar = 100 μm; bottom right panel: intracellular distribution of FITC-labeled CpG-Stat3 siRNA; scale bar = 25 μm. Results represent two independent experiments. (c, d) Repeated local peritumoral treatment with CpG-Stat3 siRNA significantly reduces Stat3 mRNA and protein in immune cells within TDLNs. Total RNA and protein were isolated from CD11c+ DCs, CD19+ B cells and CD11b+c− myeloid cells accumulated in TDLNs (inguinal) using cells pooled from 4–9 mice. (c) Shown are quantitative real-time PCR results on Stat3 expression, combining 3–4 independent experiments ± SEM; control CpG-Luc siRNA was set as 100%. (d) Total and phospho-Stat3 levels are reduced by CpG-Stat3 siRNA conjugates. Representative results of Western blot analysis for tyrosine-phosphorylated or total Stat3 and β-actin from one of two independent experiments are shown. Full-length blots are presented in Supplementary Figure 13d. (e) Two weeks after B16 tumor challenge, luciferase-overexpressing mice were injected peritumorally with CpG-Luc siRNA or CpG-scrambled RNA every day for 3 d. The level of luciferase activity was assessed in CD11b+ and CD4+ cells isolated from TDLNs; shown are representative results from one of 3 independent experiments using 3 mice/group/experiment.
We next evaluated the gene silencing and antitumor effects of CpG-Stat3 siRNAs in vivo. Peritumoral injections of the CpG-Stat3 siRNA resulted in relatively effective gene silencing in DCs, macrophages and B cells in TDLNs, compared to control CpG-Luc siRNA, as measured by quantitative real-time PCR (Fig. 3c). Stat3 inhibition in CD11c+ DCs isolated from TDLNs was confirmed at protein level (Fig. 3d). Furthermore, quantitative real-time PCR and Western blotting indicated Stat3 silencing in the total TDLNs (Supplementary Fig. 10). We also used CpG-Luc siRNA conjugate to confirm that CpG-siRNA conjugates are able to reduce protein expression specifically within myeloid cells in vivo. Mice over-expressing firefly luciferase under control of the β-actin promoter39 were challenged with tumor cells, followed by repeated peritumoral injections of CpG-Luc siRNA. Results from these experiments indicated effective inhibition of luciferase activity in CD11b+ myeloid cells but not in CD4+ lymphocytes within TDLNs (Fig. 3e).
Both DCs and macrophages in the tumor microenvironment are known to promote immune tolerance11,17. Our previous work demonstrated that Stat3 is persistently-activated in myeloid cells in the tumor milieu and that genetic ablation of Stat3 in the myeloid compartment elicits potent antitumor immunity10. Furthermore, both CpG and LPS treatment activates Stat338,25,40, which acts as a negative feedback mechanism to constrain Th1 immune responses. Therefore, we assessed whether the CpG-Stat3-siRNA conjugates could reverse the immunosuppressive effects imposed by the tumor-microenvironment and at the same time allow effective antitumor immunity induced by TLR9 triggering. Local treatment with CpG-Stat3 siRNA oligonucleotides inhibited growth of subcutaneously growing B16 melanoma (3–5 mm in diameter at the initial treatment). In contrast, treatment with unconjugated CpG-ODN plus Stat3 siRNA, or CpG-scrambled RNA construct, or GpC-Stat3 siRNA had significantly less antitumor effects (Fig. 4a). The antitumor effects of CpG-Stat3 siRNA were confirmed by using two alternative Stat3 siRNA sequences (data not shown). To directly show that the antitumor effects induced by CpG-Stat3 siRNA were mainly mediated by immune cells, we performed in vivo experiments with antibody-mediated depletion of CD8/CD4 T cells and NK cells. Without CD8+ and CD4+ immune cell populations (possibly also the cross-priming CD11c+CD8+ DCs), the effects of CpG-Stat3 siRNA treatment were partially reduced (Fig. 4b). The therapeutic effects of CpG-Stat3 siRNA were abrogated in the absence of NK cells (Fig. 4b).
Figure 4.
