Abstract
Stimulation of iNKT cells is highly dependent on the structures of the glycolipids presented by CD1d. Furthermore, antigen processing and CD1d loading in lysosomes play central roles in controlling the stimulatory properties of glycolipid antigens. Previously, we determined that substitution at C6″ on α-galactosylceramides did not significantly impact stimulatory properties; however, it was not known if substitution at this position influenced lysosomal processing of oligoglycosylceramides. We have prepared a series of mono- and di-galactosylceramides to observe the impact of C6″ substitution on glycosidase truncation of these glycolipids. We found that substitution did not significantly impact glycosidase activity or loading into CD1d.
Keywords: glycosylation, glycosphingolipid synthesis, stimulatory activity, IL-2 release, NKT cells
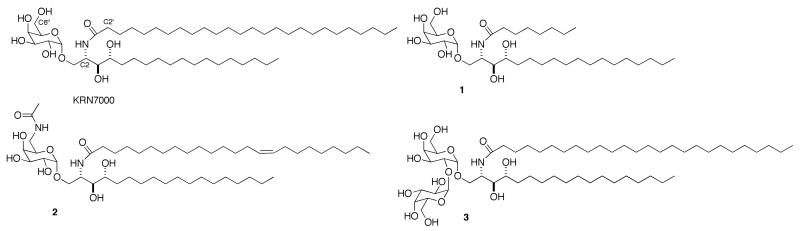
Natural killer T (NKT) cells are a subset of T cells and have been characterized as regulatory T cells.1 The most prevalent and best studied subset of NKT cells expresses an invariant T cell Vα14/24 (mouse/human) receptor and are referred to as invariant NKT cells (iNKT) cells. Through this invariant T cell receptor, iNKT cells recognize bacterial and endogenous glycolipid antigens presented by CD1d, an antigen presentation protein related to major histocompatibility complex (MHC) proteins. The hallmark response of the iNKT cells is the rapid production and release of large quantities of cytokines, including proinflammatory cytokines (e.g., IFN-γ, IL-2) and immuno-modulatory cytokines (e.g., IL-4). These cytokines play critical roles in inducing a series of events leading to the activation of innate and adaptive immune cells. The most well-studied ligand for iNKT cells is a glycolipid, KRN7000 (Figure 1) developed through anti-tumor structure-activity studies of glycolipids from the marine sponge Agelas mauritianus.2 In further structure activity studies using cytokine release from NKT cells as an end point, we have determined that shortening the acyl chain in KRN7000 to eight carbons (from 26) yields a compound (1 in Figure 1) that loads effectively into CD1d without dependence on lipid-transfer proteins that are required for loading most glycolipids into CD1d.3,4 We have also found that replacement of the hydroxyl group at C6″ (see Figure 1 for glycosylceramide numbering) with an acetylamide gives a compound, 2, that is a more potent stimulator of NKT cells than KRN7000 (both in vitro and in vivo).5
Figure 1.
Structure of glycolipids KRN7000, 1, 2, and 3.
An important aspect of glycolipid presentation by CD1d and recognition by iNKT cells is lysosomal processing of glycolipids. Lysosomes in antigen presenting cells contain an array of glycosidases that can truncate oligoglycosylceramides to monoglycosylceramides. Work by Prigozy, et al.6 demonstrated that diglycosylceramides with 1-2 or 1-3 glycosidic linkages (e.g., 3 in Figure 1) require lysosomal processing and truncation to KRN7000 for presentation by CD1d and stimulation of iNKT cells. CD1d is cycled between lysosomes, where it is loaded with glycolipids, and the cell surface, where it presents bound glycolipids to iNKT cells. Deletion of the peptide sequence required for this cycling yields “tail-deleted” CD1d (tdCD1d), which does not effectively cycle between the cell surface and lysosomes.7 Consequently, glycolipids that are processed in the lysosome are not loaded into CD1d, and tdCD1d is useful for observing dependence on lysosomal glycolipid processing.6,7
The rationale for preparing glycolipids substituted at C6″ was to use groups appended at this position to monitor glycolipid trafficking, processing, and presentation of glycolipids.8 We have found that fluorophores appended at C6″ do not significantly alter stimulation of iNKT cells.9 However, we have not determined if C6″ substitution affects lysosomal processing. To this end, we prepared a series of diglycosylceramides including one with C6″ substitution. For this series, we used the C8 acyl chain in 1 to remove possible influences of lipid transfer proteins10,11 and thereby only focus on activities of lysosomal glycosidases.