Both local and systemic treatments with CpG-Stat3 siRNA inhibit tumor growth. (a) Mice with subcutaneous (s.c) B16 tumors were treated by peritumoral injections of CpG-Stat3 siRNA, GpC-Stat3 siRNA, CpG-scrambled RNA, combination of equimolar amounts of uncoupled CpG and Stat3 siRNA or PBS only every other day, starting 6 d after challenge with 1×105 B16 cells, n = 5–6. Statistically significant differences between CpG-Stat3 siRNA- and CpG-scrambled RNA-treated groups are indicated by asterisks. Similar results were obtained in 3 independent experiments. (b) Tumor growth inhibition by CpG-Stat3 siRNA involves NK cell- and T cell-mediated immunity. Mice with established B16 tumors were depleted of NK cell or CD4/CD8 lymphocytes prior to CpG-Stat3 siRNA treatment; shown are means ± SEM, P < 0.0001 (from two-way ANOVA test). (c, d) Local treatment with CpG-Stat3 siRNA reduces growth of C4 melanoma (c) and CT26 colon carcinoma (d). Tumor cells were injected (s.c.) into C3H or BALB/c mice, respectively. Mice with established tumors were treated by peritumoral injections of CpG-Stat3 siRNA, CpG-Luc siRNA, CpG alone or PBS every other day, starting 7d after challenge with 1×105 tumor cells. Statistically significant differences between the CpG-siRNA-treated groups are indicated by asterisks. (e) C57BL/6.CEA mice were challenged s.c. with 1×105 of MC38.CEA cells and treated as described above using CpG-Stat3 siRNA or CpG-Luc siRNA starting from d 11 after tumor challenge, P < 0.0001 by two-way ANOVA (n = 4 for each group). (f) Systemic treatment with CpG-Stat3 siRNA reduces Stat3 expression in DCs within TDLNs (cervical). Samples pooled from 6 mice/group were analyzed by real-time PCR. Shown is the average level of Stat3 expression in CpG-Stat3 siRNA-treated mice from one of two independent experiments analyzed in triplicates ± SEM, relative to control CpG-scrambled RNA, which was set as 100%. (g) Systemic treatment with CpG-Stat3 siRNA reduces the number of B16 tumor metastasis. Mice were i.v. injected with B16 cells and treated with CpG alone, CpG-Stat3 siRNA or CpG-scrambled RNA i.v. injections every other day starting from two days post tumor challenge. Lung colonies were enumerated 15 d later when control mice become moribund. Shown are mean numbers of colonies ± SEM (n = 7), P = 0.0054 by two-way ANOVA test. Representative photos of lung tissues excised from mice inoculated and treated as described above. The in vivo data are representative of two independent experiments.
We confirmed that local treatment with CpG-Stat3 siRNA can reduce growth of other tumors independently of their genetic background. CpG-Stat3 siRNA oligonucleotides inhibited growth of both a poorly immunogenic variant of K1735 melanoma, C441, and CT26 colon carcinomas in C3H and BALB/c mice, respectively (Fig. 4c,d). Furthermore, CpG-Stat3 siRNA treatment of the carcinoembryonic antigen (CEA) transgenic C57BL/6 mice bearing MC38 colon carcinomas expressing CEA led to tumor regression in some mice (Fig. 4e). To assess in vivo effects of the CpG-Stat3 siRNA on tumor cells, we stained B16 tumor tissues with fluorescent antibody specific to activated caspase-3. Data from these analysis showed that B16 tumors received CpG-Stat3 siRNA had undergone more extensive apoptosis relative to the other three treatment groups (Supplementary Fig. 11). We also investigated the possibility that CpG-Stat3 siRNA could be systemically delivered to achieve gene silencing and antitumor effects. Intravenous injection (i.v.) of CpG-Stat3 siRNA (0.78 nmol) reduced Stat3 expression in DCs within TDLNs relative to CpG-scrambled RNA (Fig. 4f). Systemic delivery of CpG-Stat3 siRNA to inhibit metastasis was tested in an experimental B16 lung metastasis model. Relatively small amounts of the oligonucleotide (< 1 mg/kg) were used for the systemic injection, which led to significant reduction in the number of lung metastasis (Fig. 4g). Lower antitumor effect due to CpG-scrambled RNA and CpG ODN alone was also observed. Thus, maximal antitumor effects required conjugation of the CpG TLR9 ligand with a functional Stat3 siRNA.
Modulation of the tumor immunologic milieu by the CpG-Stat3 siRNA
To further assess the role of immune modulation in the observed antitumor effects mediated by CpG-Stat3 siRNA conjugate treatment, we analyzed changes in Th1 cytokine/chemokines and co-stimulatory molecule expression by DCs in the TDLNs. Lack of Stat3 in DCs has been shown to upregulate expression of Th1 cytokines/chemokines10,20,42,43, which can be greatly amplified by CpG treatment25. As shown in Fig. 5a, injection of the CpG-Stat3 siRNA at tumor site resulted in upregulation of several Th1 cytokines and chemokines, which were shown to be upregulated by Stat3 ablation10,20,42,43. It has also been documented that DCs with low expression levels of co-stimulatory molecules mediate immune tolerance44, which is one of the proposed mechanisms for tumor immune evasion induced by Stat3 activation in tumor-associated DCs10. We therefore analyzed expression of co-stimulatory molecules by DCs enriched from TDLNs. Results from these analyses indicated that CpG-Stat3 siRNA reduced the number of the DCs with low expression of co-stimulatory molecules, including MHC class II, CD80 and CD40, which was accompanied by a modest increase in expression of these co-stimulatory molecules (Supplementary Fig. 12). Stat3 ablation in myeloid cells followed by local treatment has been shown to induce potent antitumor innate immune responses that involve neutrophils10. We therefore assessed whether CpG-Stat3 siRNA conjugate treatment could lead to neutrophil-associated tumor cell apoptosis. Co-staining B16 tumor tissue sections with antibodies specific to activated caspase-3 and neutrophils revealed that CpG-Stat3 siRNA treatment-induced massive tumor cell apoptosis (activated caspase 3-positive), which was associated with an increase in tumor-infiltrating neutrophils (Fig. 5b). An increase in neutrophils in tumor after CpG-Stat3 siRNA treatment was further confirmed by flow cytometry (Fig. 5c).
Figure 5.
In vivo administration of CpG-Stat3 siRNA induces innate immunity. (a) Immunostimulatory cytokine/chemokine gene expression was analyzed in DCs enriched from TDLN cell suspensions pooled from 4–10 mice, prepared after 3 peritumoral injections of CpG-Stat3 siRNA or CpG-Luc siRNA. Data from quantitative real-time PCRs run in triplicates were normalized to GAPDH expression. The averaged results from 4 independent in vivo experiments were combined and analyzed for statistical significance using unpaired t-test with unequal variance. Shown are mean values ± SEM; CpG-Luc siRNA was set as a baseline (100%). (b) Frozen sections of tumor tissues isolated from mice after indicated treatments were stained with antibodies specific to neutrophils (green) and activated caspase-3 (red) and analyzed by fluorescent microscopy. Shown are results representative of two independent experiments; scale bar = 200 μm. (c) Single cell suspensions prepared from tumors pooled from 3–6 mice were analyzed by flow cytometry for the presence of Gr1+CD11b− neutrophils. Shown are means ± SEM combined from three independent experiments.
The ratio of effector to regulatory T cells within the tumor microenvironment is considered indicative of the effect of adaptive immune responses on tumor progression and metastasis45. We investigated the numbers of tumor infiltrating T cell populations in subcutaneously growing B16 tumors treated locally for 2 weeks with CpG-Stat3 siRNA, CpG-scrambled RNA control or PBS only. Although CpG-Stat3 siRNA treatment did not induce significant changes in overall CD4+ T cell numbers within the tumors, as shown by flow cytometric analysis (Fig. 6a), the percentage of CD4+/FoxP3+ Treg cells within all CD4+ T cells dropped from approximately 60% to 25% after peritumoral injections of CpG-Stat3 siRNA (Fig. 6b). We observed an increase in the infiltration of total CD8+ T cells in the tumor stroma, although CpG-scrambled RNA control treatment also led to some recruitment of CD8+ T cells (Fig. 6c). These effects probably result from TLR9-mediated immunostimulation of tumor-infiltrating antigen presenting cells. Fluorescent immunostaining of frozen tumor tissues with anti-CD8 antibody confirmed data generated by the flow cytometric analysis that CpG-Stat3 siRNA treatment caused increased CD8+ T cell infiltration in tumors (data not shown). Activation of tumor antigen-specific CD8+ T cells is believed to be critical for immune-mediated antitumor effects. We therefore examined the ability of CpG-Stat3 siRNA treatment to generate CD8+ T cells specific for the B16 tumor antigen, TRP2. ELISPOT assays to determine IFNγ production by T cells isolated from TDLNs in response to recall stimulation with TRP2 peptide antigen indicated that in vivo CpG-Stat3 siRNA administration indeed induced antigen-specific CD8+ T cells (Fig. 6d).
Figure 6.
Targeting Stat3 using CpG-siRNA augments innate and adaptive antitumor immunity. Effects of in vivo CpG-siRNA treatment on immune cell populations within tumor. (a, b, c, d) Single cell suspensions prepared from tumors pooled from 3–6 mice/group were analyzed by flow cytometry for the presence of CD4+ (a), CD4+FoxP3+ (b) and CD8+ (c). Representative dot plots (left panels in b, c) and means ± SEM combining three independent experiments (right panels in b, c) are shown. (d) Local treatments with CpG-Stat3 siRNA generate tumor antigen-specific immune responses. IFNγ ELISPOT assays were performed using cell suspensions prepared from 4 pooled TDLNs per each treatment group; presented are the results from one of two independent experiments. Bars represent average numbers of TRP2-specific dots ± SEM from triplicate samples; P-values from one-way ANOVA test for statistical significance are indicated.