The glycolipids prepared to determine the effects of C6″ substitution on lysosomal processing are shown in Figure 2. Comparison of iNKT cell stimulation of 1 and 4 with wild-type CD1d and tdCD1d provides a measure of the influences of lysosomal processing on these glycolipids. Glycolipid 1 would be expected to load into CD1d well and stimulate iNKT cells comparably with wild-type and tdCD1d. However, the requirement for processing would make 4 less stimulatory with tdCD1d. If the substitution at C6″ does not significantly influence lysosomal trafficking, then the same trend would be expected. That is, 6 should lose stimulatory properties with tdCD1d.
Figure 2.
Structures of diglycosyl and C6″ appended glycolipids used in the study of lysosomal processing of C6″ substituted glycolipids.
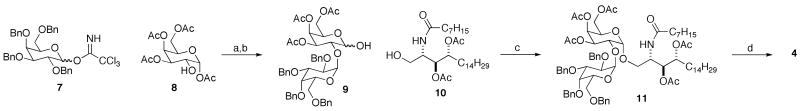
The synthesis of disaccharide 4 is described in Scheme 1. A Schmidt coupling12 of 7 and 8 gave the corresponding disaccharide, and the anomeric hydroxyl group was revealed after deacylation with the mono-acetate salt of ethylene diamine. The disaccharide was coupled with the appropriately protected ceramide using Gin's method,13 and deprotection gave 4.
Scheme 1.
Preparation of diglycosylceramide 4.
Reagents (yields in parentheses): a) TMSOTf, CH2Cl2 (40%). b) ethylene diamine, AcOH, THF (75%). c) diphenylsulfoxide, Tf2O, tri-t -butylpyrimidine, CH2Cl2 (71%). d) H2, Pd/C, THF, methanol; MeONa, methanol (56%).
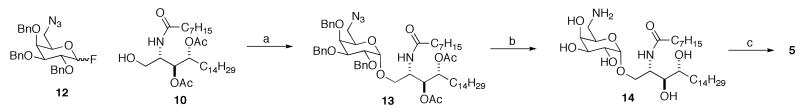
The preparation of C6″ modified galactosylceramide 5 is detailed in Scheme 2. The synthesis borrows from work with fluorophore labeled galactosylceramides.9 Coupling of C6 azido galactose 12 with ceramide gave 13. Dissolving metal deprotection also caused reduction of the azide to the amine, and subsequent acylation gave 5.
Scheme 2.
Preparation of C6″ N-acyl amino substituted galactosylceramide 5.
Reagents (yields in parentheses): a) AgClO4, SnCl2, CH2Cl2 (54%). b) Na°, NH3 (40%). c) AcOH, HOBt, DCC (90%).
For the preparation of 6 (Scheme 3), C6 azido acceptor 15 was used, and a pathway similar to that used for the synthesis of 4 was followed, except that deprotection was achieved using dissolving metal conditions. The acyl group was installed as described in the preparation of 5.
Scheme 3.
Preparation of C6″ N-acyl amino substituted digalactosylceramide 6.
Reagents (yields in parentheses): a) TMSOTf, CH2Cl2 (37%). b) ethylene diamine, AcOH, THF(64%). c) diphenylsulfoxide, Tf2O, tri-t -butylpyrimidine, CH2Cl2 (53%). d) Na/NH3, (27%). e) AcOH, DCC, HOBt, (22%).
The iNKT cell stimulatory properties of glycolipids 1, 4, 5 and 6 were determined using iNKT cell hybridoma and dendritic cells as antigen-presenting cells. Dendritic cells generate a well-established lysosome and effectively truncate many oligoglycosylceramides to stimulatory compounds. As shown in Figure 3A, all four of the glycolipids stimulated cytokine release. The monoglycosylceramides are not dependent on processing and can load into CD1d directly, and as expected they proved to be more potent than the diglycosylceramides. Notably, C6″ did not play a significant role in stimulatory properties. To further establish the dependency of the diglycosylceramides for lysosomal processing, dendritic cells with tdCD1d were used as antigen presenting cells. Because tdCD1d does not effectively sample the lipids in lysosomes, it was expected that stimulation with 4 and 6 would be decreased with tdCD1d. As shown in Figure 3B, this trend was observed, while the monoglycosylceramides were less dependent on CD1d sampling of lysosomes.
Figure 3.
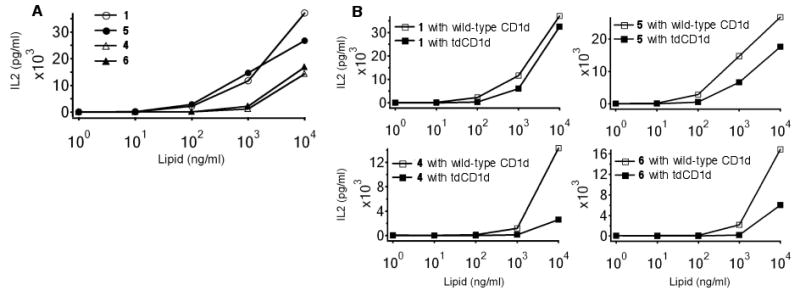
Comparison of stimulatory activity on mouse Vα14 iNKT hybridoma DN3.2.D3 by murine bone marrow-derived dendritic cells in the presence of different concentrations of glycosylceramides 1, 4, 5, and 6. Indicated are the mean release amounts of IL-2 (pg/mL) in cell culture supernatants determined by ELISA. A: Wild-type CD1d. B: Comparison of stimulation with wild-type CD1d and tdCD1d.