DISCUSSION
We have developed a new strategy for targeted siRNA delivery together with immune activation by covalently linking TLR oligonucleotide agonists to siRNAs. These conjugates encompass three activities in a single molecule: targeting to immune cells, which include DCs, macrophages, and B cells, TLR activation and immune checkpoint silencing. In addition to TLR9, several other intracellular TLRs, such as TLR3, TLR7 and TLR8, also recognize/interact with nucleic acids, suggesting a broad application of this approach using various ligands for these TLRs to deliver siRNAs into different immune cells. TLRs are important for stimulating DC maturation, antigen uptake and presentation, leading to CTL activation and CD4+ T helper cell differentiation. Therefore, TLR agonist-siRNA approaches can further stimulate desired immune responses for treating diseases such as cancer and infections. Although it has been established that binding to TLR9 is necessary for CpG-mediated immune activation, it remains to be fully explored how CpG ODN enter cells36. Our results indicated that cellular uptake of both CpG ODN and the CpG-siRNA constructs can occur in the absence of TLR9. However, TLR9, at least in mouse cells, is essential for CpG-siRNA-mediated gene silencing. While the exact underlying mechanism(s) remains to be determined, it is possible that triggering TLR9 could effect either endosomal release of CpG-siRNA into the cytoplasm, or/and its intracellular processing.
Although TLR9 is expressed in different types of mouse DCs, its expression is more selective in humans. Nevertheless, while the highest levels of constitutive TLR9 expression is observed on human plasmacytoid DCs and B cells, it is also expressed at lower levels on human monocytes and macrophages26. These immune cells can serve as antigen-presenting cells and induce innate, adaptive or humoral immunity27,29,46. Furthermore, it has been demonstrated recently that adding triphosphate to the 5′ of siRNA can potentiate the antitumor effects of siRNA by stimulating antitumor immune responses, likely through intracellular RNA sensors such as RIG-I or MDA-54. It is therefore conceivable to incorporate triphosphate to the CpG-siRNA to further amplify antitumor immunity. In addition, a critical role of tumor stromal macrophages and B cells in promoting tumor development has been well documented47,48. Importantly, Stat3 and several other molecules produced by the tumor myeloid population, and possibly tumor-associated B cells, are critical for tumor immunosuppression17, and Stat3 activity in the myeloid compartment (possibly B cells as well) promotes Stat3 activity in tumor cells and endothelial cells in the tumor, enhancing tumor cell growth/survival12,21,22,23. In addition to Stat3, other oncogenic molecules produced by the tumor myeloid/B cell compartment are also critical in promoting cancer growth and resistance to various therapies. Therefore, being able to target the tumor stromal myeloid cells/B cells through CpG-siRNA conjugate molecules is highly desirable for cancer therapies. In addition to normal immune cells, several types of tumor cells, including those of B cell origin, and some solid tumor cells, are also positive for TLR949. Our preliminary results suggested the feasibility of CpG-siRNA approach to silence genes in TLR9+ tumor cells. For example, treating human TLR9+ tumors in NOD/SCID/IL-2RγKO mice with CpG-Stat3 siRNA resulted in tumor cell apoptosis and tumor growth inhibition (Kortylewski and Yu, unpublished data). More experiments remain to be performed to optimize this approach for potential clinical use.
Our results indicated that the gene silencing effects by CpG-siRNA in cultured cells requires high concentrations of the conjugates and are suboptimal relative to in vivo treatment. Work is underway to determine the possible cause(s) of this difference, which might include serum-associated degradation of CpG-siRNAs or reduction of the overall silencing effect in rapidly dividing cell populations. It is possible that in vivo repeated treatments allow accumulative gene silencing effects, and the crosstalk between various cells in the tumor microenvironment could lead to secondary inhibitory effects on Stat3 activity23. The half-life of the constructs at present is limited. Although the CpG ODN in the construct is phosphothioated, which should resist serum degradation, the siRNA in the chimeric construct is unmodified and negatively charged. Chemically modifying the siRNA to prolong serum stability and to neutralize the negative charge of the siRNA to facilitate endosomal release may improve the efficacy of TLR agonist-siRNA approach. Although more studies are necessary to further understand the mechanism(s), and to improve CpG-siRNA gene silencing effects, our results raise the possibility to use oligonucleotide TLR agonists for siRNA delivery into tumor-associated myeloid cells and B cells, and possibly certain tumor cells, to inhibit expression of tumor-promoting/immunosuppressive molecules while activating TLR(s) for immune activation.
Methods
Cells
Murine B16 melanoma, CT26 colon carcinoma and A20 B cell lymphoma lines were purchased from American Type Culture Collection. Mouse dendritic DC2.4 cells were originally from Dr. Kenneth Rock (University of Massachusetts Medical School, MA). Highly metastatic clone of K1735 melanoma (C4) was kindly provided by Drs. S. Huang and J. Fidler of M. D. Anderson Cancer Center (Houston, TX). The stably transduced A20-Luc cell line was provided by Dr. Defu Zheng (City of Hope, Duarte, CA). The generation of transgenic C57BL/6.CEA mice and MC-38.CEA cell line was previously described50.
Oligonucleotide design and synthesis
The phosphothioated oligodeoxyribonucleotide(ODN) and antisense strands (AS) of siRNAs were linked using 6 units of C3 carbon chain linker, (CH2)3 (Glen Research, Sterling, VA). The resulting constructs were hybridized with complementary sense strands (SS) of siRNAs to create chimeric ODN-siRNA constructs used in the study (deoxynucleotides are shown underlined). Sequences of single stranded constructs are listed below; the target Stat3 sequence is found between bases 1898 and 1922 of the mouse Stat3 gene (accession number NM_213659).
Mouse Stat3 siRNA (SS)
5′ CAGGGUGUCAGAUCACAUGGGCUAA 3′
CpG1668-mouse Stat3 siRNA(AS)
5′ TCCATGACGTTCCTGATGCT-linker-UUAGCCCAUGUGAUCUGACACCCUGAA 3′
GpC-mouse Stat3 siRNA (AS)
5′ TCCATGAGCTTCCTGATGCT-linker-UUAGCCCAUGUGAUCUGACACCCUGAA 3′
Scrambled RNA (SS)
5′ UCCAAGUAGAUUCGACGGCGAAGTG 3′
CpG1668-scrambled RNA (AS)
5′ TCCATGACGTTCCTGATGCT-linker-CACUUCGCCGUCGAAUCUACUUGGAUU 3′
The sequence of firefly luciferase-specific 25/27mer siRNA (Luc1 R 25D/27), used for the CpG1668-Luc siRNA conjugate molecule, was published34. The correct formation of siRNA duplex was confirmed by in vitro Dicer cleavage assays. 0.5 μg of each ODN-siRNA construct was subjected to processing by 1U of Dicer (Ambion) in 37°C for 1.5 h, resolved with 15% polyacrylamide/7.5M urea gel and results of the dicing reaction were visualized with SYBR Gold staining (Invitrogen).
Quantitative real-time PCR
Total RNA was extracted from cultured or primary cells using RNeasy kit (Qiagen). After cDNA synthesis using iScript cDNA Synthesis kit (Bio-Rad), samples were analyzed using pairs of primers specific for Stat3, TNF, IL-6, IP-10, RANTES, p35/IL-12, p40/IL-12 and GAPDH mRNAs (SuperArray Bioscience Corporation). Sequence-specific amplification was detected by fluorescent signal of SYBR Green (Bio-Rad) by using Chromo4 Real-time PCR Detector (Bio-Rad).
Transient transfections
B16 cells were transiently transfected with 15 nM CpG-linked or uncoupled Stat3 siRNA and scrambled RNA using Lipofectamine 2000 (Invitrogen). 48 h later cells were lysed and used for Western blot analysis.
Electromobility shift assay (EMSA) and Western blot analysis
EMSA and Western blot analysis to detect Stat3 DNA-binding and protein expression were performed as described previously18. The protein levels of Stat3 detected by Western blotting were later quantified by densitometry using AlphaEase FC software (Alpha Innotech).
Luciferase reporter gene assay
A20-Luc cells incubated with CpG-RNAs for 48 h or primary cells isolated from TDLNs of Luc+ mice treated with CpG-Luc siRNA were lysed and luciferase activities were determined using the Luciferase Assay System (Promega) after normalization to the protein content of the sample.
In vivo experiments
Mouse care and experimental procedures were performed under pathogen-free conditions in accordance with established institutional guidance and approved protocols from Research Animal Care Committees of the City of Hope. For s.c. tumor challenge, we injected 1 × 105 B16, C4 or CT26 tumor cells into 7–8 weeks C57BL/6, C57BL/6.CEA, C3H or Balb/C mice, respectively. After tumors reached average size of ca. 5 mm, mice were injected peritumorally with 0.78 nmol of CpG1668 ODN alone, in combination with Stat3 siRNA or CpG/GpC ODN linked to various double stranded RNA (dsRNA) sequences described above. Tumor growth was monitored every other day. For the analysis of cellular and molecular mechanisms of CpG/GpC-dsRNAs effects, mice were killed after 2–3 weeks of treatment. For experiments on silencing of luciferase activity in vivo, Luc+ mice (originally kindly provided by Dr. Christopher H. Contag, Stanford University School of Medicine, CA) were challenged with tumor and treated with 3 daily injections of CpG-Luc siRNA or CpG-scrambled RNA. Lymph nodes as well as tumor specimens were harvested. For B16 lung metastasis model, mice were challenged with 0.5 × 105 B16 tumor cells (i.v.) and two d later, after tumors were established, were treated systemically with 0.78 nmol (ca. 1 mg/kg) of CpG1668 ODN or various CpG-dsRNAs. Intravenous injections were repeated every other day for two weeks. Lungs were harvested, fixed and the number of B16 tumor colonies was manually counted. The level of Stat3 silencing was assessed by real-time PCR in DCs isolated from tumor-draining inguinal (for s.c. tumor model) or cervical (for metastasis model) lymph nodes. For immune cell depletion, mice were pretreated with anti-CD8 plus anti-CD4 antibodies (clone 2.43 and GK1.5, respectively, depleting 99% and 98% of CD8 and CD4 cells, respectively) or anti-asialo-GM1 antibodies (Wako, depleting min. 79% of NK cells) before tumor inoculation then injected weekly.