Comparisons of the iNKT cell stimulatory properties of mono- and diglycosylceramides with and without substitution at C6″ indicate that substitution at this position does not play a significant role in stimulatory properties of these glycolipids or their abilities to act as substrates for lysosomal glycosidases. Small changes in glycosylceramide structure can change iNKT cell stimulatory properties dramatically: α-galactosylceramides (e.g., KRN7000 and 1) are potent stimulators while β-galactosylceramide is only very weakly active or inactive; α-glucosylceramide is much less potent than α-galactosylceramide. However, the C6″ position appears to be well suited for manipulation without significantly impacting glycolipid processing and presentation. We have shown that fluorophores of modest size (e.g., a danyl group) can be appended at C6″ without significantly altering NKT cell responses,9 and the acetamide at C6″ in 6 does not interfere with processing. It is anticipated that larger groups, such as fluorophores of modest size, appended at this position with not impact processing; however, this has not yet been demonstrated. Furthermore, it is not yet well understood how substitution at this position affects glycolipid trafficking and lipid-transfer protein interactions, and studies are ongoing to determine impacts on these interactions.
Supplementary Material
Acknowledgments
We gratefully acknowledge generous financial support from the National Institutes of Health (NIAID AI053725). JM is a CRI Fellow and was supported by a grant from LRI. SF is a Swiss National Science Foundation/SSMBS Fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bendelac A, Savage PB, Teyton L. Annu Rev Immunol. 2007;25:297. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 2.Morita M, Motoki K, Akimoto K, Natori T, Sakai T, Sawa E, Yamaji K, Koezuka Y, Kobayashi E, Fukushima H. J Med Chem. 1995;38:2176. doi: 10.1021/jm00012a018. [DOI] [PubMed] [Google Scholar]
- 3.Goff RD, Gao Y, Mattner J, Zhou DP, Yin N, Cantu C, Teyton L, Bendelac A, Savage PB. J Am Chem Soc. 2004;126:13602. doi: 10.1021/ja045385q. [DOI] [PubMed] [Google Scholar]
- 4.Zajonc DM, Cantu C, Mattner J, Zhou DP, Savage PB, Bendelac A, Wilson IA, Teyton L. Nat Immunol. 2005;6:810. doi: 10.1038/ni1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y, Goff RD, Zhou D, Mattner J, Sullivan BA, Khurana A, Cantu C, Altman JD, Teyton L, Bendelac A, Savage PB. J Immun Methods. 2006;312:34. doi: 10.1016/j.jim.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Prigozy TI, Naidenko O, Qasba P, Elewaut D, Brossay L, Khurana A, Natori T, Koezuka Y, Kulkarni A, Kronenberg M. Science. 2001;291:664. doi: 10.1126/science.291.5504.664. [DOI] [PubMed] [Google Scholar]
- 7.Chiu YH, Park SH, Benlagha K, Forestier C, Jayawardena-Wolf J, Savage PB, Teyton L, Bendelac A. Nat Immunol. 2002;3:55. doi: 10.1038/ni740. [DOI] [PubMed] [Google Scholar]
- 8.Sagiv Y, Hudspeth K, Mattner J, Schrantz N, Stern RK, Zhou D, Savage PB, Teyton L, Bendelac A. J Immunol. 2006;177:26. doi: 10.4049/jimmunol.177.1.26. [DOI] [PubMed] [Google Scholar]
- 9.Zhou XT, Forestier C, Goff RD, Li CH, Teyton L, Bendelac A, Savage PB. Org Let. 2002;4:1267. doi: 10.1021/ol025565+. [DOI] [PubMed] [Google Scholar]
- 10.Zhou D, Cantu CC, Sagiv Y, Kulkarni AB, Qi X, Morales C, Grabowski GA, Benlagha K, Savage PB, Bendelac A, Teyton L. Science. 2004;303:523. doi: 10.1126/science.1092009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schrantz N, Sagiv Y, Liu Y, Savage PB, Bendelac, Teyton L. J Exp Med. 2007;204:841. doi: 10.1084/jem.20061562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt RR, Michel J. Angew Chem IEE. 1980;19:731. [Google Scholar]
- 13.Garcia BA, Poole JL, Gin DY. J Am Chem Soc. 1997;119:7597. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.