Flow cytometry and ELISA
We prepared single-cell suspensions of spleen, lymph node or tumor tissues by mechanic dispersion followed by collagenase D/DNase I treatment as described before10. For extracellular staining of immune markers 1 × 106 of freshly prepared cells suspended in PBS/2% FCS/0.1% w/v sodium azide was preincubated with FcγIII/IIR-specific antibody to block non-specific binding and stained with different combinations of fluorochrome-coupled antibodies to CD11c, I-Ab (MHCII), CD40, CD80, B220, CD11b, Gr1, CD3, CD8 or CD4 (BD Biosciences). Prior to intracellular staining with antibodies to TLR9 (eBioscience), Dicer (Santa Cruz) or FoxP3 (eBioscience), various immune cell subsets were fixed in paraformaldehyde and permeated in methanol. Fluorescence data were collected on FACScalibur (Beckton Dickinson) and analyzed using FlowJo software (Tree Star).
ELISPOT assay
5 × 105 cells isolated form TDLNs of CpG- or CpG-siRNAs-treated mice, were seeded into each well of 96-well filtration plate in the presence or absence of 10 μg/ml of TRP2 peptide. After 24 h of incubation at 37°C, peptide-specific IFNγ-positive spots were detected according to manufacturer’s procedure (Cell Sciences), scanned and quantified using Immunospot Analyzer from Cellular Technology Ltd.
Fluorescent, confocal and intravital two-photon microscopy
For immunofluorescent staining, we fixed the flash-frozen tumor specimens in formaldehyde and permeabilized with methanol before antibody staining. For confocal microscopy, cultured cells were fixed with 2% paraformadehyde for 20 m, permeabilized in PBS/0.1 % Triton X-100/1 mM MgCl2, and 0.1 mM CaCl2 for 5 m and quenched in 50 mM NH4Cl for 5 m before blocking in 1% BSA for 1 h. Samples were stained with antibodies to CD11b (BD Biosciences), neutrophils (7/4, Cedarlane), active caspase-3 (Cell Signaling), TLR9 (eBiosciences), Dicer (Santa Cruz) and detected with Alexa488- or Alexa555-coupled secondary antibodies (Invitrogen). After staining the nuclei with DAPI (Vector) or Hoechst 33342, slides were mounted and analyzed by fluorescent or confocal microscopy. The confocal imaging was carried out using a 63x water immersion objective on cLSM510Meta confocal microscope (Zeiss). For intravital two-photon imaging, B16 tumor-bearing mice received single intratumoral injection of 0.78 nmol FITC-labeled CpG-Stat3 siRNA, followed by retroorbital injection of dextran-rhodamine (Invitrogen) and Hoechst 33342 prior to imaging 1 h later. Mice were anesthetized and intravital two-photon microscopy was carried out using equipment and software from Ultima Multiphoton Microscopy Systems.
Statistical analysis
Unpaired t-test with equal or unequal variance (specifically for the analysis of cytokine expression in Fig. 5a) was used to calculate two-tailed P value to estimate statistical significance of differences between two treatment groups in the whole study. One-way ANOVA followed by Newman-Keuls test was applied for comparison of multiple treatment groups. Two-way ANOVA plus Bonferroni posttest were applied to assess statistical significance of differences in tumor growth kinetics between multiple treatment groups. Statistically significant P values were indicated in figures and/or legends and labeled as follows: ***; P < 0.001; **, P < 0.01 and *, P < 0.05. Data were analyzed using GraphPad Prism version 4.0 software (GraphPad).
Supplementary Material
Acknowledgments
We are grateful to the Light Microscopy Imaging and Flow Cytometry Cores, and Animal Facilities of Beckman Research Institute at City of Hope Medical Center for technical support and assistance; Dr Xuejun Li of Department of Biomedical Informatics for consultation on statistical analyses. We thank Dr C. H. Contag of Stanford University for providing the original luciferase mice. This work is funded in part by grants from Board of Governors for City of Hope Medical Center, Harry Lloyd Charitable Trust and Keck Foundation, in addition to NIH grants (R01CA122976, R01CA115815, R01CA115674, P50CA107399).
Footnotes
AUTHOR CONTRIBUTIONS
H. Y. and M. K. conceived the project, designed majority of the experiments, analyzed the data and wrote the paper. M. K. also carried out many of the key experiments and is instrumental for the design of the CpG-siRNA construct. P. S. contributed to the design of the construct and synthesized all the CpG-siRNA constructs. A. H. performed imaging and EMSA experiments. L. W. did A20 tumor cell experiments, luciferase CpG-siRNA experiments in vivo. C. K. tested siRNA sequences and carried out all the real-time PCR experiments. M. K. (Kujawski) and Y. L. performed some in vivo experiments. H. L. and C. Y. did Western blot analysis. A. S. and J. D. tested CpG-siRNA in B cell malignant cells. H. S. suggested the in vitro Dicer experiment. A. R. contributed to the MC38-CEA tumor experiment. J. R. and S. F. provided helpful discussion. J. R. is also helpful for the siRNA design. D. P. contributed to immunological experimental design, provided insightful discussion and assisted in writing the mansucript. R. J. contributed to the concept of the project, and the Stat3 siRNA design.
References
- 1.Song E, et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 2.McNamara JO, 2nd, et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 3.Kumar P, et al. Transvascular delivery of small interfering RNA to the central nervous system. Nature. 2007;448:39–43. doi: 10.1038/nature05901. [DOI] [PubMed] [Google Scholar]
- 4.Poeck H, et al. 5′-Triphosphate-siRNA: turning gene silencing and Rig-I activation against melanoma. Nat Med. 2008;14:1256–1263. doi: 10.1038/nm.1887. [DOI] [PubMed] [Google Scholar]
- 5.Li BJ, et al. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque. Nat Med. 2005;11:944–951. doi: 10.1038/nm1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zimmermann TS, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 7.Bui JD, Schreiber RD. Cancer immunosurveillance, immunoediting and inflammation: independent or interdependent processes? Curr Opin Immunol. 2007;19:203–208. doi: 10.1016/j.coi.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Koebel CM, et al. Adaptive immunity maintains occult cancer in an equilibrium state. Nature. 2007;450:903–907. doi: 10.1038/nature06309. [DOI] [PubMed] [Google Scholar]
- 9.Shankaran V, et al. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410:1107–1111. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 10.Kortylewski M, et al. Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat Med. 2005;11:1314–1321. doi: 10.1038/nm1325. [DOI] [PubMed] [Google Scholar]
- 11.Zou W. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat Rev Cancer. 2005;5:263–274. doi: 10.1038/nrc1586. [DOI] [PubMed] [Google Scholar]
- 12.Kujawski M, et al. Stat3 mediates myeloid cell-dependent tumor angiogenesis in mice. J Clin Invest. 2008;118:3367–3377. doi: 10.1172/JCI35213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu CL, et al. Enhanced DNA-binding activity of a Stat3-related protein in cells transformed by the Src oncoprotein. Science. 1995;269:81–83. doi: 10.1126/science.7541555. [DOI] [PubMed] [Google Scholar]
- 14.Bromberg JF, et al. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 15.Yu H, Jove R. The STATs of cancer--new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 16.Darnell JE., Jr Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 17.Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7:41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- 18.Wang T, et al. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat Med. 2004;10:48–54. doi: 10.1038/nm976. [DOI] [PubMed] [Google Scholar]
- 19.Kortylewski M, Yu H. Role of Stat3 in suppressing anti-tumor immunity. Curr Opin Immunol. 2008;20:228–233. doi: 10.1016/j.coi.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kortylewski M, et al. Regulation of the IL-23 and IL-12 balance by Stat3 signaling in the tumor microenvironment. Cancer Cell. 2009;15:114–123. doi: 10.1016/j.ccr.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bollrath J, et al. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell. 2009;15:91–102. doi: 10.1016/j.ccr.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 22.Grivennikov S, et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15:103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee H, et al. Persistently activated Stat3 maintains constitutive NF-kappaB activity in tumors. Cancer Cell. 2009;15:283–293. doi: 10.1016/j.ccr.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, et al. IL-17 is pro-carcinogenic through an IL-6/Stat3 signaling pathway. J Exp Med. 2009;206:1457–64. doi: 10.1084/jem.20090207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kortylewski M, et al. Toll-like receptor 9 activation of signal transducer and activator of transcription 3 constrains its agonist-based immunotherapy. Cancer Res. 2009;69:2497–2505. doi: 10.1158/0008-5472.CAN-08-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Immunol. 2004;5:987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 27.Kanzler H, Barrat FJ, Hessel EM, Coffman RL. Therapeutic targeting of innate immunity with Toll-like receptor agonists and antagonists. Nat Med. 2007;13:552–559. doi: 10.1038/nm1589. [DOI] [PubMed] [Google Scholar]
- 28.Barchet W, Wimmenauer V, Schlee M, Hartmann G. Accessing the therapeutic potential of immunostimulatory nucleic acids. Curr Opin Immunol. 2008;20:389–395. doi: 10.1016/j.coi.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 29.Krieg AM. Toll-like receptor 9 (TLR9) agonists in the treatment of cancer. Oncogene. 2008;27:161–167. doi: 10.1038/sj.onc.1210911. [DOI] [PubMed] [Google Scholar]
- 30.Klinman DM, Currie D, Gursel I, Verthelyi D. Use of CpG oligodeoxynucleotides as immune adjuvants. Immunol Rev. 2004;199:201–216. doi: 10.1111/j.0105-2896.2004.00148.x. [DOI] [PubMed] [Google Scholar]
- 31.Klinman DM, Yi AK, Beaucage SL, Conover J, Krieg AM. CpG motifs present in bacteria DNA rapidly induce lymphocytes to secrete interleukin 6, interleukin 12, and interferon gamma. Proc Natl Acad Sci U S A. 1996;93:2879–2883. doi: 10.1073/pnas.93.7.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krieg AM, et al. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature. 1995;374:546–549. doi: 10.1038/374546a0. [DOI] [PubMed] [Google Scholar]
- 33.Kim DH, et al. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 34.Rose SD, et al. Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res. 2005;33:4140–4156. doi: 10.1093/nar/gki732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hemmi H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 36.Latz E, et al. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat Immunol. 2004;5:190–198. doi: 10.1038/ni1028. [DOI] [PubMed] [Google Scholar]
- 37.Chendrimada TP, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samarasinghe R, et al. Induction of an anti-inflammatory cytokine, IL-10, in dendritic cells after toll-like receptor signaling. J Interferon Cytokine Res. 2006;26:893–900. doi: 10.1089/jir.2006.26.893. [DOI] [PubMed] [Google Scholar]
- 39.Cao YA, et al. Shifting foci of hematopoiesis during reconstitution from single stem cells. Proc Natl Acad Sci U S A. 2004;101:221–226. doi: 10.1073/pnas.2637010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benkhart EM, Siedlar M, Wedel A, Werner T, Ziegler-Heitbrock HW. Role of Stat3 in lipopolysaccharide-induced IL-10 gene expression. J Immunol. 2000;165:1612–1617. doi: 10.4049/jimmunol.165.3.1612. [DOI] [PubMed] [Google Scholar]
- 41.Xie TX, et al. Stat3 activation regulates the expression of matrix metalloproteinase-2 and tumor invasion and metastasis. Oncogene. 2004;23:3550–3560. doi: 10.1038/sj.onc.1207383. [DOI] [PubMed] [Google Scholar]
- 42.Takeda K, et al. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity. 1999;10:39–49. doi: 10.1016/s1074-7613(00)80005-9. [DOI] [PubMed] [Google Scholar]
- 43.Welte T, et al. STAT3 deletion during hematopoiesis causes Crohn’s disease-like pathogenesis and lethality: a critical role of STAT3 in innate immunity. Proc Natl Acad Sci U S A. 2003;100:1879–1884. doi: 10.1073/pnas.0237137100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dhodapkar MV, Steinman RM, Krasovsky J, Munz C, Bhardwaj N. Antigen-specific inhibition of effector T cell function in humans after injection of immature dendritic cells. J Exp Med. 2001;193:233–238. doi: 10.1084/jem.193.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bui JD, Uppaluri R, Hsieh CS, Schreiber RD. Comparative analysis of regulatory and effector T cells in progressively growing versus rejecting tumors of similar origins. Cancer Res. 2006;66:7301–7309. doi: 10.1158/0008-5472.CAN-06-0556. [DOI] [PubMed] [Google Scholar]
- 46.Klinman D, Shirota H, Tross D, Sato T, Klaschik S. Synthetic oligonucleotides as modulators of inflammation. J Leukoc Biol. 2008;84:958–964. doi: 10.1189/jlb.1107775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sica A, Bronte V. Altered macrophage differentiation and immune dysfunction in tumor development. J Clin Invest. 2007;117:1155–1166. doi: 10.1172/JCI31422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan TT, Coussens LM. Humoral immunity, inflammation and cancer. Curr Opin Immunol. 2007;19:209–216. doi: 10.1016/j.coi.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 49.Spaner DE, Foley R, Galipeau J, Bramson J. Obstacles to effective Toll-like receptor agonist therapy for hematologic malignancies. Oncogene. 2008;27:208–217. doi: 10.1038/sj.onc.1210905. [DOI] [PubMed] [Google Scholar]
- 50.Clarke P, Mann J, Simpson JF, Rickard-Dickson K, Primus FJ. Mice transgenic for human carcinoembryonic antigen as a model for immunotherapy. Cancer Res. 1998;58:1469–1477. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.